Abstract
The profile of selective modulation of hepatic cytochrome P450 (P450) gene expression caused by infection with the murine intestinal pathogen Citrobacter rodentium has been well characterized in multiple genetic backgrounds; yet, the mechanisms underlying this modulation are still not entirely understood. Although several studies have addressed the roles of cytokines from the innate immune system, the influence of the adaptive immune system is not known. To address this deficiency, we used mice harboring the severe combined immune deficiency (SCID) spontaneous mutation, which lack mature T and B lymphocytes and are unable to mount an acquired immune response. Female C57BL/6 (B6) and SCID mice were infected orally with C. rodentium and assessed for bacterial colonization/translocation and P450 and flavin monooxygenase-3 (Fmo3) expression levels after 7 days. SCID mice showed similar patterns of colonic bacterial colonization and a similar degree of colonic mucosal hypertrophy compared with infected B6 mice, but SCID mice displayed 6-fold greater bacterial translocation to the liver. In the SCID mice, Cyp4a10 and Cyp2b9 down-regulations were partially and fully blocked, respectively, whereas the regulation of other P450s and Fmo3 was similar in both strains. In the C3H genetic background, the SCID mutation also blocked the down-regulation of Cyp3a11, Cyp3a25, Cyp2d22, and Cyp2c29. The results clearly dissociate bacterial translocation to the liver from hepatic drug-metabolizing enzyme regulation and suggest a possible role of T cells, T-cell cytokines, or other proteins regulated by such cytokines in the selective regulation of a limited subset of hepatic P450 enzymes during C. rodentium infection.
Introduction
Infections and inflammatory diseases are associated with significant alterations in the hepatic expressions and activities of drug-metabolizing enzymes and transporters, including numerous cytochrome P450 (P450) enzymes, UDP-glucuronosyltransferases, sulfotransferases, glutathione transferases, ATP-binding cassette family efflux transporters, and solute carrier family influx transporters (Aitken et al., 2006). Changes in P450-mediated drug clearance, associated with inflammation, have been demonstrated in humans with various infections including influenza and HIV (Chang et al., 1978; Jones et al., 2010), as well as in patients with cancer and heart failure (Frye et al., 2002; Rivory et al., 2002). In both humans and laboratory animals, most P450 enzymes studied are down-regulated (Aitken et al., 2006), primarily by pretranslational mechanisms.
We have studied the attaching and effacing murine enteric pathogen Citrobacter rodentium (CR) as a model for pathogen and host interactions leading to modified host P450 expression. CR infection is associated with a profound down-regulation of mouse liver Cyp4a mRNA and protein expression, as well as of flavin monooxygenase 3 (Fmo3) in the liver (Richardson et al., 2006; Chaluvadi et al., 2009a; Zhang et al., 2009). Drug-metabolizing P450s of the Cyp3a gene subfamily, as well as Cyp2b9, are down-regulated to a lesser extent, whereas other 1-, 2-, and 3-family P450 enzymes are less affected and display some variability in response between experiments. Cyp4f18 and Cyp2d9 mRNAs are induced (Richardson et al., 2006). The magnitudes of the responses are strain-dependent: both Toll-like receptor 4 (TLR4)-expressing C3H/HeOuJ and TLR4-deficient C3H/HeJ mice display more profound intestinal pathology and greater mortality than C57BL/6 (B6) mice (Vallance et al., 2003), and this is accompanied by greater effects on the P450 enzymes in the C3H strains (Richardson et al., 2006; Nyagode et al., 2010), although the enzyme specificity pattern is maintained.
The mechanisms that underlie hepatic gene regulation in the CR model are not well understood. Proinflammatory cytokines are capable of down-regulating many P450 enzymes in hepatocyte cultures and as such are thought to mediate most of the effects of inflammation on P450 expression (Aitken et al., 2006). In the CR model of infection, genetic ablation of tumor necrosis factor-α (TNFα) receptor-1, interleukin (IL)-6, or interferon-γ (IFNγ) each significantly attenuated the down-regulation of specific Cyp3a gene products with IL-6 or IFNγ-knockout effects possibly occurring via negative regulation of TNFα (Nyagode et al., 2010; Kinloch et al., 2011). However, the absence of IL-6, IL-1 receptor-1, TNFα receptor-1, or IFNγ did not attenuate the down-regulation of the genes that are most sensitive to CR infection, i.e., Cyp4a10, Cyp4a14, and Fmo3 (Nyagode et al., 2010; Kinloch et al., 2011). Thus, alternative mechanisms for the regulation of these genes must be sought. The involvement of cells of the adaptive immune system has not been tested.
Several groups have described significantly different immune responses and pathogenicity in T- and B-cell-deficient mice infected with CR (Vallance et al., 2003; Sherman and Kalman, 2004). Severe combined immune deficiency (SCID) mice are homozygous for the SCID spontaneous mutation, and recombination activating gene 1 (RAG1) mice are homozygous-null for RAG1. Both of these models are deficient in mature T and B lymphocytes. Both models are associated with increased or prolonged CR colonization of the colon, although the effect of T- and B-lymphocyte deficiency on tissue pathology and mortality seems to depend on the genetic background and/or the age of the mice (Vallance et al., 2002, 2003; Simmons et al., 2003). Thus, tissue pathology was reduced or shortened, and mortality was delayed in juvenile (3–4 weeks old) SCID mice in a C3H background (Vallance et al., 2003). Tissue pathology was reduced or shortened, and mortality was increased in young RAG1 mice on a C57BL/6 background (Vallance et al., 2002), whereas adult RAG1 mice (C57BL/6) had greatly prolonged tissue pathology, higher bacterial loads in blood, spleen, and liver, but minimal mortality out to 8 weeks of infection (Simmons et al., 2003).
Thus, to better understand the features of CR infection and the accompanying colonic inflammation that influence P450 expression in the liver, we studied hepatic P450 expression in livers of CR-infected SCID mice, on both the C57BL/6 and the C3H background.
Materials and Methods
Bacteria.
Wild-type (WT) CR (51116) were obtained from the American Type Culture Collection (Manassas, VA) and grown in Luria broth. After dilution in phosphate-buffered saline (PBS), the nominal concentration of the bacteria was calculated spectrophotometrically.
Chemicals, Animals, and Treatments.
All chemicals and reagents, unless otherwise specified, were obtained from Sigma-Aldrich (St. Louis, MO). Female B6.CB17-Prkdcscid/SzJ (SCID-B6) and C3SnSmn.CB17-Prkdcscid/J (SCID-J) mice, as well as their respective control strains C57BL/6J (B6) and C3H/HeJ, were obtained from The Jackson Laboratory (Bar Harbor, ME). The mice were given a 1-week acclimatization period in the animal facility and were 9 weeks old at the time of infection. Mice were housed in groups of four to six per cage and were given access to 20% sucrose (control) or 20% sucrose inoculated with CR (infected) as the sole source of drinking water for 24 h while food was withheld. Actual doses were determined from retrospective plating on MacConkey agar, which were determined to be on average 2 × 108 cells/mouse in the B6 experiment and 1.7 × 108 cells/mouse in the C3H experiment. Mice were sacrificed 7 days after administration of sucrose or bacteria. Livers were stored at −80°C for RNA preparation or homogenized in PBS for colony counts. Colon samples were homogenized in PBS for colony counts.
Determination of Tissue Bacterial Loads.
Measurement of colony-forming units of CR was performed as described previously (Chaluvadi et al., 2009a).
Colonic Hyperplasia.
Paraffin-embedded colon tissue sections were stained with hematoxylin and eosin. Three measurements of well oriented crypts were taken in the distal colon for each mouse, using a Zeiss 200M microscope (Carl Zeiss, Inc., Thornwood, NY) with a 20× NA1.4 lens, and Slidebook (Intelligent Imaging Innovations, Denver, CO).
Cytokine Analysis.
Serum samples were assayed for the cytokines IL-2, IL-4, IL-6, IFNγ, and TNFα using the Milliplex mouse cytokine/chemokine kit (Millipore Corporation, Billerica, MA) following the manufacturer's protocol as described previously (Chaluvadi et al., 2009a). Data were analyzed using MasterPlex software 1.2 (Hitachi Software Engineering America, Ltd., San Francisco, CA).
Real-Time Reverse-Transcriptase Polymerase Chain Reaction.
Total liver RNA was prepared using RNA-Bee isolation reagent following the manufacturer's protocol (Tel-Test, Inc., Friendswood, TX). Using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA), purified total RNA was reverse-transcribed according to the manufacturer's protocol. Real-time reverse-transcriptase polymerase chain reaction was performed using the ABI Prism 7000 Sequence Detection System and SYBR Green to determine the expression of mRNAs of interest in mouse liver as described previously (Richardson and Morgan, 2005; Chaluvadi et al., 2009a). The value obtained from each target gene was normalized to the expression of glyceraldehyde-3-phosphate dehydrogenase by the ΔΔCt method (Livak and Schmittgen, 2001). The expression level in control samples was arbitrarily set at 1. Product dissociation curves were checked routinely to ensure primer specificity.
Western Blotting.
Western blotting and chemiluminescent detection were used to measure relative levels of P450 proteins in mouse hepatic microsomes (Chaluvadi et al., 2009a). Antibodies to rat CYP2B1 and CYP4A1 were generously provided by Dr. James Halpert (University of California, San Diego College of Pharmacy, San Diego, CA) and Dr. Gordon Gibson (University of Surrey, Guildford, UK), respectively. Antibodies to guanine nucleotide binding protein β (Gβ T-20) were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Proteins were detected using the SuperSignal West Pico chemiluminescent substrate kit (Thermo Fisher Scientific, Waltham, MA). The intensity of chemiluminescent protein bands was visualized by fluorography on X-ray film and was scanned, and the protein bands were quantified by densitometry. All assays were performed within a linear range, and bands' intensities were normalized to Gβ, a loading control for microsomal samples (Caro et al., 2009).
Statistical Analysis.
Data are presented as mean values ± S.E.M. Equality of variance was assessed by Levene's test, and differences between groups were compared by two-tailed t test (SPSS, Inc., Chicago, IL). The level of significance was set at p < 0.05.
Results
Effect of the SCID Genotype on Pathogenesis, Bacterial Burden, and Serum Cytokines during CR Infection.
All B6 mice infected with CR or given control sucrose solution had lost approximately 5 to 7% of their body weight by 24 h of the start of the experiment, presumably because of the withdrawal of food during that period (data not shown). All mice began to regain weight on the second day. Control B6 and SCID-B mice continued to gain weight throughout the experiment. Infected B6 and SCID-B mice tended to gain weight until days 4 to 5 (although less so than uninfected mice), after which they lost weight so that their final weights were 93 to 94% of their starting values (data not shown). Infected C3H mice had lost 15% of their body weights at sacrifice, whereas infected SCID-J mice lost only 5%.
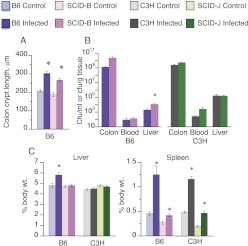
Colon crypt lengths, a measure of the degree of mucosal inflammation and hypertrophy, were significantly elevated in both B6 and SCID-B mice, and there was no notable difference in the size of this effect between the strains (Fig. 1A). Crypt height data are not available for the C3H strains because of sample loss. There was a trend toward increased colonic burdens of CR in SCID-B mice (Fig. 1B); the mean of the SCID-B mice was 20-fold higher than that of the B6 mice, although this did not achieve significance (p = 0.077). SCID-J mice showed a similar but smaller trend. The densities of CR in the livers of B6 and C3H mice were approximately 1 million-fold less than those in their colons (Fig. 1B). Liver CR burden was 6-fold higher in the SCID-B mice than in the B6 mice (Fig. 1B), whereas there was no significant difference between the C3H and SCID-J strains. The SCID genotype had no effect on the amount of CR in the blood in either background. CR infection resulted in a 21% increase in liver weights in B6 mice, but not in the SCID-B animals or in the C3H or SCID-J strains (Fig. 1C). Spleen weights of uninfected SCID-B and SCID-J mice were approximately half the weights of their WT counterparts (Fig. 1C). Infection only increased spleen weight by 1.6-fold in SCID-B compared with a 2.7-fold increase in the B6 animals. However, the fold increase in spleen weight due to infection was similar in the C3H and SCID-J mice.
Fig. 1.
Effect of SCID genotype on mucosal hypertrophy, bacterial colonization, and translocation in CR infection. Nine-week-old female C57BL/6J (B6; n = 6), B6.CB17-Prkdcscid/SzJ (SCID-B6), C3H/HeJ (C3H), or C3SnSmn.CB17-Prkdcscid/J (SCID-J) mice were infected with CR in sucrose solution as described in the text and were sacrificed 7 days later. Control animals received sucrose solution only. A, crypt lengths in the distal colons were measured by microscopy. B, bacterial loads were measured and expressed per gram of tissue in the colon and liver or per ml in the blood. C, spleen and liver weights are expressed per gram of body weight. Data are presented as the means ± S.E.M. n = 6 for all the mice with a B6 background and n = 5 for all the mice with a C3H background except for the C3H control group, which had four animals. *, significantly different from uninfected mice of the same genotype; p < 0.05 Student's t test.
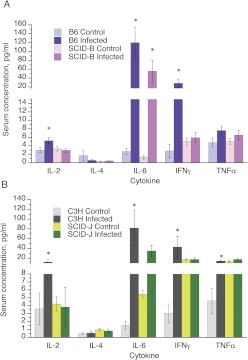
Of the cytokines measured, IL-2, IFNγ, and IL-6 were each elevated in the sera of the infected B6 and C3H mice (Fig. 2). TNFα was elevated in the sera of C3H mice, and a similar but nonsignificant trend was observed in the B6 mice (Fig. 2). The increases in IL-2, IFNγ, and TNFα were not apparent in the SCID mice of either background (Fig. 2). However, the lack of effect on IFNγ and TNFα in the SCID-J mice appeared to be due to much higher levels of these cytokines in the uninfected animals. In contrast, serum IL-6 was still ∼40-fold elevated in the SCID-B and SCID-J mice, and although the mean levels tended to be lower than in the WT strains, this was within the variability of the data. Serum IL-4 levels were not different from control in any mouse strain (Fig. 2).
Fig. 2.
Effect of SCID genotype on plasma cytokine profiles during CR infection. Mice were infected as described in Fig. 1, and sera were harvested on day 7 for analysis of cytokine concentrations as described. A, data from B6 and SCID-B mice. B, data from C3H and SCID-J mice. Data are presented as the means ± S.E.M. *, significantly different from uninfected mice of the same genotype; p < 0.05 Student's t test. The number of animals in each group was the same as that in Fig. 1.
Effect of the SCID Genotype (C57BL/6 Background) on Hepatic P450 and Fmo3 Expression.
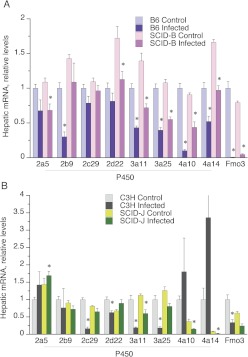
During CR infection, the majority of P450 enzymes studied to date are either down-regulated or unaffected, whereas Cyp2d9, Cyp3a13, and Cyp4f18 are induced (Chaluvadi et al., 2009a; Nyagode et al., 2010). The effects of the SCID mutation in a B6 background on the hepatic response to CR are shown in Fig. 3A for unaffected or down-regulated genes and in Fig. 4A for genes that are normally induced by the infection.
Fig. 3.
Effect of SCID genotype on down-regulation of hepatic mRNAs during CR infection. Mice were infected as described in Fig. 1, and livers were harvested on day 7 for analysis of gene expression by RT-QPCR. A, results from B6 and SCID-B mice. B, results from C3H and SCID-J mice. Data are presented as the means ± S.E.M. *, significantly different from uninfected mice of the same genotype, p < 0.05 Student's t test. The number of animals in each group was the same as that in Fig. 1.
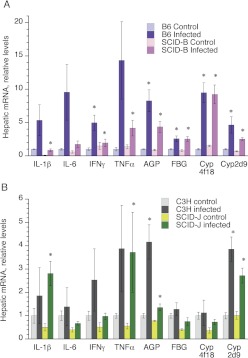
Fig. 4.
Effect of SCID genotype on induction of hepatic mRNAs during C. rodentium infection. Mice were infected as described in Fig. 1, and livers were harvested on day 7 for analysis of gene expression by reverse-transcriptase quantitative polymerase chain reaction. A, results from B6 and SCID-B mice. B, results from C3H and SCID-J mice. Data are presented as the means ± S.E.M. *, significantly different from uninfected mice of the same genotype; p < 0.05 Student's t test. The number of animals in each group was the same as that in Fig. 1.
Most of the P450 genes studied, as well as Fmo3, had similar levels of basal expression in the two strains. The exceptions were Cyp2d22, Cyp3a11, and Cyp4a14, each of which were expressed at 1.4- to 1.7-fold higher levels in the SCID-B mice (Fig. 3). The mRNA levels of Cyp3a11, Cyp3a25, Cyp4a14, Cyp2a5, and Fmo3 were all down-regulated to a similar extent in both strains, when values in infected mice were calculated as a percentage of those in the uninfected mice of the same strain (Fig. 3A). The mRNAs for Cyp2d9, Cyp2f18, and the acute-phase protein fibrinogen were also induced to a similar extent in both strains (Fig. 3B).
The down-regulation of Cyp2b9 mRNA was blocked, and that of Cyp4a10 was attenuated in the SCID-B mice (Fig. 3A). Induction of the mRNA for the acute-phase protein α1-acid glycoprotein appeared to be attenuated as well (Fig. 4A). IFNγ was the only cytokine mRNA that was significantly induced in the liver during CR infection (Fig. 4A), and the effect was greatly attenuated in the SCID-B mice. The other cytokines IL-6, IL-1β, and TNFα showed similar trends, but their induction in the B6 mice was not significant, because of high variability. Finally, Cyp2d22 mRNA showed significant down-regulation only in the SCID-B mice (Fig. 3A).
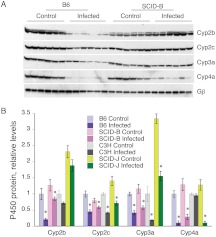
In infected B6 mice, Cyp2b, Cyp2c, Cyp3a, and Cyp4a protein levels were down-regulated to 21, 45, 42, and 10% of control levels, respectively, and the effects on Cyp2b, Cyp2c, and Cyp4a were attenuated (67, 73, and 20% of control, respectively) in the SCID-B strain (Fig. 5). Cyp3a was similarly down-regulated in both strains (Fig. 5).
Fig. 5.
Effect of SCID genotype in a B6 background on regulation of hepatic P450 proteins during CR infection. B6, SCID-B, C3H, or SCID-J mice were infected as described in Fig. 1, and livers were harvested on day 7 for preparation of microsomes. Relative levels of P450 proteins were assayed by Western blotting. A, Western blot of B6 and SCID-B mouse microsomes. The Gβ loading control is shown for the Cyp4a blot, although each blot was probed with Gβ antibody and normalized to its own loading control. B, quantification of the data. Data are presented as the means ± S.E.M. and are normalized to the Gβ signal for each blot. *, significantly different from uninfected mice of the same genotype; p < 0.05 Student's t test. The number of animals in each group was the same as that in Fig. 1.
Effect of the SCID Genotype (C3H Background) on the Responses to CR Infection.
Several differences could be discerned between the patterns of hepatic gene regulation in infected C3H mice (Figs. 3B and 4B) compared with B6 mice (Figs. 3A and 4A). C3H mice showed more profound down-regulation of Cyp3a11 and Cyp3a25 mRNAs during CR infection (Fig. 3B) than that seen in the B6 strain (Fig. 3A), and these effects were greatly attenuated in the SCID-J mice (Fig. 3B). Likewise, CR infection down-regulated Cyp2c29 and Cyp2d22 mRNAs in the C3H strain (Fig. 3B), whereas the effects were not significant in the B6 strain or in the SCID-J mice (Fig. 3, A and B). In contrast, Fmo3 and Cyp2b9 were less sensitive to down-regulation in the C3H mice than in the B6 mice (Fig. 3), and Cyp4f18 was induced only in the B6 strain (Fig. 4). The SCID mutation still had little effect on Fmo3 down-regulation in the C3 background (Fig. 4B).
In contrast to the B6 background, basal levels of Cyp4a10 and Cyp4a14 mRNAs were much lower in the SCID-J mice than in their C3H counterparts (Fig. 4B). Cyp4a10 and Cyp4a14 mRNAs tended to be induced by infection rather than down-regulated in the C3H mice, although this was not significant because of great variability (Fig. 4B). Despite the already relatively low Cyp4a mRNA levels in the SCID-J mice, both Cyp4a10 and Cyp4a14 mRNAs were down-regulated in the infected mice of this strain (Fig. 4B).
In infected C3H mice, Cyp2c and Cyp3a proteins levels were down-regulated to 42 and 19% of control levels, respectively, and the effect on Cyp3a protein was attenuated to 50% of control in the SCID-J strain (Fig. 5). Cyp4a and Cyp2b proteins were not significantly affected by infection in the C3H strain, whereas Cyp4a proteins were down-regulated to 22% of control in the SCID-J animals.
Discussion
This work shows that the lack of mature T and B cells in a C57BL/6 mouse background attenuates the down-regulation or induction of select hepatic P450, acute-phase proteins, and cytokine mRNAs during CR infection while failing to affect the regulation of others. Colonization of the colon by CR is associated with massive infiltration of the lamina propria with T cells and macrophages. These T cells are of both T-helper (Th) 1 (expressing IL-1, TNFα, IL-12, and IFNγ) (Higgins et al., 1999) and Th17 phenotypes (expressing IL-17 and IL-22) (Mangan et al., 2006). Th17 cells trigger antimicrobial responses such as the release of antimicrobial peptides, but they also favor pathogen colonization, possibly by preferential killing of other gut bacteria (Liu et al., 2009). Hyperplasia is associated with an increased expression of keratinocyte growth factor (KGF) in colonic tissue (Higgins et al., 1999). The T-cell infiltration, cytokine production, and epithelial cell proliferation are very similar to what occurs in inflammatory bowel disease (Higgins et al., 1999). As noted above, the effect of T- and B-cell depletion on infection varies with the model used, the genetic background, and the age of the animals. The various studies suggest that T cells are important for bacterial translocation across the epithelial cell barrier (Vallance et al., 2003), generation of CR-directed antibodies (Simmons et al., 2003), and epithelial cell hyperplasia.
Effects of the SCID mutation in a C57BL/6 background have not been reported. We chose to study this genetic background because we have extensively characterized the time course of hepatic gene regulation in relation to intestinal pathology and colonization in this strain (Chaluvadi et al., 2009a), as well as the roles of cytokines and Kupffer cells (Nyagode et al., 2010; Kinloch et al., 2011). In B6 mice infected with CR, the time course or hepatic P450 up- or down-regulation correlates with that of colonic colonization, with a maximum at 7 to 10 days of infection and resolution of the responses by 15 days. Colonic hypertrophy persists until 15 days of infection, and the down-regulation of Fmo3 persists at least until 21 days (Zhang et al., 2009). Therefore, we chose the 7-day time point for the present study.
Although not significant because of high variability, the tendency toward higher bacterial colonization of the colon in SCID-B mice is in general agreement with previous work in young SCID-J or RAG1 mice and in adult RAG1 mice (Vallance et al., 2002, 2003; Simmons et al., 2003). The number of CR cells in the liver was increased 6-fold in our SCID-B mice, whereas liver translocation of the organism was not reported in the previous studies. Despite this effect, the SCID-B mutation actually abrogated the increase in liver weight caused by the infection in WT B6 mice. Furthermore, no effect of the SCID mutation on liver translocation was observed in the SCID-J mice.
The dichotomous effect of T- and B-cell ablation on P450 and Fmo3 regulation in B6 mice further emphasizes that multiple mechanisms regulate different gene products in the liver in this model (Aitken et al., 2006). Only a few P450 enzymes or acute-phase proteins had their regulation blocked or attenuated in SCID-B mice, i.e., Cyp2b9 mRNA (blocked), Cyp4a10 mRNA, Cyp4a protein, Cyp2b protein, and α1-acid glycoprotein mRNA (attenuated). Translocation of CR to the liver can be ruled out as a cause of the observed effects, because it was increased in the SCID-B mice. There was a trend for induction of hepatic cytokine mRNAs to be attenuated in the SCID-B mice, suggesting that reduced cytokine production in the liver could be responsible for the observed effects on the P450s. However, the observed cytokine mRNA responses were small and variable, and we have shown previously that down-regulation of Cyp4a10 and other P450 mRNAs can occur in the absence of significant hepatic cytokine mRNA induction (Chaluvadi et al., 2009a,b) or Kupffer cell depletion (Kinloch et al., 2011). In addition, neither Cyp4a10 nor Cyp4a14 down-regulations were attenuated in IL-6, IFNγ, TNF receptor 1, or IL-1 receptor 1 knockout mice (Nyagode et al., 2010; Kinloch et al., 2011).
In the current study, we found that SCID-B and B6 mice had very similar plasma patterns of proinflammatory cytokines produced mainly by the innate immune system, i.e., IL-6 and TNFα. Cytokines produced largely by T cells, IL-2, and IFNγ were attenuated in the sera of the infected SCID-B mice. Thus, the current results suggest that some P450s such as Cyp4a10 and Cyp2b9 could be regulated by T-cell cytokines such as IL-2, IL-12, IL-17, and IL-22. In addition, the attenuation of Cyp4a10 and Cyp2b9 down-regulation paralleled the effects of infection on liver weight. The reason for the increased liver weight is not known, but we speculate that it could be due to increased production of KGF in colonic tissue (Higgins et al., 1999). Although causality has not been proven between T cells and KGF production, cytokines including KGF (which are growth factors for intestinal epithelial cells) are likely responsible for intestinal hypertrophy seen in CR infection (Sherman and Kalman, 2004). KGF is a powerful mitogen for rat hepatocytes (Itoh et al., 1993; Housley et al., 1994), although its effects on hepatic P450 expression have not been studied.
The majority of the P450 mRNAs studied were regulated similarly in B6 and SCID-B mice, suggesting that T cells and T-cell cytokines are not directly involved in their regulation in this genetic background. Translocation of CR to the liver is unlikely to be a cause of the regulation of these genes, for the same reasons as discussed for Cyp2b9 and Cyp4a10. These genes are also unaffected by whatever factor causes hepatomegaly during infection, because they exhibited the same response in SCID-B mice in the absence of hepatomegaly. The results are consistent with our previously described contributions of IL-6, IFNγ (Nyagode et al., 2010), and TNFα (Kinloch et al., 2011) to in vivo regulation of Cyp3as in this model. Finally, Cyp2d22 regulation correlated with bacterial translocation to the liver, and it is possible that such translocation could be affecting this gene.
The differential regulation of Cyp4a10 and Cyp4a14 in SCID-B mice is the first time we have seen their noncoordinate regulation in the CR model (Richardson et al., 2006; Chaluvadi et al., 2009a; Nyagode et al., 2010; Kinloch et al., 2011). The Cyp4a protein levels in the liver of B6 mice correlated with Cyp4a10 regulation and not Cyp4a14, suggesting either that Cyp4a10 is the dominant Cyp4a protein in mouse liver or that these antibodies to rat CYP4A1 preferentially recognize Cyp4a10 in mouse liver microsomes.
As mentioned under Introduction, pathology, mortality, and down-regulation of P450 gene expression in the liver are higher in C3H strains than in other mouse strains. Therefore, we were interested to study the effect of the SCID mutation in this genetic background. Since P450 regulation during CR infection was not dependent on the presence of functional TLR4 (Richardson et al., 2006), we used C3H/HeJ mice as the control strain as recommended by the supplier. The effects of infection on Cyp4a mRNA expression in the C3H/HeJ strain were opposite to what we observed previously in this strain and the TLR4-competent C3H/HeOu strain (Richardson et al., 2006), and this was also reflected in a lack of down-regulation of Cyp4a proteins. Further analysis revealed that the mice with high Cyp4a mRNA levels were losing weight most rapidly at the time of sacrifice. Because Cyp4a enzymes are induced by starvation via the peroxisome proliferator-activated receptor-α (Kroetz et al., 1998), we propose that the observed mRNA induction was due to reduced food intake secondary to the enhanced pathogenicity of the organism in this strain.
The substantial decreases in Cyp3a11 and Cyp2c29 mRNAs observed in the C3H/HeJ mice are in agreement with the effects reported previously (Richardson et al., 2006). The down-regulations of both mRNAs, as well as of Cyp3a25 and Cyp2d22, were greatly attenuated in the SCID-J mice. In line with the arguments above, this suggests that T-cell cytokines and/or growth factors such as KGF might be responsible for the down-regulation of these P450s under conditions favoring highly pathogenic effects. On the other hand, Cyp2c and Cyp3a proteins were down-regulated similarly in both C3H and SCID-J mice. This suggests that the antibodies may recognize additional related protein species in mouse liver microsomes.
In conclusion, the current results suggest that T-cell-derived factors, or molecules regulated by T-cell cytokines, may contribute to the regulation of a subset of hepatic P450 mRNAs and proteins including Cyp4a10 and Cyp2b9. These mechanisms may extend to other genes such as Cyp3a11, Cyp3a25, Cyp2d22, and Cyp2c29 when conditions (e.g., genetic background of the host) favor high pathogenicity.
Acknowledgments
We thank Daniel Kalman for the use of the microscopy facilities and Dean P. Jones and Yongliang Liang (Clinical Biomarkers Laboratory, Department of Medicine, Emory University) for equipment used to quantify serum cytokines.
This work was supported by the National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant R01-DK072372].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- P450
- cytochrome P450
- CR
- Citrobacter rodentium
- Fmo
- flavin monooxygenase
- TLR
- Toll-like receptor
- TNF
- tumor necrosis factor
- IL
- interleukin
- IFNγ
- interferon-γ
- SCID
- severe combined immune deficiency
- RAG
- recombination activating gene
- PBS
- phosphate-buffered saline
- WT
- wild type
- Th
- T helper
- KGF
- keratinocyte growth factor.
Authorship Contributions
Participated in research design: Nyagode, Watkins, Kinloch, and Morgan.
Conducted experiments: Nyagode, Watkins, and Kinloch.
Performed data analysis: Nyagode, Watkins, Kinloch, and Morgan.
Wrote or contributed to the writing of the manuscript: Nyagode, Watkins, Kinloch, and Morgan.
References
- Aitken AE, Richardson TA, Morgan ET. (2006) Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol 46:123–149 [DOI] [PubMed] [Google Scholar]
- Caro AA, Evans KL, Cederbaum AI. (2009) CYP2E1 overexpression inhibits microsomal Ca2+-ATPase activity in HepG2 cells. Toxicology 255:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaluvadi MR, Kinloch RD, Nyagode BA, Richardson TA, Raynor MJ, Sherman M, Antonovic L, Strobel HW, Dillehay DL, Morgan ET. (2009a) Regulation of hepatic cytochrome P450 expression in mice with intestinal or systemic infections of citrobacter rodentium. Drug Metab Dispos 37:366–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaluvadi MR, Nyagode BA, Kinloch RD, Morgan ET. (2009b) TLR4-dependent and -independent regulation of hepatic cytochrome P450 in mice with chemically induced inflammatory bowel disease. Biochem Pharmacol 77:464–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KC, Bell TD, Lauer BA, Chai H. (1978) Altered theophylline pharmacokinetics during acute respiratory viral illness. Lancet 1:1132–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RF, Schneider VM, Frye CS, Feldman AM. (2002) Plasma levels of TNF-alpha and IL-6 are inversely related to cytochrome P450-dependent drug metabolism in patients with congestive heart failure. J Card Fail 8:315–319 [DOI] [PubMed] [Google Scholar]
- Higgins LM, Frankel G, Douce G, Dougan G, MacDonald TT. (1999) Citrobacter rodentium infection in mice elicits a mucosal Th1 cytokine response and lesions similar to those in murine inflammatory bowel disease. Infect Immun 67:3031–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housley RM, Morris CF, Boyle W, Ring B, Biltz R, Tarpley JE, Aukerman SL, Devine PL, Whitehead RH, Pierce GF. (1994) Keratinocyte growth factor induces proliferation of hepatocytes and epithelial cells throughout the rat gastrointestinal tract. J Clin Invest 94:1764–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Suzuki M, Mitsui Y. (1993) Keratinocyte growth factor as a mitogen for primary culture of rat hepatocytes. Biochem Biophys Res Commun 192:1011–1015 [DOI] [PubMed] [Google Scholar]
- Jones AE, Brown KC, Werner RE, Gotzkowsky K, Gaedigk A, Blake M, Hein DW, van der Horst C, Kashuba AD. (2010) Variability in drug metabolizing enzyme activity in HIV-infected patients. Eur J Clin Pharmacol 66:475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinloch RD, Lee CM, van Rooijen N, Morgan ET. (2011) Selective role for tumor necrosis factor-α, but not interleukin-1 or Kupffer cells, in down-regulation of CYP3A11 and CYP3A25 in livers of mice infected with a noninvasive intestinal pathogen. Biochem Pharmacol 82:312–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroetz DL, Yook P, Costet P, Bianchi P, Pineau T. (1998) Peroxisome proliferator-activated receptor alpha controls the hepatic CYP4A induction adaptive response to starvation and diabetes. J Biol Chem 273:31581–31589 [DOI] [PubMed] [Google Scholar]
- Liu JZ, Pezeshki M, Raffatellu M. (2009) Th17 cytokines and host-pathogen interactions at the mucosa: dichotomies of help and harm. Cytokine 48:156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Mangan PR, Harrington LE, O'Quinn DB, Helms WS, Bullard DC, Elson CO, Hatton RD, Wahl SM, Schoeb TR, Weaver CT. (2006) Transforming growth factor-beta induces development of the T(H)17 lineage. Nature 441:231–234 [DOI] [PubMed] [Google Scholar]
- Nyagode BA, Lee CM, Morgan ET. (2010) Modulation of hepatic cytochrome P450s by Citrobacter rodentium infection in interleukin-6- and interferon-{gamma}-null mice. J Pharmacol Exp Ther 335:480–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson TA, Morgan ET. (2005) Hepatic cytochrome P450 gene regulation during endotoxin-induced inflammation in nuclear receptor knockout mice. J Pharmacol Exp Ther 314:703–709 [DOI] [PubMed] [Google Scholar]
- Richardson TA, Sherman M, Antonovic L, Kardar SS, Strobel HW, Kalman D, Morgan ET. (2006) Hepatic and renal cytochrome P450 gene regulation during Citrobacter rodentium infection in wild-type and Toll-like receptor 4 mutant mice. Drug Metab Dispos 34:354–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivory LP, Slaviero KA, Clarke SJ. (2002) Hepatic cytochrome P450 3A drug metabolism is reduced in cancer patients who have an acute-phase response. Br J Cancer 87:277–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman MA, Kalman D. (2004) Initiation and resolution of mucosal inflammation. Immunol Res 29:241–252 [DOI] [PubMed] [Google Scholar]
- Simmons CP, Clare S, Ghaem-Maghami M, Uren TK, Rankin J, Huett A, Goldin R, Lewis DJ, MacDonald TT, Strugnell RA, et al. (2003) Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect Immun 71:5077–5086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Deng W, Jacobson K, Finlay BB. (2003) Host susceptibility to the attaching and effacing bacterial pathogen Citrobacter rodentium. Infect Immun 71:3443–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallance BA, Deng W, Knodler LA, Finlay BB. (2002) Mice lacking T and B lymphocytes develop transient colitis and crypt hyperplasia yet suffer impaired bacterial clearance during Citrobacter rodentium infection. Infect Immun 70:2070–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Chaluvadi MR, Reddy R, Motika MS, Richardson TA, Cashman JR, Morgan ET. (2009) Hepatic flavin-containing monooxygenase gene regulation in different mouse inflammation models. Drug Metab Dispos 37:462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]