Abstract
Signal transduction typically begins by ligand-dependent activation of a concomitant partner which is otherwise in its resting state. However, in cases where signal activation is constitutive by default, the mechanism of regulation is unknown. The Arabidopsis thaliana heterotrimeric Gα protein self-activates without accessory proteins, and is kept in its resting state by the negative regulator, AtRGS1 (Regulator of G protein Signaling 1), which is the prototype of a seven transmembrane receptor fused with an RGS domain. Endocytosis of AtRGS1 by ligand-dependent endocytosis physically uncouples the GTPase accelerating activity of AtRGS1 from the Gα protein, permitting sustained activation. Phosphorylation of AtRGS1 by AtWNK8 kinase causes AtRGS1 endocytosis, required both for G protein-mediated sugar signaling and cell proliferation. In animals, receptor endocytosis results in signal desensitization, whereas in plants, endocytosis results in signal activation. These findings reveal how different organisms rearrange a regulatory system to result in opposite outcomes using similar phosphorylation-dependent endocytosis.
Keywords: AGB1, Arabidopsis, heterotrimeric G protein, RGS1, receptor endocytosis
Many cells employ coordinated mechanisms to control the amplitude, duration and spatial distribution of G-protein mediated responses to extracellular signals that activate 7-transmembrane (7TM) G-protein coupled receptors. One such mechanism is 7TM receptor internalization, which physically removes the receptor from the cell surface to desensitize cells from continuous stimulation of the G protein complex by the activated receptor 1, 2. Here we report identification of a previously unknown mechanism of signal control where internalization of a 7TM Regulator of G signaling (RGS) protein leads to sustained signaling.
Recently, we discovered that the model plant organism, Arabidopsis thaliana, employs signal regulation mechanisms distinct from those identified in animals 3, 4. In contrast to animals, the Arabidopsis Gα protein self-activates without the aid of a receptor because its rate of guanine nucleotide exchange is about 100 times faster than its rate of GTP hydrolysis 3, 4. Consistent with this self-activating property, the Arabidopsis genome encodes no canonical 7TM, G protein-coupled receptors (GPCR) 5, 6. However, the Arabidopsis genome encodes a 7TM domain fused to an RGS protein, AtRGS1, that stimulates the rate-limiting GTPase activity of the Arabidopsis Gα subunit, AtGPA1 3, 7, 8.
AtRGS1 and its cognate heterotrimeric G protein complex are required for normal glucose sensing, cell proliferation, cell elongation and development 8–12. Genetic evidence suggested that D-glucose or a sugar metabolite regulates AtRGS1 activity toward AtGPA1 3, 8, although direct evidence for glucose binding to RGS1 is lacking and the molecular basis of how D-glucose and AtRGS1 control G protein signaling is not known.
Here we show that D-glucose causes AtRGS1 endocytosis. This sugar-dependent re-localization physically uncouples the inhibitory activity of AtRGS1 from the plant Gα protein, leaving the Gα protein constitutively active at the cell surface for “sustained” signaling. These results reveal how land plants employ RGS protein internalization to sense energy status and regulate growth and development. The presence of 7TM-RGS proteins in fungi and single-cell eukaryotes suggests this mechanism is utilized in other organisms.
RESULTS
AtRGS1 internalizes in response to D-glucose
Mutations in Arabidopsis G proteins confer altered responsiveness to glucose 8, 13, 14. Because AtRGS1 contains a predicted 7TM domain reminiscent of GPCRs, localizes to the cell surface, and interacts with the plant Gα protein in a glucose-dependent manner, AtRGS1 was proposed to be a glucose receptor or co-receptor in G protein-mediated glucose sensing 3, 8, 13, 15, 16.
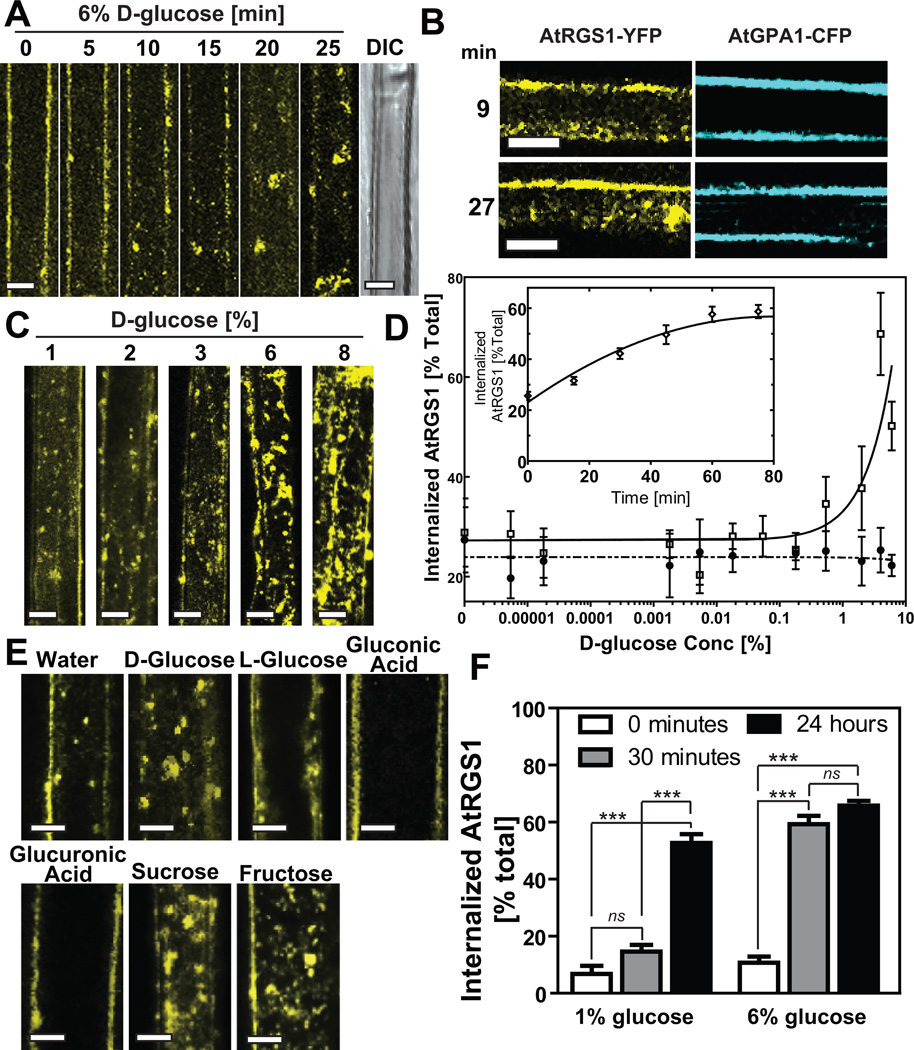
In animals, ligand-induced 7TM receptor endocytosis desensitizes cells to the ligand by reducing the amount of receptor at the cell surface 1. To determine the effect of the candidate ligand on AtRGS1 internalization, epidermal cells expressing AtRGS1-YFP were treated with several concentrations of D-glucose, and the subcellular localization of AtRGS1 was captured over time (Fig. 1 and Supplementary Fig. S1A). The maximum steady-state level of internalized AtRGS1 at steady state varied between 60%-90%, depending on expression levels, and was reached within 60 min. 3-D reconstruction revealed that the observed change in AtRGS1 was due to internalization as opposed to clustering on the plasma membrane (Supplemental Movie S1). AtRGS1 internalization showed glucose dose dependency (Fig. 1C, Fig. 1D) and structural stereo-specificity in that D- but not L-glucose caused internalization (Fig. 1E). Likewise, two similar structures, gluconic and glucuronic acids (Fig. 1E) did not affect AtRGS1 localization. Three analogous sugars, mannose, fructose and sucrose, each able to yield glucose through metabolism17–19, induced AtRGS1 internalization (Fig. 1E). General reciprocity was observed between dose-dependence and time-dependence of AtRGS1 internalization; 1% D-glucose induced internalization (Fig. 1C, D), but required 6 hr to reach maximum achieved by the acute dose of 6% in 30 minutes (Fig. 1F).
Figure 1. AtRGS1 internalizes in response to sugar.
AtRGS1-YFP internalized by glucose. (A) AtRGS1-YFP and (B) AtGPA1-CFP localization after treatment with 6% glucose in an Arabidopsis hypocotyl epidermal cell. Differential interference contrast (DIC) shows that 30 min of glucose does not disrupt cell integrity (last in series, panel A). (C) Dose-dependent internalization of AtRGS1. Arabidopsis cells stably expressing AtRGS1-YFP imaged after treatment with varying concentrations of glucose for 30 min. (D) Quantitation of dosage response of AtRGS1 (open square) and AtRGS1(E320K) mutant (GAP dead; close circle) with increasing glucose concentrations. At the 30 min time point, YFP fluorescence was measured by subtracting internalized RGS1-YFP fluorescent signal from total cell fluorescence. A point mutation that inhibits AtRGS1 interaction with AtGPA1, AtRGS1(E320K), disrupts AtRGS1-YFP internalization. Error bars = SEM, n = 5. (D Inset) Quantitation of the glucose dosage response of AtRGS1-YFP internalization imaged at 30 min post-glucose treatment. Error bars = SEM, n = 5. (E) Sugar specificity of AtRGS1 internalization. Several sugar and sugar analogs (6% of each) were applied to seedlings expressing AtRGS1-YFP for 30 min prior to imaging as described in Methods. (F) RGS1-YFP reciprocity of time and dose dependence. AtRGS1 seedlings stably expressing AtRGS1-YFP were treated without or with 1% or 6% D-glucose. After 30 min or 24 hr treatment, internalized AtRGS1 was quantified. Error = SEM, n = 5. Labels with ns has no statistical difference (P > 0.05), *** mean highly significantly different (P < 0.001). All scale bars = 10 µm. Quantitation of fluorescence is described in Methods.
To eliminate the possibility that glucose caused a general and nonspecific sweep of membrane proteins from the plasma membrane, we tested a 7TM domain protein, AtMLO6 20, 21 (Supplementary Fig. S1B, bottom pair) and showed that this plasma membrane 7TM protein does not internalize with glucose. These results show that the effect of D-glucose on AtRGS1 internalization is specific, as well as time- and dose-dependent.
A critical observation was that while AtRGS1-YFP internalized by glucose, the cognate Gα, CFP-AtGPA1 co-expressed in the same cell, did not (Fig. 1B). No internalization of CFP-AtGPA1, was observed at any tested glucose dose including the 6% acute treatments even monitored over extended observation times using both stably- (Fig. 1B) or transiently-transformed cells (Supplementary Fig. S1B, center pair). Taken with the AtRGS1 data above, we conclude that glucose causes physical separation of the plant Gα subunit from AtRGS1.
Animal GPCRs are internalized via the endosomal pathway 22–24. To determine AtRGS1 localization after internalization, we measured AtRGS1 co-localization with various compartmental markers in epidermal pavement cells. AtRGS1 co-trafficked with the endosomal dye, FM4-64 (Supplementary Fig. S2A), endosomal markers that reside in the early to late endosomes (Supplementary Fig. S2). Co-expression of AtRGS1-YFP with RFP-tagged compartment markers was also performed using tobacco cells because their larger size provides the required spatial resolution for co-localization (Supplementary Fig. S2B). Of particular interest was the observed 100% co-localization of AtRGS1 with SYP23 (Supplementary Fig. S2B), a late endosomal syntaxin because AtRGS1 and SYP23 are known to physically interact 25. AtRGS1-YFP did not co-localize with the mitochondrial-RFP marker (Mt-rk; Supplementary Fig. S2B) 26. These results indicate that AtRGS1-YFP internalization occurs through the endocytic pathway and hereafter we refer to the observation of AtRGS1 internalization as endocytosis.
Free Gβ, AGB1, is essential, but not sufficient, for AtRGS1 endocytosis
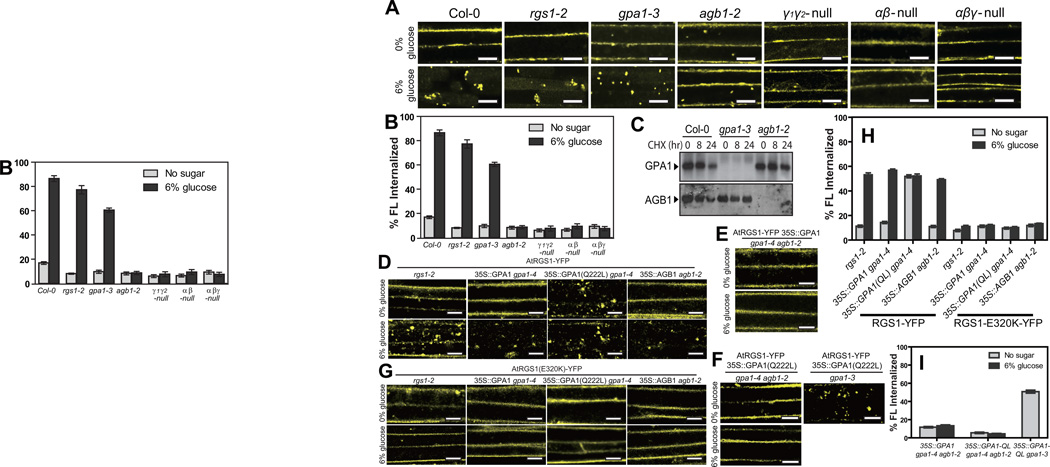
D-glucose-induced endocytosis of AtRGS1-YFP was reduced 40% in the absence of the Gα subunit, AtGPA1 (gpa1-3), and completely abrogated in the absence of the Gβ subunit AGB1 (Fig. 2 A,B). Loss of AtGPA1 or AGB1 individually, did not dramatically affect the steady-state levels of the respective protein-binding partner (Fig. 2C), suggesting that both G protein components, particularly the Gβ subunit, directly mediate glucose-stimulated AtRGS1 endocytosis.
Figure 2. AGB1 is essential for AtRGS1 internalization.
(A) AtRGS1-YFP was transiently expressed in an informative set of G-protein mutants and treated with 6% glucose. Internalization of AtRGS1-YFP was imaged in Col-0, rgs1-2, gpa1-3, agb1-2, gpa1-4 agb1-2 double null mutants (αβ-null) and gpa1-4 agb1-2 agg1-2 agg2-1 quadruple null mutants (αβγ-null) without and with 6% glucose. (B) Quantitation of percent AtRGS1-YFP fluorescence (FL) internalized in epidermal cells transiently expressed before and after glucose stimulation. Error bars = SEM, n = 5. (C) Seedlings of Col-0, gpa1-3 and agb1-2 were treated with 200 mM cycloheximide (CHX). Relative steady-state levels of AtGPA1 and AGB1 protein in the seedling were analyzed by immunoblot analysis with anti-AtGPA1 and anti-AGB1 antisera. (D) AtRGS1-YFP was transiently expressed in rgs1-2 null mutants, 35S::AtGPA1, 35S::AtGPA1(Q222L) (active mutant) or 35S::AGB1 lines and treated with 6% glucose. “35S::” represents a constitutive promoter from the Cauliflower Mosaic virus used for ectopic overexpression. (E) AtRGS1-YFP and 35S::AtGPA1 were transiently expressed in a gpa1-4 agb1-2 double mutant and treated with 6% glucose. (F) Both AtRGS1-YFP and 35S::AtGPA1(Q222L) were transiently expressed in the gpa1-4 agb1-2 double mutant and treated with 6% glucose (left panels). AtRGS1-YFP and 35S::AtGPA1(Q222L) were also transiently expressed in the gpa1-3 mutant (right panel). (G) AtRGS1(E320K)-YFP was transiently expressed in rgs1-2 null mutants, 35S::AtGPA1, 35S::AtGPA1(Q222L) (active mutant) or 35S::AGB1 lines then treated with 6% glucose prior to imaging.(H) Quantitation of percent AtRGS1-YFP and AtRGS1-E320K-YFP fluorescence internalized in epidermal cells before and after 6% glucose stimulation. Error bars = SEM, n = 5. The genetic background is indicated: rgs1-2, ectopic expression of AtGPA1 (35S-GPA1 in gpa1-4 null background), ectopic expression of constitutively-active AtGPA1 (35S::GPA1(Q222L) in gpa1-4 null background), and ectopic expression of AGB1 (35S::AGB1 in agb1-2 null background). (I) Quantitation of percent of AtRGS1-YFP fluorescence in epidermal cells transiently expressing the AtGPA1 in the gpa1/agb1 double mutant (35S::AtGPA1 in gpa1-4 agb1-2 null background), constitutively-active AtGPA1 (35S::GPA1-QL) in the gpa1/agb1 double mutant), and 35S::GPA1-QL in the gpa1-3 mutant). Error = SEM, n = 5.All scale bars = 10 µm. GPA1-QL represents GPA1(Q222L). Quantitation of fluorescence is described in Methods.
The complete loss of AtRGS1-trafficking in the agb1-2 background was genetically complemented by ectopic expression of AGB1 (Fig. 2D and 2H). Ectopic overexpression of wild type AtGPA1 did not significantly affect AtRGS1 trafficking (Fig. 2D). However, over-expression of the constitutively-active AtGPA1 mutant (GTPase-deficient AtGPA1(Q222L)) in the absence of the wild type Gα subunit triggered AtRGS1 endocytosis without glucose (Fig. 2D, 2H, and 2I). This induction by AtGPA1(Q222L) of AtRGS1 endocytosis required AGB1 (Fig. 2E, F). Taken together, these results indicate that the Gβγ dimer is required for AtRGS1 endocytosis induced by D-glucose and that the activated Gα subunit plays a role for AtRGS1 endocytosis. While these results suggest that the freed Gβ subunit is necessary, its release from the complex alone, an expectation for the Gα null mutants, is insufficient for AtRGS1 endocytosis.
Physical coupling of AtRGS1 and AtGPA1 is essential for AtRGS1 endocytosis
A charge reversal at glutamate 320 of AtRGS1 uncouples interaction between AtRGS1 and AtGPA1, therefore abrogates acceleration of GTP hydrolysis activity 3. To determine if AtRGS1 interaction with AtGPA1 is required for AtRGS1 endocytosis, we measured glucose-induced internalization of the AtRGS1(E320K) mutant protein. We found that AtRGS1(E320K)-YFP did not internalize under conditions that promoted endocytosis of the wild-type protein (Fig. 1D, 2D vs. 2G, and 2H). To distinguish between a loss of interaction or failure to accelerate GTP hydrolysis, we combined the AtGPA1(Q222L) mutation with the AtRGS1(E320K) mutation. Although constitutively active AtGPA1(Q222L) caused glucose-independent internalization of wild-type AtRGS1, AtGPA1(Q222L) failed to trigger AtRGS1(E320K) internalization under any condition (Fig. 2D, G). These results indicate that physical interaction between AtGPA1 and AtRGS1 is required for AtRGS1 endocytosis, but AtGPA1 cycling between inactive and active states is dispensable for initiating the AtRGS1 internalization process. One interpretation is that without AtRGS1 interaction with AtGPA1, the heterotrimer forms, thus sequestering AGB1, shown to be essential for AtRGS1 endocytosis (Fig. 2 A, B).
AtWNK8 is an AtRGS1 kinase
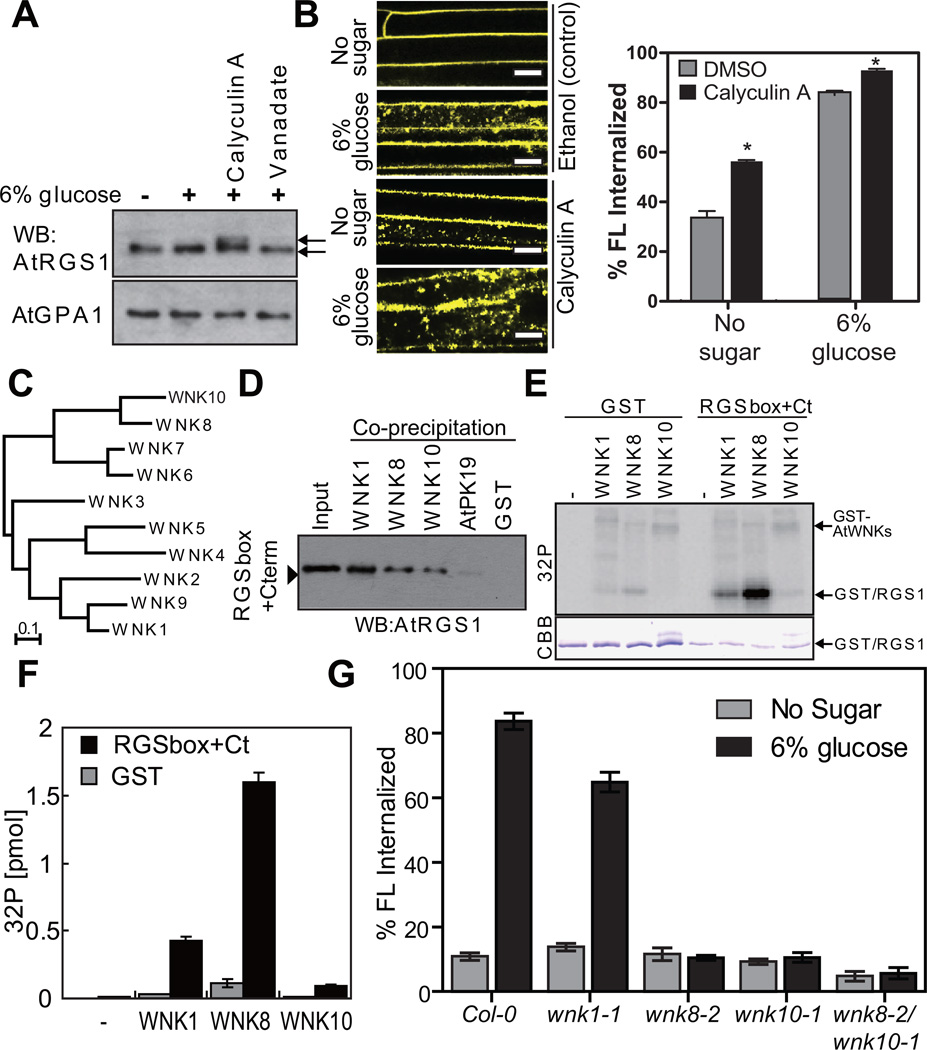
Upon sustained ligand occupancy, mammalian GPCRs are phosphorylated by G protein receptor kinases (GRKs). Thisphosphorylation event is critical for assembly of the trafficking complex and concomitant receptor endocytosis 1, 27. Thus, we hypothesized that AtRGS1 phosphorylation is required for AtRGS1 endocytosis. When calyculin A, a Ser/Thr phosphatase inhibitor 28 was applied to Arabidopsis seedlings, a band shift for AtRGS1, but not AtGPA1, was observed using SDS-PAGE (Fig. 3A), suggesting the phosphorylation of AtRGS1 in vivo. Likewise, calyculin A accelerated receptor endocytosis even in the absence of glucose suggesting tonic cycling (Fig. 3B; P<0.01). In contrast, the mobility of AtRGS1 and AtGPA1 were not changed by the tyrosine phosphatase inhibitor, sodium orthovanadate (Fig. 3A). These data suggest that AtRGS1 phosphorylation on Ser/Thr residues promotes endocytosis.
Figure 3. In vivo and In vitro function of AtWNK8.
(A) In vivo phosphorylation of AtRGS1. Seedlings expressing AtRGS1-TAP were pretreated with 100 nM calyculin A and 10 mM sodium orthovanadate for 3 h followed by 6% D-glucose stimulation for 90 min. AtRGS1-TAP or AtGPA1 in seedling lysates was separated on a 12.5% Anderson’s gel and detected by immunoblot with peroxidase anti-peroxidase or anti-AtGPA1 antibody. (B) Four-day-old AtRGS1-YFP expressing seedlings were treated with phosphatase inhibitors, calyculin A, for 2 h followed by 6% glucose treatment or not (No glucose) for 1 h prior to imaging epidermal cells. Scale bars = 10 µm. Error = SEM, n = 5. (C) Phylogenetic tree of the AtWNK-family kinases. Full-length amino acid sequences were aligned with CLUSTAL W implemented in CLC Genomics Workbench using the following settings; Gap open penalty, 10; Gap extension penalty 1. The neighbor joining tree (1000 bootstrap replicate) was created with the aligned sequences. (D) In vitro binding between AtRGS1 and AtWNKs. Recombinant RGSbox+Cterm was tested for interaction with GST (negative control) or GST-AtWNKs using glutathione-Sepharose, and detected by immunoblot analysis using an anti-AtRGS1 antibody. (E) In vitro phosphorylation of AtRGS1 by AtWNK kinases. Recombinant GST or His-RGSbox+Cterm was incubated with GST-AtWNKs in reaction buffer containing γ32P-ATP. Proteins were separated on SDS-PAGE. (F) Radioactivity incorporated into the GST/RGS1 bands. Phosphorylation levels of three independent experiments were quantified in (E). Error bars = SEM. (G) Quantitation of sugar-induced AtRGS1 internalization in AtWNK-null mutants. Seedlings of Col-0, wnk1-1, wnk8-1, wnk8-2 or wnk10-2 transiently expressing AtRGS1-YFP were treated with 6% D-glucose for 30 min. WNK# denotes AtWNK members in panels C-F. Error bars = SEM, n = 5. Quantitation of fluorescence is described in Methods.
AtRGS1 has many di-serines at its C terminus, analogous to sequence found in vertebrate GPCRs that are phosphorylated, a requisite for endocytosis in response to their ligands 1, 27. Although the Arabidopsis genome encodes ten-fold more kinases than the human genome 29, none of these plant kinases have the GRK architecture typified by an N-terminal RGS-homology domain 30. Therefore, we searched ab initio for candidate AtRGS1-kinases among candidate AtRGS1-interacting proteins that were identified from yeast complementation screens 25, 31. We found several AtRGS1-interacting kinases (Supplementary Fig. S3A) and confirmed in vivo interactions using bimolecular fluorescence complementation (BiFC, Supplementary Fig. S3B). While all four kinases assigned to the G protein interactome interacted with AtRGS1, only AtWNK8, among these, phosphorylated a recombinant AtRGS1 substrate-RGSbox+Cterm (284–459 aa) under these in vitro conditions (Supplemental 3A).
AtWNK8 is one of eleven WITH NO LYSINE (WNK) family Ser/Thr kinases in Arabidopsis (Fig. 3C). This kinase family has the catalytic lysine at the expected location in the catalytic center at an unusual position in the linear sequences 32. To address AtRGS1 specificity for WNK family members, we measured interactions using yeast two-hybrid (Supplementary Fig. S3C), BiFC (Supplementary Fig. S3B) and in vitro co-precipitation assays (Fig. 3D). We included AtWNK1, the most divergent from AtWNK8, and AtWNK10, the most similar to AtWNK8 in sequence (Fig. 3C). AtRGS1 interacted with all three representative WNK family kinases in all three assays, but AtRGS1 was phosphorylated by AtWNK8 the most under these experimental conditions (Fig. 3E, F; c.f. AtRGS1 phosphorylation to the respective AtWNK autophosphorylation band). Although AtWNK1 and AtWNK10 have lower specific activities, these kinases are clearly able to phosphorylate AtRGS1 under these in vitro conditions.
Having identified kinases that phosphorylate AtRGS1 in vitro, we next determined how these kinases affected AtRGS1 endocytosis in vivo using genetic ablation (alleles shown in Supplemental Fig. S3E and S3F). Toward this end, we measured AtRGS1 trafficking in the absence of the selected WNK kinases with and without 6% D-glucose (Fig. 3G). AtRGS1 was internalized in Col-0 and wnk1 cells, but not in the absence of AtWNK8 (wnk8-1 and wnk8-2 alleles) or AtWNK10 (wnk10-2) or both AtWNK8 and AtWNK10 (wnk8-1/wnk10-1). These results shown in Fig. 3G indicate that AtWNK8 and AtWNK10, but not WNK1, are AtRGS1 kinases that drive AtRGS1 endocytosis.
AtRGS1 endocytosis requires C-terminal phosphorylation
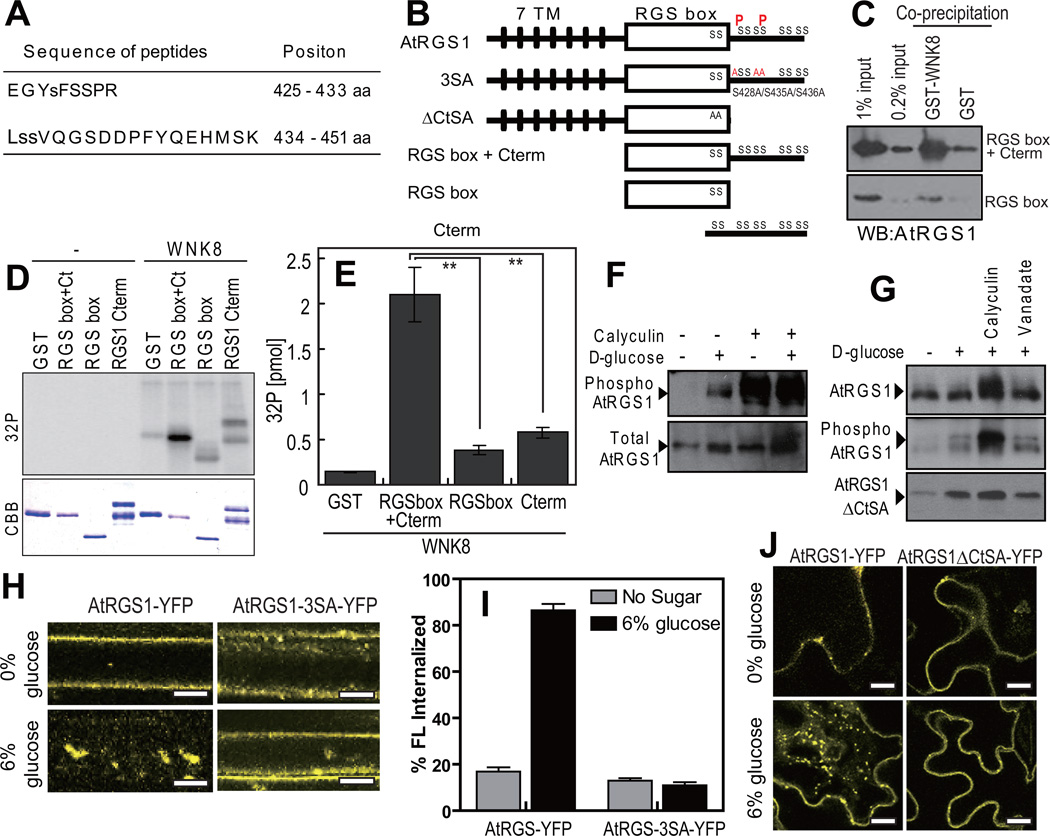
AtRGS1 has eleven serines within its fifty most C-terminal residues that are candidate phosphorylation sites. Our in vitro phosphorylation of RGSbox+Cterm showed ~2 mole 32PO4 incorporated per mole of substrate (Supplementary Fig. S4A). LS/MS/MS analysis of recombinant RGSbox+Cterm phosphorylated by AtWNK8 identified two phosphorylation sites: Ser428 and Ser435 or Ser436 (Fig. 4A, 4B and Supplementary Fig. S4C, 4D) in the AtRGS1 C-terminus. To validate the tandem MS results, we determined if AtWNK8 phosphorylates in vitro an AtRGS1 substrate lacking the C-terminal domain (AtRGS1-RGS box; 284–416 aa) and the isolated C-terminal domain (AtRGS1-Cterm; 400–459 aa). AtWNK8 preferentially phosphorylated AtRGS1 substrates that contained both the RGS and the C-terminal domains (Fig. 4D,E) and this result correlated with stronger interactions in co-precipitation assays using purified proteins (Fig. 4C). In BiFC experiments, cYFP-AtWNK8 specifically interacted with AtRGS1-nYFP but not with the mutant lacking the C-terminal domain (ΔCt; 1–416 aa, Fig. 4B; (Supplementary Fig. S4E), although both AtRGS1-cYFP and the ΔCt mutant interacted with cYFP-AtGPA1 as a positive control. These results indicate that the C-terminal domain on AtRGS1 is necessary for phosphorylation and interaction with AtWNK8 and that the RGS box plays a structural role in these processes. Because our MS analyses failed to identify phosphopeptides from the extreme C-terminus of AtRGS1 (non-highlighted sequence in Supplementary Fig. S4B), we do not exclude additional phosphorylation sites in this region. However, since we showed that the specific activity is 2 moles phosphate per mole of AtRGS1 C-terminal fragment (Supplementary Fig. S4A), additional phosphorylation sites would be mutually exclusive to the ones identified.
Figure 4. Phosphorylation and function of the carboxyl terminus of AtRGS1.
(A) Phosphorylated peptides of AtRGS1 isolated and identified by tandem mass spectrometry. Recombinant RGSbox+Cterm was phosphorylated by AtWNK8, trypsinized and subjected to LC-MS/MS as described in the Methods. (B) Schematic model of AtRGS1 mutants. Transmembrane regions are shown as black lines, AtRGS1 box as a white box and the identified phosphorylation sites are denoted “P”. (C) In vitro binding between AtRGS1 truncated mutants and AtWNK8. Recombinant RGSbox+Cterm or RGSbox of AtRGS1 was tested for interaction with GST (negative control) or GST-AtWNK8. Inputs and precipitated proteins were analyzed by immunoblot analysis using an anti-AtRGS1 antibody. (D) In vitro phosphorylation of AtRGS1 by AtWNK8. Recombinant His-tagged RGSbox+Cterm, RGSbox, or GST-AtRGS1-Cterm plus γ32P-ATP were incubated with (WNK8) or without (−) GST-AtWNK8. Radiolabelled (32P) proteins were separated by SDS-PAGE and detected as described in the Methods. (E) The levels of phosphorylation were quantified. Error bars = SEM, n = 3. **, P < 0.01. (F) In vivo phosphorylation of AtRGS1. Seedlings expressing AtRGS1-TAP were pretreated with 100 nM calyculin A for 3 h followed by 6% D-glucose for 90 min. Phosphorylation of AtRGS1 was detected by immunoblot analysis using an anti-phospho-AtRGS1 antibody or peroxidase anti-peroxidase (Total RGS1). (G) In vivo phosphorylation of AtRGS1 or the ΔCtSA mutant. TAP-tagged AtRGS1 or AtRGS1-ΔCtSA seedlings were pretreated with 100 nM calyculin A or 10 mM sodium orthovanadate for 3 h followed by 6% D-glucose stimulation for 90 min. AtRGS1 or AtRGS1-ΔCtSA lysates was separated by 12.5% Anderson’s gel and subjected to immunoblot analysis using anti-phospho-AtRGS1 antibody or peroxidase anti-peroxidase (Total RGS1, RGS1ΔCtSA). (H) Internalization of AtRGS1-YFP and AtRGS1-3×SA-YFP. AtRGS1-YFP and AtRGS1-3×SA-YFP were transiently expressed, followed by 30 min treatment in 6% glucose prior to imaging. Scale bars = 10 µm. (I) Quantitation of AtRGS1-YFP and AtRGS1-3×SA-YFP fluorescence internalization. Error = SEM, n = 5. (J) Internalization of full-length and carboxyl-terminal truncated mutant of AtRGS1. AtRGS1-YFP or AtRGS1-ΔCtSA was expressed in tobacco leaves and treated with 6% D-glucose for 30 min. WNK8 denotes AtWNK8. Scale bars = 20 µm. Quantitation of fluorescence is described in Methods.
We generated phospho-specific AtRGS1 antibodies for immunoblot analysis. In Arabidopsis seedlings lysates, AtRGS1-TAP (tandem affinity protein tagged) was phosphorylated upon D-glucose treatment (Fig. 4F). The phosphatase inhibitor, calyculin A, increased AtRGS1 phosphorylation, even without D-glucose (Fig. 4F and 4G), suggesting that AtRGS1 undergoes steady-state phosphorylation and dephosphorylation in plant cells.
Since phosphorylation of animal GPCRs is critical for their endocytosis, we engineered a C-terminal truncation mutant (AtRGS1-ΔCtSA) and a full-length AtRGS1-3SA mutant (S428A/S435A/S436A) in which three phosphorylation sites identified were mutated to alanine residues (Fig. 4B), and assayed for D-glucose dependent endocytosis. While the wild-type AtRGS1-YFP was localized to the plasma membrane and endocytosed upon D-glucose treatment, the AtRGS1-3SA mutant (Fig. 4H and 4I) or AtRGS1-ΔCtSA (Fig. 4J) remained at the plasma membrane with D-glucose in both stable and transient expression assays. While steady-state levels of phosphorylated full-length AtRGS1 increased with phosphatase inhibition by calyculin A, no equivalent change was observed for the AtRGS1-ΔCtSA mutant (Fig. 4G). These results indicate that the carboxyl-terminus on AtRGS1 is the only phosphorylation region and that carboxyl-terminal phosphorylation is required for AtRGS1 endocytosis.
Gβγ subunit interacts with AtWNK8 on the plasma membrane
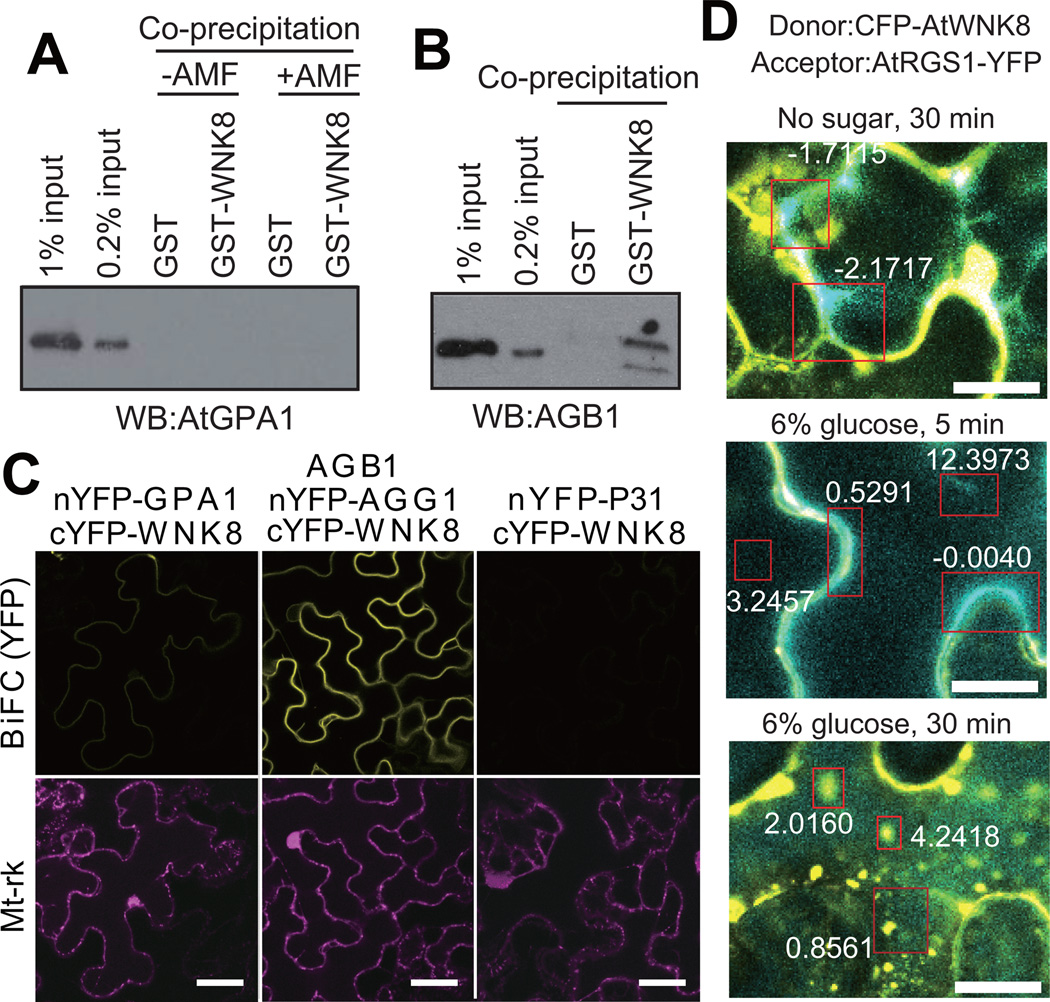
AtWNK8 is known to interact with the β-propeller protein AtRACK1 25, which has a similar structure to the Gβ protein, AGB1 33. Thus, we hypothesized that AtWNK8 directly interacted with AGB1 as well. We found, using purified components, that AtWNK8 directly interacted with AGB1/AGG1 in co-precipitation experiments, but AtWNK8 did not interact with AtGPA1 (Fig. 5A, B). Using BiFC analysis, AGB1/AGG1 strongly interacted with AtWNK8 at the plasma membrane in both stable and transient assays (Fig. 5C and Supplementary Fig. S5), although the majority of WNK8 was cytoplasmic (Fig. 5D). This indicates that the rate-limiting factor is the level of available Gβγ dimer at the plasma membrane.
Figure 5. AtWNK8 physically interacts with the G-protein βγ subunit.
(A, B) In vitro binding between AtWNK8 and heterotrimeric G protein. Inactive, GDP-bound AtGPA1, AtGPA1 activated by aluminum tetrafluoride (AMF: 50 µM GDP, 30 µM AlCl3, 10 mM MgCl2 and 5 mM NaF) or AGB1/AGG1 was co-precipitated with GST or GST-AtWNK8. Proteins were subjected to immunoblot analysis with anti-AtGPA1 or anti-AGB1 antisera. The two different amounts of input protein (0.2% or 1% of total) were loaded as reference. (C) In vivo binding between AtWNK8 and heterotrimeric G protein. nYFP-tagged AtGPA1, AGG1 with AGB1, or P31 was co-transformed with cYFP-tagged AtWNK8 and mitochondrial marker (Mt-rk, transformation control) into tobacco leaves. Fluorescence complementation of split YFP and expression of RFP were observed by confocal fluorescence microscopy. Scale bars = 50 µm. (D) CFP-AtWNK8 associates with AtRGS1-YFP. Acceptor photobleaching of CFP-AtWNK8 and AtRGS1-YFP transiently expressed in tobacco in no sugar and 6% D-glucose for the indicated times. Bleached zones are in red boxes and numbers denote the net FRET value. WNK8 denotes AtWNK8. The method for determining the FRET efficiency indicated by the respective boxes is described in the Methods. Scale bars = 20 µm.
FRET analysis showed that AtWNK8 association with AtRGS1 increased upon treatment with 6% D-glucose (Fig. 5D). Within minutes of stimulation, the AtWNK8 associated with AtRGS1 at the plasma membrane (5 min.; P<0.05) followed by the endosomes (30 min.; P<0.05). The FRET results suggest that the association of AtWNK8 and AtRGS1 is triggered by induction of glucose causing initial interaction at the plasma membrane (Fig 5D, 5 min. time point) continued with movement into endocytic compartments (Fig 5D, 30 min. time point).
AtWNK8/10 is required for activation of G protein signaling
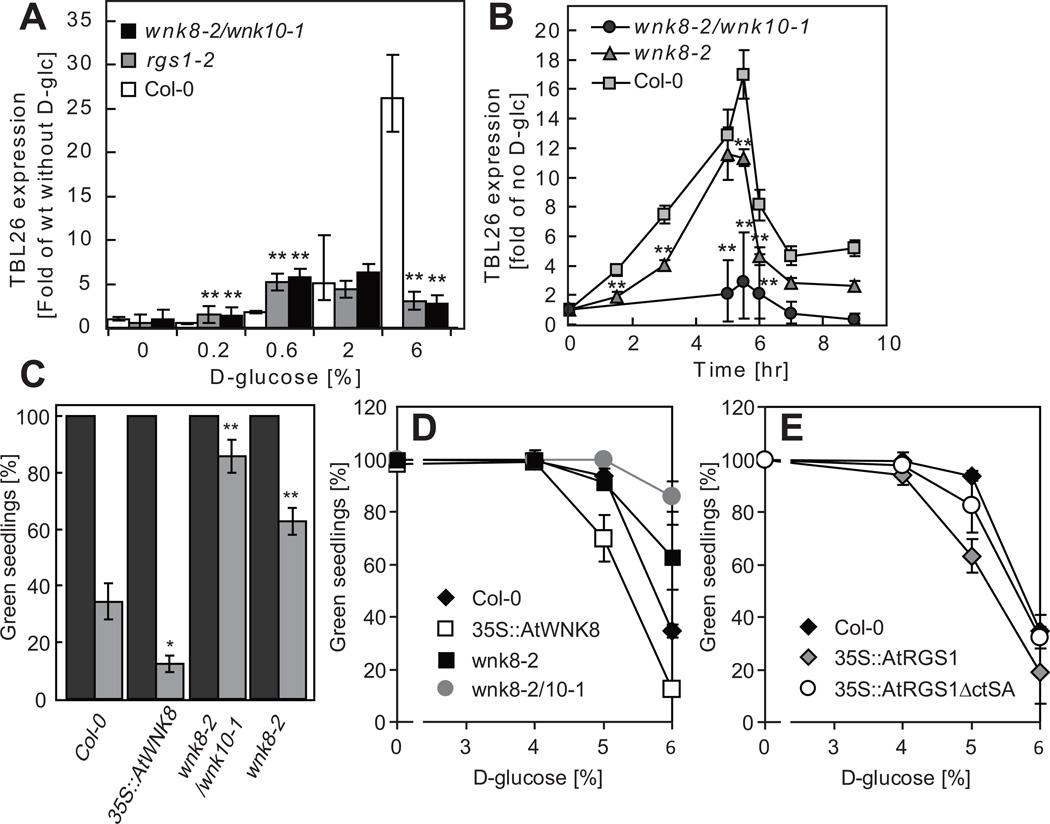
Glucose/fructose signaling through AtRGS1 is known to activate transcription of the TBL26 gene (At4G01080) 16 and therefore glucose-induced TBL26 expression is used as a reporter of AtRGS1-dependent, sugar signaling. We found that TBL26 expression in cells lacking AtRGS1 or both AtWNK8 and AtWNK10 was at the basal level, only 20% of the expression detected in wild-type cells treated with 6% D-glucose (Fig. 6A). These results were consistent with our earlier finding that AtRGS1 is endocytosed in 6% D-glucose, and minimally or not at all at lower concentration and less duration of D-glucose (Fig. 1). In the rgs1-2 or wnk8-2/wnk10-1 double mutant cells, acute D-glucose treatment failed to stimulate TBL26 expression at the wild type level (Fig. 6A). Loss of WNK8 attenuated TBL26 expression but loss of both WNK8 and 10 completely abrogated sugar signaling as reported by TBL26 expression (Fig 6A and B). Sugar-stimulated AtRGS1 endocytosis correlated with TBL26 expression.
Figure 6. wnk8 mutant expression and phenotypes.
(A) Glucose signaling as reported by gene expression analysis in wnk mutants. Dose-dependent TBL26 expression in Col-0, rgs1-2 or wnk8-2/wnk10-1. 7-day old seedlings were starved for 2 days and then treated with various concentration of D-glucose for 3 h. TBL26 transcript levels were normalized with that of Col-0 without D-glucose stimulation. Data shows mean ± S.D. (n = 3) from a representative experiment. (B) Time course of TBL26 expression and the reset timing. Seedlings were starved for 2 days and either stimulated with 6% D-glucose for 5 h or not (by moving to ½ X MS media lacking sugars). TBL26 expressions at each time period were normalized with the expression without D-glucose stimulation. Data shows mean ± S.D. (n = 3) from a representative experiment. (C, D, E) Physiological sugar-related phenotype of wnk and rgs1 mutants. Vernalized seeds of wild type col-0 or the indicated mutants were grown on ½ X MS plates containing 0%, 3%, 4%, 5% and 6% D-glucose at 23° C under continuous light (50 µmol s−1 m−2) for 10 days. The average number of seedlings having green cotyledons were determined and presented with SEM. The histogram in C provides the average plus SEM of 0% glucose (black) or 6% D-glucose (gray). Error bars = SEM. Pairwise Student’s t test was used to compare values to the Col-0. *, P < 0.05; **, P < 0.01. “35S::” represents a constitutive promoter from the Cauliflower Mosaic virus used here for ectopic overexpression of the indicated open reading frame. n (the number of independent experiments) = 2 for 0%, 4 for 6% or 3 for the other concentration of D-glucose.
Previously, we identified a subset of genes that were induced by D-glucose in an AtRGS1-independent manner 16. To determine the role of AtWNK8 in expression of these additional genes, we quantified their induction following a brief acute treatment with D-glucose. We found that induction of AtRGS1-dependent genes (TBL26 and three Jacalin-like) were attenuated in rgs1-2, gpa1-4, agb1-2, wnk8-2 or wnk10-1 (Supplementary Fig. S6A). In contrast, AtRGS1-independent genes (At4G23600 and At5G48850) were similarly induced in all genotypes (Supplementary Fig. S6A). Likewise, the time course of D-glucose stimulation and reset timing after starvation was unaffected in wnk8 null mutants (Fig. 6B and Supplementary Fig. S6B). Together, these results suggest that sugar signals are mediated by several different mechanisms, and AtWNK8 and AtWNK10 influence only the AtRGS1-dependent sugar signals. Moreover, these results suggest that AtWNK-mediated endocytosis of AtRGS1 is critical for activating G protein-mediated sugar signaling.
AtRGS1 endocytosis activates cell division
G-protein signaling is involved in cell proliferation and elongation in Arabidopsis seedlings 9, 10, 13. After 2 days of etiolated growth, gpa1 or agb1 seedlings have shorter hypocotyls than wild type seedlings because of a reduction in cell proliferation 9, 10. On the other hand, rgs1 plants and plants that express the constitutively active Gα, AtGPA1(Q222L), have longer hypocotyls 8. A visual sugar phenotype of the G mutants is growth arrest and this also is a cell division phenomenon. We used the standard “green seedling” assay in conjunction with our sugar signaling assay34 to illustrate the role of the WNK kinases. Fig. 6A shows that over a range of glucose, sugar signaling is greatly attenuated in the rgs1 single and wnk8-2/10-1 double mutants (combination of a different set of wnk8 and wnk10 alleles gave the same results). Loss of WNK8 alone slightly reduces signaling over time compared to wild type (Fig 6B). Sugar signaling as reported by TBL26 gene expression correlates with the growth behavior on acute level of D-glucose. Growth arrest on 6% glucose was more than 60% for wild type yet only 15% for the wnk8-2/10-1 double mutant and 40% for the wnk8-2 single mutant (Fig 6C). This was observed over a range of glucose concentrations (Fig 6D). Consistent with the loss-of-function mutations, ectopic over expression of either WNK8 or AtRGS1 confers the opposite effect (Fig. 6C vs. 6D for WNK8 vs. 6E for AtRGS1). The increased sensitivity to glucose conferred by AtRGS1 expression requires the C-terminus and/or the serines targeted for phosphorylation by WNK8 (Fig 6E). The role of glucose-induced AtRGS1 endocytosis fits the context of sugar-induced growth arrest and also suggests that sugar signaling for this growth behavior originates, at least in part, at the endosome.
DISCUSSION
xSignaling through theheterotrimeric G protein complex is one mechanism by which plants sense sugar levels and regulate cell proliferation and cell elongation accordingly 3, 8–11, 13, 15, 16. Unlike in humans, where the serum glucose levels are held nearly constant by a feedback mechanism involving glucose uptake and metabolism, both the apoplastic and symplastic glucose levels in plants swing over 4 orders of magnitude 35. Diurnal changes in the rate of glucose production from photosynthesis account for most of this variation in glucose concentration, but also important are long term changes such as the positions and strengths of glucose sinks such as roots, meristems, and fruits 36. Yeast, like plants, adopt growth strategies based over wide swings in glucose levels and do so, in part, through a G protein coupled signaling pathway 37–39. However, the level of glucose must be sustained for the cell to commit to an altered growth strategy.
While the rate-limiting step for G protein activation in humans is 7TM receptor-catalyzed nucleotide exchange, this process is spontaneous for the Arabidopsis Gα protein3, 7. Plant cells must therefore utilize accessory control to keep the complex in the basal state until self-activation is permitted. The protein prototype 7TM-GAP, AtRGS1, serves as a receptor or co-receptor for glucose (and/or glucose metabolites) and accelerates the intrinsic hydrolysis of its cognate Gα subunit, AtGPA1 3, 8 to keep the complex in its basal state 40.
WNKs may play a role in circadian and developmental timing in Arabidopsis 41–43 and in abiotic stress 44, 45 but until now, no specific biochemical pathway was known although recently, human WNK1 was shown to coordinate signaling through PIP2 and Gq-coupled pathways by an interesting dual mechanism 46. Of particular interest here is that WNK1 promotes clathrin-dependent endocytosis of renal ion transporters and potassium channels 47, suggesting by comparison to AtWNK8 that WNK’s regulatory role in membrane protein trafficking is its primordial function.
In animals, internalization of 7TM GPCRs desensitizes cells to the receptor agonists (Fig. 7A–C), however recent data indicate that, in some cases, signaling subsequent to endocytosis occurs at the endosome 2, 48. The results from our epistasis analyses here prompt us to propose that AtRGS1 mediates sugar signaling both via the endosome and via G protein at the plasma membrane. For example, for many sugar phenotypes, the loss-of-function alleles of the G protein complex (gpa1, agb1 or agg1/agg2) and AtWNK8 display the opposite phenotype as the rgs1 mutants, suggesting that AtWNK8-mediated AtRGS1 endocytosis drives G protein activation at the plasma membrane. In contrast, null alleles of gpa1,agb1, rgs1 and wnk8/10 all abrogate signaling as measured by TBL26 (and several others) expression, implying that internalized AtRGS1 transmits signals from the endosome.
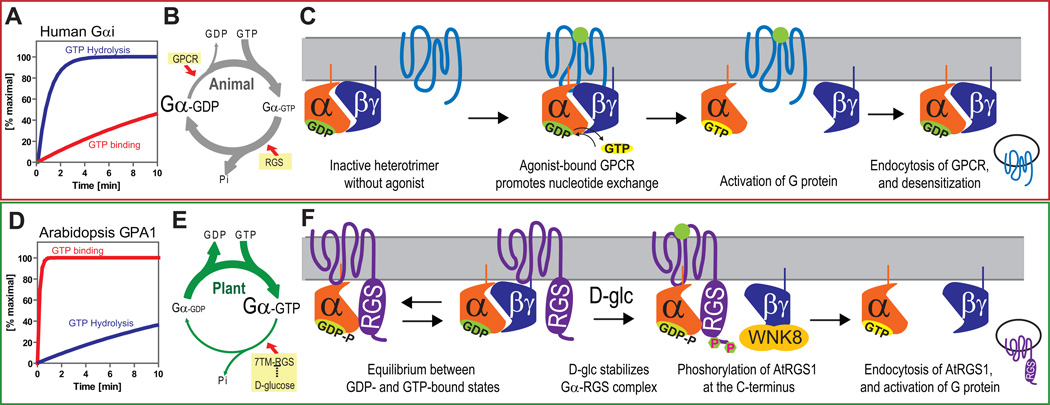
Figure 7. Model of sustained G protein activation in Arabidopsis; comparison of activation mechanisms between fast and slow nucleotide exchanging G proteins.
(A, D) Rate of guanine nucleotide exchange of human G protein is slower than that of GTP hydrolysis. However, Arabidopsis AtGPA1 rapidly releases GDP, while GTP hydrolysis is slow. (B, E) Based on these intrinsic properties, human G proteins require GPCRs to form the active GTP-bound state. In contrast, AtGPA1 requires a constitutively active 7TM-RGS protein, AtRGS1, to keep the inactive GDP-bound state. Genetic evidence suggests that D-glucose inhibits AtRGS1 to activate the Arabidopsis G protein pathway. (C, F) Many human GPCRs are endocytosed after phosphorylation and subsequent endocytosis causes desensitization of G protein signaling. In Arabidopsis, in the absence of glucose, Gα subunit binding is in equilibrium between the RGS and Gβ dimerization interfaces, both shared on the Gα subunit. Glucose shifts the equilibrium toward the Gα-RGS dimer increasing the time the Gβγ remains free from the heterotrimer. The free Gβγ dimer recruits AtWNK8 to phosphorylate AtRGS1 at its C terminus. Phosphorylation is requisite for AtRGS1 endocytosis. Endocytosis causes uncoupling of AtGPA1 from its inhibitor, AtRGS1 and subsequent sustained activation of G protein signaling.
Here we show that glucose-induced endocytosis of AtRGS1 is achieved by AtWNK8-mediated phosphorylation of at least 2 C-terminal serines. Although we show the elements, order, and mechanism of this process, one question remains. Specifically, what causes the instantaneous G protein activation that leads to the sustained activation mechanism we elucidate here? Fig. 7 illustrates a model that summarizes the key findings and our interpretation: In the absence of glucose stimulation, the plant self-activating Gα protein (Fig. 7 D, E) cycles between the GTP and GDP bound states with its binding partners, Gβγ and AtRGS1 (Fig. 7E,F). The GTPase accelerating activity of AtRGS1 promotes inactive heterotrimer formation while the self-activating property of AtGPA1 promotes association with AtRGS1 3. The newly-freed Gβγ dimer recruits AtWNK8 to the plasma membrane to phosphorylate the C-terminus of AtRGS1. Once phosphorylated, AtRGS1 undergoes endocytosis and the Gα protein is physically uncoupled from its negative regulator, left to self-activate at the plasma membrane. Each signaling component, both endosomal AtRGS1 and plasma membrane-delimited Gα and Gβγ, consequently relays signals for a sustained response to the sugar signal. The initial mechanism of Arabidopsis G protein activation is still unknown, but we previously reported that the D-glucose stimulation increased AtGPA1 association with AtRGS1 3. Because the RGS domain and Gβ have overlapping interaction interfaces on the Gα protein surface 49, 50, increased association between AtGPA1 and AtRGS1 would preclude heterotrimer formation and favor Gβγ release (Fig. 7F). An alternative hypothesis is that glucose may directly inhibit the GAP activity of AtRGS1 thus permitting Gα self-activation and heterotrimer dissociation. While glucose has no effect in vitro on the GAP activity of the RGSbox+Cterm of AtRGS1 (Supplementary Fig. S7C, D), it remains possible that sugar directly regulates GAP activity in the full-length protein or indirectly by conformation changes promoting GAP activity. Both hypotheses are consistent with the glucose-induced change in FRET efficiency between AtGPA1 and AtRGS1 3.
In conclusion, sustained plant G protein activation is regulated in an opposite manner as in animals (Fig. 7). Despite the contrasting mechanisms of G protein activation between plant and animal cells, there is an intriguing similarity in the requirement for phosphorylation in GPCR endocytosis. While we elucidated the critical role of C terminal phosphorylation of AtRGS1 for its endocytosis and provide a mechanism for sustained G protein-dependent signaling, we raised questions of whether trafficking mechanisms of mammals and of Arabidopsis 7TM proteins (e.g. GPCRs vs. 7TM-RGS) share evolutionary history, and how Arabidopsis and mammals independently combined similar functional compatibilities from different gene products to evolve receptor-trafficking mechanisms.
METHODS
Plant materials
The gpa1-3, agb1-2, gpa1-4 agb1-2 double, gpa1-4 agb1-2 agg1-1 agg2-1 quadruple, rgs1-1 and rgs1-2 mutants were described previously 8–11. The 35S::AtGPA1-Q222L gpa1-4, 35S::AtGPA1 gpa1-4, 35S::AGB1 agb1-2 were previously generated 9, 10. The 35S::AtWNK8 was provided by Dr. Thomas Eulgem (UC-Riverside). wnk1, wnk8-1, wnk8-2, wnk10-1 and wnk10-2 mutants were obtained from the ABRC stock center (SALK_015778, SALK_103318, SALK_024887, SALK_012899, and SALK_071328C). All DNA-insertion lines were made homozygous and the T-DNA insertions were confirmed by PCR of genomic DNA with the following primer sets;
wnk8-1_RP: TGCCATGAATTCAGGAGTACC,
wnk8-1_LP: AAAGATCCTTCTGGCCGTTAC,
wnk8-2_RP: TACTCCTGAATTCATGGCACC,
wnk8-2_LP: CAGCAGATCTTGGAAGGACTG,
wnk1_RP: CGCAAGACATTCTTCGAATTC,
wnk1_LP: GGGAATCAAGGAGAGGTCAAG,
wnk10_RP: TGCTCTTCTGCTAAAAGCAGC,
wnk10_LP: GGGTCCATTCCTCTCTCTCAG, and
T-DNA_LB: ATTTTGCCGATTTCGGAAC (Supplementary Fig. S3E).
Transient expression in plants
Transient expression in tobacco was as described by Sparkes, et al 51 with the following modifications to the infiltration buffer (10 mM MgCl2, 10 mM MES, 200 µM acetosyringone). Agrobacterium tumefacien was incubated in infiltration solution for at least 4 h. Images were typically captured 3 days after inoculation to obtain the lowest detectable expression. Transient expressions in Arabidopsis seedlings were as described by Grefen, et al. 52. Equal volumes of the A. tumefacien (O.D.600=1.0) harboring the plasmids were mixed just prior to the dipping step in the Arabidopsis transformation protocol.
BiFC was performed using 4–5 week-old tobacco leaves that were infected with A. tumefacien to express nYFP- and cYFP-tagged proteins as described previously 16 except with the following modifications. pENTR clones of the genes of interest were subcloned into one or more of the BiFC vectors pBatTL-sYFP-N or pBatTL-sYFP-C (for C-terminal tagged nYFP and cYFP halves, respectively) and pCL112_JO or pCL113_JO (for N-terminal tagged nYFP and cYFP halves, respectively). An internal positive transformation control (mitochondrial RFP marker; Mt-rk obtained from Arabidopsis Biological Resource Center (CD3-991)) was included to address the issue of transgene silencing (p19 gene silencing suppressor, mt-rk and two BiFC halves).
Plasmids, Recombinant Proteins, and Antibodies
cDNA for AtWNK1 was obtained from Dr. Norihito Nakamichi, and cDNAs for AtWNK8 and AtWNK10 were obtained from the Arabidopsis Biological Resource Center. AtWNK coding sequences were subcloned into pDEST15 (N-terminal GST), pCL113_JO (N-terminal split-cYFP), pACTGW-attR and pB7WGC2 (N-terminal CFP). Ara7, Rha1, RabA5d, VTI11 and SYP23 were obtained from the ABRC and subcloned into pEarleyGate101 (C-terminal YFP-HA), pEarleyGate102 (C-terminal CFP), and pEarleyGate104 (N-terminal YFP) plasmids. AtRGS1 (encoding 1–459 aa) and its C-terminal truncated mutant (encoding 1–416 aa, designated as AtRGS1-ΔCt) were subcloned to pEarleyGate205 (C-terminal TAP), pEarleyGate101 and pBAT-TL-B-sYFP-N (C-terminal split-nYFP). Coding sequences for cytoplasmic region of AtRGS1 were synthesized by Celtek Bioscience (Nashville, TN, USA) with codons optimized for expression in E. coli. AtRGS1-coding sequences RGSbox+Cterm (284–459 aa), RGSbox (284–416 aa) and Cterm (400–459 aa) of AtRGS1 were subcloned into pENTR-TEV-TOPO, pAS-attR and pDEST17 (N-terminal His) or pDEST15. AtRGS1-ΔCtSA (1–416 aa, S405A/S406A) was generated with QuickchangeTM by direct mutagenesis. Recombinant GST- or His-tagged proteins were expressed in E. coli (ArcticExpress RP, Agilent Technologies) with 0.5 mM IPTG at 12°C, solubilized in buffer A (50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 5 mM 2-Mercaptoethanol, 1 mM PMSF and 1 µg/ml leupeptin) with 0.25 mg/ml lysozyme and 0.2 % NP-40, purified from the soluble fraction using glutathione-Sepharose 4B (GE Healthcare) or TALON Metal Affinity Resin (Clonetech), washed with buffer A containing 500 mM NaCl and 0.1% sodium cholate, and eluted with 20 mM glutathione or 500 mM imidazole, respectively. For His-tagged proteins, 5 mM imidazole was included in crude extracts to reduce nonspecific binding. The purified proteins were dialyzed in buffer B (20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 1 mM EDTA, 1 mM DTT and 1 mM PMSF).
The AtGPA1 and AGB1 antibodies were described previously 10, 53. The AtRGS1 and phosphor-specific AtRGS1 antisera were raised with the His-tagged RGS1box+Cterm protein or the phosphorylated AtRGS1 peptide (424–440 aa): CKEGY-pS-FSSPRL-pS-pS-VQGS (pS; phosphorylated-serine) (YenZym Antibodies). The anti-phospho-AtRGS1 antibody was purified by affinity chromatography using the phosphorylated AtRGS1 peptide conjugated to affinity matrix. The purified antibody was further cleaned up with unphosphorylated-AtRGS1 peptide (424–440 aa) to remove phosphorylation-independent AtRGS1 antibody. The specificity of the antibody against phosphorylated- and unphosphorylated-AtRGS1 peptides was checked with ELISA. Scans of blots used in figures here are shown in Supplemental Figure S8.
Microscopies
Vertical optical sections (Z stacks) of hypocotyl epidermal cells of dark grown seedlings located approximately. 3–4 mm from the cotyledon were imaged using a Zeiss LSM710 confocal laser scanning microscope equipped with a Plan-NeoFluor 20×/0.5 objective and a C-Apochromat 40×/1.20 water immersion objective. YFP, CFP and RFP were excited by a 514nm and 458nm Argon laser and a 560nm diode laser, respectively, and their respective emissions were detected at 526–569, 460–520 and 565–621 nm by a photomultiplier detector. The digital images were analyzed with Zen software (Zeiss).
Some fluorescence images of tobacco transients and Arabidopsis seedlings were captured by using an Olympus IX81 inverted microscope (Olympus America, Merville, NY) with a Hamamatsu CCD C4742-80-12AG (Hamamatsu Photonics, Bridgewater, NJ) and analyzed using Slidebook 5.0 (3i). Filter sets used were YFP (excitation, 500/20 nm; emission, 535/30 nm; same filter for GFP-tag), CFP (excitation, 436/20 nm; emission, 480/40 nm).
FRET measurements in CFP/YFP double labeled cells was determined by the acceptor photobleaching method. The intracellular region of interest (ROI) varied in size depending on the substructure analyzed but fell into two categories: (i) initial regions at or adjacent the plasma membrane, and (ii) regions inside the cell or not adjacent to the plasma membrane. The laser intensity and duration was optimized to achieve at least 80% YFP photobleaching without affecting donor emission quantum yield. CFP and YFP were excited by the 458 nm and 514 nm lines of an Argon laser, respectively, Emissions were detected at 460–520 and 526–569 nm. Pre- (Ipre.λ) and post-bleaching (Ipost.λ) fluorescence intensities of both CFP and YFP were obtained and the FRET efficiencies in the ROI were calculated using the following equation:
The digital images and FRET analysis were analyzed with Zen software (Zeiss).
Quantitation and Statistics
Fluorescence quantitation was obtained using the software ImageJ 54. Randomly selected hypocotyl images from at least five (5) whole Z-section image stacks from 3 independent experiments were selected for quantification. Images were thresholded for fluorescence signal prior to selecting regions. Boxed regions of fluorescence from within cells were selected and subtracted from the total hypocotyl fluorescence of the hypocotyl in the image (fluorescence not in the hypocotyl was excluded from measurements). Boxed regions were chosen distant from the cell periphery as determined in the bright field/DIC images.
Statistical comparison of mean fluorescence was performed using Analysis of Variance to confirm differences in treatments and genotypes followed by mean analysis by Tukey’s test. Typically, a minimum of 5 Z-sections of the root were randomly selected and used for quantitation. There is little variation between different Z sections of cells in the scan range which was typically 20–30 nm from root surface.
In vitro Co-precipitations
Seventy five pmol of His-RGSbox+Cterm and 50 pmol of GST-AtWNK kinases were mixed with 10 µl of glutathione-Sepharose 4B (GE Healthcare) in 250 µl of buffer C (50 mM MES (pH 6.0), 10 mM MgCl2, 5 mM MnCl2, 2 mM DTT, 1 mM PMSF and 1 µg/ml leupeptin). The tubes were rotated for 60 min at 4°C, and the resin was washed with buffer C with 0.1% NP-40 four times. The co-precipitated or input proteins were visualized by immunoblot analysis with anti-AtRGS1 antisera and Coomassie Brilliant Blue (CBB) staining. To test binding with AtGPA1, the same procedure was followed except that 50 µM GDP with or without aluminum tetrafloride (30 µM AlCl3, 10 mM MgCl2 and 5 mM NaF) was added into buffer C.
Yeast Two Hybrid
pAS-RGSbox+Cterm, RGSbox or Cterm of AtRGS1 and pACTGW-AtWNK1, AtWNK8 or AtWNK10 were transformed into AH109 yeast cells. The colonies grown on SD (-LW) plates were inoculated into SD (-LW) liquid media and cultured overnight at 30°C. The β-galactosidase activity of yeast was determined per the manufacturer’s instruction (Clonetech).
Physiological Assay
Approximately 40 seeds of Arabidopsis were sterilized, vernalized and sown on ½ X MS plates containing 0%, 3%, 4%, 5% and 6% D-glucose. The plates were incubated at 23°C under continuous light (50 µmol s−1 m−2) for 10 days for 0%, 3%,5% and 6%. Number of green seedlings was divided by number of germinated-seeds. Score was presented with SEM.
In vitro and in vivo Phosphorylation Assays
Seventy-five pmol of His-RGSbox+Cterm, -RGSbox or GST-AtRGS1-Cterm was incubated with 0.05 pmol of GST, GST-AtWNK1, GST-AtWNK8 or GST-AtWNK10 in 15 µl of buffer C containing 0.2 mM [γ-32P]ATP for 6 h at room temperature. The reactions were terminated by adding Laemmli sample buffer and separated by SDS-PAGE. Incorporation of 32P into the separated proteins was visualized with a phosphor screen (Molecular Dynamics).
Approximately 50 sterilized and stratified seeds were grown in ½ X MS liquid media with 1% sucrose at 23°C, at 100 rpm under low light (50µE) condition. After 7 days culture, seedlings were starved of sugars with ½ X MS media lacking sucrose for 2 days in a dark chamber, and then stimulated with 6% D-glucose, 100 nM calyculin A and 10 mM sodium orthovanadate, as indicated. The seedlings were frozen, powdered with mortar and pestle then lysed with buffer D (150 mM Tris-HCl (pH 7.5), 300 mM NaCl, 20% glycerol, 5 mM DTT, 2 mM Na3VO4, 10 mM NaF, 20 mM β-glycerophosphate, 1 mM PMSF, 2 µg/ml leupeptin and 1% ASB-14). After 2 h incubation at 4°C, the lysates were centrifuged and the supernatants were subjected to SDS-PAGE and immunoblot analysis. The phosphorylated AtRGS1 was detected with anti-phospho-RGS1 antibody. Total AtRGS1-TAP amount was determined with peroxidase anti-peroxidase soluble complex (PAP). To observe a band shift of AtRGS1-TAP, AtRGS1-ΔCtSA-TAP or AtGPA1, total extracts were separated with 12.5% Anderson’s gel 55 and subjected to SDS-PAGE and immunoblot analyses using PAP or anti-AtGPA1 antisera.
Quantitative Real Time PCR
Approximately 150 seeds were grown in ½ X MS liquid media with 1% glucose at 23°C with low light (50µE). After 7 days culture, seedlings were starved with ½ XMS media for 2 days in the dark. Then, seedlings were stimulated with 6% sugar for 3 h in dark condition. The mRNA and cDNA were prepared with RNAeasyTM (Qiagen) and Superscript IIITM (Invitrogen) per the manufacturer’s instruction. Expression of TBL26 and TUB4 were analyzed by real-time PCR with SYBRgreenTM or Taqman-probes using the following primer and probe sets. TBL26-fw; CGCCATCGAACCTTCGTCAAATTC, TBL26-rv; TCGTCCATTCAATAGGCAGTTCTGA, TBL26-Taqman;CTCCCGGAAACGTTTCATCAGCAAAG, TUB4-fw; AGAGGTTGACGAGCAGATGA, TUB4-rv; ACCAATGAAAGTAGACGCCA, TUB4-Taqman; CCCAAACAACGTCAAGTCCAGTGTCTGT.
Mass Spectrometry
Phosphorylated protein bands were excised from 10% polyacrylamide gels (Biorad) and digested as described previously 56. The trypsinized peptides were analyzed by MALDI-TOF/TOF and by LC-ESI-MS/MS. A 90-min gradient was used and MS analysis was performed with an LTQ Orbitrap XL mass spectrometer (ThermoFisher Scientific). Peptide masses were queried using the MASCOT algorithm against the nonredundant NCBI database and AtRGS1 specific database with possible phosphorylations. Based on MASCOT results, two phosphorylated peptides (EGYSFSSPR and LSSVQGSDDPFYQEHMSK) were identified. Based on manual inspection of the spectra, the phosphorylation sites were defined.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. David Smalley for determining the phosphorylation sites of AtRGS1, Dr. Norihito Nakamichi, Dr. Thomas Eulgem, and Dr. Hong Ma for supplying experimental materials, Kaitlin Williamson and Melissa Mathew for assistance in protein purification and gene cloning, Ariko Urano for assisting in experiments and Dr. Henrik Dohlman for helpful discussions. This work was supported by grants from the NIGMS (R01GM065989) and NSF (MCB-0723515 and MCB-0718202) to A.M.J. The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through the grant DE-FG02-05er15671 to A.M.J. funded the genotyping of the materials in this study.
Abbreviations
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GPCR
G protein-coupled receptor
- GRK
G protein-coupled receptor kinase
- GST
glutathione S-transferase
- RGS
regulators of G protein signaling
- TAP
tandem affinity purification
- WNK
With No K-Lysine kinase
Footnotes
Accession Numbers
| wnk1 | SALK_015778 |
| wnk8-1 | SALK_103318 |
| wnk8-2 | SALK_024887 |
| wnk10-1 | SALK_012899 |
| wnk10-2 | SALK_071328 |
AUTHOR CONTRIBUTIONS
A.M.J. and J.H. made the initial observation of glucose-dependent AtRGS1 internalization. J.H., J.G. and J.P.T. characterized the sugar dependency of endocytosis of wild type and mutant AtRGS1 (Fig. 1). N.P. quantitated endocytosis of AtRGS1 in mutant genotypes and performed FRET analyses and co-localization of AtRGS1 with compartment markers. J.C.J. assisted in the initial intrinsic tryptophan fluorescence measurement. J.Y. assisted in the real-time PCR experiments. D.U. designed most of the experiments, performed all the biochemical and physiological analyses, performed some of the BiFC experiments and analyses, performed all the gene expression profiling, and constructed all the expression vectors. A.M.J., D.U. and N.P. wrote the manuscript. All authors edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Kohout TA, Lefkowitz RJ. Regulation of G protein-coupled receptor kinases and arrestins during receptor desensitization. Mol Pharmacol. 2003;63:9–18. doi: 10.1124/mol.63.1.9. [DOI] [PubMed] [Google Scholar]
- 2.Hanyaloglu AC, von Zastrow M. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu Rev Pharmacol Toxicol. 2008;48:537–568. doi: 10.1146/annurev.pharmtox.48.113006.094830. [DOI] [PubMed] [Google Scholar]
- 3.Johnston CA, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proc Natl Acad Sci U S A. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano D, et al. G Protein Activation without a GEF in the Plant Kingdom. PLoS Genet. 2012;8:e1002756. doi: 10.1371/journal.pgen.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gookin TE, Kim J, Assmann SM. Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice, and poplar: computational prediction and in-vivo protein coupling. Genome Biol. 2008;9:R120. doi: 10.1186/gb-2008-9-7-r120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriyama EN, Strope PK, Opiyo SO, Chen Z, Jones AM. Mining the Arabidopsis thaliana genome for highly-divergent seven transmembrane receptors. Genome Biol. 2006;7:R96. doi: 10.1186/gb-2006-7-10-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones JC, et al. The crystal structure of a self-activating G protein alpha subunit reveals its distinct mechanism of signal initiation. Sci Signal. 2011;4:ra8. doi: 10.1126/scisignal.2001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JG, et al. A seven-transmembrane RGS protein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- 9.Ullah H, et al. The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ullah H, et al. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–2069. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- 11.Trusov Y, et al. Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakravorty D, et al. An atypical heterotrimeric G protein gamma subunit is involved in guard cell K(+) channel regulation and morphological development in Arabidopsis thaliana. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04638.x. [DOI] [PubMed] [Google Scholar]
- 13.Chen JG, Gao Y, Jones AM. Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 2006;141:887–897. doi: 10.1104/pp.106.079202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JG, Jones AM. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 2004;389:338–350. doi: 10.1016/S0076-6879(04)89020-7. [DOI] [PubMed] [Google Scholar]
- 15.Booker KS, Schwarz J, Garrett MB, Jones AM. Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigston JC, et al. D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett. 2008;582:3577–3584. doi: 10.1016/j.febslet.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pego JV, Smeekens SC. Plant fructokinases: a sweet family get-together. Trends Plant Sci. 2000;5:531–536. doi: 10.1016/s1360-1385(00)01783-0. [DOI] [PubMed] [Google Scholar]
- 18.Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J Exp Bot. 2003;54:525–531. doi: 10.1093/jxb/erg055. [DOI] [PubMed] [Google Scholar]
- 19.Valpuesta V, Botella MA. Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends Plant Sci. 2004;9:573–577. doi: 10.1016/j.tplants.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, et al. Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol. 2006;60:583–597. doi: 10.1007/s11103-005-5082-x. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z, et al. Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell. 2009;21:1972–1991. doi: 10.1105/tpc.108.062653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hislop JN, von Zastrow M. Role of ubiquitination in endocytic trafficking of G-protein-coupled receptors. Traffic. 2011;12:137–148. doi: 10.1111/j.1600-0854.2010.01121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hislop JN, Zastrow M. In: Regulated Membrane Trafficking and Proteolysis of GPCRs The G Protein-Coupled Receptors Handbook. Devi LA, editor. Humana Press; 2005. pp. 95–105. [Google Scholar]
- 24.Marchese A, Paing MM, Temple BR, Trejo J. G protein-coupled receptor sorting to endosomes and lysosomes. Annual review of pharmacology and toxicology. 2008;48:601–629. doi: 10.1146/annurev.pharmtox.48.113006.094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klopffleisch K, et al. Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol Syst Biol. 2011;7:532. doi: 10.1038/msb.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 27.Moore MJ, Soltis PS, Bell CD, Burleigh JG, Soltis DE. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc Natl Acad Sci U S A. 2010;107:4623–4628. doi: 10.1073/pnas.0907801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stubbs MD, et al. Purification and properties of Arabidopsis thaliana type 1 protein phosphatase (PP1) Biochim Biophys Acta. 2001;1550:52–63. doi: 10.1016/s0167-4838(01)00267-9. [DOI] [PubMed] [Google Scholar]
- 29.Seki M, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- 30.Day PW, Wedegaertner PB, Benovic JL. Analysis of G-protein-coupled receptor kinase RGS homology domains. Methods Enzymol. 2004;390:295–310. doi: 10.1016/S0076-6879(04)90019-5. [DOI] [PubMed] [Google Scholar]
- 31.Lalonde S, et al. A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol. 2010;1:24. doi: 10.3389/fphys.2010.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang CL, Cha SK, Wang HR, Xie J, Cobb MH. WNKs: protein kinases with a unique kinase domain. Exp Mol Med. 2007;39:565–573. doi: 10.1038/emm.2007.62. [DOI] [PubMed] [Google Scholar]
- 33.Chen JG, et al. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006;57:2697–2708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- 34.Grigston JC, et al. D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett. 2008;582:3577–3584. doi: 10.1016/j.febslet.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deuschle K, et al. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell. 2006;18:2314–2325. doi: 10.1105/tpc.106.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lalonde S, Wipf D, Frommer WB. Transport mechanisms for organic forms of carbon and nitrogen between source and sink. Annu Rev Plant Biol. 2004;55:341–372. doi: 10.1146/annurev.arplant.55.031903.141758. [DOI] [PubMed] [Google Scholar]
- 37.Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 2004;16:293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a G alpha subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 39.Versele M, de Winde JH, Thevelein JM. A novel regulator of G protein signalling in yeast, Rgs2, downregulates glucose-activation of the cAMP pathway through direct inhibition of Gpa2. Embo J. 1999;18:5577–5591. doi: 10.1093/emboj/18.20.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones JC, Temple BR, Jones AM, Dohlman HG. Functional reconstitution of an atypical G protein heterotrimer and regulator of G protein signaling protein (RGS1) from Arabidopsis thaliana. J Biol Chem. 2011;286:13143–13150. doi: 10.1074/jbc.M110.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murakami-Kojima M, Nakamichi N, Yamashino T, Mizuno T. The APRR3 component of the clock-associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant Cell Physiol. 2002;43:675–683. doi: 10.1093/pcp/pcf084. [DOI] [PubMed] [Google Scholar]
- 42.Park HY, et al. EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol. 2011;52:1376–1388. doi: 10.1093/pcp/pcr084. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, et al. The plant WNK gene family and regulation of flowering time in Arabidopsis. Plant Biol (Stuttg) 2008;10:548–562. doi: 10.1111/j.1438-8677.2008.00072.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Suo H, Zhuang C, Ma H, Yan X. Overexpression of the soybean GmWNK1 altered the sensitivity to salt and osmotic stress in Arabidopsis. J Plant Physiol. 2011 doi: 10.1016/j.jplph.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 45.Kumar K, Rao KP, Biswas DK, Sinha AK. Rice WNK1 is regulated by abiotic stress and involved in internal circadian rhythm. Plant Signal Behav. 2011;6:316–320. doi: 10.4161/psb.6.3.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An SW, et al. WNK1 Promotes PIP2 Synthesis to Coordinate Growth Factor and GPCR-G(q) Signaling. Curr Biol. 2011;21:1979–1987. doi: 10.1016/j.cub.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang CL, Yang SS, Lin SH. Mechanism of regulation of renal ion transport by WNK kinases. Curr Opin Nephrol Hypertens. 2008;17:519–525. doi: 10.1097/MNH.0b013e32830dd580. [DOI] [PubMed] [Google Scholar]
- 48.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular Mechanism of β-Arrestin-Biased Agonism at Seven-Transmembrane Receptors. Annu Rev Pharmacol Toxicol. 2011;52:179–197. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4--activated G(i alpha1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 50.Lambright DG, et al. The 2.0 A crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 51.Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 52.Grefen C, et al. A ubiquitin-10 promoter-based vector set for fluorescent protein tagging facilitates temporal stability and native protein distribution in transient and stable expression studies. Plant J. 2010;64:355–365. doi: 10.1111/j.1365-313X.2010.04322.x. [DOI] [PubMed] [Google Scholar]
- 53.Friedman EJ, et al. Acireductone dioxygenase 1 (ARD1) is an effector of the heterotrimeric G Protein {beta} subunit in Arabidopsis. J Biol Chem. 2011;286:30107–30118. doi: 10.1074/jbc.M111.227256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 55.Anderson CW, Baum PR, Gesteland RF. Processing of adenovirus 2-induced proteins. J Virol. 1973;12:241–252. doi: 10.1128/jvi.12.2.241-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanna SL, Sherman NE, Kinter MT, Goldberg JB. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology. 2000;146(Pt 10):2495–2508. doi: 10.1099/00221287-146-10-2495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.