Abstract
Ricin A chain (RTA) depurinates the sarcin/ricin loop (SRL) of 28S ribosomal RNA and inhibits protein synthesis in mammalian cells. In yeast, the ribosomal stalk facilitates the interaction of RTA with the ribosome and subsequent depurination. Despite homology between the stalk structures from yeast and humans there are notable differences. The human ribosomal stalk contains two identical heterodimers of P1/P2 bound to P0, while the yeast stalk consists of two different heterodimers, P1α/P2β and P2α/P1β, bound to P0. RTA exhibits higher activity towards mammalian ribosomes than ribosomes from other organisms, suggesting that the mode of interaction with ribosomes may vary. Here we examine whether the human ribosomal stalk proteins facilitate the interaction of RTA with human ribosomes and subsequent depurination of the SRL. Using siRNA-mediated knockdown of P1/P2 expression in human cells, we demonstrate that the depurination activity of RTA is lower when P1 and P2 protein levels are reduced. Ribosomes from P1/P2-depleted cells have a reduced ability to bind RTA by Biacore analysis, which correlates with reduced depurination activity both in vitro and inside cells. RTA interacts directly with recombinant human P1/P2 dimer, further demonstrating the importance of the human P1/P2 proteins in enabling RTA to bind and depurinate human ribosomes.
Keywords: ricin, ribosome, P protein, sarcin/ricin loop, ribosomal stalk
INTRODUCTION
Ricin, a naturally occurring plant protein found in castor beans (Ricinus communis), is highly toxic to mammalian cells and is a concern for bioterrorism [1]. Due to its toxicity to cancer cells, ricin has also been used in the design of novel cancer therapeutics [1]. Ricin is a ribosome inactivating protein (RIP), which consists of a 32 kDa enzymatically active A subunit (RTA) and a 34 kDa galactose/N-acetylgalactosamine-binding B subunit (RTB) coupled by a single disulfide bond [2]. Following endocyctic uptake, a limited portion of ricin proceeds to the trans-Golgi network, where it undergoes retrograde trafficking to the endoplasmic reticulum (ER) [3]. In the ER lumen, a resident disulfide isomerase facilitates reductive separation of RTA and RTB [4]. RTA is able to dislocate from the ER into the cytosol and interact with ribosomes [5]. RTA is an N-glycosidase, which specifically cleaves an adenine (A4324 in rat ribosomes) residue of the universally conserved sarcin/ricin loop (SRL) of the 28S rRNA, a process termed depurination [6]. The SRL region is critical for ribosome function as it facilitates binding and activation of translational GTPases that regulate protein synthesis [7]. Currently, there is no FDA approved vaccine or a therapeutic that can protect against ricin or related RIPs, such as Shiga toxins, which share similar modes of action. Therefore, understanding interactions of ricin with human ribosomes is critical for developing therapeutics against intoxication.
In a yeast model, we have shown that RTA binds to a component of the large ribosomal subunit known as the stalk, to depurinate the SRL [8, 9]. The ribosomal stalk regulates protein synthesis by promoting translation factor GTPase activity and binding to the ribosome [10–12]. The stalk structure is universally conserved among all domains of life. However, the acidic phosphoproteins (P-proteins) that comprise the stalk display species-specific adaptation, which suggests that they may confer distinct functional features to the ribosomes of each species [13–15]. While there is significant homology between eukaryotic P-proteins, the bacterial proteins represent a distinct evolutionary group [16]. The human ribosomal stalk consists of two identical dimers (P1/P2) bound to P0 in a pentameric configuration P0-(P1/P2)2. In contrast, the yeast ribosomal stalk is a pentameric protein complex comprised of a single P0 protein bound by two different heterodimers of acidic ribosomal P1/P2 proteins, P1α/P2β and P1β/P2α [17]. Human P1 shares 46.5% and 40.4% amino acid sequence identity with yeast P1α and P1β, respectively, while human P2 shares 53% and 56% amino acid sequence identity with yeast P2α and P2β, respectively. The P1/P2 proteins form stable, functionally relevant heterodimers [18], with the P1 proteins anchoring the dimers at two independent sites on the P0 protein [19, 20]. The N-terminal domains of P1/P2 proteins are responsible for dimerization and binding to P0 via the P1 proteins, while the C-terminal domains are mobile in the cytosol and interact with translational GTPases [21, 22]. A distinctive feature of P-proteins is a cluster of acidic and hydrophobic amino acids at their C-termini, and the last 11 residues (SDDDMGFGLFD) are almost identical in all organisms. This highly conserved peptide, appears to be involved in stalk activity during protein synthesis [23, 24] and interacts with several RIPs, including trichosanthin [25, 26] and Shiga toxin 1 in vitro [27, 28]. However, since the ribosomal P proteins are part of a P1/P2 complex and are not found individually in free form in the cytoplasm [29], the significance of these interactions for ribosome depurination inside cells is not well understood.
Although RTA interacts with yeast stalk proteins, which contain C-termini that are highly conserved among eukaryotes, RTA differs in its activity towards ribosomes from different organisms. Mammalian ribosomes are most sensitive to the action of RTA, yeast ribosomes are less sensitive, while prokaryotic ribosomes are resistant [30, 31]. Additionally, RTA is 23,000 times more active on rat liver ribosomes than on plant ribosomes [32], despite the fact that plant P-proteins contain the almost identical C-terminal 11 residues. These observations suggest that the mode of interaction between RTA and ribosomes may vary among different species. Here we examine the interaction of RTA with human ribosomes to determine whether P-proteins are important for RTA activity in human cells. We present the first evidence that the ribosomal stalk is required for ribosome binding and depurination by ricin in human cells.
RESULTS
Effect of P-protein depletion in human cells on depurination activity of ricin
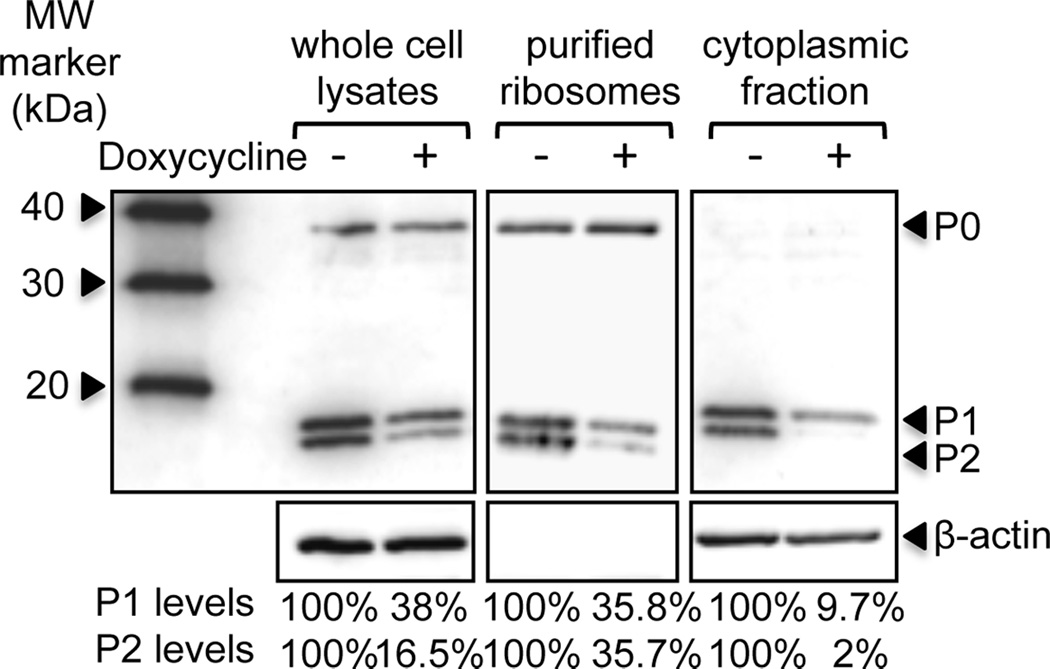
To examine the role of the human stalk proteins in ricin activity we used human embryonic kidney cells (HEK293T) stably transfected with a doxycycline-inducible construct, which produces siRNA specific for P2 mRNA [33]. To knock-down P2 expression, P2 siRNA-transfected cells were treated with 0.1µg/ml doxycycline for 96 h, using a previously established protocol to effectively reduce P-protein levels without affecting cell viability [33]. In response to doxycycline treatment, cellular and ribosomal P2 protein levels typically decreased by ~84% and ~64%, respectively (Fig. 1). Since P1/P2 complex is found free in the cytoplasm and can exchange with ribosome bound P1/P2 [29], we analyzed P1/P2 levels in the cytoplasmic fraction. Cytoplasmic P2 levels were reduced by 98% (Fig. 1). P2 depletion induced a concomitant decrease in P1 protein levels (Fig. 1) due to instability of P1 in the absence of P2 in human cells [33]. In contrast, the amount of P0 was unaffected [33].
Fig. 1.
siRNA-mediated silencing of P-protein expression in HEK293T cells. HEK293T cells stably transected with doxycycline-inducible P2 RNAi were treated with 0.1µg/ml doxycycline for 96 h to knockdown expression of P2 protein. Whole cell lysates, purified ribosomes and cytoplasmic fractions were prepared post-treatment and separated by SDS-PAGE. Proteins were transferred to nitrocellulose, and probed with monoclonal antibody against the conserved C-termini of P-proteins (3BH5). P1 and P2 protein levels are expressed as a percentage relative to levels in undepleted cells. Levels were calculated from the band intensities, which were normalized to either P0 (whole cell lysates and purified ribosomes) or ß-actin (cytoplasmic fraction). Representative blots from 2–3 independent experiments are shown.
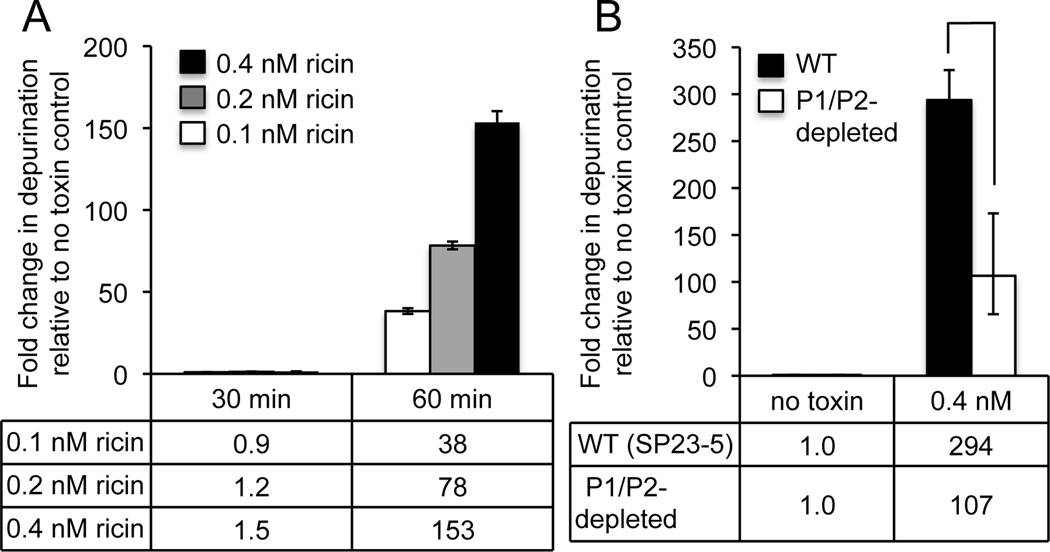
Preliminary experiments were performed to establish the relative sensitivity of HEK293T cells to ricin. Cells were treated with 0.1–0.4 nM ricin over a time-course of 0–90 minutes and depurination was measured using a previously described qRT-PCR assay [34, 35]. A dose-dependent increase in depurination could be detected by 60 minutes with increasing concentrations of ricin (Fig. 2A). To determine if depletion of P1/P2 affected depurination of ribosomes by ricin, undepleted and P1/P2-depleted HEK293T cells were treated with 0.4 nM ricin for 1 h (Fig. 2B). Treatment of WT cells with ricin yielded a high level of depurination (294–fold increase). In comparison, depurination levels were markedly lower in P1/P2-depleted cells (P < 0.05, one-tailed paired t test), with only a 107-fold change in depurination relative to the no toxin control. Similar analysis of parental HEK293T cells that lack P2-siRNA confirmed that the reduction in depurination was P2-siRNA-dependent and not related to doxycycline treatment (Fig. S1). Despite only partial depletion of P1/P2 proteins, a 2.7-fold reduction in depurination was observed, clearly implicating P1/P2 proteins in contributing to ricin-dependent depurination of human ribosomes.
Fig. 2.
Depurination activity of ricin inside human cells. (A) HEK293T cells were treated with either 0.1, 0.2 or 0.4 nM ricin for 0–60 minutes and depurination levels measured by qRT-PCR analysis using the comparative cycle threshold (ΔΔCt) method. Data indicate the fold increase in depurination relative to untreated (no toxin) cells. The mean ± SD of triplicate reactions is shown. (B) P2 RNAi-transfected HEK293T cells were pre-treated with or without doxycycline (0.1µg/ml for 96h), followed by treatment with 0.1 nM or 0.4 nM ricin for 60 minutes. Data indicate the fold increase in depurination relative to untreated (no toxin) cells and represent the mean ± SD of triplicate reactions from two independent experiments. * indicates difference was significant (P < 0.05), as analyzed by a Student’s paired, one-tailed t test.
Effect of P-protein depletion on the binding of RTA to mammalian ribosomes
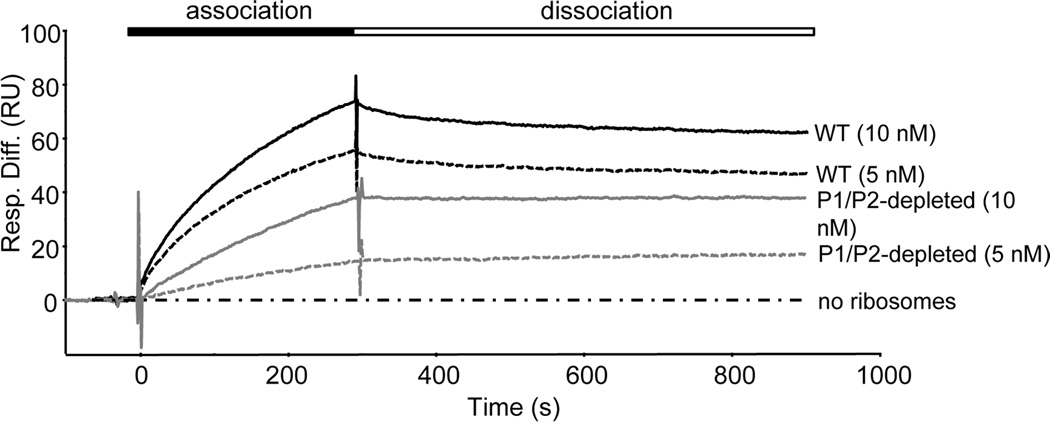
We sought to investigate the molecular basis for the decrease in toxin activity in P1/P2-depleted cells by examining the ability of RTA to bind to depleted ribosomes. The HEK293T cells stably transfected with inducible P2 siRNA were treated with or without doxycycline and monomeric salt-washed ribosomes were isolated for comparison. Depleted ribosomes typically displayed a 64% reduction in P1/P2 levels (Fig 1A). The interaction between RTA and P1/P2-depleted and control ribosomes was examined by surface plasmon resonance (SPR) with a Biacore 3000, as previously described [8, 36]. For these experiments, His-tagged RTA was immobilized on to the surface of a Ni2+–nitrilotriacetic acid (NTA) chip and an analyte of purified monomeric ribosomes was passed over the surface. The N-terminal His-tagged EGFP was immobilized on a reference channel and analyzed in parallel as a negative control. WT ribosomes at 10 nM and 5 nM exhibited rapid association with RTA during the first 30–50s of the association period, followed by a slower association phase, reaching 73 and 55 resonance units (RU), respectively. Similarly, a brief, rapid dissociation phase was followed by a very slow dissociation phase (Fig. 3). In contrast, only slow association and dissociation phases were observed with the P1/P2-depleted ribosomes, reaching only 38 RU at 10 nM and 14 RU at 5 nM. Negligible dissociation of P1/P2-depleted ribosomes and RTA was observed over a 10-minute period. These results showed that both association and dissociation of RTA with ribosomes were reduced when P1/P2 proteins were depleted.
Fig. 3.
Analysis of the interaction of RTA with WT and P1/P2-depleted human ribosomes measured using surface plasmon resonance. N-terminal His-tagged RTA was coupled to an NTA chip on the Biacore 3000 and used as a ligand for WT or P1/P2-depleted ribosomes from HEK293T cells. As a control for non-specific binding, the interaction of ribosomes with N-terminal His-tagged EGFP coupled to a reference surface was measured in parallel and subtracted from the signal obtained for the target surface with RTA. Ribosomes (5 or 10 nM) or running buffer were passed over the target and the reference surface.
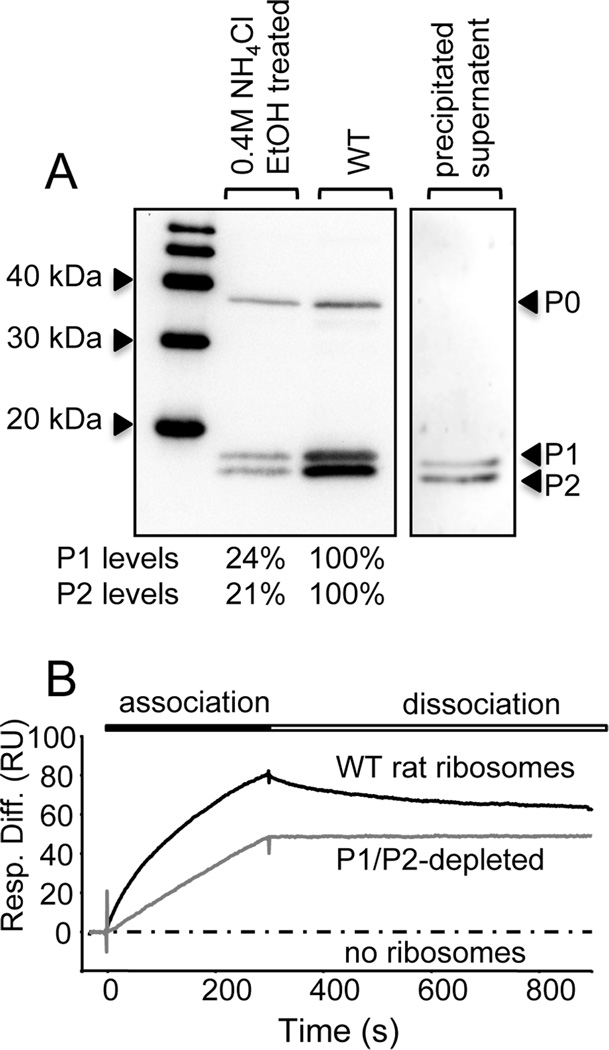
We sought to verify these results using an independent means of depletion by exploiting the unique acidic nature of ribosomal P-proteins. P1/P2 proteins can be efficiently and selectively depleted from ribosomes by NH4Cl-ethanol treatment [37]. P-proteins from rat liver ribosomes show greater than 97% sequence identity to human P1 and P2, and the last 11 C-terminal residues are identical to the human P-proteins. Since ribosomes can be isolated from rat liver at a higher yield than from cultured cells, rat liver ribosomes were chemically treated with 0.4 M NH4Cl and ethanol, as described in the Experimental procedures. The resultant ribosomes displayed a 76–79% reduction in P1/P2 levels compared to WT rat liver ribosomes (Fig. 4A). SPR analysis demonstrated that as observed with human ribosomes (Fig. 3), WT rat ribosomes also exhibited heterogeneous interaction profiles with RTA, consisting of an initial rapid association phase, followed by a slower association phase, reaching 80 RU after 5 minutes (Fig. 4B). Although there was a brief, rapid decrease in binding in the dissociation phase, the ribosomes dissociated slowly from RTA (Fig. 4B). The interaction profile of P1/P2-depleted rat ribosomes and RTA did not exhibit the same heterogeneity as WT ribosomes. P1/P2-depleted rat ribosomes displayed significantly slower rate of association with RTA, reaching only 48 RU after 5 minutes, and negligible dissociation over a time-course of 10 minutes. Overall these results demonstrate that depletion of P1/P2 proteins diminishes binding of RTA to mammalian ribosomes.
Fig. 4.
Depletion of P1 and P2 proteins from rat ribosomes and its effect on ribosome binding by RTA. (A) Ribosomes were treated with NH4Cl (0.4M) followed by the addition of ethanol (50% [v/v]) to selectively remove P1and P2. Following high speed centrifugation, the ribosomal pellet and the supernatant containing stripped P1/P2 proteins were collected and analyzed by western blotting with a monoclonal antibody against the conserved C-termini of P-proteins. The indicated P1/P2 protein levels in depleted ribosomes are expressed as a percentage the P1/P2 protein levels in untreated samples, normalized to P0 levels. (B) The interaction of RTA with WT and P1/P2-depleted rat ribosomes measured by surface plasmon resonance. N-terminal His-tagged RTA was coupled to an NTA chip on the Biacore 3000 and ribosomes (5 nM) were passed over the surface for 5 min and dissociation was monitored for 10 min. The interaction of ribosomes with a control ligand (EGFP) was analyzed in parallel and subtracted from the signal obtained for the target surface (RTA), to account for non-specific binding.
Interaction of RTA with the human P1/P2 heterodimer
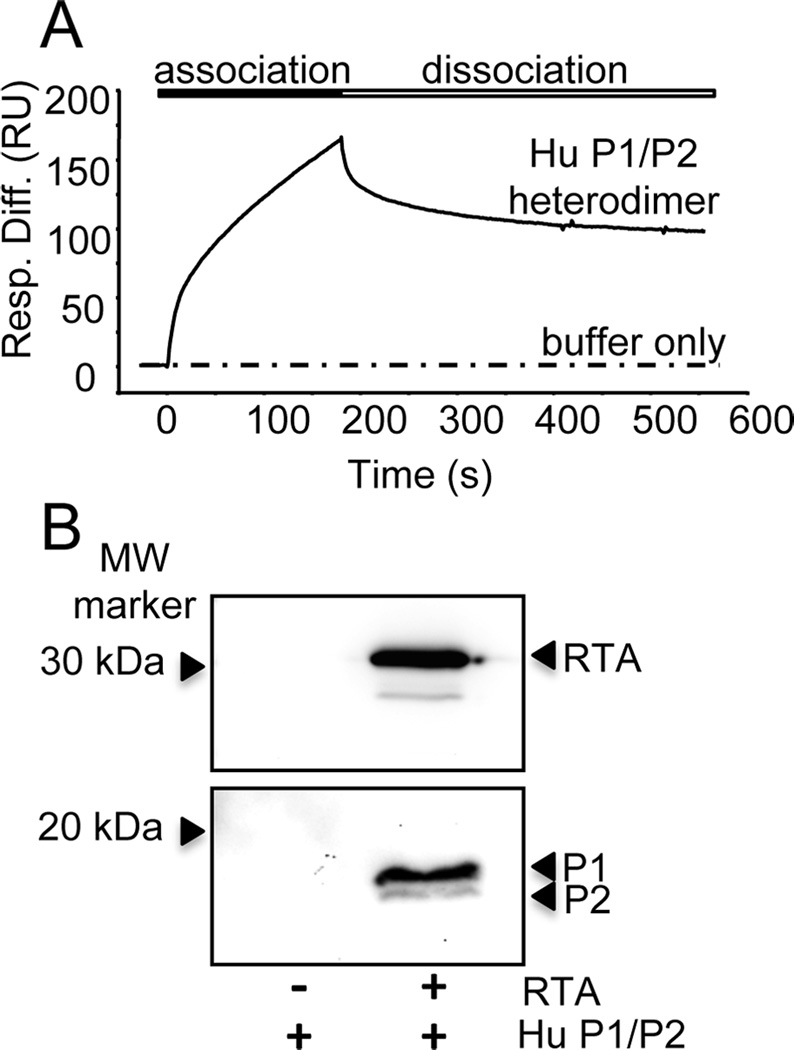
The decreased ability of RTA to bind to P1/P2-depleted ribosomes suggested that RTA interacts with the P1/P2 heterodimer. To verify this, we reconstituted the human P1/P2 heterodimer after expressing each protein in E. coli and examined the interaction of recombinant human P1/P2 heterodimer with RTA by SPR. RTA was immobilized on a carboxymethylated dextran chip (CM5) via amine coupling and recombinant human P1/P2 heterodimer [38] was passed over the surface. Human P1/P2 rapidly associated with RTA, while no significant interaction could be detected with a control protein (EGFP) (Fig. 5A). To confirm this interaction we used co-immunoprecipitation (co-IP). P1/P2 was incubated with RTA and the complexes were pulled-down with polyclonal anti-RTA antibodies. Immunoprecipitated complexes were separated by SDS-PAGE and analyzed by immunoblotting with monoclonal antibodies to either RTA or to the conserved C-termini of P-proteins (Fig. 5B). The human P1/P2 heterodimer was pulled-down only in the presence of RTA and not when it was incubated with anti-RTA antibodies alone (Fig. 5B). These results demonstrate that RTA interacts directly with the human P1/P2 heterodimer.
Fig. 5.
Interaction of RTA with recombinant human P1/P2 heterodimer. (A) The interaction of RTA with human P1/P2 heterodimer by surface plasmon resonance. RTA was amine-coupled to a CM5 chip on the Biacore T200. Recombinant human P1/P2 heterodimers (50 nM) were passed over the surface at 60 µl/min for 3 min and dissociation was monitored for 5 min. The interaction of RTA with a control ligand (EGFP) was analyzed in parallel and subtracted from the signal obtained for the target surface (RTA), to account for non-specific binding. (B) Co-immunoprecipitation of RTA with human P1/P2 heterodimer. P1/P2 heterodimer (50 pmol) was incubated RTA (500 pmol) and then mixed with polyclonal rabbit anti-RTA bound to Protein A-agarose. The agarose was washed three times with PBS and remaining proteins were eluted with SDS-PAGE sample buffer and analyzed by western blotting. The top panel shows the membrane probed with monoclonal anti-RTA (PB10) and the bottom panel shows the membrane probed with a monoclonal antibody against the conserved C-termini of P-proteins (3BH5).
Effect of P-protein depletion on the in vitro depurination activity of RTA
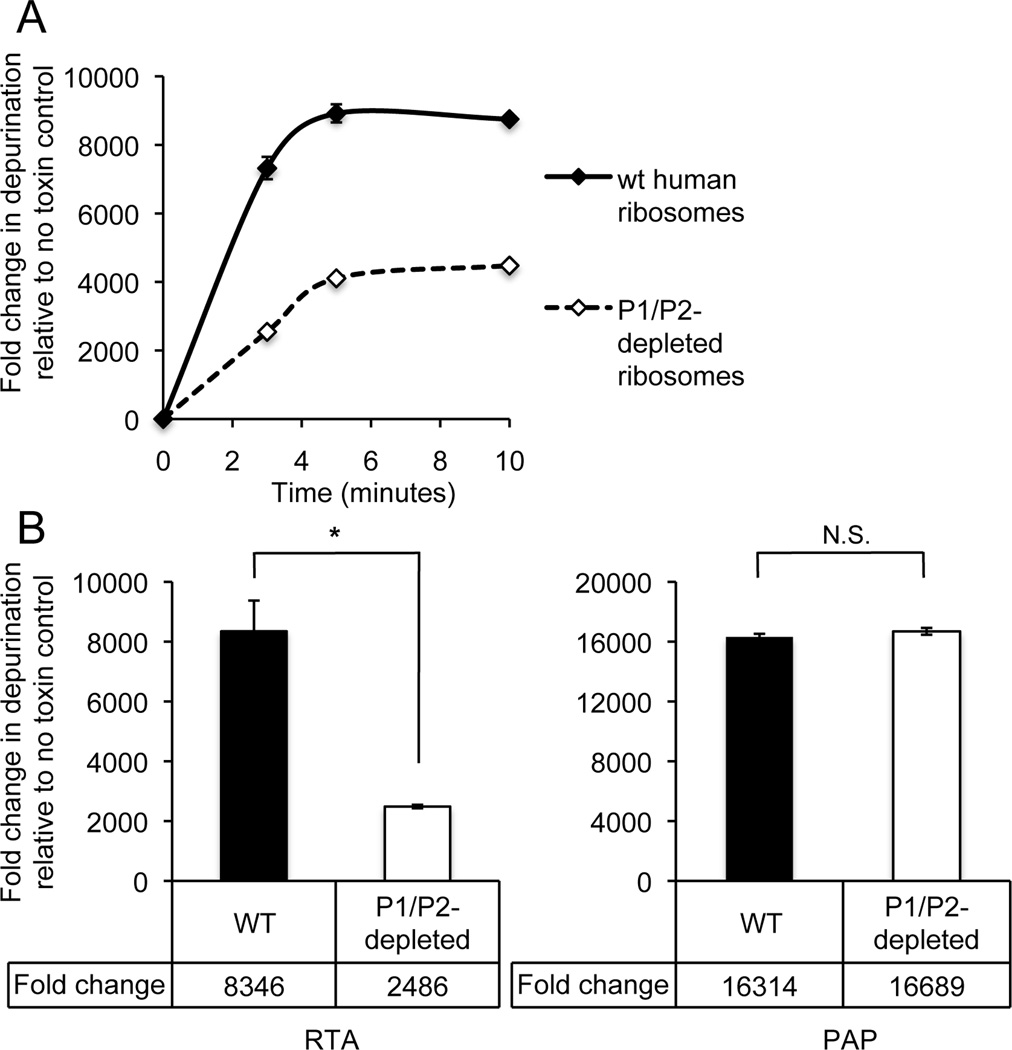
Our data indicated that not only does RTA directly interact with the human P1/P2 heterodimer but that this interaction is critical for RTA to bind to human ribosomes. Therefore, it follows that decreased binding to P1/P2-depleted ribosomes may result in decreased depurination activity of the toxin in vitro; thereby account for the decreased depurination we observed inside depleted cells. To test this, we isolated ribosomes from WT (HEK293T) and P1/P2-depleted HEK293T cells expressing P2-siRNA. Ribosomes were treated in vitro with 0.1 nM RTA over a time course of 0–10 minutes and depurination level was measured by qRT-PCR [34, 35]. Depurination occurred rapidly and both reactions reached plateau at 5 minutes post-treatment (Fig. 6A). At 3 minutes post-treatment when the reaction was linear, P1/P2-depleted ribosomes exhibited >3-fold lower depurination levels (P < 0.05, two-tailed unpaired t test) compared to WT ribosomes, indicating that P1/P2 depletion reduced depurination activity of RTA (Fig. 6B).
Fig. 6.
The effect of P1/P2-depletion on ribosome depurination by RTA and PAP in vitro. (A) WT and P1/P2-depleted human ribosomes (2 pmol) were treated with RTA (0.1nM) for a time course 0–10 minutes. (B) WT and P1/P2-depleted ribosomes (2 pmol) treated with either RTA (0.1nM) or PAP (0.1 nM) for 3 minutes. rRNA was extracted, converted to cDNA, and depurination was measured by qRT-PCR using the comparative CT method (ΔΔCT). Data indicate the fold increases in depurination relative to untreated (no toxin) WT ribosomes and represent the mean ± SD of triplicate reactions from two independent experiments. * indicates P < 0.05 and N.S. indicates difference was not significant (P > 0.05), as analyzed by a Student’s unpaired, two-tailed t test.
We previously showed that pokeweed antiviral protein (PAP) from Phytolacca americanabinds to ribosomal protein L3 to access the SRL [9, 39]. Since PAP does not require the stalk proteins for ribosome depurination, we examined depurination of P1/P2 depleted ribosomes by PAP in parallel as a control. Notably, the level of PAP-dependent depurination was not significantly reduced by P1/P2 depletion of the ribosomes (P > 0.05, two-tailed unpaired t test) (Fig. 6B). These data demonstrate that P1/P2–depletion affects RTA depurination activity in vitro in a way that is specific to RTA and has no affect on the depurination activity of PAP.
DISCUSSION
Previously, chemical cross-linking in mammalian cells indicated that internalized ricin is proximal to the stalk protein P0 and the ribosomal protein L9 [40]. RTA was reported to interact with a short peptide corresponding to the conserved C-terminal region of the human ribosomal stalk proteins, P0, P1 and P2 by pull-down analysis [27]. However, the significance of the proximity of RTA to P0 (L10e) and L9, or the ability of RTA to interact with this peptide was not investigated in these studies. We previously showed that the interaction of RTA with the stalk structure of the yeast ribosome is important for depurination [8, 9, 36]. There are discrete differences in the structural and functional aspects of the stalk from yeast and mammalian cells. RTA is more active on mammalian ribosomes than on yeast ribosomes. Here we have examined if ribosomal stalk proteins facilitate the interaction of RTA with human ribosomes and ribosome depurination in human cells. We demonstrate that depurination activity of RTA is lower when P1 and P2 protein levels are reduced in human cells. Ribosomes from P1/P2 depleted cells have reduced ability to bind RTA. RTA interacts directly with purified human P1/P2 dimer. P1/P2 depletion of human ribosomes reduces depurination by RTA, but has no effect on depurination by a related RIP, PAP. These results demonstrate for the first time the importance of the stalk proteins for RTA to bind to human ribosomes and depurinate the SRL, both in vitro and inside human cells, and are relevant to human intoxication with ricin.
It is possible that depletion of P1/P2 proteins may affect the ribosome structure. Examination of PAP, in parallel to ricin, allowed us to investigate this possibility. PAP shares the same active site structure as RTA with conserved catalytic residues [41] and depurinates the SRL of 28S rRNA at the same position as RTA [42, 43]. PAP binds to ribosomal protein L3 to access the SRL in yeast [39], and its depurination activity has been shown to be P1/P2-independent [9, 39, 44]. In the current study, PAP-dependent depurination did not vary significantly between P1/P2-depleted and undepleted human ribosomes, suggesting that any conformational changes in ribosomes that may have occurred as a result of P1/P2 depletion, did not affect the ability of PAP to depurinate the SRL. These results provide evidence that PAP may not depend on the P1/P2 proteins to depurinate human ribosomes.
RTA has previously been reported to interact with the conserved C-terminal region of the ribosomal stalk proteins (P0, P1 and P2) in vitro [27], suggesting a potential role for P0 in facilitating RTA:ribosome binding. Although we were unable to achieve complete depletion of P1 and P2, our data indicates that P0 does not efficiently facilitate ribosome binding and depurination by RTA in the absence of P1/P2. This may be because RTA is unable to interact with native P0 inside cells or because the conformation of P0 is altered in the absence of P1/P2, precluding binding of RTA. Indeed, previous analysis of the yeast stalk structure suggested that P0 was unable to fold correctly in the absence of P1α/P2β [18]. Nonetheless, although RTA may bind to the C-terminal region of the stalk proteins in vitroour results suggest that protein conformation and the overall architecture of the stalk structure play an important role in determining whether the C-termini of the P-proteins are accessible to RTA. Ultimately our data indicates that, a stalk structure comprised of P0 on human ribosomes is unable to interact efficiently with RTA in the absence of P1/P2.
In addition to examining the effect of P1/P2-depletion on ricin depurination activity in human cells, we attempted to compare the relative toxicity of ricin for P1/P2-depleted and undepleted cells by observing changes membrane permeability using propidium iodide. In the undepleted HEK293T cells, ricin treatment in excess of 48 hours was required to induce cell death. However, at this time, 6 days after induction of P2 siRNA, the P1/P2 depletion alone reduced viability. When depleted cells were also treated with toxin for 48 h, we did not observe any further reduction in viability. However, the reduction in viability due to P1/P2 depletion alone confounded the interpretation of these results, making it difficult to draw conclusions. Nonetheless, despite only partial depletion of P1/P2 proteins, a marked reduction in depurination was observed, clearly implicating P1/P2 proteins in contributing to ricin-dependent depurination of human ribosomes.
The depletion studies suggested that RTA may interact with the human P1/P2 heterodimer directly. Subsequently, using Biacore-based SPR and co-immunoprecipitation analysis we demonstrated direct binding of RTA to recombinant human P1/P2. We have previously compared kinetics of the interaction of RTA with WT and mutant yeast ribosomes lacking P1/P2 proteins [8]. This study suggested two types of interactions between RTA and WT ribosomes, a saturable fast interaction that requires an intact stalk and a non-saturable slow interaction that does not. Based on these results, we proposed a two-step model. First, the slow reaction, which consists of nonspecific electrostatic interactions, concentrates RTA on the ribosome surface. This facilitates diffusion of RTA towards the stalk where the faster, more specific interactions occur [8]. In the present study, the inability to obtain a homogenous preparation of mammalian (human and rat) ribosomes devoid of P1/P2 proteins precluded similar kinetic analyses. Nonetheless, the binding curves observed with RTA and WT mammalian ribosomes exhibited similar heterogeneity, with an initial fast association followed by a slower association rate. Moreover, the initial rapid association of WT mammalian ribosomes with RTA was P1/P2-dependent, since this was not observed with depleted ribosomes. Mutant ribosomes exhibited significantly slower association/dissociation rates with an apparent loss of heterogeneity of the observed binding curves. Overall these data are consistent with a two-step binding model.
Notably, depletion of P1/P2 proteins reduced not only the association of RTA with human ribosomes, but also its dissociation. The lack of dissociation of RTA from P1/P2 depleted human ribosomes may be significant if dissociation of RTA from the stalk is required for the subsequent depurination of the SRL. It seems unlikely that dissociation of RTA is affected by an altered interaction with the SRL in P1/P2 depleted ribosomes. A previous study showed that RTA binds to depurinated ribosomes in the same pattern as undepurinated ribosomes by SPR and the interaction of RTA with ribosomes was not inhibited by increasing concentrations of adenine [45]. It is more likely that the effect on dissociation and association observed in our study represents an effect on the interaction of RTA with the stalk, and not with the SRL. The dynamic properties of the stalk may be important not only for recruiting RTA to ribosomes, but also for delivering RTA to the SRL by undergoing conformational changes, as proposed for the interaction of the stalk proteins with the translation factors [10].
Although the SRL is the universal substrate for RIPs, it is now apparent that ribosomal proteins play an important role in facilitating ribosome binding and subsequent depurination [46]. Notably, despite targeting the same substrate, RIPs differ in their abilities to inactivate fungal, protozoan, plant, insect and prokaryotic ribosomes [46]. While, PAP exhibits comparable activity on both bacterial and eukaryotic ribosomes, RTA displays higher activity on rat liver ribosomes than on plant or yeast ribosomes, and no activity on bacterial ribosomes [30, 32, 47, 48]. A possible explanation for these differences is that the interaction of RIPs with ribosomes is influenced by their interactions with ribosomal proteins. PAP depurinates the SRL by binding to ribosomal protein L3 in yeast [39], which is highly conserved between prokaryotic and eukaryotic ribosomes [9]. We show here that RTA interacts with the human ribosomal stalk, which clearly displays species-specific adaptation. In particular the bacterial proteins which comprise the stalk structure are regarded as functionally analogous rather than homologous to the eukaryotic and archaeal proteins [16]. Furthermore, although there is significant homology between eukaryotic P-proteins, additional isoforms of P1 and P2 proteins are present in yeast and in plants, suggesting that species-dependent differences in the stalk structure may account for the differences in the activity of RTA on ribosomes from different organisms.
In summary, our data highlights the importance of the human P1/P2 proteins for ricin-dependent depurination in human cells and suggests that inhibition of ricin binding to human stalk proteins may be an attractive target for therapeutic intervention. Expression of a truncated form of the ribosomal protein L3 in transconferred resistance to PAP in a plant model [49]. Our results demonstrating that RTA binds to the P1/P2 heterodimer in human cells, is an important step towards the identification of potential inhibitors of RTA:ribosome interactions and the development of novel therapeutics. A detailed understanding of the interaction of RTA with the human stalk complex will aid in this pursuit.
EXPERIMENTAL PROCEDURES
Reagents
Ricin (RCA II, RCA60) was obtained from Vector labs (#L-1090). PAP was a generous gift of Dr James Irvin. Recombinant His-tagged RTA [8], His-tagged EGFP [8] and antibody 3BH5 specific to the C-terminal domain of stalk P-proteins have been previously described [50]. The monoclonal anti-RTA antibody (PB10) was a generous gift of Dr. Nicholas Mantis.
Tissue culture
The HEK (human embryonic kidney)-293T cell line and its derivative (clone SP23-5), which was obtained by stable transfection with an inducible siRNA specific for P2, have been previously described [33]. Cells were maintained in Dulbecco's Modified Eagle Medium supplemented with fetal bovine serum (10%). Induction P2 siRNA was achieved by treatment with 0.1µg/ml doxycycline for 96 h, with media renewal every 48h. Cell confluence was approximately 2×104/cm2 at the time of ricin addition.
Ribosome isolation
Cytoplasmic extracts were isolated as described by Bommer et al. [51], with modifications. Briefly, cultured cells or rat liver were homogenized in Buffer A (20 mM HEPES-KOH pH 7.6, 5mM Mg(CH3COO)250 mM KCl, 10% glycerol, 1mg/ml Heparin, 1mM PMSF, 1mM, DTT, mammalian protease inhibitor cocktail (P8340, Sigma-Aldrich) and either 1% or 2% Triton X-100 for cultured cells and rat liver respectively. Extracts were centrifuged at 30,000 ×g for 20 minutes to remove nuclei and mitochondria and subsequent purification of salt-washed monomeric ribosomes was performed as described by Chiou et al. [9]. Ribosomes were resuspended in Buffer C (50 mM HEPES-KOH pH 7.6, 5mM Mg(CH3COO)250 mM NH4Cl, 25% glycerol, 1mM PMSF, 1mM, DTT).
Purification of human P1/P2 dimer
Expression, purification of recombinant human P-proteins and heterodimer preparation was performed as described previously [38]. Briefly, the proteins were expressed in E. coli BL21(DE3) cells and purified using a procedure described for yeast P-proteins [52]. The heterocomplex was prepared according to a denaturation/renaturation procedure established for the yeast complex [53, 54].
Chemical depletion of ribosomal P-proteins from isolated ribosomes
NH4Cl-ethanol treatment of ribosomes was performed as described previously [37]. Briefly, ribosomes were resuspended at a concentration of 4 mg/ml in 10 mM imidazole-HCL buffer pH 7.4, containing 20 mM MgCl21 mM DTT and 0.4 M NH4Cl, followed by the addition of ethanol to 50% (v/v) final concentration, to selectively remove ribosomal proteins P1 and P2. The ribosomes were pelleted by high-speed centrifugation and the pellet containing P1/P2-depleted ribosomes was washed and resuspended in Buffer C. For western blot analysis of stripped proteins, the supernatants containing stripped P1/P2 proteins were precipitated by the addition 10–20% TCA and centrifuged at 10,000×g for 20 minutes. Protein pellets were washed in the presence of decreasing amounts of TCA (5% then 1%), resuspended in 2×SDS sample buffer (0.125 M Tris-HCl, pH 6.8, 4% [w/v] SDS, 20% [v/v] glycerol, 10% [v/v] β-mercaptoethanol, 0.04% [w/v] Bromophenol blue) and the pH adjusted with NaOH as required.
RNA isolation
Total RNA was extracted from cell cultures using an RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany), as per the manufacturer’s instructions. rRNA from ribosomes was extracted with phenol/chloroform as described previously [9].
cDNA synthesis and quantitative real-time polymerase chain reaction (qRT-PCR) for detection of 28S rRNA depurination
To detect depurination, a qRT-PCR assay was used that exploits the inherent property of reverse transcriptase to incorporate a deoxyadenosine at the position opposite a depurinated site, such that the resulting T to A transversion on the cDNA strand can be detected by sequence-specific primers [34, 35]. 200 ng of total RNA or rRNA from experimental samples was converted to cDNA using random primers (Applied Biosystems) and the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer’s protocol. cDNAs were diluted 1/40 and used as templates for qRT-PCR. Reactions were performed using an ABI StepOnePlus™ RT-PCR system (Applied Biosystems) using JumpStart Taq Ready Mix (Sigma) with SYBR Green I dye (Invitrogen) and ROX reference Dye (Invitrogen). Primers were at a final concentration of 0.2 mM. Thermal cycling conditions were 95°C for 10 min, then 40 repetitions of 95°C (15 s) and 60°C (30 s), followed by a melt curve analysis. The resulting cycle threshold values were recorded for depurinated 28 rRNA templates and normalized to total 28S rRNA. The relative amount of depurinated 28S rRNA in untreated and RTA-treated samples at each time point were compared using the comparative cycle threshold (ΔΔCt) method.
Surface plasmon resonance analysis of the interaction between RTA and mammalian ribosomes or human P1/P2 heterodimer
The Biacore 3000 (GE Healthcare) was used to analyze the interaction between RTA and either ribosomes or the human P1/P2 heterodimers as previously described with modifications [8, 36]. His-tagged ligand (RTA or EGFP) was bound to a Ni2+–nitrilotriacetic acid (NTA) sensor chip and monomeric ribosomes were used as an analyte. Running buffer was comprised of 0.01 M HEPES, 0.15 M NaCl, 10 mM MgOAc, 50 µM EDTA, 0.005 % Surfactant P20, pH 7.4. Sensor chips were first washed with a regeneration solution (0.01 M HEPES, 0.15 M NaCl, 0.35 M EDTA, 0.005 % surfactant P20, pH 8.3) and a 0.3% SDS solution. Running buffer containing 500 µM NiCl2 was injected for 1 min. After the extra washing step, the 10×His-tagged RTA prepared in the running buffer at 50 nM was injected at a flow rate of 10 µl per minute to generate a resonance signal of 1000 Resonance Units (RU). As a control, 10×His-tagged EGFP prepared in the running buffer was immobilized on a reference surface to generate the same RU. Ribosomes were then passed through both the sensor (RTA) and the reference (EGFP) surfaces. Initially series of different flow rates were analyzed to identify an optimal flow rate to eliminate mass transfer limitations (data not shown). Ultimately, ribosomes were analyzed at various concentrations at a flow rate of 30 µl per minute for 5 minutes to monitor the association. The dissociation was recorded at the same flow rate for another 10 minutes. The signal from the reference surface was subtracted from the signal from sensor surface to correct for nonspecific binding. The sensor and the reference surfaces were regenerated by injecting the regeneration solution for 1 minute followed by 1 minute of 0.3% SDS solution and 1 minute of running buffer at a flow rate of 50 µl per minute. For each experiment, a buffer sample without ribosomes was injected under the same conditions and the sensorgram generated from the buffer control was subtracted from the sensorgrams obtained at each ribosome concentration to correct for differences due to the sample injection process. At least 5 different concentrations were analyzed for each ribosome preparation.
Heterodimer interactions were performed as described for the ribosome interactions except that RTA was immobilized on a CM5 chip (Biacore) via amine coupling to 2524 RU. As a control, EGFP was also immobilized on a reference surface at equivalent RU. The dimers (50 nM) were passed over the surface at 60 µl/min for 3 min and a dissociation of 5 min was observed. All interactions were analyzed at 25°C. The sensorgram was analyzed using the BIAevaluation software 4.1.
Co-immunoprecipitation of RTA and the human P1/P2 heterodimer
Co-immunoprecipitation was performed as described previously [36]. Briefly, protein A-agarose was incubated with polyclonal rabbit anti-RTA serum overnight at 4 °C in Biacore running buffer, and then washed three times with the same buffer. Human P1/P2 heterodimer (50 pmol) were mixed with 500 pmol 10×His-tagged RTA in Biacore running buffer and incubated overnight at 4 °C. Protein A-agarose was incubated with RTA-heterodimer complexes at 4 °C for 3 h. The agarose was washed three times with PBS and proteins were eluted with SDS-PAGE sample buffer.
In vitro depurination assay
Assays were performed as previously described [9] with modifications. Briefly, 2 pmol of ribosomes were incubated with the indicated concentrations of RTA or PAP in RIP buffer (60 mM KCl, 10 mM MgCl2and 10 mM Tris-HCl pH 7.4) in a total volume of 100 µl at 37°C for incubation times up to 10 min. rRNA was extracted, converted to cDNA and assayed for depurination by qRT-PCR.
Immunoblot analysis
Cell pellets, cytoplasmic fractions, purified ribosomes or co-immunoprecipitated complexes were separated by SDS-PAGE (15%), electroblotted onto a 0.2 µm nitrocellulose membrane (Bio-Rad) and probed with monoclonal antibody 3BH5 against the C-terminal domain of stalk P-proteins, anti-ß-actin monoclonal antibody or anti-RTA monoclonal antibody, as indicated. Membranes were incubated with chemiluminescent substrate (Bio-Rad) and images were captured using a Chemidoc XRS (Bio-Rad). Net band intensities (mean pixel intensity by number of pixels) were determined using Quantity One imaging software (Bio-Rad) and were normalized to P0. Changes in protein levels were determined by comparison to the level for the undepleted controls.
Statistical analysis
Values are expressed as mean ± SD. Analysis of significance were calculated in Microsoft Excel using either unpaired or paired t tests, with one- or two-tailed P values as appropriate.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jim Irvin for the generous gift of PAP and Nicholas Mantis for the kind gift of anti-RTA antibody (PB10). We gratefully acknowledge Jennifer Nielsen-Kahn for critically reading the manuscript. This work was supported by the National Institutes of Health [grant AI072425 to NET] and Fogarty International Center [grant TW008418 to NET and MT], the Spanish Ministry of Science and Innovation [grant BFU 2009-09738 to JPGB], and an Institutional grant to the Center of Molecular Biology Severo Ochoa from Fundacion Areces to JPGB.
Abbreviations
- PAP
pokeweed antiviral protein
- qRT-PCR
quantitative real-time polymerase chain reaction
- RIP
ribosome inactivating protein
- RTA
ricin A-chain
- RTB
ricin B-chain
- SRL
sarcin/ricin loop
- SPR
surface plasmon resonance
- NTA
nitrilotriacetic acid
- co-IP
co-immunoprecipitation
Footnotes
SUPPORTING INFORMATION
The following supplementary material is available:
Fig. S1. Effect of doxycycline treatment alone on ricin-dependent depurination in parental HEK293T cells.
REFERENCES
- 1.Olsnes S. The history of ricin, abrin and related toxins. Toxicon. 2004;44:361–370. doi: 10.1016/j.toxicon.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Lord JM, Roberts LM, Robertus JD. Ricin: structure, mode of action, and some current applications. FASEB J. 1994;8:201–208. [PubMed] [Google Scholar]
- 3.Spooner RA, Lord JM. How Ricin and Shiga Toxin Reach the Cytosol of Target Cells: Retrotranslocation from the Endoplasmic Reticulum. Curr Top Microbiol Immunol. 2012;357:19–40. doi: 10.1007/82_2011_154. [DOI] [PubMed] [Google Scholar]
- 4.Spooner RA, Watson PD, Marsden CJ, Smith DC, Moore KAH, Cook JP, Lord JM, Roberts LM. Protein disulphide-isomerase reduces ricin to its A and B chains in the endoplasmic reticulum. Biochem J. 2004;383:285–293. doi: 10.1042/BJ20040742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wesche J, Rapak A, Olsnes S. Dependence of ricin toxicity on translocation of the toxin A-chain from the endoplasmic reticulum to the cytosol. J Biol Chem. 1999;274:34443–34449. doi: 10.1074/jbc.274.48.34443. [DOI] [PubMed] [Google Scholar]
- 6.Endo Y, Tsurugi K. RNA N-glycosidase activity of ricin A-chain. Mechanism of action of the toxic lectin ricin on eukaryotic ribosomes. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 7.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The Mechanism for Activation of GTP Hydrolysis on the Ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X-P, Chiou J-C, Remacha M, Ballesta JPG, Tumer NE. A two-step binding model proposed for the electrostatic interactions of ricin A chain with ribosomes. Biochemistry. 2009;48:3853–3863. doi: 10.1021/bi802371h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou J-C, Li X-P, Remacha M, Ballesta JPG, Tumer NE. The ribosomal stalk is required for ribosome binding, depurination of the rRNA and cytotoxicity of ricin A chain in Saccharomyces cerevisiae. Mol Microbiol. 2008;70:1441–1452. doi: 10.1111/j.1365-2958.2008.06492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diaconu M, Kothe U, Schlünzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 11.Tchórzewski M. The acidic ribosomal P proteins. Int J Biochem Cell Biol. 2002;34:911–915. doi: 10.1016/s1357-2725(02)00012-2. [DOI] [PubMed] [Google Scholar]
- 12.Wahl MC, Möller W. Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci. 2002;3:93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- 13.Cardenas D, Revuelta-Cervantes J, Jimenez-Diaz A, Camargo H, Remacha M, Ballesta JPG. P1 and P2 protein heterodimer binding to the P0 protein of Saccharomyces cerevisiae is relatively non-specific and a source of ribosomal heterogeneity. Nucleic Acids Res. 2012;40:4520–4529. doi: 10.1093/nar/gks036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krokowski D, Tchórzewski M, Boguszewska A, McKay AR, Maslen SL, Robinson CV, Grankowski N. Elevated copy number of L-A virus in yeast mutant strains defective in ribosomal stalk. Biochem Biophys Res Commun. 2007;355:575–580. doi: 10.1016/j.bbrc.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Szick K, Springer M, Bailey-Serres J. Evolutionary analyses of the 12-kDa acidic ribosomal P-proteins reveal a distinct protein of higher plant ribosomes. Proc Natl Acad Sci USA. 1998;95:2378–2383. doi: 10.1073/pnas.95.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grela P, Bernadó P, Svergun D, Kwiatowski J, Abramczyk D, Grankowski N, Tchórzewski M. Structural relationships among the ribosomal stalk proteins from the three domains of life. J Mol Evol. 2008;67:154–167. doi: 10.1007/s00239-008-9132-2. [DOI] [PubMed] [Google Scholar]
- 17.Ballesta JP, Remacha M. The large ribosomal subunit stalk as a regulatory element of the eukaryotic translational machinery. Prog Nucleic Acid Res Mol Biol. 1996;55:157–193. doi: 10.1016/s0079-6603(08)60193-2. [DOI] [PubMed] [Google Scholar]
- 18.Krokowski D, Tchórzewski M, Boguszewska A, Grankowski N. Acquisition of a stable structure by yeast ribosomal P0 protein requires binding of P1A-P2B complex: in vitro formation of the stalk structure. Biochim Biophys Acta. 2005;1724:59–70. doi: 10.1016/j.bbagen.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 19.Grela P, Helgstrand M, Krokowski D, Boguszewska A, Svergun D, Liljas A, Bernadó P, Grankowski N, Akke M, Tchórzewski M. Structural characterization of the ribosomal P1A-P2B protein dimer by small-angle X-ray scattering and NMR spectroscopy. Biochemistry. 2007;46:1988–1998. doi: 10.1021/bi0616450. [DOI] [PubMed] [Google Scholar]
- 20.Krokowski D, Boguszewska A, Abramczyk D, Liljas A, Tchórzewski M, Grankowski N. Yeast ribosomal P0 protein has two separate binding sites for P1/P2 proteins. Mol Microbiol. 2006;60:386–400. doi: 10.1111/j.1365-2958.2006.05117.x. [DOI] [PubMed] [Google Scholar]
- 21.Jose MP, Santana-Roman H, Remacha M, Ballesta JP, Zinker S. Eukaryotic acidic phosphoproteins interact with the ribosome through their amino-terminal domain. Biochemistry. 1995;34:7941–7948. doi: 10.1021/bi00024a019. [DOI] [PubMed] [Google Scholar]
- 22.Bargis-Surgey P, Lavergne J-P, Gonzalo P, Vard C, Filhol-Cochet O, Reboud J-P. Interaction of elongation factor eEF-2 with ribosomal P proteins. Eur J Biochem. 1999;262:606–611. doi: 10.1046/j.1432-1327.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- 23.Santos C, Ballesta JP. The highly conserved protein P0 carboxyl end is essential for ribosome activity only in the absence of proteins P1 and P2. J Biol Chem. 1995;270:20608–20614. doi: 10.1074/jbc.270.35.20608. [DOI] [PubMed] [Google Scholar]
- 24.Nomura N, Honda T, Baba K, Naganuma T, Tanzawa T, Arisaka F, Noda M, Uchiyama S, Tanaka I, Yao M, Uchiumi T. Archaeal ribosomal stalk protein interacts with translation factors in a nucleotide-independent manner via its conserved C terminus. Proc Natl Acad Sci USA. 2012;109:3748–3753. doi: 10.1073/pnas.1112934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan DSB, Chu L-O, Lee K-M, Too PHM, Ma K-W, Sze KH, Zhu G, Shaw P-C, Wong K-B. Interaction between trichosanthin, a ribosome-inactivating protein, and the ribosomal stalk protein P2 by chemical shift perturbation and mutagenesis analyses. Nucleic Acids Res. 2007;35:1660–1672. doi: 10.1093/nar/gkm065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Too PH-M, Ma MK-W, Mak AN-S, Wong Y-T, Tung CK-C, Zhu G, Au SW-N, Wong K-B, Shaw P-C. The C-terminal fragment of the ribosomal P protein complexed to trichosanthin reveals the interaction between the ribosome-inactivating protein and the ribosome. Nucleic Acids Res. 2009;37:602–610. doi: 10.1093/nar/gkn922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCluskey AJ, Poon GMK, Bolewska-Pedyczak E, Srikumar T, Jeram SM, Raught B, Gariépy J. The catalytic subunit of Shiga-like toxin 1 interacts with ribosomal stalk proteins and is inhibited by their conserved C-terminal domain. J Mol Biol. 2008;378:375–386. doi: 10.1016/j.jmb.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 28.McCluskey AJ, Bolewska-Pedyczak E, Jarvik N, Chen G, Sidhu SS, Gariépy J. Charged and hydrophobic surfaces on the a chain of Shiga-like toxin 1 recognize the C-terminal domain of ribosomal stalk proteins. PLoS ONE. 2012;7:e31191. doi: 10.1371/journal.pone.0031191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boguszewska A, Tchórzewski M, Dukowski P, Winiarczyk S, Grankowski N. Subcellular distribution of the acidic ribosomal P-proteins from Saccharomyces cerevisiae in various environmental conditions. Biol Cell. 2002;94:139–146. doi: 10.1016/s0248-4900(02)01192-9. [DOI] [PubMed] [Google Scholar]
- 30.Sturm MB, Schramm VL. Detecting ricin: sensitive luminescent assay for ricin A-chain ribosome depurination kinetics. Anal Chem. 2009;81:2847–2853. doi: 10.1021/ac8026433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartley MR, Legname G, Osborn R, Chen Z, Lord JM. Single-chain ribosome inactivating proteins from plants depurinate Escherichia coli 23S ribosomal RNA. FEBS Lett. 1991;290:65–68. doi: 10.1016/0014-5793(91)81227-y. [DOI] [PubMed] [Google Scholar]
- 32.Harley SM, Beevers H. Ricin inhibition of in vitro protein synthesis by plant ribosomes. Proc Natl Acad Sci USA. 1982;79:5935–5938. doi: 10.1073/pnas.79.19.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Azorin F, Remacha M, Ballesta JPG. Functional characterization of ribosomal P1/P2 proteins in human cells. Biochem J. 2008;413:527–534. doi: 10.1042/BJ20080049. [DOI] [PubMed] [Google Scholar]
- 34.Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17:201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melchior WB, Tolleson WH. A functional quantitative polymerase chain reaction assay for ricin, Shiga toxin, and related ribosome-inactivating proteins. Anal Biochem. 2010;396:204–211. doi: 10.1016/j.ab.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Li X-P, Grela P, Krokowski D, Tchórzewski M, Tumer NE. Pentameric organization of the ribosomal stalk accelerates recruitment of ricin a chain to the ribosome for depurination. J Biol Chem. 2010;285:41463–41471. doi: 10.1074/jbc.M110.171793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez-Madrid F, Reyes R, Conde P, Ballesta JP. Acidic ribosomal proteins from eukaryotic cells. Effect on ribosomal functions. Eur J Biochem. 1979;98:409–416. doi: 10.1111/j.1432-1033.1979.tb13200.x. [DOI] [PubMed] [Google Scholar]
- 38.Grela P, Sawa-Makarska J, Gordiyenko Y, Robinson CV, Grankowski N, Tchórzewski M. Structural properties of the human acidic ribosomal P proteins forming the P1-P2 heterocomplex. J Biochem. 2008;143:169–177. doi: 10.1093/jb/mvm207. [DOI] [PubMed] [Google Scholar]
- 39.Hudak KA, Dinman JD, Tumer NE. Pokeweed antiviral protein accesses ribosomes by binding to L3. J Biol Chem. 1999;274:3859–3864. doi: 10.1074/jbc.274.6.3859. [DOI] [PubMed] [Google Scholar]
- 40.Vater CA, Bartle LM, Leszyk JD, Lambert JM, Goldmacher VS. Ricin A chain can be chemically cross-linked to the mammalian ribosomal proteins L9 and L10e. J Biol Chem. 1995;270:12933–12940. doi: 10.1074/jbc.270.21.12933. [DOI] [PubMed] [Google Scholar]
- 41.Monzingo AF, Collins EJ, Ernst SR, Irvin JD, Robertus JD. The 2.5 A structure of pokeweed antiviral protein. J Mol Biol. 1993;233:705–715. doi: 10.1006/jmbi.1993.1547. [DOI] [PubMed] [Google Scholar]
- 42.Irvin JD. Pokeweed antiviral protein. Pharmacology & Therapeutics. 1983;21:371–387. doi: 10.1016/0163-7258(83)90061-x. [DOI] [PubMed] [Google Scholar]
- 43.Endo Y, Tsurugi K, Lambert JM. The site of action of six different ribosome-inactivating proteins from plants on eukaryotic ribosomes: the RNA N-glycosidase activity of the proteins. Biochem Biophys Res Commun. 1988;150:1032–1036. doi: 10.1016/0006-291x(88)90733-4. [DOI] [PubMed] [Google Scholar]
- 44.Ayub MJ, Smulski CR, Ma K-W, Levin MJ, Shaw P-C, Wong K-B. The C-terminal end of P proteins mediates ribosome inactivation by trichosanthin but does not affect the pokeweed antiviral protein activity. Biochem Biophys Res Commun. 2008;369:314–319. doi: 10.1016/j.bbrc.2008.01.170. [DOI] [PubMed] [Google Scholar]
- 45.Honjo E, Watanabe K, Tsukamoto T. Real-time kinetic analyses of the interaction of ricin toxin A-chain with ribosomes prove a conformational change involved in complex formation. J Biochem. 2002;131:267–275. doi: 10.1093/oxfordjournals.jbchem.a003098. [DOI] [PubMed] [Google Scholar]
- 46.Tumer NE, Li X-P. Interaction of Ricin and Shiga Toxins with Ribosomes. Curr Top Microbiol Immunol. 2012;357:1–18. doi: 10.1007/82_2011_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor S, Massiah A, Lomonossoff G, Roberts LM, Lord JM, Hartley M. Correlation between the activities of five ribosome-inactivating proteins in depurination of tobacco ribosomes and inhibition of tobacco mosaic virus infection. Plant J. 1994;5:827–835. doi: 10.1046/j.1365-313x.1994.5060827.x. [DOI] [PubMed] [Google Scholar]
- 48.Endo Y, Tsurugi K. The RNA N-glycosidase activity of ricin A-chain. Nucleic Acids Symp Ser. 1988:139–142. [PubMed] [Google Scholar]
- 49.Di R, Tumer NE. Expression of a truncated form of ribosomal protein L3 confers resistance to pokeweed antiviral protein and the Fusarium mycotoxin deoxynivalenol. Mol Plant Microbe Interact. 2005;18:762–770. doi: 10.1094/MPMI-18-0762. [DOI] [PubMed] [Google Scholar]
- 50.Vilella MD, Remacha M, Ortiz BL, Mendez E, Ballesta JP. Characterization of the yeast acidic ribosomal phosphoproteins using monoclonal antibodies. Proteins L44/L45 and L44' have different functional roles. Eur J Biochem. 1991;196:407–414. doi: 10.1111/j.1432-1033.1991.tb15831.x. [DOI] [PubMed] [Google Scholar]
- 51.Bommer U, Burkhardt N, Jünemann R, Spahn C, Triana-Alonso F, Nierhaus K. In: Subcellular Fractionation—A Practical Approach. G J, R D, editors. Washington, DC: IRL Press; 1997. [Google Scholar]
- 52.Tchórzewski M, Boguszewska A, Abramczyk D, Grankowski N. Overexpression in Escherichia coli purification, and characterization of recombinant 60S ribosomal acidic proteins from Saccharomyces cerevisiae . Protein Expr Purif. 1999;15:40–47. doi: 10.1006/prep.1998.0997. [DOI] [PubMed] [Google Scholar]
- 53.Tchórzewski M, Boguszewska A, Dukowski P, Grankowski N. Oligomerization properties of the acidic ribosomal P-proteins from Saccharomyces cerevisiae: effect of P1A protein phosphorylation on the formation of the P1A-P2B hetero-complex. Biochim Biophys Acta. 2000;1499:63–73. doi: 10.1016/s0167-4889(00)00108-7. [DOI] [PubMed] [Google Scholar]
- 54.Tchórzewski M, Krokowski D, Boguszewska A, Liljas A, Grankowski N. Structural Characterization of Yeast Acidic Ribosomal P Proteins Forming the P1A–P2B Heterocomplex. Biochemistry. 2003;42:3399–3408. doi: 10.1021/bi0206006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.