Summary
In animal gonads, PIWI proteins and their bound 23–30 nt piRNAs guard genome integrity by the sequence specific silencing of transposons. Two branches of piRNA biogenesis, namely primary processing and ping-pong amplification, have been proposed. Despite an overall conceptual understanding of piRNA biogenesis, identity and/or function of the involved players are largely unknown. Here, we demonstrate an essential role for the female sterility gene shutdown in piRNA biology. Shutdown, an evolutionarily conserved cochaperone collaborates with Hsp90 during piRNA biogenesis, potentially at the loading step of RNAs into PIWI proteins. We demonstrate that Shutdown is essential for both primary and secondary piRNA populations in Drosophila. An extension of our study to previously described piRNA pathway members revealed three distinct groups of biogenesis factors. Together with data on how PIWI proteins are wired into primary and secondary processing, we propose a unified model for piRNA biogenesis.
Highlights
► The cochaperone Shutdown is an essential piRNA biogenesis factor ► Primary and secondary piRNA biogenesis feed into a common biogenesis step ► Piwi and Aub, but not AGO3 are loaded with primary piRNAs
Introduction
PIWI interacting RNAs (piRNAs) are a class of animal small RNAs. They are bound by PIWI family proteins and guide the sequence specific silencing of selfish genetic elements such as transposable elements (TEs). Defects in the piRNA pathway lead to TE derepression, genomic instability and ultimately sterility (Malone and Hannon, 2009; Siomi et al., 2011).
In Drosophila, most piRNAs are generated from two sources; on the one hand, these are piRNA cluster transcripts that originate from discrete genomic loci and serve as reservoirs of TE sequences; on the other hand, these are RNAs derived from active TEs that engage—together with cluster transcripts—in a piRNA amplification loop called the ping-pong cycle (Brennecke et al., 2007; Gunawardane et al., 2007; Senti and Brennecke, 2010).
Two modes of piRNA biogenesis exist: (1) during primary piRNA biogenesis, a single stranded RNA is processed into pre-piRNAs, which are loaded onto PIWI proteins and are subsequently 3′ trimmed and methylated, yielding mature piRNA induced silencing complexes (piRISCs) (Kawaoka et al., 2011; Senti and Brennecke, 2010). (2) piRISCs with active slicer activity can trigger secondary piRNA biogenesis, where a new piRNA is formed out of the sliced target RNA. In the presence of corresponding sense and antisense precursor RNAs, secondary piRNA biogenesis acts as the ping-pong amplification loop. The two piRNAs engaged in ping-pong have opposite orientation and exhibit a characteristic ten nucleotide 5′ overlap (ping-pong signature; Brennecke et al., 2007; Gunawardane et al., 2007).
Primary and secondary piRNA biogenesis co-occur in germline cells, complicating the genetic and mechanistic dissection of these processes. However, somatic cells of the gonad also harbor a piRNA pathway and this is based exclusively on primary piRNA biogenesis (Lau et al., 2009; Li et al., 2009; Malone et al., 2009; Saito et al., 2009). The Drosophila ovary is therefore ideally suited to identify and characterize factors required for either primary or secondary piRNA biogenesis or both.
Somatic support cells of the Drosophila ovary express Piwi as the only PIWI family protein. Primary piRNA biogenesis is thought to take place in peri-nuclear Yb-bodies, where the RNA helicases Armitage (Armi) and Yb as well as the TUDOR domain protein Vreteno (Vret) accumulate. In addition to these three factors, the putative mitochondria-localized nuclease Zucchini (Zuc) and the RNA helicase Sister of Yb (SoYb) are essential for piRNA biogenesis in the soma (Haase et al., 2010; Handler et al., 2011; Olivieri et al., 2010; Qi et al., 2011; Saito et al., 2010; Zamparini et al., 2011). Formation of mature Piwi-RISC triggers its nuclear import, while failure in piRNA biogenesis results in destabilization of presumably unloaded Piwi (Haase et al., 2010; Olivieri et al., 2010; Qi et al., 2011; Saito et al., 2010). Mature Piwi-RISC triggers TE silencing by an unknown mechanism that requires Piwi’s nuclear localization but not its slicer activity (Saito et al., 2010; Klenov et al., 2011).
With the exception of Yb, the above mentioned biogenesis factors are also essential in germline cells for the formation of Piwi-RISC. Germline cells, however, express two additional PIWI proteins, Aubergine (Aub) and Argonaute 3 (AGO3), which localize to the cytoplasm and are enriched in peri-nuclear nuage. Aub and AGO3 are the main players in the ping-pong cycle. Several factors with essential or modulatory roles in the ping-pong cycle have been identified. These are the RNA helicases Spindle-E and Vasa and the TUDOR domain proteins Krimper, Tejas (Tej), Qin and Tudor (Anand and Kai, 2011; Lim and Kai, 2007; Malone et al., 2009; Nishida et al., 2009; Patil and Kai, 2010; Zhang et al., 2011).
The analysis of piRNA populations from wild-type and piRNA pathway mutant ovaries indicated that Piwi is mainly a recipient of primary piRNAs, while Aub and AGO3 are predominantly or exclusively recipients of secondary piRNA biogenesis (Brennecke et al., 2007; Gunawardane et al., 2007; Li et al., 2009; Malone et al., 2009). Given this, three major questions arise: (1) Are primary and secondary piRNA biogenesis processes genetically and mechanistically separate or do common factors act in both processes? (2) In which processing step do identified piRNA biogenesis factors act? (3) How are the three PIWI family proteins wired into piRNA biogenesis? In other words, are certain PIWI proteins only receiving primary or only secondary piRNAs?
Here, we show that the female sterility gene shutdown (Munn and Steward, 2000; Schüpbach and Wieschaus, 1991) encodes a piRNA biogenesis factor. We show that Shu is required for all piRNA populations in ovaries and that it acts downstream of known piRNA biogenesis factors. A comparison of Shu to several other pathway factors led to the definition of three major groups of piRNA biogenesis factors. In combination with data on how PIWI proteins are wired into piRNA biogenesis, we propose a model that accounts for the distinct association of piRNA subpopulations with specific PIWI proteins in Drosophila.
Results
The Cochaperone Shutdown Is an Essential piRNA Pathway Factor
In Drosophila, mutations in piRNA pathway factors result in sterility. In many cases, however, oogenesis proceeds and mutant females are laying eggs. These exhibit defects in egg asymmetry and fail to develop. Loss of egg asymmetry is triggered by sustained activation of the Chk2 DNA damage checkpoint that is presumably activated by uncontrolled TE activity (Klattenhoff et al., 2007). Based on the characteristic mutant egg morphology, several piRNA pathway genes were identified before piRNAs were described (e.g., Schüpbach and Wieschaus, 1991). We searched the literature for female sterility mutants with egg polarity defects yet no reported link to piRNA biology. One of the candidates was shutdown (shu), a gene encoding a cochaperone of the immunophilin class that harbors a peptidyl-prolyl-cis/trans-isomerase domain (PPIase) and a TPR domain (Munn and Steward, 2000). Notably, the orthologous mouse FKBP6 gene is essential for male fertility (Crackower et al., 2003) and was identified in PIWI immune precipitates (Vagin et al., 2009).
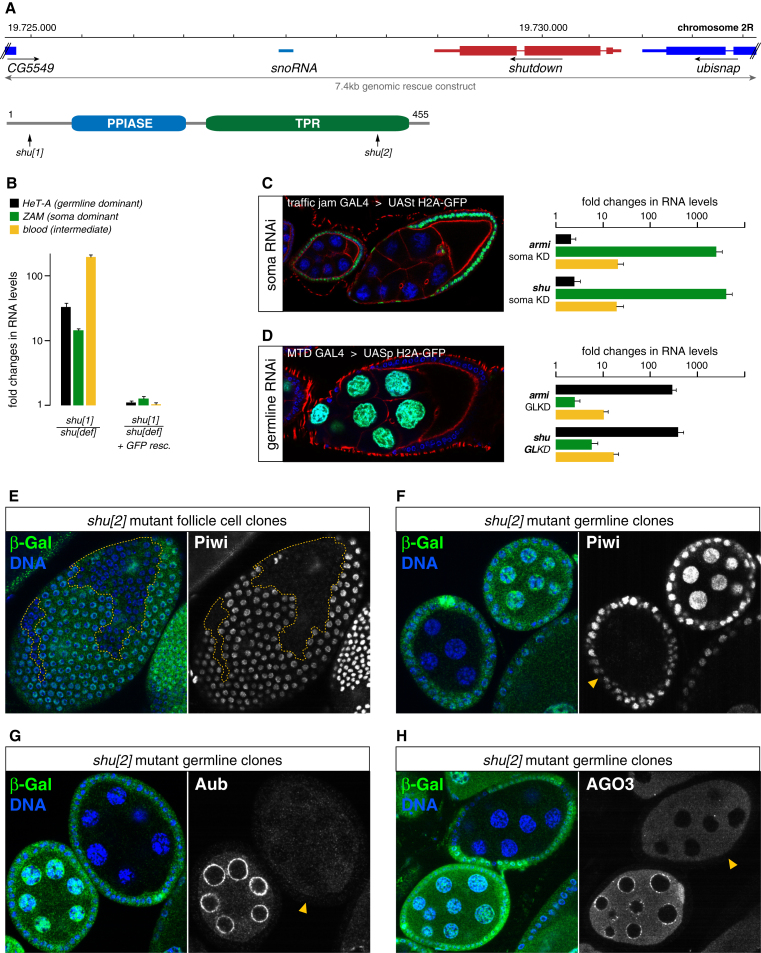
Unlike piRNA pathway mutants that only affect the germline branch and that allow overall oogenesis to occur, shu mutant ovaries are rudimentary and resemble mutants that affect also the somatic piRNA pathway (not shown). We compared TE RNA levels in shu mutant ovaries to those of heterozygous controls and observed derepression of TEs known to be active either in germline or in somatic support cells (Figure 1B). Sterility, ovary morphology and TE repression were rescued by a genomic construct harboring the shu locus with an additional N-terminal GFP cassette (Figures 1A and 1B). To support a role for Shu in ovarian somatic and germline cells we used tissue specific RNAi based on the traffic jam GAL4 driver (soma) or the nanos GAL4 driver (germline) (Handler et al., 2011; Ni et al., 2011; Olivieri et al., 2010). Similar to germline knockdowns (GLKD) of the piRNA biogenesis factor Armi, GLKD of Shu resulted in derepression of the germline controlled TE HeT-A, while soma knockdowns resulted in derepression of the soma controlled element ZAM. The blood element was derepressed in both knockdowns, in agreement with this TE being targeted by soma and germline piRNAs (Malone et al., 2009).
Figure 1.
Shutdown Is an Essential piRNA Pathway Factor
(A) Shown is the genomic shu locus, the extent of the rescue construct and the domain composition of the 455 amino acid Shu protein (arrows indicate the two nonsense mutations in shu[1] and shu[2]).
(B) Fold changes in steady state RNA levels of indicated TEs in shu mutant and GFP-shu-rescued ovaries (normalized to shu heterozygotes; n = 3; error bars indicate StDev.).
(C and D) Immunofluorescence images show soma (C) and germline (D) specificity of tj-Gal4 and MTD-Gal4 (crossed to UAS H2A-GFP (green); DNA (blue); actin (red)). Bar diagrams show fold changes in steady state RNA levels of indicated TEs upon soma (C) or germline (D) specific knockdown of armi or shu (n = 3; StDev.).
(E) Confocal section of a follicle epithelium with a mitotic shu[2] clone (marked by absence of β-Gal (green)) stained for DNA (blue) and Piwi (monochrome). The dashed yellow line marks the clone boundary.
(F–H) Confocal sections of two egg chambers with one being mutant for shu[2] in the germline (absence of β-Gal (green); yellow arrowhead in monochrome panels) stained for DNA (blue), Piwi (F), Aub (G) or AGO3 (H).
We next analyzed localization and levels of PIWI proteins in cells lacking Shu using mosaic clones. shu mutant cells exhibited a near loss of Piwi (Figures 1E and 1F). Similarly, levels and nuage accumulation of Aub were severely affected (Figure 1G). AGO3 levels were moderately decreased and residual AGO3 accumulated in cytoplasmic foci rather than in nuage (Figure 1H). Western blot analysis from shu-GLKD ovaries confirmed the strong reductions in Piwi and Aub levels (Figure S1). The only other known piRNA pathway factor with similar requirements for all PIWI proteins is the TUDOR domain containing protein Vret (Handler et al., 2011; Zamparini et al., 2011).
Shutdown Is Required for the Biogenesis of All piRNA Populations
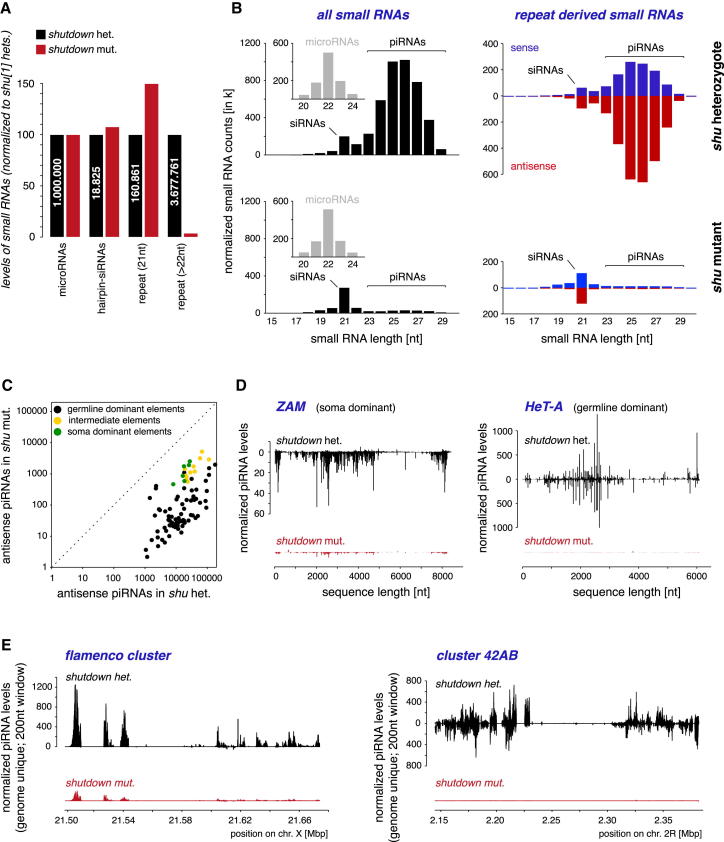
Reductions in PIWI levels such as in the case of shu mutants have been linked to defects in piRNA biogenesis. We sequenced 18–30 nt RNAs from ovaries of shu mutants and heterozygous controls. Both libraries were normalized to one million miRNA reads. In support of this normalization, levels of endogenous hairpin derived siRNAs were similar in both libraries (Figure 2A). Remarkably, while repeat derived siRNAs (21 nt) were slightly elevated in shu mutants, repeat derived piRNAs were nearly ablated (∼30-fold reduction; Figures 2A and 2B). An analysis of the 93 Repbase TEs with more than 1000 sequenced antisense piRNAs in wild-type ovaries revealed severe reductions of piRNA levels for all TEs (Figure 2C). We noted a stronger impact on germline dominant elements (∼150-fold reduction) compared to intermediate (∼23-fold) and soma dominant elements (∼15-fold; Figures 2C and 2D). We attribute this to the distorted morphology of shu mutant ovaries, which likely contain a relatively greater proportion of somatic cells than wild-type ovaries. A similar trend was observed for piRNAs mapping uniquely to piRNA clusters; the soma dominant cluster flamenco was strongly, but less affected than germline dominant clusters such as cluster 42AB (Figure 2E). Also gene-derived piRNAs required Shu as piRNAs mapping to the 3′UTR of traffic jam, a prominent source of piRNAs in Drosophila, were nearly lost (not shown; Robine et al., 2009; Saito et al., 2009).
Figure 2.
Shutdown Is Required for the Biogenesis of All Ovarian piRNA Populations
(A) Shown are normalized levels of indicated ovarian small RNAs from shu[1] heterozygous and shu[1]/shu[def] flies.
(B) To the left, length profiles of normalized small RNAs split into miRNAs (small panels) and remaining RNAs (siRNAs and piRNAs) are shown. To the right, length profiles of repeat derived small RNAs (red antisense; blue sense) are shown; top panels show shu heterozygotes, bottom panels shu mutants.
(C) Scatter plot (log scale) showing levels of antisense piRNAs mapping to soma dominant (green), intermediate (yellow) or germline dominant (black) TEs in shu het. or shu mut. libraries.
(D) normalized piRNA profiles (sense up; antisense down) from shu het. (black) or shu mut. (red) libraries mapping to indicated TEs.
(E) normalized profiles of genome unique piRNAs (sense up; antisense down) from shu het. (black) or shu mut. (red) libraries mapping to indicated piRNA clusters (200nt walking window).
Shutdown Acts Downstream of Known piRNA Biogenesis Factors
Current models postulate that PIWI proteins are loaded with longer pre-piRNA molecules. Subsequently, an unknown ‘trimmer’ activity generates the 3′ end (Kawaoka et al., 2011). This predicts a hierarchy of factors that (1) direct precursor RNAs to the biogenesis machinery, that (2) generate pre-piRNA molecules, that (3) load pre-piRNAs into PIWI proteins and that (4) catalyze piRNA 3′ end maturation.
Thus, piRNA biogenesis is a dynamic process and underlying protein-protein and protein-RNA interactions must be transient, challenging the molecular dissection of the biogenesis pathway. A hierarchy of piRNA biogenesis factors can, however, be delineated based on their localization to Yb-bodies in somatic cells or to nuage in germline cells, the proposed sites of piRNA biogenesis (Handler et al., 2011; Lim and Kai, 2007; Olivieri et al., 2010; Saito et al., 2010).
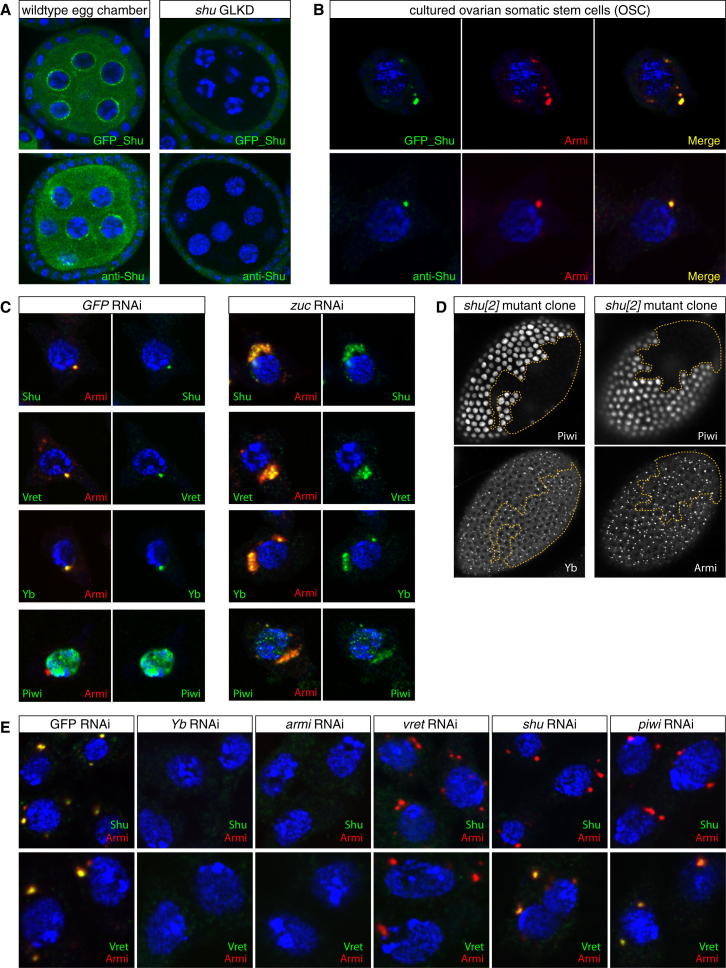
Based on the GFP-Shu rescue construct and antibody stainings, Shu localized to the cytoplasm of ovarian germline and somatic cells and was enriched in nuage (Figure 3A). In follicle cells, Shu levels were considerably lower. We therefore turned to cultured ovarian somatic cells (OSCs; Niki et al., 2006; Saito et al., 2009). Here, GFP-Shu as well as endogenous Shu was cytoplasmic and typically enriched in Yb-bodies (Figure 3B). The primary piRNA biogenesis factors Yb, Armi and Vret colocalize in Yb-bodies (Figure 3C) and a genetic hierarchy exists (Yb → Armi → Vret; Handler et al., 2011; Olivieri et al., 2010; Saito et al., 2010). Comparable to observations in mouse (Watanabe et al., 2011), loss of the essential biogenesis factor Zuc led to clustering of mitochondria (Figure S2). Under those conditions, Yb, Armi and Vret decorated most mitochondria and defined a “giant” Yb-body that also accumulated Shu and—presumably unloaded—Piwi (Figure 3C; Olivieri et al., 2010; Saito et al., 2010). We conclude that Shu is a bona fide Yb-body factor.
Figure 3.
Shutdown Acts Downstream of Known piRNA Biogenesis Factors
(A) Shown are confocal sections of egg chambers expressing GFP-shu (top) or stained for endogenous Shu (bottom) from wild-type flies (left) or shu-GLKD. Signal in somatic follicle cells is unaffected.
(B) IF analysis of OSCs expressing GFP-Shu (top) or stained for Shu (bottom). Yb-bodies are identified via anti-Armi (red) and the right panels indicate colocalization of Shu and Armi in yellow. DNA in all panels stained with DAPI (blue).
(C) IF analysis of OSCs stained for DNA (blue), Armi (red) and indicated proteins (green) in control knockdowns (left) or zuc knockdowns (right). Colocalization of red and green appears yellow in the merge.
(D) Shown are confocal sections of the follicular epithelium with mitotic shu[2] clones stained for Piwi (top) and Yb or Armi (bottom). Clone borders shown with dashed yellow line.
(E) IF analysis of OSCs knocked down for indicated factors stained for DNA (blue), Armi (red) and Shu (green; top panels) or Vret (green; bottom panels). All panels show the merge (colocalization of red and green appears yellow).
We tested Shu’s requirement for Yb-body formation. In clones of shu mutant follicle cells, Yb and Armi accumulated in discrete foci, indistinguishable from surrounding control cells, arguing that Yb-body formation does not require Shu (Figure 3D). Based on knockdown experiments in OSCs, Yb-body localization of Vret (though dependent on Armi and Yb), was only mildly affected by loss of Shu (Figure 3E). On the other hand, Shu localization to Yb-bodies required all known biogenesis factors (Figure 3E). Notably, knockdown of Piwi also caused delocalization of Shu from Yb-bodies. This suggests the possibility that unloaded Piwi recruits Shu. Indeed, low levels of Piwi colocalize with Shu in Yb-bodies and Piwi copurified with Shu and Armi in IP-experiments (Figure S2).
Taken together, Shu localizes to sites of piRNA biogenesis, potentially interacts with unloaded PIWI proteins and acts downstream of known biogenesis factors.
Shutdown’s TPR Domain Is Essential for Its Function and Shutdown and Hsp83 Colocalize with Unloaded AGO3
Heat shock protein 90 (Hsp90) is required for loading siRNA duplexes into Argonaute proteins (Iki et al., 2010; Iwasaki et al., 2010; Miyoshi et al., 2010). Recently, plant Cyclophilin 40 (Cyp40) has been shown to be an important Hsp90 cochaperone for siRNA loading into AGO1 (Iki et al., 2011). Cyp40 shares the same domain organization with Shu and the C-terminal TPR domain was shown to be essential for Hsp90 interaction and Cyp40 function. We therefore tested whether Shu cooperates with Hsp83 (Drosophila Hsp90) during piRNA biogenesis.
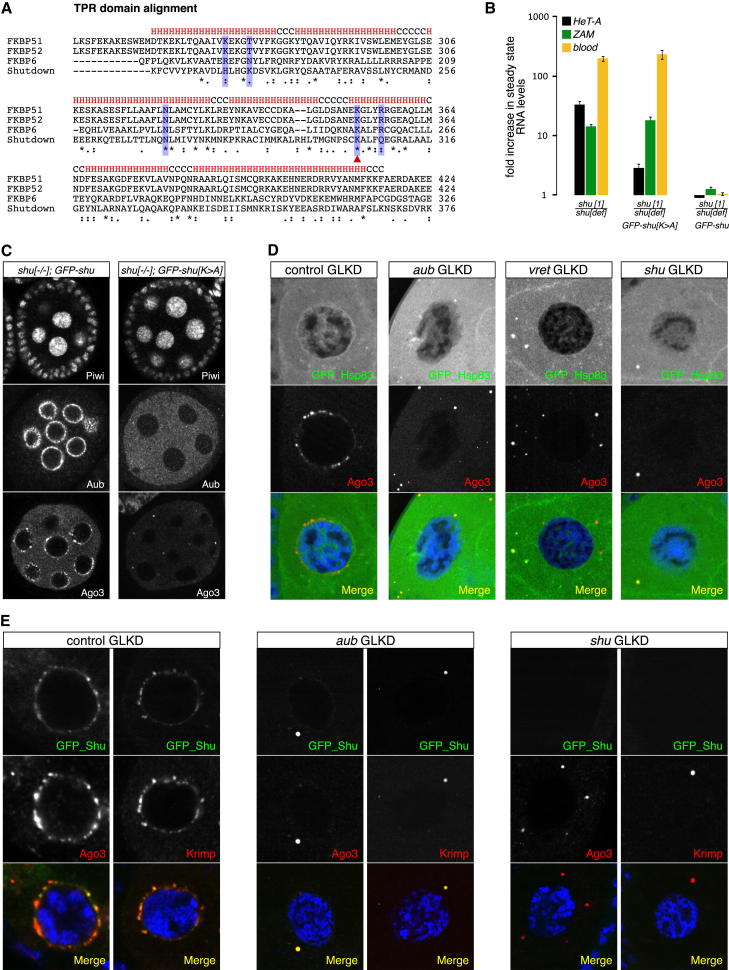
TPR domains interact with the C-terminal MEEVD peptide of Hsp90 (Scheufler et al., 2000). The key amino acid residues for an Hsp90 interaction are present in the Shu TPR domain (Figure 4A). We mutated Lys 304 into Ala (K304A) in the GFP tagged rescue construct and tested whether shu[K304A] could rescue shu loss of function alleles. The wild-type shu-GFP-rescue construct fully restored fertility and PIWI protein localization. In contrast, shu[K304A] flies were sterile, lacked Aub/AGO3 nuage localization and showed elevated TE levels (Figures 4B and 4C). Essentially identical results were obtained upon mutating Gln 308 into Glu (Figure S3). In both cases Piwi localization and thus presumably primary piRNA biogenesis was not severely affected although ZAM, a soma specific TE was moderately derepressed. It is possible that the introduced TPR mutations do not result in a complete loss of Shu’s function and secondary piRNA biogenesis is more sensitive toward loss of Shu activity.
Figure 4.
Shutdown Cooperates with Hsp83 and Colocalizes with Unloaded AGO3
(A) CLUSTAL alignment of the TPR domains from indicated proteins. α-helical segments indicated with ‘H’; residues critical for an Hsp90 interaction highlighted with blue; the red arrowhead indicates the mutated Lys 304.
(B) Shown are fold changes in RNA levels (normalized to shu het.) of indicated TEs in ovaries from indicated genotypes (n = 3; StDev).
(C) Confocal sections of shu mutant egg chambers expressing wild-type (left) or K304A mutant (right) GFP-Shu stained for Piwi, Aub or AGO3.
(D) Confocal sections of nurse cells from indicated GLKDs stained for GFP-Hsp83, (green), AGO3 (red) and DNA (blue).
(E) Confocal sections of nurse cells from indicated GLKDs stained for GFP-Shu (green), AGO3 or Krimp (red) and DNA (blue).
To support a role of Hsp83 in piRNA biogenesis, we determined its localization in wild-type and piRNA biogenesis defective ovaries using a genomic GFP-tagged Hsp83 construct. In wild-type nurse cells, Hsp83 localized uniformly to the cytoplasm and weakly to the nucleus (Figure 4D). Hsp83 was enriched in a peri-nuclear rim, potentially reflecting nuage accumulation. Strikingly, upon GLKD of aub, vas, vret or shu, Hsp83 colocalized with AGO3 in cytoplasmic foci (Figure 4D). We isolated equivalent amounts of AGO3 from shu-GLKD and control ovaries and found no association of AGO3 with piRNAs upon loss of Shu (Figure S3). This suggests that unloaded AGO3 that localizes to cytoplasmic foci interacts with Hsp83.
In support of its proposed interaction with Hsp83, Shu also localized to the cytoplasmic AGO3 foci in every knockdown that disrupted Aub/AGO3 nuage localization (shown for aub-GLKD in Figure 4E). In all cases, AGO3-Shu foci were also positive for the piRNA biogenesis factor Krimp. Thus, loss of secondary piRNA biogenesis results in the accumulation of the three nuage factors AGO3, Krimp and Shu in large cytoplasmic foci, which also accumulate Hsp83. These foci depended on Krimper (see below), but also formed in the absence of Shu (Figures 4D and 4E). This indicated that the cochaperone Shu is not required for the recruitment of Hsp83 to AGO3 foci, analogous to the Cyp40/AGO1/Hsp90 biology in plants (Iki et al., 2011).
Taken together, our genetic and cell biological data point toward an important role for the Hsp83 machinery in piRNA biogenesis. Indeed, Piwi has been found in a complex with Hsp83 in embryo lysate (Gangaraju et al., 2011) and Hsp83 has been demonstrated to affect phenotypic variation at least in part via the piRNA pathway (Specchia et al., 2010).
A Controlled System to Compare the Impact of Biogenesis Factors on piRNA Populations
The analysis of small RNAs from shu mutant ovaries indicated that Shu is required for all piRNA biogenesis branches. However, this conclusion and also comparisons to other pathway factors were hampered by the distorted morphology of shu mutant ovaries. We therefore turned to robust germline knockdown systems (Ni et al., 2011; Handler et al., 2011) and analyzed the resulting impacts on piRNAs. This offered two advantages: First, overall ovarian morphology was unaffected as somatic cells that maintain the stem cell niche were unperturbed (full mutants of piwi, shu, armi, zuc or vret have rudimentary ovaries). Second, the genetic background of the analyzed flies was nearly identical, minimizing variations in TE load and piRNA cluster composition.
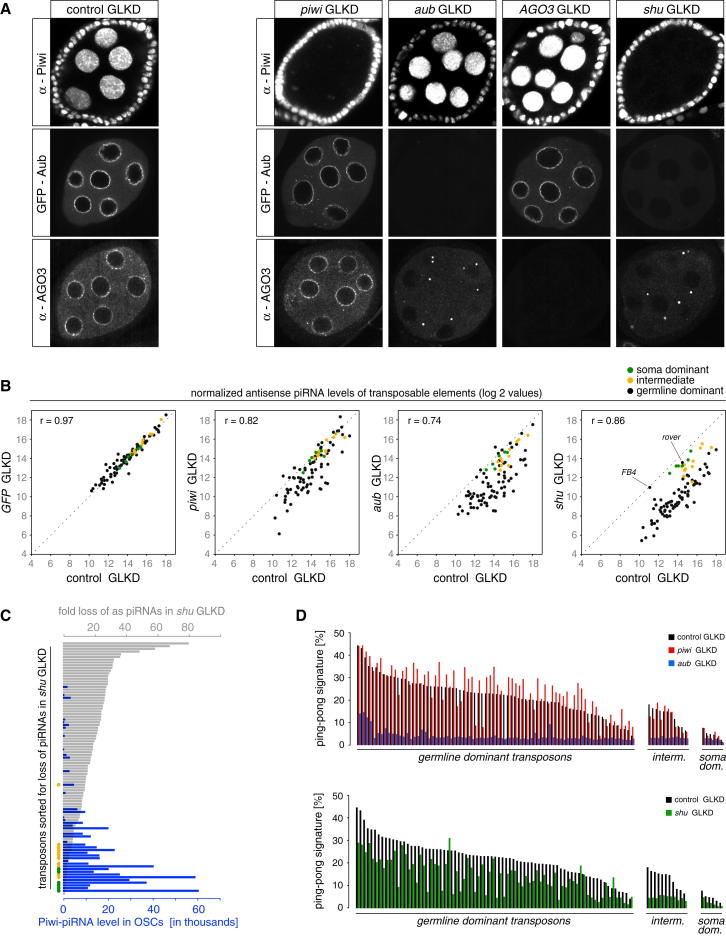
To benchmark this approach, we analyzed knockdowns for the three PIWI proteins. Immunofluorescence analysis indicated a near complete loss of germline Piwi/Aub/AGO3 levels in the respective knockdowns (Figure 5A), which was further supported by western analysis (Figure S1). As Aub antibody stainings were variable, we used a functional GFP tagged aub rescue construct. Loss of Piwi had no detectable impact on Aub or AGO3 localizations; loss of Aub did not affect Piwi, but resulted in cytoplasmic AGO3 bodies; and loss of AGO3 had no impact on localizations of Piwi or Aub (Figure 5A). These results are in line with reports showing that Piwi is dispensable for ping-pong, that Aub is an essential ping-pong factor and that loss of AGO3 permits auto ping-pong of Aub, though more pronounced effects of AGO3 loss on Piwi and Aub localization have been reported (Li et al., 2009; Malone et al., 2009).
Figure 5.
Probing the Importance and Role of Biogenesis Factors in the Germline
(A) Shown are confocal sections of egg chambers from indicated GLKDs stained for Piwi, Aub or AGO3.
(B) Scatter plots showing log2 values of normalized antisense piRNA levels mapping to soma dominant (green), intermediate (yellow) or germline dominant (black) TEs in indicated knockdowns (Pearson correlations based on germline dominant TEs).
(C) Shown is the fold-loss of piRNAs (gray bars) in shu-GLKD versus control-GLKD mapping to Repbase TEs (sorted for fold loss). Blue bars indicate levels of Piwi bound piRNAs for the same TEs. Bullets indicate soma-dominant (green), intermediate (yellow) and germline dominant (black) TEs.
(D) Shown are ping-pong signatures of germline dominant, intermediate and soma dominant TEs in control-GLKD (black bars) compared to piwi-, and aub-GLKDs (top) or shu-GLKD (bottom).
We then performed shu-GLKD and, in agreement with our previous results, localization of all PIWI proteins was disrupted (Figure 5A). We sequenced 18–30 nt small RNAs from ovaries of two control knockdowns (control-GLKD [MTD x w[1118]strain] and GFP-GLKD) as well as from piwi, aub and shu-GLKDs. All libraries were normalized to one million miRNA reads and the levels of antisense 23–30 nt RNAs (piRNAs) were determined for the set of annotated Repbase elements (Figure 5B; Tables S1 and S2).
piRNA levels in the two control libraries were very similar (Figure 5B; Pearson correlation 0.97), arguing for the reproducibility of the approach. Knockdowns of Piwi or Aub resulted in significant reductions of piRNAs mapping to a subset of TEs, with defects being more pronounced in aub-GLKD ovaries (Figure 5B). As we only affected the piRNA pathway in germline cells, we divided TEs into soma dominant, intermediate and germline dominant (Malone et al., 2009). As expected, piRNA levels of soma dominant elements were unaffected and those of intermediate elements showed only slightly lower piRNA levels compared to controls (Figure 5B).
Knockdown of Shu resulted in a remarkably uniform reduction of piRNAs mapping to germline elements (Pearson correlation 0.86; see Figure S4 for the two outliers rover and FB4) and in a minor or no reduction of piRNAs mapping to intermediate or soma dominant elements, respectively (Figure 5B). The universal requirement of Shu for piRNA biogenesis allowed us to test the previous classification of TEs into soma-dominant, intermediate and germline-dominant (Malone et al., 2009). When sorted for the extent of piRNA loss in shu-GLKD ovaries, soma and intermediate elements ranked at the end and a clear anti-correlation resulted with levels of OSC piRNAs (Figure 5C).
We next calculated ping-pong signatures for all TEs (Table S3). The two control libraries showed a strong correlation of 0.97 (Pearson). We sorted elements according to their classification and their ping-pong signature in control knockdowns and plotted the respective ping-pong signatures for the piwi and aub GLKD libraries (Figure 5D). In agreement with data from loss of function mutants (Malone et al., 2009), Piwi was not required for ping-pong (signatures were slightly enhanced), while Aub was uniformly required for it. In shu-GLKD, we observed a moderate to strong reduction in ping-pong signatures for nearly all elements (Figure 5D), arguing that the residual piRNA populations undergo low-level primary and secondary piRNA biogenesis. Knockdown of shu apparently affects primary and secondary piRNA biogenesis in a very uniform manner.
Primary and Secondary piRNA Biogenesis Require Distinct Factors and Feed into a Common Biogenesis Step
Numerous piRNA biogenesis factors have been identified. These include Armi, the Tdrd12 family (Yb, SoYb and BoYb), Zuc, Vas, Spindle-E (Spn-E), Krimp, Tej, Qin and Vret. We tested most of these in the GLKD assay for impacts on PIWI proteins and sequenced small RNAs from a subset of these knockdowns.
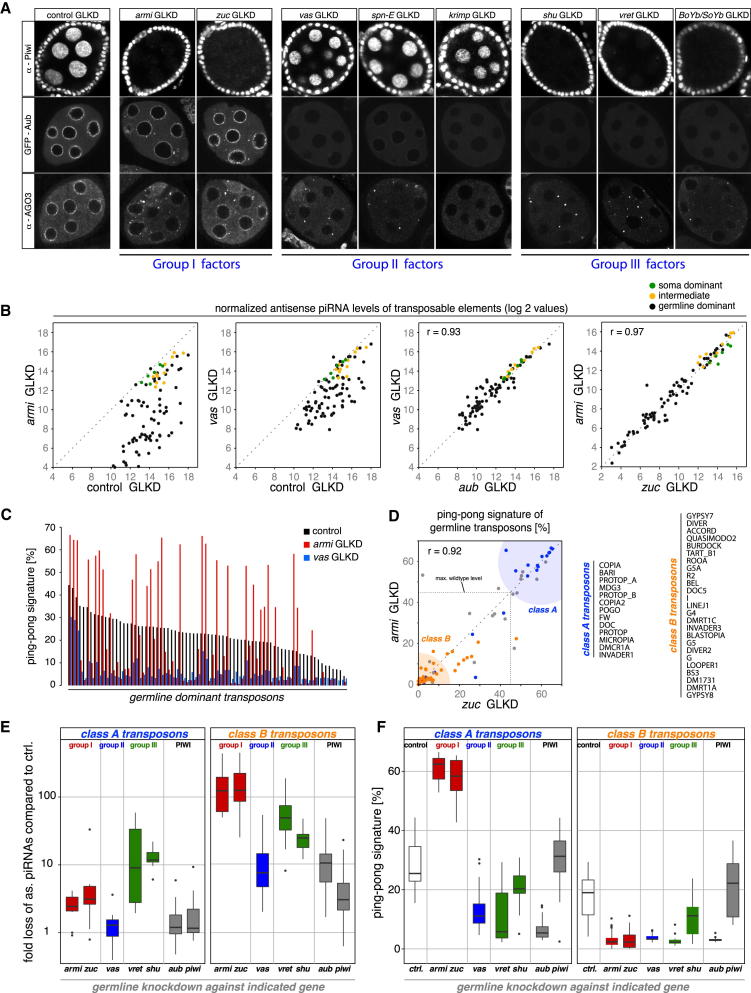
The localization patterns of PIWI proteins in these knockdowns defined three major groups (Figure 6A): Group I factors (Armi, Zuc) were required for Piwi levels and its nuclear accumulation, but had no or only limited impact on Aub and AGO3. Both factors are essential piRNA biogenesis factors in follicle cells supporting a role in primary piRNA biogenesis (Haase et al., 2010; Olivieri et al., 2010; Saito et al., 2010). Group II factors (Vas, Spn-E, Krimp) had no significant impact on Piwi levels, but were required for Aub/AGO3 biology; both proteins were delocalized from nuage, and AGO3 formed cytoplasmic foci (AGO3 foci were always positive for Krimp but were absent in krimp-GLKDs; Figure 6A). No group II factor is required for primary piRNA biogenesis in follicle cells (Malone et al., 2009; Olivieri et al., 2010), suggesting specific roles in germline specific secondary piRNA biogenesis. Group III factors (Shu, Vret, BoYb+SoYb) were required for all three PIWI proteins and resembled a combination of group I and II (Figure 6A). Consistent with their impact on Piwi, group III factors are also required for primary piRNA biogenesis in follicle cells (Handler et al., 2011; Zamparini et al., 2011). Our data support the proposed existence of factors that are specific for the primary (group I) or secondary (group II) biogenesis pathways (Malone et al., 2009) and further indicate that both branches feed into a common biogenesis step that requires group III factors.
Figure 6.
Three Groups of piRNA Biogenesis Factors
(A) Shown are confocal sections of egg chambers stained for Piwi, Aub or AGO3 from indicated GLKDs, which defines three groups of biogenesis factors.
(B) Scatter plots showing log2 values of normalized antisense piRNA levels mapping to soma dominant (green), intermediate (yellow) or germline dominant (black) TEs in the indicated GLKDs (Pearson correlations based on all TEs).
(C) Shown are ping-pong signatures of germline dominant TEs in control-GLKD (black bars) compared to armi- (red), and vas-GLKDs (blue).
(D) Scatter plot showing the correlation (r = Pearson) of ping-pong signatures between armi- and zuc-GLKD libraries. The dashed line indicates the maximum value of any element in control GLKD. Elements with piRNA reductions < 5-fold are in blue, those with > 50-fold loss are in orange, all others in gray. Class A and class B TEs are listed to the right.
(E) Box plots showing the fold loss of antisense piRNAs mapping to class A (left) or class B (right) TEs in the indicated GLKD libraries compared to the control GLKD library.
(F) Box plots showing the ping-pong signatures of class A (left) or B (right) TEs in the control GLKD library (white) or the indicated GLKD libraries.
To support this classification of biogenesis factors at a molecular level, we sequenced small RNAs from GLKDs of armi, zuc (group I) and vas (group II) and compared these to shu or vret knockdown libraries (group III). Consistent with the knockdowns being germline specific, piRNAs mapping to soma elements were not affected and those mapping to intermediate elements were mildly affected in all cases (Figure 6B). Knockdown of the group II gene vas led to decreases in germline piRNA profiles that were highly correlated to the decreases in aub knockdowns (Figure 6B; Pearson correlation 0.93), arguing that Vas is tightly linked to Aub biology. In contrast, knockdown of armi resulted in a very different outcome. While many elements exhibited basically a collapse of piRNA levels, several elements were not or only mildly affected in the armi-GLKD library. A nearly identical impact on piRNA populations was observed in the zuc-GLKD library (Figure 6B; armi/zuc Pearson correlation: 0.93).
An even more striking contrast between armi/zuc knockdowns and vas knockdown was observed when ping-pong signatures were analyzed (Figure 6C). Consistent with previous findings (Malone et al., 2009), vas knockdowns resulted in a strong reduction of ping-pong signatures for nearly all elements. In contrast, knockdown of armi split the set of TEs essentially into two classes. While several elements lost ping-pong signatures, numerous elements exhibited strongly increased signatures (Figure 6C). Highly similar results were obtained from zuc knockdowns (Figure 6D; Pearson correlation: 0.92). Ping-pong signatures correlated with piRNA levels: While most elements with increased ping-pong signatures were only mildly affected in their piRNA levels (Figure 6D; elements with < 5-fold reductions in piRNAs colored blue), elements that lost ping-pong signals typically exhibited > 50-fold reductions in piRNA levels (orange). We specified elements with increased ping-pong signatures and moderate piRNA reductions in armi/zuc-GLKD as class-A elements and those with collapsed piRNA populations and ping-pong signatures as class-B elements (listed in Figure 6D).
We extended the analysis of class-A and B TEs to all libraries and calculated loss of piRNA populations as well as ping-pong signatures. Figures 6E and 6F show box-plots of this data and Figure S5 depicts the piRNA profiles of two representatives (copia for class-A and I-element for class-B) in detail. Several conclusions can be drawn from this analysis:
(1) Levels of class-A element piRNAs were remarkably resistant against knockdown of group I or group II biogenesis factors (Figure 6E). Strikingly, despite comparable piRNA levels, the ping-pong signatures (Figures 6F and S5) either doubled (group I) or collapsed (group II). Loss of group I or group II factors therefore split the population of primary and secondary piRNAs for class-A elements. In agreement with group III factors being required for both piRNA biogenesis branches, only shu- or vret-GLKD resulted in severely reduced piRNA levels (Figures 6E and S5).
(2) piRNA populations of class-B elements were highly sensitive toward loss of any biogenesis factor, with the group II factor Vas having the weakest impact (Figures 6E and S5). Similarly, ping-pong signatures essentially collapsed upon knockdown of any factor with Piwi being the only exception (Figures 6F and S5).
(3) The behavior of class-A elements indicated that group I factors are not required for ping-pong per se. As secondary piRNA biogenesis must be triggered by an input piRNA and must therefore depend at some point on primary piRNA biogenesis, we speculate that maternally deposited piRNAs (Brennecke et al., 2008) allow class-A elements to engage in ping-pong in the absence of zygotic primary piRNA biogenesis. It is unclear why class-B elements do not behave similarly. Different temporal expression patterns of piRNA clusters with distinct TE content may lead to gaps in the availability of precursor RNA molecules that are required for continuous ping-pong.
Primary piRNA Biogenesis Factors Load Both Piwi and Aub Proteins, but Not AGO3
piRNA populations mapping to class-B elements are highly sensitive toward loss of the primary biogenesis (group I) factors Armi or Zuc. The only well established recipient of primary piRNAs in Drosophila is Piwi. Of all knockdowns, however, loss of Piwi had the lowest impact on class-B elements and ping-pong signatures were even slightly enhanced (Figures 6E and 6F). Thus, group I factors must also feed primary piRNAs into another PIWI protein. Indeed, loss of Aub had a pronounced impact on class-B element piRNAs (Figure 6E). However, due to Aub’s involvement in ping-pong, this result was inconclusive.
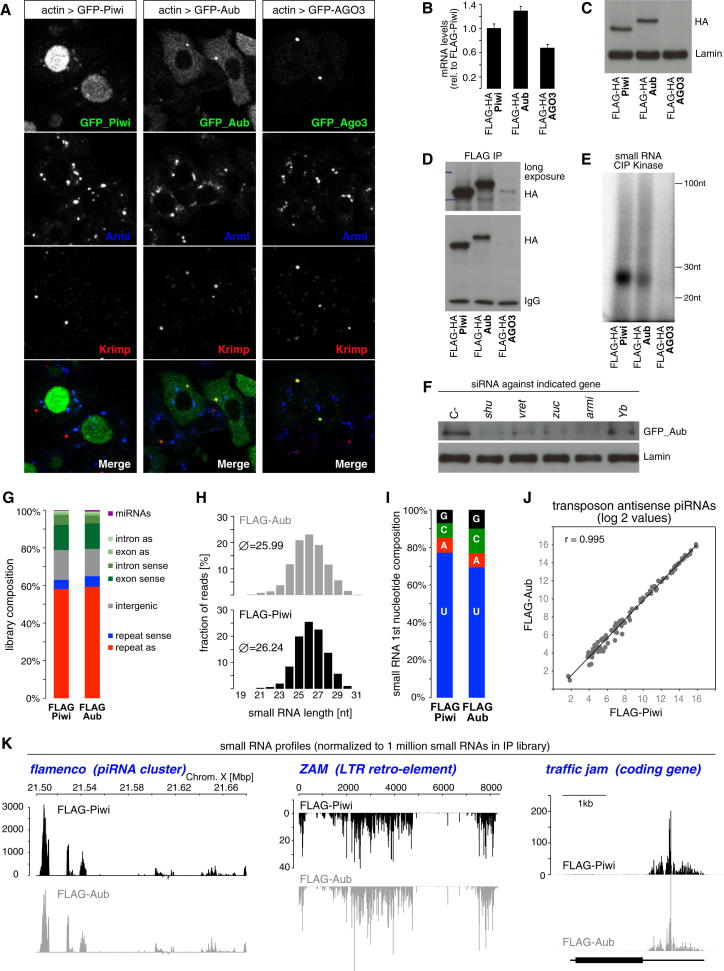
We therefore turned to the OSC system that harbors a primary pathway feeding into Piwi. Importantly, OSCs do not express Aub or AGO3 (Lau et al., 2009; Saito et al., 2009). We expressed GFP- or FLAG/HA-tagged Piwi, Aub or AGO3 in OSCs and determined their subcellular localization and expression levels (Figures 7A–7C). As expected, tagged Piwi localized to nuclei and the faint cytoplasmic accumulations of Piwi colocalized with Armi in Yb-bodies (Figure 7A). Levels of tagged Aub were comparable to Piwi, but Aub localized to the cytoplasm (Figures 7A–7C). Aub was weakly enriched in Yb-bodies but interestingly accumulated in a discrete cytoplasmic dot that stained positive for Krimp, which is endogenously expressed in OSCs but not required for primary piRNA biogenesis (Malone et al., 2009). In contrast, tagged AGO3 (despite comparable mRNA levels; Figure 7B) was hardly detectable by western analysis (Figure 7C) and the residual AGO3 protein localized to Krimp bodies (Figure 7A) that never overlapped with Yb-bodies. We immunoprecipitated all three tagged PIWI proteins and tested for small RNA loading by CIP-Kinase labeling of copurified small RNAs (Figures 7D and 7E). Remarkably, this indicated that Aub was loaded efficiently with primary piRNAs.
Figure 7.
Piwi and Aub, but Not AGO3 Can Be Loaded by the Somatic Primary piRNA Biogenesis Pathway
(A) Shown are IF images of OSCs transfected with the indicated GFP-tagged PIWI proteins (green) stained for Armi (blue) and Krimp (red). The bottom row shows the overlay of all channels.
(B) Shown are relative levels of indicated FLAG/HA-PIWI mRNAs isolated from transfected cells (n = 3; error bars indicate StDev).
(C) Shown are protein levels of FLAG/HA-PIWI proteins isolated from transfected cells in comparison to Lamin levels.
(D) IP-western analysis indicating efficient immunoprecipitation of tagged Piwi and Aub, but not AGO3 from lysate of transfected OSCs.
(E) CIP-Kinase experiment indicating efficient loading of tranfected Piwi and Aub in OSCs.
(F) Western blot indicating that ectopically expressed GFP-Aub is destabilized in OSCs upon knockdown of piRNA biogenesis factors.
(G) Annotation of small RNAs isolated from indicated IPs from OSCs.
(H) Size profile of small RNAs isolated from indicated IPs.
(I) First nucleotide bias of small RNAs isolated from indicated IPs.
(J) Scatter plot of antisense piRNAs isolated from indicated IPs mapping to Repbase TEs.
(K) Profiles of small RNAs isolated from indicated IPs mapping uniquely to the flamenco cluster (left; 200 nt walking window) to the ZAM TE (middle) or to the traffic jam locus (right). All plots were normalized to one million small RNAs in the respective library.
To probe the identity of Aub bound OSC RNAs we sequenced small RNAs from FLAG-Piwi and FLAG-Aub immunoprecipitates. The two RNA populations were mirror images of each other in terms of library composition (Figure 7G), piRNA levels mapping to TEs (Figure 7J; Pearson correlation 0.995) as well as in terms of piRNA profiles mapping to clusters, TEs or genes (Figure 7K). The only slight differences were in terms of the uridine bias at the first nucleotide (Figure 7I; Piwi: 77%; Aub: 69%) and the length distribution (Figure 7H; Piwi: 26.2 nt; Aub: 26.0 nt on average). Both parameters also differ in piRNAs bound by Piwi or Aub in ovaries and follow the same trend (Brennecke et al., 2007). Thus, the rudimentary piRNA pathway in OSCs is fully capable of loading the germline specific Aub protein.
Ectopic expression of AGO3 did not yield efficient AGO3-piRISC formation (Figure 7C and 7E). Based on the following observations, we propose that primary piRNA biogenesis in OSCs is incompatible with AGO3 loading: (1) accumulating evidence suggests that PIWI proteins are degraded if not loaded with piRNAs. Indeed, AGO3 protein levels in OSCs were very low despite comparable mRNA levels. Moreover, GFP-Aub levels in OSCs were significantly reduced upon knockdown of any biogenesis factor including shu (Figure 7F) and Aub levels were nearly undetectable in shu-GLKD ovaries (Figure S1). (2) residual AGO3 colocalized with Krimp in a cytoplasmic body. This body also accumulated Shu, which was never found in Krimp bodies of un-transfected OSCs (not shown). siRNA mediated knockdown of Krimp in OSCs transfected with AGO3 did not result in higher AGO3 levels. Thus, AGO3 behaves in OSCs just as in germline cells that are defective in AGO3-piRISC formation. We conclude that the core genetic requirements for primary Piwi- and Aub-piRISC formation are likely identical and that the primary pathway is either incompatible with AGO3 loading or that AGO3 is actively prevented to be a substrate for primary piRISC formation.
Taken together, our analyses of piRNA populations from group I-III germline knockdowns as well as the analysis of PIWI protein biology in OSCs strongly argue for a very simple model (Figure S6): In somatic cells, a primary biogenesis machinery consisting of group I and group III factors loads Piwi with primary piRNAs. In germline cells, the same set of core factors is required to funnel primary piRNAs into Piwi and Aub but not AGO3. While it is unclear to which extent Piwi participates in ping-pong, Aub efficiently triggers secondary piRNA biogenesis via group II and group III factors and this is the only possibility to load AGO3 with piRNAs, which in turn triggers biogenesis of an Aub bound piRNA.
Discussion
The outcome of this work is threefold: (1) The cochaperone Shutdown is essential for the biogenesis of all Drosophila piRNA populations. (2) Three major groups of piRNA biogenesis factors can be distinguished. (3) Piwi and Aub but not AGO3 are loaded with primary piRNAs, explaining how the cell maintains highly specific piRNA populations in the three PIWI proteins.
The Role of the Cochaperone Shutdown in piRNA Biogenesis
A remarkable feature of the shu mutant phenotype is that piRNA populations for every TE collapse. This already points to a common piRNA biogenesis step downstream of the primary and secondary branches. Our epistatic analysis in somatic follicle cells is consistent with Shu acting at a late step in piRNA biogenesis: Shu is not required for the localization of any known biogenesis factor to Yb-bodies. On the other hand, Shu’s localization to Yb-bodies depends on all other biogenesis factors and even on Piwi, arguing that unloaded Piwi recruits Shu to the Yb-body. Similarly, Shu colocalizes with nonloadable AGO3 in OSCs as well as in ovaries defective of ping-pong in discrete foci that also contain and are dependent on Krimp. Thus, in wild-type and in biogenesis mutants, Shu appears to colocalize with unloaded PIWI proteins.
Shu’s C-terminal TPR domain falls into the class of Hsp90 binders (Scheufler et al., 2000) and Hsp90 is important for small RNA loading into Argonaute proteins (Iki et al., 2010; Iwasaki et al., 2010; Miyoshi et al., 2010). In addition, the plant cochaperone Cyp40 interacts with Hsp90 via its TPR domain and is a critical cofactor for small RNA loading into AGO1 (Iki et al., 2011). Our genetic and localization data support an analogous role for Shu and Hsp90 during small RNA loading into PIWI proteins. Clearly, in vitro assays will be crucial to dissect the precise order of events and the molecular role of Shu, especially its PPIase domain.
Three Groups of piRNA Biogenesis Factors in Drosophila
A major challenge in the field is to assemble piRNA biogenesis factors into pathways that explain the stereotypic populations of piRNAs in vivo. We took advantage of efficient germline specific knockdowns to study the impact of several factors on piRNA populations. Based on levels and localization of PIWI proteins as well as on piRNA populations obtained from several pathway factor knockdowns, we propose three major groups of piRNA biogenesis factors.
Group I factors are required for primary piRNA biogenesis but dispensable for secondary biogenesis. In fact, piRNAs that initiated ping-pong in group I knockdowns were amplified and ping-pong signatures of such TEs were strongly increased, presumably as primary piRNAs that do not feed into ping-pong were absent.
Group II factors are specific for ping-pong, as primary piRNA biogenesis feeding into Piwi was unaffected. An alternative explanation that we cannot exclude is that some or all group II genes are required specifically for Aub biology (primary and secondary) per se. This would similarly leave Piwi bound piRNAs intact and would lead to a collapse in ping-pong. Given our data on Aub loading in OSCs, we favor, however, a model where the primary biogenesis machineries that feed Aub and Piwi are very similar.
Finally, group III factors are required for the biogenesis of Piwi/Aub/AGO3 bound piRNAs. The prototypic member of this group is Shu. Loss of Shu affects essentially all piRNA populations to the same extent. We note that analysis of piRNA populations from vret mutants indicated a role for this group III factor in primary biogenesis but not ping-pong (Handler et al., 2011; Zamparini et al., 2011). The distorted tissue composition of vret mutant ovaries coupled with perdurance of maternal Vret protein or RNA may underlie this discrepancy. The existence of group III factors predicts that primary and secondary piRNA biogenesis feed into a final piRISC maturation step that requires a set of common factors for all PIWI proteins. Given that piRNA biogenesis—irrespective of the source of the precursor RNA—requires an RNA loading step as well as a 3′ trimming step, the existence of group III factors suggests itself.
The three proposed groups serve as a rough classification of biogenesis factors. Clearly, at a molecular level, the precise role of each factor within the biogenesis process will vary considerably. Of note, the classification of group I and group II genes extends to the mouse piRNA pathway. The Armi and Zuc orthologs MOV10L1 and PLD6 are required for primary piRNA biogenesis (Watanabe et al., 2011; Zheng et al., 2010), whereas mouse VAS and TDRD9 (mouse Spn-E) were reported to be dispensable for primary biogenesis but are required for secondary biogenesis pathway (Kuramochi-Miyagawa et al., 2010; Shoji et al., 2009).
Wiring of PIWI Proteins into piRNA Biogenesis Pathways
Our data indicate that Aub is not only loaded via ping-pong, but also via primary piRNA biogenesis. We also postulate that Aub and Piwi proteins are wired into primary piRNA biogenesis processes in a very similar manner, meaning that they require the same or highly overlapping core factors (e.g., Armi or Zuc). In agreement with this, ectopically expressed Aub is loaded in OSCs that harbor a fully functional primary pathway but lack critical ping-pong factors such as Vas. We showed that the genetic requirements for Aub loading in OSCs are identical to those for Piwi. We extrapolate from this that the core primary biogenesis machinery that loads Piwi in the soma also loads Piwi and Aub in the germline. Analyses of piRNA populations from armi versus piwi or aub-GLKDs support a model where Armi and Zuc are required for the biogenesis of both Piwi and Aub bound primary piRNAs. We do not exclude the possibility that—despite a similar biogenesis machinery—populations of primary piRNAs in Aub and Piwi are different. For example, differences in subcellular localizations of PIWI proteins as well as piRNA precursor RNAs might result in such differences.
In contrast to Aub, AGO3 was unstable in OSCs. Coexpression of Aub or simultaneous knockdown of krimp had no impact on AGO3 stability. We therefore conclude that primary piRNA biogenesis is incompatible with AGO3. In fact, also in the germline genetic data indicated that AGO3 depends on secondary piRNA biogenesis for being loaded (Li et al., 2009; Malone et al., 2009). Blocking AGO3′s access to the primary biogenesis machinery would allow the cell to load AGO3 with a unique class of piRNAs if it couples AGO3 loading to a precursor RNA originating from Aub-slicer mediated cleavage of an active TE. This would explain the remarkable bias of AGO3 bound piRNAs being sense and carrying an Adenosine at position ten.
Interestingly, on a primary sequence level Aub—despite its significantly different biology—is more closely related to Piwi than to AGO3. A critical question emanating from this is to which extent Piwi is participating in ping-pong, and if it does not, why. A weak, yet statistically significant, ping-pong signature has been observed between Piwi and AGO3 bound piRNAs (Brennecke et al., 2007; Li et al., 2009). This could mean that there is indeed low level of Piwi-AGO3 ping-pong. An alternative explanation is that the Piwi-AGO3 signal is a misleading computational signal: If Piwi and Aub are loaded via the same primary biogenesis machinery, initiator piRNAs for ping-pong that end up in Aub also end up in Piwi. As primary piRNA biogenesis appears to be nonrandom and preferentially processed piRNAs likely trigger ping-pong more robustly, an “artificial” AGO3/Piwi ping-pong signature might result.
What could be the molecular basis of why Piwi does not or only moderately participate in ping-pong? Either, specific features of Aub (e.g., symmetric Arginine methylation) are funneling this protein into ping-pong and similar features are absent on Piwi. Or, the mere sequestration of Piwi into the nucleus prevents Piwi from participating in ping-pong. Notably, N-terminally truncated Piwi that is still loaded but that cannot translocate into the nucleus is enriched in nuage the proposed site of secondary piRNA biogenesis (Klenov et al., 2011). A simple difference in the subcellular localization of Aub and Piwi might thus contribute to the dramatic differences of piRNA populations residing in Aub or Piwi.
Experimental Procedures
Drosophila Stocks
Flies (stocks listed in Table S4) were kept at 25°C and were aged 5 days before analysis.
Immunocytochemistry
Rabbit (rb) α-Shu was raised against peptides C-SPIQEDVLTLSKPDVKFA (western and IP) and MEENFEPYTPQLLKNP-C (IF). Other antibodies were: rb α-Piwi, α-Aub, α-AGO3 (Brennecke et al., 2007), rb α-Armi (Handler et al., 2011), mouse α-Armi (Saito et al., 2010), rb α-Vret and rb α-Yb (Handler et al., 2011), rb α-Krimp (Lim and Kai, 2007), mouse α-FLAG (SIGMA M2), mouse α-HA (16B12), mouse α-beta-Gal (Promega Z378B), mouse α-Lamin (DSHB ADL67.10). MitoTracker Red CMXRos was used according to the manufacturer’s instructions.
Cell Culture
OSCs were cultured as described in Niki et al. (2006) and transfected with Nucleofector kit V (Amaxa; program T-029). siRNAs (Table S5) were transfected 2x and cells analyzed after 96 hr.
act > GFP_Shu, act > FLAG/HA_Piwi/Aub/AGO3, act > GFP_Piwi/Aub/AGO3 plasmids were produced with the Gateway Collection (DGRC).
Immunoprecipitation
IPs were performed as in Olivieri et al. (2010).
Small RNA Cloning
Total ovarian RNA was isolated with TRIzol and small RNAs from IPs with Phenol/Chloroform. Small RNA libraries were generated as described in Brennecke et al. (2007).
piRNA Labeling
piRNAs purified from IPs were dephosphorylated (CIP) and radioactively labeled with T4 PNK.
Transposon qPCR Analysis
qPCR analysis (primers in Table S6) was carried out according to Handler et al. (2011).
Bioinformatics Analysis
Raw libraries were demultiplexed, trimmed off linker sequences and mapped to the genome (100% match; release 5). For piRNA cluster mapping we considered genome-unique mappers, for TE mappings (Repbase; Jurka et al., 2005) all mappers (up to 3 MM). Libraries were normalized to 1 Mio miRNA reads. Small RNAs mapping to rRNAs, tRNAs and snoRNAs were excluded. The calculation of TE piRNA levels was based on antisense piRNAs. Ping-pong signatures were calculated as in Malone et al. (2009).
Acknowledgments
We thank the Brennecke laboratory for support and discussions. We thank A. Stark for computational help, S. Lopez for fly injections, the CSF for deep sequencing, and M. Madalinski for antibody purifications. We thank M. Siomi, T. Kai, and the DSHB for antibodies; the DGRC for vectors; and T. Schupbach, the Bloomington stock center, the VDRC and TRiP for fly lines. Work in the Brennecke laboratory is supported by an ERC starting grant (FP7/2007-2013; #260711) from the European Union and a START grant (Y 510-B12) from the Austrian Science Fund (F.W.F.). K.S. is supported by a Marie Curie Fellowship. D.O. and J.B. conceived the experiments, D.O. conducted the experiments, R.S. and S.S. provided the small RNA database, K.A.S. provided genetic tools, J.B. and R.S. performed the computational analysis, and J.B., K.A.S., and D.O. wrote the paper.
Published online: August 16, 2012
Footnotes
Supplemental Information includes six figures and seven tables and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2012.07.021.
Accession Numbers
Small RNA libraries were deposited at GEO (accession number GSE38728).
Supplemental Information
References
- Anand A., Kai T. The tudor domain protein kumo is required to assemble the nuage and to generate germline piRNAs in Drosophila. EMBO J. 2011;31:870–882. doi: 10.1038/emboj.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crackower M.A., Kolas N.K., Noguchi J., Sarao R., Kikuchi K., Kaneko H., Kobayashi E., Kawai Y., Kozieradzki I., Landers R. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science. 2003;300:1291–1295. doi: 10.1126/science.1083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangaraju V.K., Yin H., Weiner M.M., Wang J., Huang X.A., Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 2011;43:153–158. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane L.S., Saito K., Nishida K.M., Miyoshi K., Kawamura Y., Nagami T., Siomi H., Siomi M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- Haase A.D., Fenoglio S., Muerdter F., Guzzardo P.M., Czech B., Pappin D.J., Chen C., Gordon A., Hannon G.J. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010;24:2499–2504. doi: 10.1101/gad.1968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D., Olivieri D., Novatchkova M., Gruber F.S., Meixner K., Mechtler K., Stark A., Sachidanandam R., Brennecke J. A systematic analysis of Drosophila TUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011;30:3977–3993. doi: 10.1038/emboj.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Nishikiori M., Jaudal M.C., Matsumoto-Yokoyama E., Mitsuhara I., Meshi T., Ishikawa M. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol. Cell. 2010;39:282–291. doi: 10.1016/j.molcel.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Iki T., Yoshikawa M., Meshi T., Ishikawa M. Cyclophilin 40 facilitates HSP90-mediated RISC assembly in plants. EMBO J. 2011;31:267–278. doi: 10.1038/emboj.2011.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Kobayashi M., Yoda M., Sakaguchi Y., Katsuma S., Suzuki T., Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kawaoka S., Izumi N., Katsuma S., Tomari Y. 3′ end formation of PIWI-interacting RNAs in vitro. Mol. Cell. 2011;43:1015–1022. doi: 10.1016/j.molcel.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C., Bratu D.P., McGinnis-Schultz N., Koppetsch B.S., Cook H.A., Theurkauf W.E. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Klenov M.S., Sokolova O.A., Yakushev E.Y., Stolyarenko A.D., Mikhaleva E.A., Lavrov S.A., Gvozdev V.A. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc. Natl. Acad. Sci. USA. 2011;108:18760–18765. doi: 10.1073/pnas.1106676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Takamatsu K., Chuma S., Kojima-Kita K., Shiromoto Y., Asada N., Toyoda A., Fujiyama A. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 2010;24:887–892. doi: 10.1101/gad.1902110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Robine N., Martin R., Chung W.J., Niki Y., Berezikov E., Lai E.C. Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res. 2009;19:1776–1785. doi: 10.1101/gr.094896.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Vagin V.V., Lee S., Xu J., Ma S., Xi H., Seitz H., Horwich M.D., Syrzycka M., Honda B.M. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell. 2009;137:509–521. doi: 10.1016/j.cell.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.K., Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Hannon G.J. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone C.D., Brennecke J., Dus M., Stark A., McCombie W.R., Sachidanandam R., Hannon G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137:522–535. doi: 10.1016/j.cell.2009.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T., Takeuchi A., Siomi H., Siomi M.C. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat. Struct. Mol. Biol. 2010;17:1024–1026. doi: 10.1038/nsmb.1875. [DOI] [PubMed] [Google Scholar]
- Munn K., Steward R. The shut-down gene of Drosophila melanogaster encodes a novel FK506-binding protein essential for the formation of germline cysts during oogenesis. Genetics. 2000;156:245–256. doi: 10.1093/genetics/156.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J.Q., Zhou R., Czech B., Liu L.P., Holderbaum L., Yang-Zhou D., Shim H.S., Tao R., Handler D., Karpowicz P. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki Y., Yamaguchi T., Mahowald A.P. Establishment of stable cell lines of Drosophila germ-line stem cells. Proc. Natl. Acad. Sci. USA. 2006;103:16325–16330. doi: 10.1073/pnas.0607435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K.M., Okada T.N., Kawamura T., Mituyama T., Kawamura Y., Inagaki S., Huang H., Chen D., Kodama T., Siomi H., Siomi M.C. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J. 2009;28:3820–3831. doi: 10.1038/emboj.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri D., Sykora M.M., Sachidanandam R., Mechtler K., Brennecke J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010;29:3301–3317. doi: 10.1038/emboj.2010.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil V.S., Kai T. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr. Biol. 2010;20:724–730. doi: 10.1016/j.cub.2010.02.046. [DOI] [PubMed] [Google Scholar]
- Qi H., Watanabe T., Ku H.Y., Liu N., Zhong M., Lin H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 2011;286:3789–3797. doi: 10.1074/jbc.M110.193888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine N., Lau N.C., Balla S., Jin Z., Okamura K., Kuramochi-Miyagawa S., Blower M.D., Lai E.C. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr. Biol. 2009;19:2066–2076. doi: 10.1016/j.cub.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Inagaki S., Mituyama T., Kawamura Y., Ono Y., Sakota E., Kotani H., Asai K., Siomi H., Siomi M.C. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461:1296–1299. doi: 10.1038/nature08501. [DOI] [PubMed] [Google Scholar]
- Saito K., Ishizu H., Komai M., Kotani H., Kawamura Y., Nishida K.M., Siomi H., Siomi M.C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010;24:2493–2498. doi: 10.1101/gad.1989510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- Schüpbach T., Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti K.A., Brennecke J. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 2010;26:499–509. doi: 10.1016/j.tig.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M., Tanaka T., Hosokawa M., Reuter M., Stark A., Kato Y., Kondoh G., Okawa K., Chujo T., Suzuki T. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev. Cell. 2009;17:775–787. doi: 10.1016/j.devcel.2009.10.012. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Specchia V., Piacentini L., Tritto P., Fanti L., D’Alessandro R., Palumbo G., Pimpinelli S., Bozzetti M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G.J., Aravin A.A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Chuma S., Yamamoto Y., Kuramochi-Miyagawa S., Totoki Y., Toyoda A., Hoki Y., Fujiyama A., Shibata T., Sado T. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell. 2011;20:364–375. doi: 10.1016/j.devcel.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamparini A.L., Davis M.Y., Malone C.D., Vieira E., Zavadil J., Sachidanandam R., Hannon G.J., Lehmann R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development. 2011;138:4039–4050. doi: 10.1242/dev.069187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Xu J., Koppetsch B.S., Wang J., Tipping C., Ma S., Weng Z., Theurkauf W.E., Zamore P.D. Heterotypic piRNA Ping-Pong requires qin, a protein with both E3 ligase and Tudor domains. Mol. Cell. 2011;44:572–584. doi: 10.1016/j.molcel.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K., Xiol J., Reuter M., Eckardt S., Leu N.A., McLaughlin K.J., Stark A., Sachidanandam R., Pillai R.S., Wang P.J. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. USA. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.