Abstract
Objective
To identify pathogenic mutations responsible for autosomal recessive retinitis pigmentosa in 5 consanguineous Pakistani families.
Methods
Affected individuals in the families underwent a detailed ophthalmological examination that consisted of fundus photography and electroretinography. Blood samples were collected from all participating family members, and genomic DNA was extracted. A genome-wide linkage scan was performed, followed by exclusion analyses among our cohort of nuclear consanguineous families with microsatellite markers spanning the TULP1 locus on chromosome 6p. Two-point logarithm of odds scores were calculated, and all coding exons of TULP1 were sequenced bidirectionally.
Results
The results of ophthalmological examinations among affected individuals in these 5 families were suggestive of retinitis pigmentosa. The genome-wide linkage scan localized the disease interval to chromosome 6p, harboring TULP1 in 1 of 5 families, and sequential analyses identified a single base pair substitution in TULP1 that results in threonine to alanine substitution (p.T380A). Subsequently, we investigated our entire cohort of families with autosomal recessive retinitis pigmentosa and identified 4 additional families with linkage to chromosome 6p, all of them harboring a single base pair substitution in TULP1 that results in lysine to arginine substitution (p.K489R). Results of single-nucleotide polymorphism haplotype analyses were suggestive of a common founder in these 4 families.
Conclusion
Pathogenic mutations in TULP1 are responsible for the autosomal recessive retinitis pigmentosa phenotype in these consanguineous Pakistani families, with a single ancestral mutation in TULP1 causing the disease phenotype in 4 of 5 families.
Clinical Relevance
Clinical and molecular characterization of pathogenic mutations in TULP1 will increase our understanding of retinitis pigmentosa at a molecular level.
Retinitis pigmentosa (RP) is a clinically and genetically heterogeneous group of inherited retinal degeneration disorders characterized by night blindness, gradual constriction of visual fields, and finally, loss of central vision.1 It primarily affects the rod photoreceptors, whereas cone receptor function is compromised as the disease progresses.2 Affected individuals often have severely abnormal or nondetectable electroretinographic responses, and as the disease progresses, melanin pigments like bone spicules appear, along with attenuated retinal vessels.2
Mutations in TULP1 have previously been identified in patients with Leber congenital amaurosis and in patients having RP with early or juvenile onset.3–8 The tub and tubbylike (TULP) gene family consists of 4 members (TUB, TULP1, TULP2, and TULP3), which have been identified in plants and in animals.9,10 TULP protein has an essential role in the development and function of the central nervous system.11 TUB and TULP proteins share a conserved C-terminal region of approximately 200 amino acid residues.12 They also have distinct tissue expression patterns: TULP1 is expressed exclusively in retina, whereas TULP2 is expressed in retina and testis.10,11,13 The N-terminal half ofTULP1protein has a nuclear localization signal, and its tubby domain exhibits DNA-binding activity that demonstrates a potential transcription factor mechanism.12 TULP1 protein controls the vesicular trafficking of photoreceptor proteins in the nerve terminal during synaptic transmission and at the inner segment during translocation of protein to the outer segment.14,15
Herein, we describe 5 consanguineous Pakistani families with multiple individuals having RP. Our objective was to identify pathogenic mutations responsible for autosomal recessive retinitis pigmentosa. Ophthalmological examination of these families is suggestive of retinitis pigmentosa with an early onset. Linkage and exclusion analyses localized the disease phenotype to chromosome 6p harboring TULP1. Bidirectional sequencing identified pathogenic mutations in TULP1 that segregated with the disease phenotype in their respective families and were not present in ethnically matched control samples.
METHODS
PATIENT ASCERTAINMENT
In total, 125 consanguineous Pakistani families with inherited retinal dystrophies were recruited to participate in a collaborative study between the National Center of Excellence in Molecular Biology, Lahore, Pakistan, and the National Eye Institute, Bethesda, Maryland. Institutional review board approval was obtained for this study from both organizations. Participating family members gave informed consent consistent with the tenets of the Declaration of Helsinki. A detailed medical history was obtained by interviewing family members. Fundus photographs were taken at the Layton Rahmatulla Benevolent Trust Hospital, Lahore. Measurements were recorded using electroretinographic equipment (LKC; Gaithersburg, Maryland). Rod responses were determined using an incident flash attenuated by −25 dB, and rod-cone responses were measured at 0 dB. Isolated cone responses were recorded at 0 dB using a 30-Hz flicker over a background illumination of 17 to 34 candela/m2. Blood samples were collected from all participants, and genomic DNA was extracted using a nonorganic method described previously.16
GENOTYPE ANALYSIS
Genome-wide linkage and exclusion analyses were performed using highly polymorphic fluorescent-labeled short tandem repeat markers. Genome-wide scan (ABI PRISM Linkage Mapping Set MD-10; Applied Biosystems, Foster City, California) was completed using a mean spacing of 10 centimorgan (cM). Multiplex polymerase chain reaction (PCR) was carried out in a thermocycler (GeneAmp PCR System 9700, Applied Biosystems). Briefly, each reaction was performed in a 5-µL mixture containing 40 ng of genomic DNA, various combinations of 10mM dye-labeled primer pairs, 0.5 µL of 10× buffer, 1mM deoxyribonucleotide triphosphate mix, 2.5mMmagnesium chloride, and 0.2 U of Taq DNA polymerase (Applied Biosystems). Initial denaturation was performed for 5 minutes at 95°C, followed by 10 cycles of 15 seconds at 94°C, 15 seconds at 55°C, and 30 seconds at 72°C and then 20 cycles of 15 seconds at 89°C, 15 seconds at 55°C, and 30 seconds at 72°C. The final extension was performed for 10 minutes at 72°C. The PCR products from each DNA sample were pooled and mixed with a high-density loading cocktail (containing HD-400 size standards, Applied Biosystems). The resulting PCR products were separated in a DNA analyzer (ABI 3100, Applied Biosystems), and genotypes were assigned using available software (GeneMapper, Applied Biosystems).
LINKAGE ANALYSIS
Two-point linkage analyses were performed using software available in the public domain (FASTLINK version of MLINK from the LINKAGE Program Package; provided by the Human Genome Mapping Project Resources Centre, Cambridge, England)17,18;maximum 2-point logarithm of odds (LOD) scores were calculated using ILINK. Autosomal recessive RP was analyzed as a fully penetrant trait with an affected allele frequency of 0.001. The marker order and distances between the markers were obtained from the Marshfield database (http://www.marshfieldclinic.org/research/pages/index.aspx) and the National Center for Biotechnology Information (Bethesda, Maryland) chromosome 6 sequence maps.
MUTATION SCREENING
Primer pairs for individual exons were designed using available software (primer3 program; available at http://primer3.sourceforge.net/). The sequences and annealing temperatures are available from the authors on request. Amplifications were performed in 25-µL reactions containing 50 ng of genomic DNA, 400nM of each primer, 250µM of deoxyribonucleotide triphosphates, 2.5mM magnesium chloride, and 0.2 U of Taq DNA polymerase in the standard PCR buffer provided by the manufacturer (Applied Biosystems). The PCR amplification consisted of a denaturation step at 96°C for 5 minutes, followed by 40 cycles, each consisting of 96°C for 30 seconds, followed by 57°C (or primer set–specific annealing temperature) for 30 seconds and 72°C for 1 minute. The PCR products were analyzed on a 2% agarose gel and purified by ethanol precipitation. The PCR primers for each exon were used for bidirectional sequencing using a reaction mix (BigDye Terminator Ready, Applied Biosystems) according to the manufacturer’s instructions. Sequencing products were precipitated and resuspended in 10 µL of formamide (Applied Biosystems) and denatured at 95°C for 5 minutes. Sequencing was performed on an automated sequencer (ABI PRISM 3100, Applied Biosystems), and results were assembled (ABI PRISM sequencing analysis software, version 3.7) and analyzed (Seq-Scape software, Applied Biosystems).
PREDICTION ANALYSIS
Evolutionary conservation of T380 and K489 in other TULP1 orthologs was examined using the University of California, Santa Cruz, genome browser (http://genome.ucsc.edu/). The degree of evolutionary conservation of the mutated amino acids and the possible effect of these substitutions on the structure of TULP1 were examined using available online tools (SIFT [http://sift.jcvi.org]) and PolyPhen [http://genetics.bwh.harvard.edu/pph]).
RESULTS
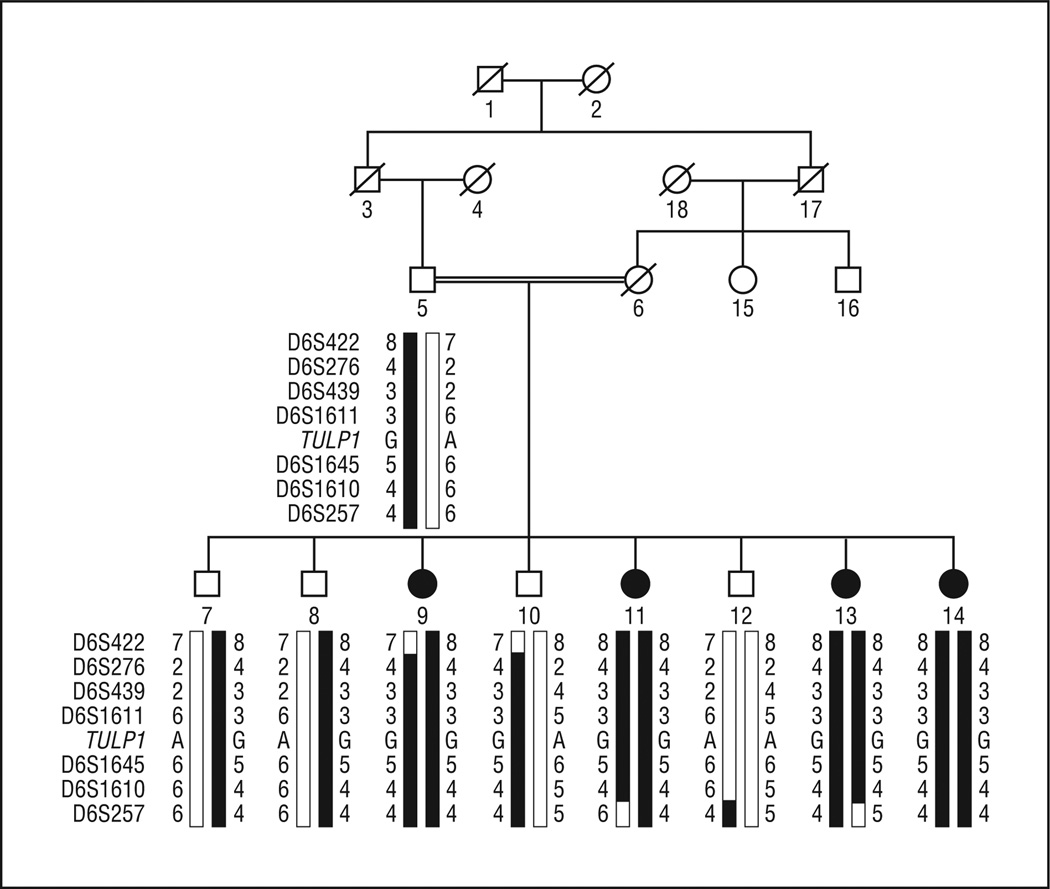
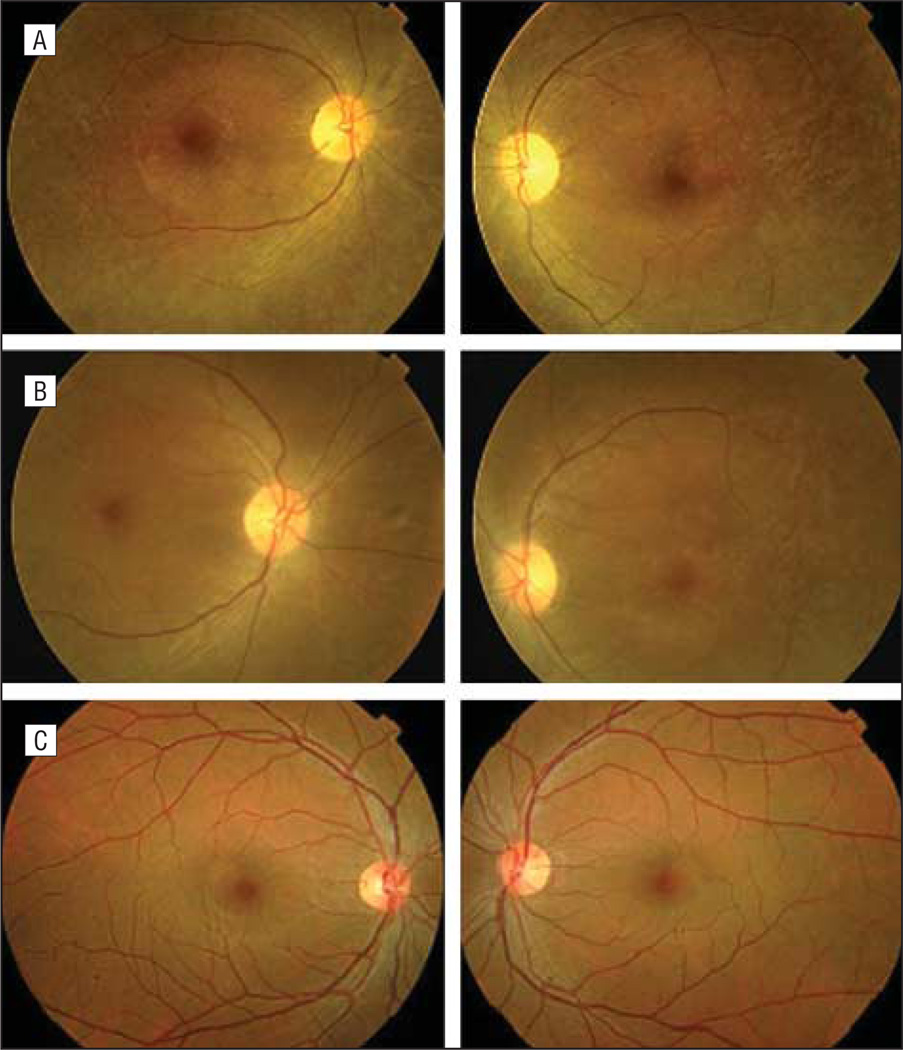
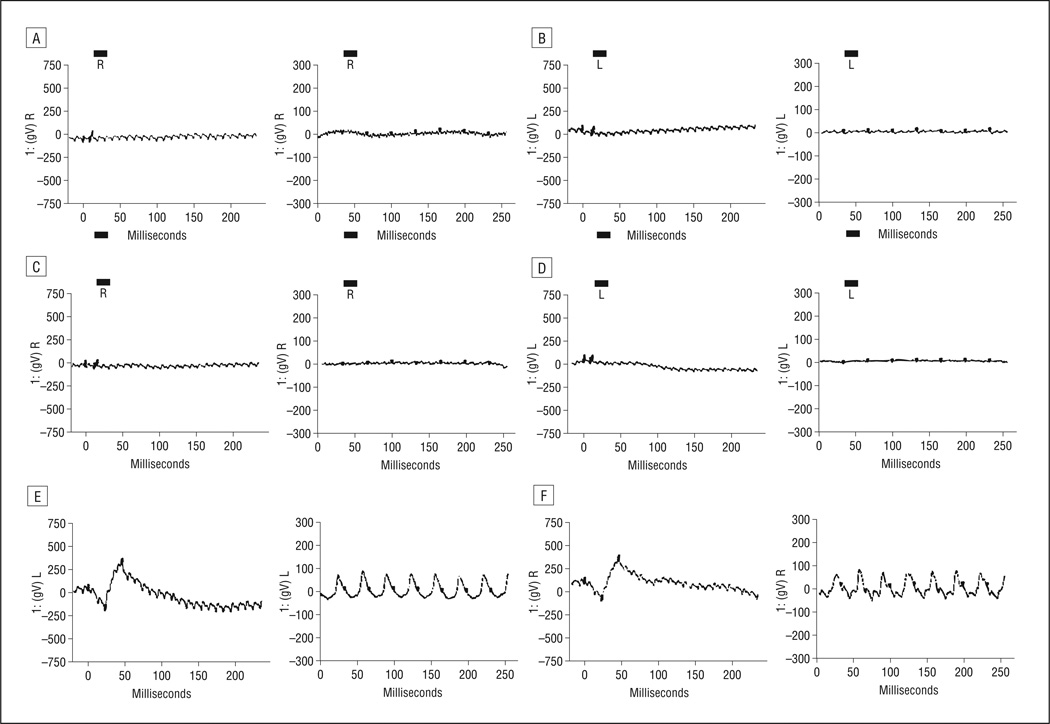
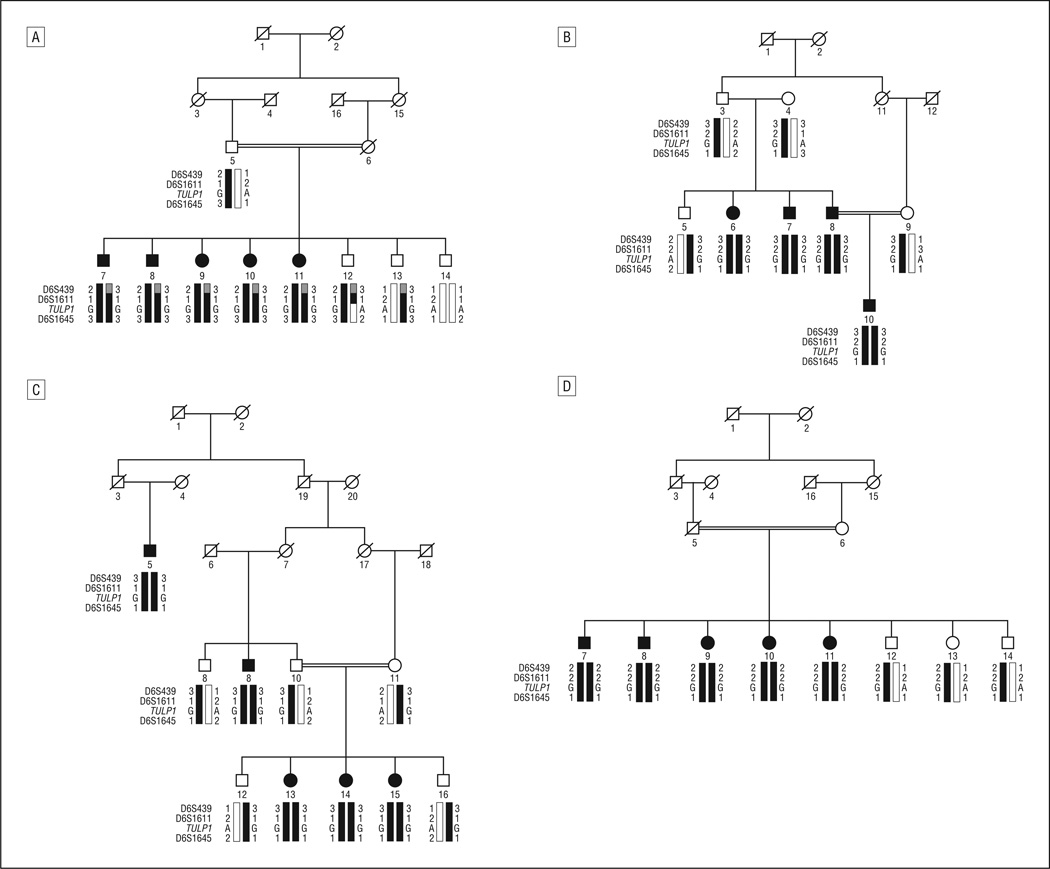
All 5 families (PKRP063, PKRP084, PKRP111, PKRP122, and PKRP171) were recruited from the Punjab province of Pakistan. Reviewed medical records of previous ophthalmological examinations suggested that all affected individuals in these families had experienced night blindness since early childhood, with progressive loss of peripheral vision and decreased visual acuity with age (Table 1). Family PKRP063 was recruited from the southern part of the Punjab province and had 9 family members, including 4 affected members who agreed to participate in the study (Figure 1). Fundus examination in PKRP063 showed attenuated retinal blood vessels and typical RP pigmentation in the midperipheral retina (Figure 2). Electroretinography in affected individuals demonstrated diminished rod and cone responses under scotopic conditions, whereas isolated cone responses using a 30-Hz flicker were absent (Figure 3).
Table 1.
Clinical Characteristics of Individuals in the 5 Pakistani Families With Pathogenic Mutations in TULP1a
| Family | Mutation | Protein Change |
Electroretinographic Findings |
|---|---|---|---|
| PKRP063 | c.A1138G | T380A | Diminished rod and cone responses under scotopic conditions, absent isolated cone response |
| PKRP122 | c.A1466G | K489R | NA |
| PKRP171 | c.A1466G | K489R | NA |
| PKRP111 | c.A1466G | K489R | NA |
| PKRP084 | c.A1466G | K489R | NA |
Abbreviation: NA, not available.
All individuals had early-onset progressive disease. Fundus examination in all individuals showed artery attenuation, pigment deposit, and pale optic disc.
Figure 1.
In PKRP063, haplotypes of 6p markers and segregation of c.1138A>G variation with the disease phenotype. A square represents a male individual; a circle, a female individual; shading, an affected individual; a double line, consanguinity; and a slash mark, a deceased individual. Haplotypes with alleles forming the risk haplotype are shaded black, and alleles not cosegregating with autosomal recessive retinitis pigmentosa are white.
Figure 2.
Fundus photographs in PKRP063 showing right eyes (left) and left eyes (right). A, Individual 9. B, Individual 11. C, Individual 12. Shown are several features associated with retinitis pigmentosa, including waxy pallor of the optic disc, attenuated arterioles, atrophy of retinal pigment epithelium, and peripheral bone spicules.
Figure 3.
Electroretinographic responses in PKRP063 showing combined rod and cone responses and isolated cone responses in right eyes (left panels) and left eyes (right panels). A and B, Individual 9. C and D, Individual 11. E and F, Individual 12. Electroretinographic recordings show no rod and cone response for affected individuals, with photopic flicker response absent as well.
A genome-wide linkage scan was completed for PKRP063, and evidence of significant linkage was only observed with markers on chromosome 6p. Two-point LOD scores of 3.17 and 3.10 (at θ = 0.00) were obtained with markers D6S276 and D6S1610, respectively (Table 2). Subsequently, additional short tandem repeat markers were selected from the Marshfield database, and genomic DNA from all family members was genotyped. Two-point LOD scores of 3.16, 3.15, and 3.01 (at θ = 0.00) were obtained with markers D6S439, D6S1611, and D6S1645, respectively.
Table 2.
Two-Point Logarithm of Odds (LOD) Scores of 6p21.3 Markersa
| Marker | cM | Mb | 0.00 | 0.01 | 0.05 | 0.09 | 0.10 | 0.20 | 0.30 | Zmaxb | θmaxc |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D6S422d | 35.66 | 20.37 | −∞ | 1.10 | 1.55 | 1.53 | 1.19 | 0.76 | 0.33 | 1.56 | 0.07 |
| D6S276d | 44.41 | 24.18 | 3.17 | 3.10 | 2.85 | 2.52 | 1.85 | 1.18 | 0.53 | 3.17 | 0.00 |
| D6S439 | 48.26 | 35.15 | 3.16 | 3.09 | 2.84 | 2.50 | 1.83 | 1.17 | 0.51 | 3.16 | 0.00 |
| D6S1611 | 47.71 | 35.37 | 3.15 | 3.08 | 2.83 | 2.50 | 1.82 | 1.14 | 0.50 | 3.15 | 0.00 |
| D6S1645 | 48.26 | 35.58 | 3.01 | 2.94 | 2.68 | 2.36 | 1.69 | 1.02 | 0.37 | 3.01 | 0.00 |
| D6S1610d | 53.81 | 39.25 | 3.10 | 3.03 | 2.81 | 2.47 | 1.83 | 1.12 | 0.51 | 3.10 | 0.00 |
| D6S257d | 79.92 | 55.91 | −∞ | −0.89 | 0.27 | 0.58 | 0.59 | 0.40 | 0.18 | 0.63 | 0.15 |
Two-point linkage analyses were performed using the FASTLINK version of MLINK from the LINKAGE Program Package (provided by the Human Genome Mapping Project Resources Centre, Cambridge, England), whereas maximum 2-point LOD scores were calculated using ILINK.
Zmax represents maximum 2-point LOD score.
θmax is the recombination fraction at which the maximum LOD score is achieved.
Marker included in genome-wide scan.
Visual inspection of the haplotypes of chromosome 6p markers supported results of the linkage analysis (Figure 1). There is a proximal recombination event at D6S422 in affected individual 9 and a distal recombination event at D6S257 in affected individuals 11 and 13. This places the disease locus in a 44.26-cM (35.54 Mb [million bases]) region on chromosome 6p, flanked by D6S422 proximally and D6S257 distally. This region harbors the previously reported RP-causing gene TULP1.9
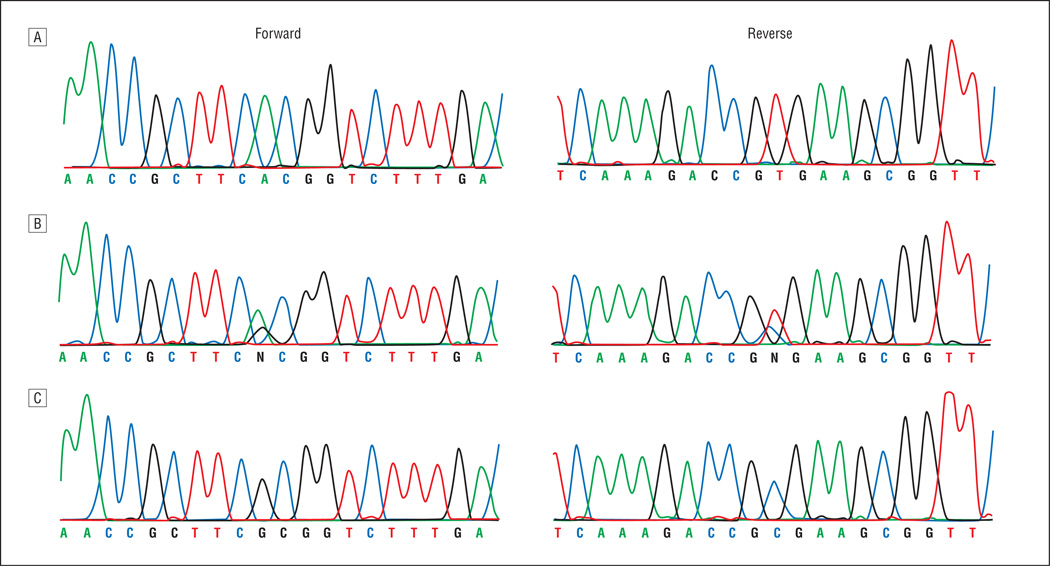
Sequencing of the coding exons of TULP1 identified a single base pair substitution (c.1138A>G), resulting in threonine to alanine substitution (p.T380A) (Figure 4). This mutation segregated with the disease phenotype in the family. All affected individuals were homozygous for the mutation, whereas unaffected individuals were heterozygous carriers or were homozygous for the wild-type allele. This mutation was not present in 192 racially/ethnically matched control chromosomes.
Figure 4.
Forward and reverse sequence chromatograms in PKRP063. A, Individual 12, harboring wild-type allele. B, Individual 7, a heterozygous carrier. C, Individual 9, homozygous for the single base pair substitution c.1138A>G, resulting in threonine to alanine substitution (p.T380A).
Identification of a causal mutation in TULP1 prompted us to investigate our cohort of nuclear families with closely spaced fluorescently labeled short tandem repeat markers spanning the TULP1 locus. We identified 4 families in whom results of linkage and haplotype analyses were suggestive of linkage to chromosome 6p (Figure 5). Maximum 2-point LOD scores (at θ = 0.00) were 3.58 with marker D6S1645 for PKRP084, 2.07 with marker D6S1645 for PKRP111, 2.56 with marker D6S439 for PKRP122, and 3.24 with marker D6S439 for PKRP171. Subsequent sequencing of the coding exons of TULP1 identified a single change (c.1466A>G) in all 4 families, which results in lysine to arginine substitution (p.K489R). All affected individuals were homozygous, whereas unaffected individuals were heterozygous carriers or homozygous for the wild-type allele. This variation was not present in 192 racially/ethnically matched control chromosomes.
Figure 5.
Haplotypes of 6p markers and segregation of c.1466A>G variation with the disease phenotype. A, In PKRP084. B, In PKRP111. C, In PKRP122. D, In PKRP171. Symbols are explained in the legend for Figure 1.
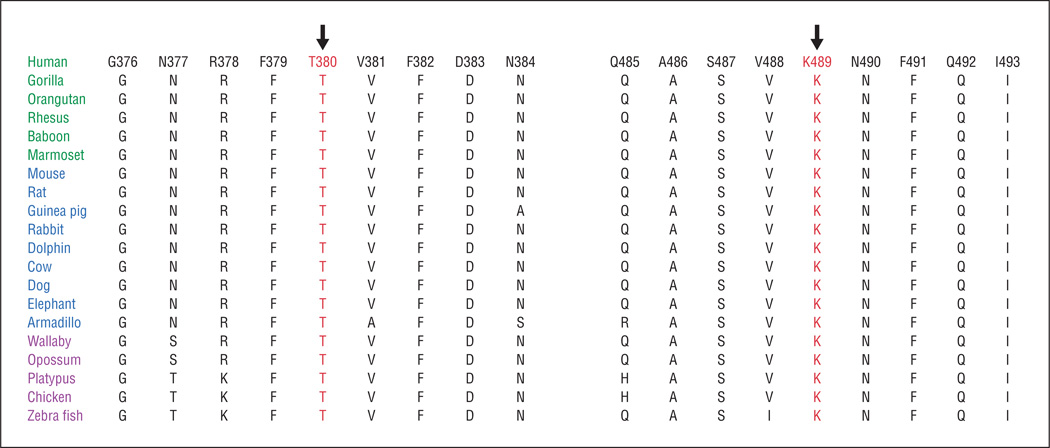
Next, we investigated the evolutionary conservation of T380 and K489 in other TULP1 orthologs. Both T380 and K489 are conserved, along with amino acid residues residing in the immediate neighborhood of both T380 and K489 (Figure 6). Position-specific independent count score differences obtained from PolyPhen suggested that T380A and K489R substitutions could potentially have a deleterious effect on TULP1 structure, with scores of 1.92 and 1.52, respectively (a position-specific independent count score difference >1.0 is probably damaging).
Figure 6.
Sequence conservation of T380 and K489 residues in TULP1 orthologs (primates are green, placental mammals are blue, and vertebrates are purple). The arrows point to amino acid residues T380 and K489, which were mutated in individuals with autosomal recessive retinitis pigmentosa.
All 5 families described herein reside in the same geographic region of Pakistan, but racially/ethnically they belong to different social castes. We were unable to identify any family relationships among these clans. Hence, the presence of common causal mutations in 4 families prompted us to investigate the ancestral relationships, if any, among these 4 families. We used single-nucleotide polymorphisms flanking the causal mutation and constructed a haplotype (CC/TGTC), which was suggestive of a common founder for the c.1466A>G variation (Table 3).
Table 3.
Single-Nucleotide Polymorphism (SNP) Haplotypes of Affected Individuals in PKRP084, PKRP111, PKRP122, and PKRP171 Harboring the p.K489R Mutation in TULP1
| SNP Haplotype | ||||||
|---|---|---|---|---|---|---|
| Family | Individual | rs7764472 | rs34126023 | rs12215920 | rs7770128 | rs12665445 |
| PKRP084 | 8 | C | C/T | G | T | C |
| PKRP111 | 8 | C | C/T | G | T | C |
| PKRP122 | 13 | C | C/T | G | T | C |
| PKRP171 | 7 | C | C/T | G | T | C |
COMMENT
Herein, we describe 5 consanguineous Pakistani families manifesting cardinal symptoms of RP with early onset. Genome-wide linkage and exclusion analyses localized the critical interval to chromosome 6p, and sequential analysis identified causal mutations in TULP1 that segregated with the disease phenotype in their respective families and were not present in racially/ethnically matched control samples. Linkage to chromosome 6p markers, identification of mutations that are predicted to be deleterious for the native protein structure, segregation of these mutations with the disease phenotype in their respective families, and the absence of mutations in racially/ethnically matched control samples suggest that these mutations are responsible for the autosomal recessive RP phenotype. To our knowledge, this is the first study associating TULP1 with autosomal recessive RP in Pakistani families.
To date, more than 25 pathogenic mutations have been reported in TULP1, with most of them in sporadic patients with autosomal recessive RP. Previously, Gu et al3 screened a large cohort of patients with autosomal recessive RP, including 155 German patients and 16 familial cases of Sardinian descent, but identified only 2 pathogenic variations in TULP1, including p.K489R. Likewise, we screened our entire cohort of 125 nuclear families with autosomal recessive RP and identified only 2 pathogenic mutations in TULP1. Although mutations in TULP1 are responsible for the RP phenotype, our results suggest that they are uncommon and that TULP1 does not contribute significantly to the genetic load of autosomal recessive RP. Identification of the pathogenic mutations in TULP1 and the clinical presentation of the disease phenotype associated with these mutations will increase our understanding of RP at a molecular level.
Acknowledgments
Funding/Support: This study was supported in part by the Higher Education Commission and the Ministry of Science and Technology, Islamabad, Pakistan.
Footnotes
Financial Disclosure: None reported.
Additional Contributions: We thank the members of the 5 families for their participation in this research.
REFERENCES
- 1.Heckenlively JR, Yoser SL, Friedman LH, Oversier JJ. Clinical findings and common symptoms in retinitis pigmentosa. Am J Ophthalmol. 1988;105(5):504–511. doi: 10.1016/0002-9394(88)90242-5. [DOI] [PubMed] [Google Scholar]
- 2.Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. Am J Ophthalmol. 1995;119(5):543–562. doi: 10.1016/s0002-9394(14)70212-0. [DOI] [PubMed] [Google Scholar]
- 3.Gu S, Lennon A, Li Y, et al. Tubby-like protein-1 mutations in autosomal recessive retinitis pigmentosa. Lancet. 1998;351(9109):1103–1104. doi: 10.1016/S0140-6736(05)79384-3. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee P, Kleyn PW, Knowles JA, et al. TULP1 mutation in two extended Dominican kindreds with autosomal recessive retinitis pigmentosa. Nat Genet. 1998;18(2):177–179. doi: 10.1038/ng0298-177. [DOI] [PubMed] [Google Scholar]
- 5.Hagstrom SA, North MA, Nishina PL, Berson EL, Dryja TP. Recessive mutations in the gene encoding the tubby-like protein TULP1 in patients with retinitis pigmentosa. Nat Genet. 1998;18(2):174–176. doi: 10.1038/ng0298-174. [DOI] [PubMed] [Google Scholar]
- 6.Paloma E, Hjelmqvist L, Bayés M, et al. Novel mutations in the TULP1 gene causing autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41(3):656–659. [PubMed] [Google Scholar]
- 7.den Hollander AI, van Lith-Verhoeven JJ, Arends ML, Strom TM, Cremers FP, Hoyng CB. Novel compound heterozygous TULP1 mutations in a family with severe early-onset retinitis pigmentosa. Arch Ophthalmol. 2007;125(7):932–935. doi: 10.1001/archopht.125.7.932. [DOI] [PubMed] [Google Scholar]
- 8.Mataftsi A, Schorderet DF, Chachoua L, et al. Novel TULP1 mutation causing leber congenital amaurosis or early-onset retinal degeneration. Invest Ophthalmol Vis Sci. 2007;48(11):5160–5167. doi: 10.1167/iovs.06-1013. [DOI] [PubMed] [Google Scholar]
- 9.North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM. Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci U S A. 1997;94(7):3128–3133. doi: 10.1073/pnas.94.7.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda S, He W, Ikeda A, Naggert JK, North MA, Nishina PM. Cell-specific expression of tubby gene family members (tub, Tulp1, 2, and 3) in the retina. Invest Ophthalmol Vis Sci. 1999;40(11):2706–2712. [PubMed] [Google Scholar]
- 11.Ikeda A, Nishina PM, Naggert JK. The tubby-like proteins, a family with roles in neuronal development and function. J Cell Sci. 2002;115(pt 1):9–14. doi: 10.1242/jcs.115.1.9. [DOI] [PubMed] [Google Scholar]
- 12.Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L. Implication of tubby proteins as transcription factors by structure-based functional analysis. Science. 1999;286(5447):2119–2125. doi: 10.1126/science.286.5447.2119. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda S, Ikeda A, Naggert JK, Nishina PM. Towards understanding the function of the tubby gene family in the retina. Adv Exp Med Biol. 2003;533:309–314. doi: 10.1007/978-1-4615-0067-4_38. [DOI] [PubMed] [Google Scholar]
- 14.Xi Q, Pauer GJ, Marmorstein AD, Crabb JW, Hagstrom SA. Tubby-like protein 1 (TULP1) interacts with F-actin in photoreceptor cells. Invest Ophthalmol Vis Sci. 2005;46(12):4754–4761. doi: 10.1167/iovs.05-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xi Q, Pauer GJ, Ball SL, et al. Interaction between the photoreceptor-specific tubby-like protein 1 and the neuronal-specific GTPase dynamin-1. Invest Ophthalmol Vis Sci. 2007;48(6):2837–2844. doi: 10.1167/iovs.06-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaul H, Riazuddin SA, Yasmeen A, et al. A new locus for autosomal recessive congenital cataract identified in a Pakistani family. Mol Vis. 2010;16:240–245. [PMC free article] [PubMed] [Google Scholar]
- 17.Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. Am J Hum Genet. 1984;36(2):460–465. [PMC free article] [PubMed] [Google Scholar]
- 18.Schäffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44(4):225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]