Abstract
Metabolites in safety testing have gained a lot of attention recently. Regulatory agencies have suggested that the kinetics of preformed and in vivo-formed metabolites are comparable. This subject has been a topic of debate. We have compared the kinetics of in vivo-formed with preformed metabolites. trans-3,5,4′-Trihydroxystilbene [trans-resveratrol (RES)] and its two major metabolites, resveratrol-3-sulfate (R3S) and resveratrol-3-glucuronide (R3G) were used as model substrates. The pharmacokinetics (PK) of R3S and R3G were characterized under two situations. First, the pharmacokinetics of R3S and R3G were characterized (in vivo-formed metabolite) after administration of RES. Then, synthetic R3S and R3G were administered (preformed metabolite) and their pharmacokinetics were characterized. PK models were developed to describe the data. A three-compartment model for RES, a two-compartment model for R3S (preformed), and an enterohepatic cycling model for R3G (preformed) was found to describe the data well. These three models were further combined to build a comprehensive PK model, which was used to perform simulations to predict in vivo-formed metabolite kinetics. Comparisons were made between in vivo-formed and preformed metabolite kinetics. Marked differences were observed in the kinetics of preformed and in vivo-formed metabolites.
Introduction
Pharmaceutical companies and regulatory agencies (Baillie et al., 2002; Hastings et al., 2003; Smith and Obach, 2005; Prueksaritanont et al., 2006; Frederick and Obach, 2010) have increasingly focused attention on drug metabolites in safety testing (MIST). Two regulatory guidelines were recently published on this topic: 1) Center for Drug Evaluation and Research (U.S.) (http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm079266.pdf) and 2) International Conference on Harmonisation (http://www.emea.europa.eu/docs/en_GB/document_library/Scientific_guideline/2009./09/WC500002720.pdf). These guidelines recommend that metabolite safety evaluation studies be performed as early as possible during the clinical development program. The recommendation is to synthesize the metabolite and to evaluate it in preclinical toxicity studies.
One major assumption that underlies metabolite toxicity evaluation studies is that the kinetic behavior of a preformed metabolite is the same as that of the metabolite formed in vivo after administration of a parent compound. This theory has been a topic of debate (Prueksaritanont et al., 2006; Pang et al., 2008; Pang, 2009). It is understood that the pharmacokinetics (PK) of a preformed metabolite depends on the absorption, distribution, metabolism, and excretion properties of the metabolite, whereas the kinetics of a metabolite generated in vivo depends on the parent as well as the metabolite (Prueksaritanont et al., 2006). Therefore, differences between the kinetic behavior of a preformed metabolite and the same metabolite generated in vivo could be the result of intrinsic differences between the disposition of the parent and its metabolite, e.g., their physicochemical properties or their interactions with transporters. Metabolites are generally more polar than their precursors. A polar preformed metabolite may experience diffusional barriers to its penetration into an eliminating organ, and hence its elimination clearance may be less than that of an in vivo-generated metabolite, whose entry into the eliminating organ is in the form of a more lipophilic parent (Pang, 1985). In addition, it is important to note that the diffusional barrier to penetration in eliminating organ pertains to the biliary excretion of substance and does not refer to renal (filtration) clearance.
As observed by Pang and coworkers (Pang et al., 2008; Pang, 2009), although preformed metabolite administration may not directly reflect the time course of the in vivo-formed metabolite, the kinetic data on the preformed metabolite can be extremely useful to develop a robust model for predictions and simulations. The data generated from preformed metabolite administration can be wisely incorporated into a comprehensive PK model of the parent metabolite to improve predictions of the behavior of formed metabolite through modeling and simulation.
In the present study, metabolite kinetics of preformed and in vivo-generated metabolites were compared with two approaches: 1) assuming similar PK of preformed versus in vivo-formed metabolite and 2) assuming dissimilar PK of preformed versus in vivo-formed metabolite. The first approach assumes that the systemic or elimination clearance of in vivo-formed and preformed metabolites are similar (Pearson and Wienkers, 2009), whereas the second approach does not make this assumption. The goal of the present study was to build a comprehensive PK model. This comprehensive PK model was used to predict in vivo formation of trans-resveratrol-3-sulfate (R3S) and trans-resveratrol-3-O-glucuronide (R3G) after trans-3,5,4′-trihydroxystilbene (RES) administration. Simulation assuming different elimination clearances of preformed and in vivo-formed metabolites was compared with simulation with the assumption that PK of preformed versus in vivo-formed metabolites are comparable.
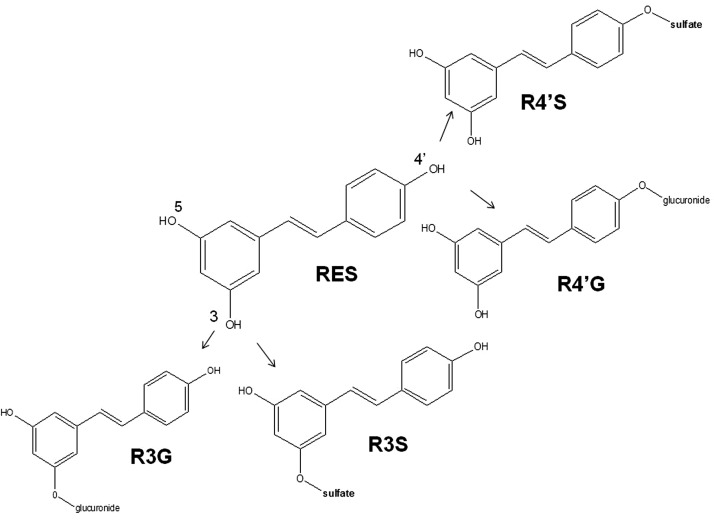
The polyphenol RES (Fig. 1) was used as a model substrate in this study. RES is almost completely metabolized into its sulfated and glucuronidated metabolites in humans as well as rodents (Meng et al., 2004; Hoshino et al., 2010). RES is useful as a model substrate for polyphenols that are heavily conjugated into phase II metabolites. Conjugated metabolites are generally more polar than phase I metabolites. We have previously reported the synthesis of pure RES metabolites resveratrol-3-sulfate and resveratrol-3-glucuronide (Fig. 1) (Iwuchukwu et al., 2012). The PK of RES and synthesized and purified R3S and R3G were characterized in mice. These data were used to develop PK models. Our models corroborate differences in the PK of preformed versus in vivo-formed metabolites (Prueksaritanont et al., 2006; Pang et al., 2008; Pang, 2009).
Fig. 1.
Structure of RES and its four monoconjugated metabolites, i.e., R3S, R4′S, R3G, and R4′G.
Materials and Methods
Chemicals.
RES was purchased from Cayman Chemical (Ann Arbor, MI). R3S, R3G, and trans-resveratrol-4′-O-glucuronide (R4′G) for calibration were purchased from Toronto Research Chemicals (North York, Canada), and they were additionally synthesized for animal dosing studies in the laboratory of Dr. Daniel J. Canney (Temple University, Philadelphia, PA) (Iwuchukwu et al., 2012). trans-Resveratrol-4′-sulfate (R4′S) was also synthesized in the laboratory of Dr. Daniel J. Canney. Other reagents were purchased from standard sources. All reagents for analytical procedures were of analytical grade.
Animals.
Male C57BL/6 mice weighing between 20 and 25 g were supplied by The Jackson Laboratory (Bar Harbor, ME) and maintained in the American Association for the Accreditation of Laboratory Animal Care-accredited University Laboratory Animal Resources of Temple University. Animals were fed a normal diet, with water continuously available, and housed in a standard 12-h dark/light cycle. Animals were acclimatized for 4 days before procedure. During the procedures, animals were not provided food for the initial 10 h of sampling to rule out any variability of gall bladder emptying in response to food. Animals had free access to water during the procedure. All animal studies were approved by the Institutional Animal Care and Use Committee.
Catheterization.
Right carotid artery cannulation was performed under anesthesia using the EZ-Anesthesia (Bethlehem, PA) apparatus with 1.5% isoflurane and 2 l/min oxygen. An incision was made to the right of midline in the neck and the right carotid artery was isolated. The right carotid artery was ligated, a small cut was made, and a medical grade vinyl catheter tubing (0.28 mm i.d. × 0.64 mm o.d.; SCI, Lake Havasu City, AZ) filled with heparin-saline (50 IU/ml; APP Pharmaceuticals, LLC, Schaumburg, IL) was inserted into the right carotid artery. The cannula was tied into place, exteriorized at the back of the neck, and the incision sutured. Animals were allowed to recover from the surgery.
Drug Administration and Blood Sampling.
RES was solubilized in 20% 2-hydroxypropyl-β-cyclodextrin in saline (Juan et al., 2010). Synthesized and purified R3G and R3S (Iwuchukwu et al., 2012) were solubilized in saline. The carotid artery cannula was used for systemic drug administration and blood sampling. Heparin-saline (20 μl, 50 IU/ml) was used to flush the cannula after systemic administration or blood sampling. RES was administered intra-arterially (i.a.) at 15 mg/kg (65.79 μM/kg). R3S was administered at 5 mg/kg (16.23 μM/kg i.a.) and R3G was administered at 3.5 mg/kg (8.67 μM/kg i.a.). These doses were selected by performing pilot studies, which gave R3S and R3G exposures [area under the plasma concentration-time curves (AUCs)] in a range comparable with R3S and R3G observed upon 15 mg/kg RES (i.a.) administration. Blood (20 μl) was serially sampled at 2.5, 5, 10, 15, 45, 90, 180, 300, 420, and 600 min. A 24-h collection was additionally made for animals with functional cannulas. A total of 3 to 5 mice per dose and time point was used. For 15 mg/kg RES, 5 mice were available for all the time points except 4 mice at 600 min and 2 mice at 24 h. For 5 mg/kg R3S, 4 mice were available for all the time points until 600 min, and for 3.5 mg/kg R3G, 4 mice were available for all the time points except 3 mice at 24 h. Blood samples were centrifuged at 14,000 rpm for 2 min, and harvested plasma was collected and stored at −80°C until liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
LC-MS/MS Analysis.
RES, R3S, R4′S, R3G, and R4′G concentrations in plasma were measured with an electrospray ionization LC-MS/MS system (API 4000; ABSciex, Foster City, CA) set in negative ion scan mode as described previously (Iwuchukwu et al., 2012). In brief, ascorbic acid (2.5 μl of a 15% solution) was added to 10 μl of plasma samples and vortexed for 1 min. Then, 30 μl of methanol containing 78 ng/ml acetaminophen [(APAP) internal standard (IS)] was added and vortexed for 1 min and centrifuged at 15,000 rpm for 15 min at room temperature. The supernatant (10 μl) was injected into the LC-MS/MS system. The chromatographic separation system consisted of a guard column (Zorbax SB-C18, 5 μm, 4.6 × 12.5 mm; Agilent Technologies, Santa Clara, CA), an analytical column (Zorbax SB-C18, 5 μm, 4.6 × 150 mm; Agilent Technologies), and a gradient mobile phase of 5 mM ammonium acetate (phase A) and methanol (phase B). The elution started with 10% B at 0 min and linearly increased to 20% B over 2 min, then to 35% B from 2 to 10 min. The gradient was further increased to 60% from 10 to 12 min and remained constant at 60% B for 17 min. Subsequently, methanol was decreased to 10% over 17 to 19 min. Flow rate of the mobile phase was 1 ml/min, and the flow from the column was split 1:3 into a ABSciex API 4000 triple-quadrupole mass spectrometer equipped with a Turbo IonSpray source operating at 450°C. The column temperature was maintained at 35°C. The column effluent was monitored at the following precursor-product ion transitions: m/z 227→185 for RES, m/z 150→107 for the IS (APAP), 403→113 for R4′G and R3G, and 307→227 for R4′S and R3S with a dwell time of 400 ms for each ion transition. The retention time was ∼5 min for the IS, ∼5.9 min for R4′G, ∼7.3 min for R3G, ∼9.2 min for R4′S, ∼10.2 min for R3S, and ∼14.2 min for RES. The lower limit of quantification (LOQ) was 3.5 ng/ml (0.012 μM) for R4′S, 2.4 ng/ml (0.008 μM) for R3S, 10 ng/ml (0.025 μM) for R4′G and R3G, and 10 ng/ml (0.044 μM) for RES. Both the accuracy and precision of the assay were greater than 85%, expressed by <15% intraday and interday error.
Pharmacokinetic Analysis.
Noncompartmental analysis.
Pharmacokinetic parameters of RES, R3G, and R3S were obtained by noncompartmental analysis with Phoenix, WinNonlin (version 6.1; Pharsight, Mountain View, CA). All concentrations and masses were expressed as molar quantities. The AUC was calculated by the linear trapezoidal method; clearance (Cl) was calculated as Cl = Doseia/AUCia0-inf; volume of distribution at steady state (Vss) was calculated as Vss = Cl × MRT0-inf; the terminal half-life (t1/2) was calculated as 0.693/k, and k was the slope of the terminal regression line, where AUCia0-inf is the area under the curve from time 0 to infinity and AUC0-t is the area under the curve from time 0 to last sampling point.
Characterization of Metabolite Kinetics.
Noncompartmental estimation.
The apparent fraction of RES, converted to R3S and R3G (fm), can be calculated assuming that R3S and R3G are formed directly and only from RES, entry of the metabolite into the eliminating organ is not diffusion limited, and elimination is not perfusion limited, with the following equations:
If metabolite elimination clearance is assumed to be the same whether preformed or in vivo formed, eqs. 1 and 2 simplify to eqs. 3 and 4, respectively (Pang and Kwan, 1983):
where AUCR3SRES is the AUC of R3S when RES is administered i.a., AUCR3GRES is the AUC of R3G when RES is administered i.a., AUCR3SR3S is the AUC of R3S when R3S is administered i.a., and AUCR3GR3G is the AUC of R3G when R3G was administered i.a.
Compartmental analysis.
Because there was wide variability in the individual animal data, most likely due to enterohepatic recycling, a naive averaged data approach was used for modeling (Gabrielsson and Weiner, 2006). This approach has been commonly used and reported by other researchers (Ogiso et al., 1998). The average concentration of each administration group at each sampling time point was used to perform the PK data analysis using SAAM II software system (version 1.2; SAAM Institute, Seattle, WA). For each dataset, the simplest compartment model was tested, and complexity was built into the model in subsequent steps. Model selection was based on goodness of fit and comparison of objective functions. Different weighting schemes were tested in SAAM II. A fractional S.D. of 0.1 was used, and data- and model-relative as well as absolute variance models were evaluated. Model-absolute variance was selected and used, because it gave parameter estimates with the lowest coefficient of variation (CV). Criteria for goodness of fit of each proposed model to the observed data were based on Akaike's Information Criterion as the objective function (Akaike, 1974). The S.E. of the parameter estimation was expressed as %CV (S.E./estimate × 100). The PK parameters for RES (model 1), R3G (model 2), and R3S (model 3) were first characterized as independent parent compounds. Linear PK were assumed at the dose levels used, and elimination solely from the central compartment was assumed. Next, a comprehensive PK model (model 4) for the formation of R3S and R3G was built combining models 1, 2, and 3. This model was used to predict the concentration of its two major in vivo-formed metabolites (R3G and R3S) when RES was administered by an i.a. bolus dose. The volume of distribution of central compartment was calculated as Vc = Dose/C0, where C0 is the initial concentration of drug in plasma. Cl calculated as product of elimination rate constant and volume of central compartment was calculated and reported. Formation clearances of R3S and R3G were calculated by multiplying the volume of central compartment of RES to corresponding formation rate constants, i.e., kf, R3S and kf, R3G. Fraction metabolized was calculated assuming that RES was completely metabolized into R3S and R3G.
Results
Noncompartmental Analysis.
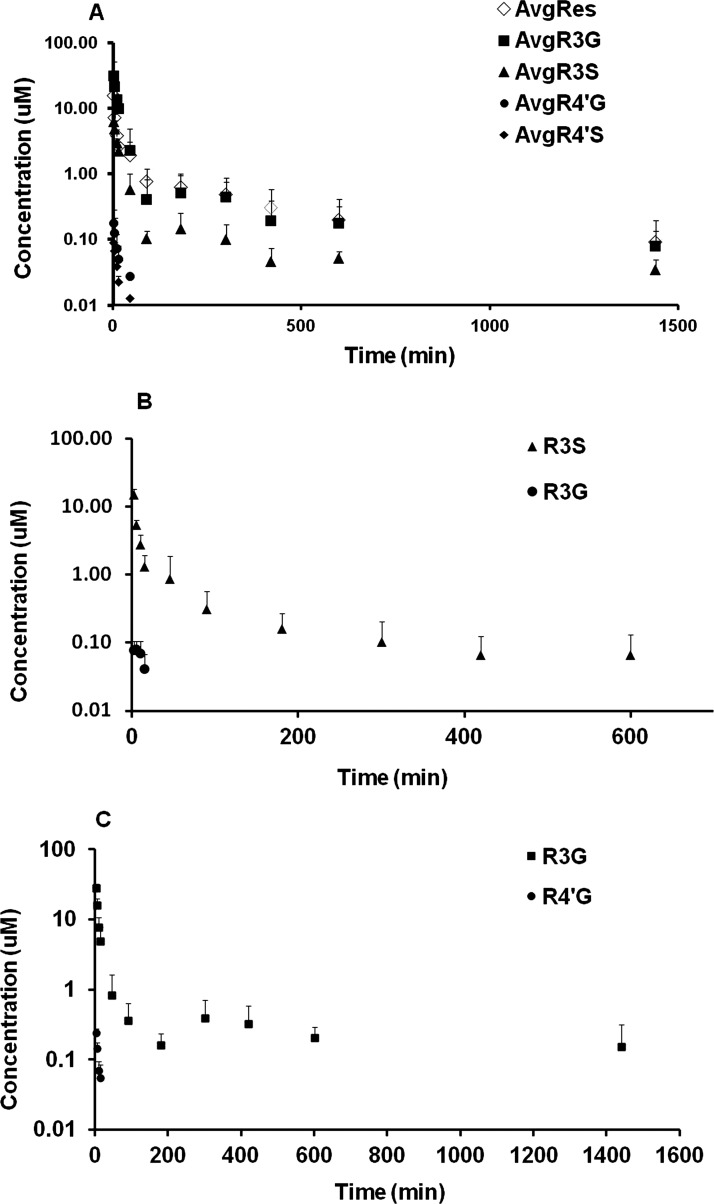
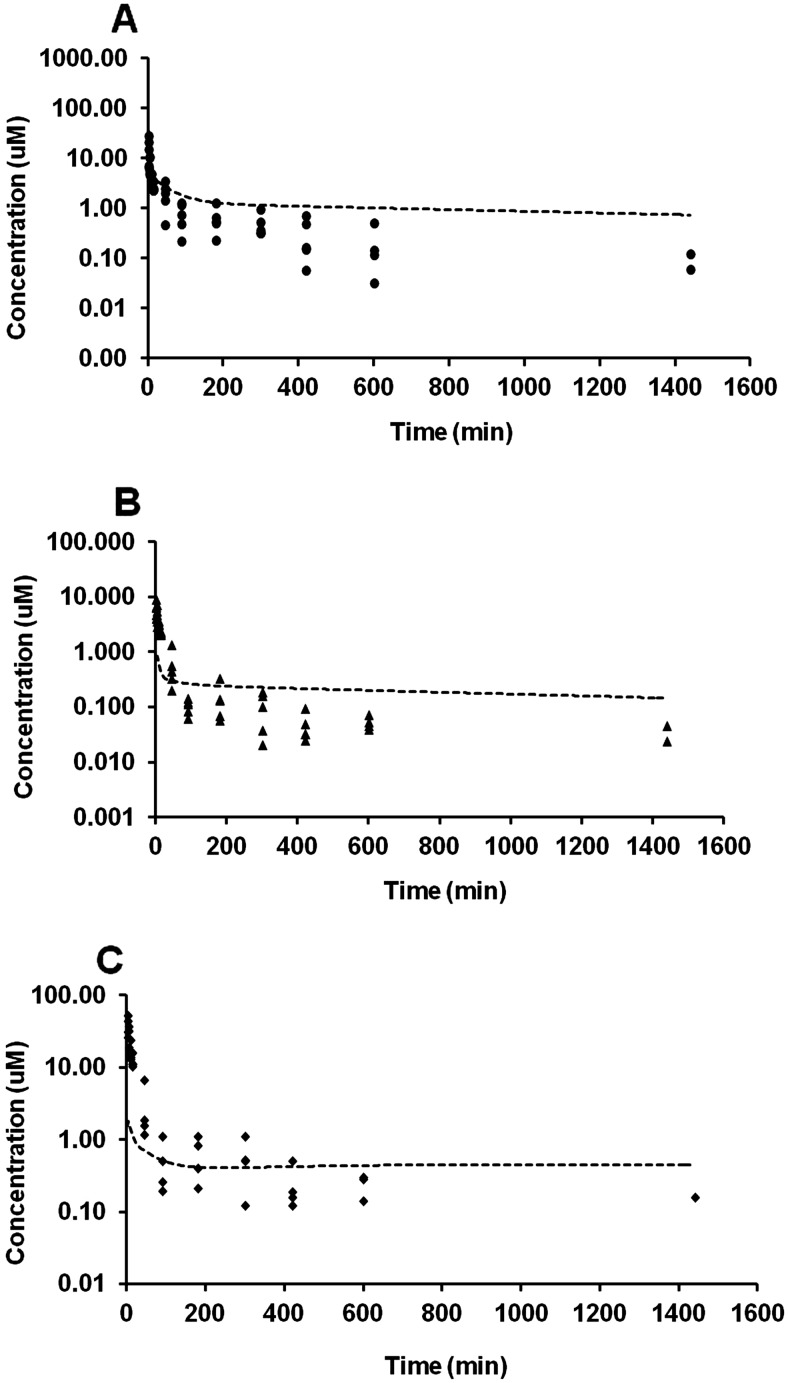
The concentration-time profiles of RES and its metabolites after administration of RES are shown in Fig. 2A. R3S and R3G were the major metabolites and R4′G and R4′S were minor metabolites based on the relative plasma exposure upon i.a. administration. Four metabolites (R3S, R3G, R4′G, and R4′S) exhibited early peak plasma concentration after i.a. RES administration. RES exhibited high clearance and high volume of distribution. Its clearance was interestingly higher compared with the normal blood flow to liver (90 ml · min−1 · kg−1) in mouse (Davies and Morris, 1993). The results of the noncompartmental pharmacokinetic analysis are summarized in Table 1.
Fig. 2.
Mean plasma concentration-time profiles after administration of RES (15 mg/kg i.a.; A), R3S (5 mg/kg i.a.; B), and R3G (3.5 mg/kg i.a.; C). Data are presented as mean ± S.D., n = 4–5.
TABLE 1.
Noncompartmental pharmacokinetic analysis upon a single dose of RES (15 mg/kg i.a.), R3S (5 mg/kg i.a.), or R3G (3.5 mg/kg i.a.)
Data are presented as mean ± S.D.
| Estimate | RES (15 mg/kg i.a.) | R3S (5 mg/kg i.a.) | R3G (3.5 mg/kg i.a.) | Units |
|---|---|---|---|---|
| n = 5 | n = 4 | n = 4 | ||
| AUC0-t | 510.01 ± 105.54 | 243.29 ± 113.10 | 650.66 ± 216.50 | min · μM |
| AUC0-inf | 591.08 ± 167.29 | 255.84 ± 124.98 | 710.10 ± 273.30 | min · μM |
| Cl | 118.77 ± 33.36 | 76.29 ± 37.07 | 13.78 ± 5.75 | ml · min−1 · kg−1 |
| Vss | 37.59 ± 23.70 | 6.37 ± 2.36 | 4.55 ± 1.07 | l/kg |
| Terminal t1/2 | 190.58 ± 69.65 | 128.08 ± 26.21 | 272.48 ± 17.07 | min |
| Cmax | 15.27 ± 9.07 | 14.79 ± 3.15 | 27.84 ± 4.70 | μM |
| Tmax | 2.5 ± 0 | 2.5 ± 0 | 2.5 ± 0 | min |
| Metabolite | R3S | RES | R3S | |
| AUC0-t | 163.87 ± 42.38 | N/A | N/A | min · μM |
| AUC0-inf | 174.94 ± 45.75 | N/A | N/A | min · μM |
| Terminal t1/2 | 201.12 ± 158.12 | N/A | N/A | min |
| Metabolite | R3G | R3G | RES | |
| AUC0-t | 857.36 ± 396.17 | 1.04 ± 0.29 | N/A | min · μM |
| AUC0-inf | 921.23 ± 457.07 | 1.59 ± 0.73 | N/A | min · μM |
| Terminal t1/2 | 264.75 ± 248.66 | 10.05 ± 3.98 | N/A | min |
| Metabolite | R4′G | R4′G | R4′G | |
| AUC0-t | 2.03 ± 1.78 | N/A | 2.08 ± 1.27 | min · μM |
| AUC0-inf | 2.61 ± 1.87 | N/A | 2.51 ± 1.41 | min · μM |
| Terminal t1/2 | 14.11 ± 5.98 | N/A | 8.64 ± 5.57 | min |
| Metabolite | R4′S | R4′S | R4′S | |
| AUC0-t | 0.83 ± 0.39 | N/A | N/A | min · μM |
| AUC0-inf | 1.07 ± 0.43 | N/A | N/A | min · μM |
| Terminal t1/2 | 8.85 ± 4.37 | N/A | N/A | min |
N/A, not applicable.
The concentration-time profiles of R3S and its metabolites after R3S administration at 5 mg/kg i.a. dose are shown in Fig. 2B. R3G was observed in the plasma after R3S administration and decreased rapidly to levels below the LOQ after 15 min. Subsequently, R3G levels were detectable at some points after 180 min in some animals. Due to paucity of data points, the R3G plasma profile could not be characterized after 15 min. The results of noncompartmental pharmacokinetic analysis are summarized in Table 1. RES was not detected after the 5 mg/kg R3S dose. The results also indicated that R3S exhibits a high clearance compared with mouse liver blood flow (Davies and Morris, 1993) and a high volume of distribution.
The concentration-time profiles of R3G and its metabolites after R3G administration at 3.5 mg/kg are shown in Fig. 2C. R4′G was observed in plasma after R3G administration. The AUC ratio of R4′G metabolite to total AUC0-inf (R3G + R4′G) was less than 1%. The results of the noncompartmental pharmacokinetic analysis are summarized in Table 1. R3G exhibits a low clearance and a high volume of distribution in mice. Re-entry peaks of R3G in the plasma, likely due to enterohepatic recirculation, were clearly observed at 5 h after i.a. administration of 3.5 mg/kg R3G.
Estimation of Metabolite Kinetics Assuming Similar Characteristics of Preformed versus In Vivo-Formed Metabolites.
Fraction of RES being metabolized into R3S and R3G using eqs. 3 and 4 (fmR3S and fmR3G) was found to be 0.168 and 0.17, respectively. The sum of apparent fm values from R3S and R3G was 0.34.
Estimation of Metabolite Kinetics Assuming Dissimilar Characteristics of Preformed versus In Vivo-Formed Metabolites.
PK modeling of RES, preformed R3S, and preformed R3G: (models 1, 2, and 3).
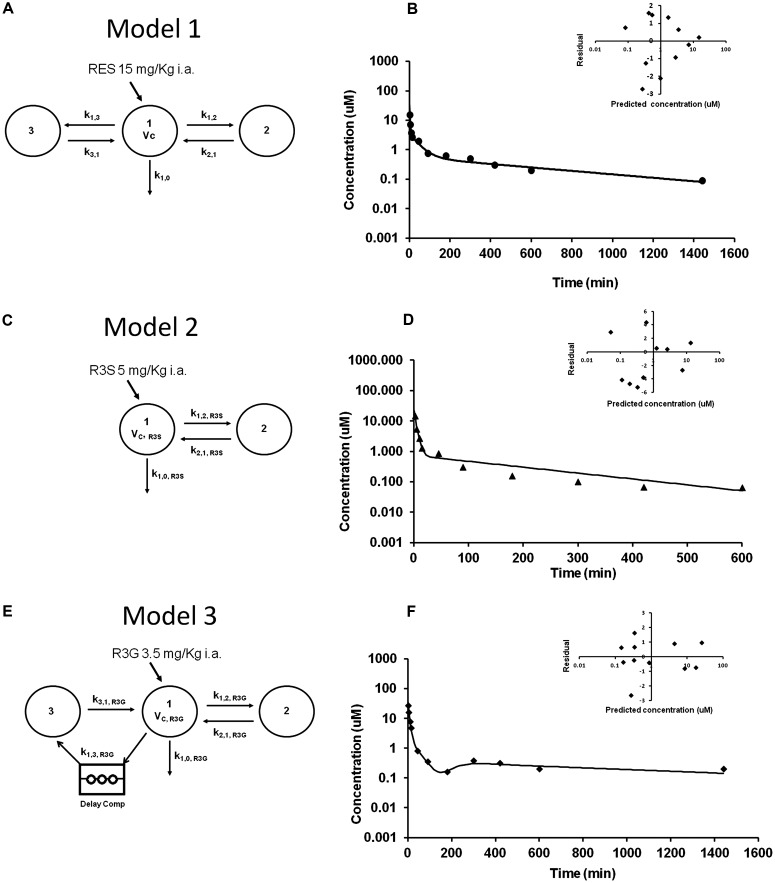
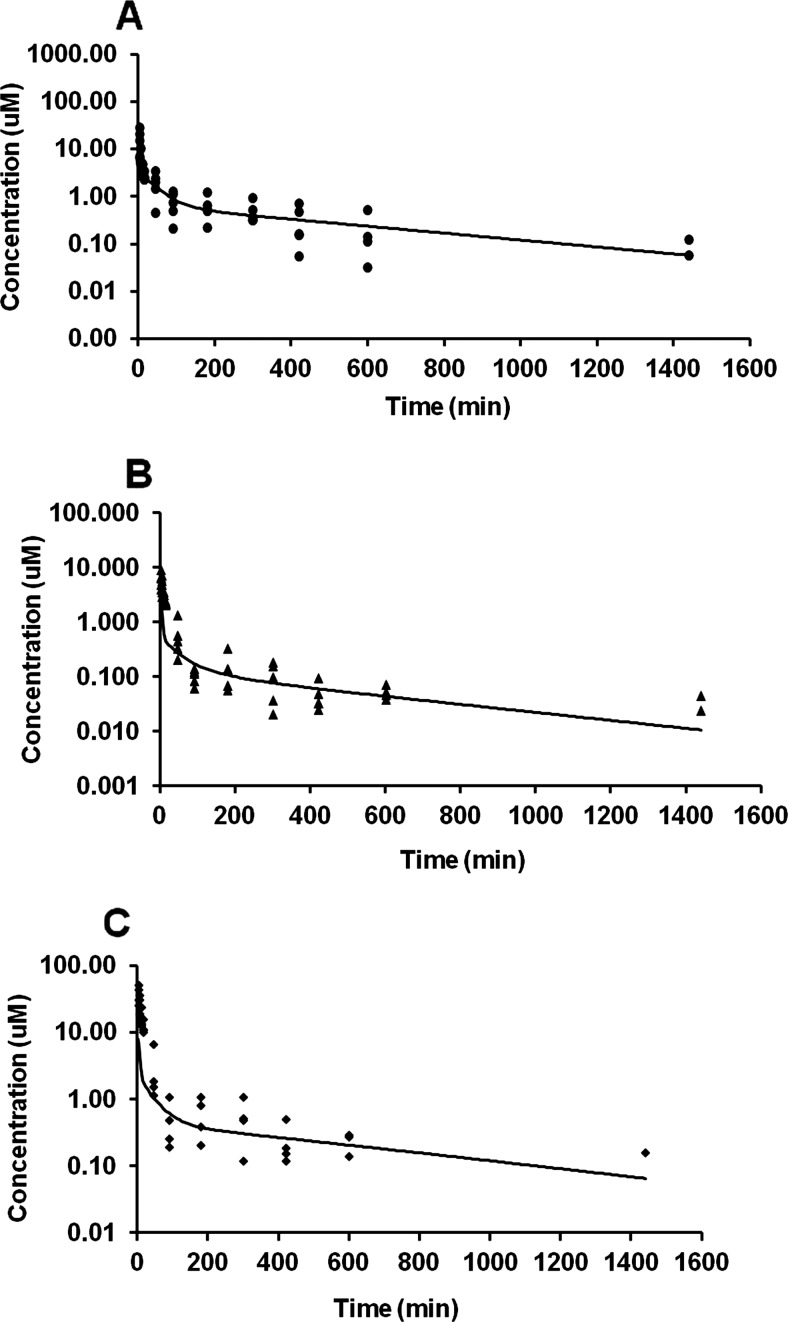
R3S and R3G are the major metabolites based on exposure when RES (15 mg/kg i.a.) was administered (Fig. 2A). R4′G and R4′S were minor metabolites and together account for only 0.2% of the total exposure of RES and metabolites combined (Table 1). We have therefore ignored these minor metabolites in the model for the sake of simplicity. One, two, and three compartment linear models were evaluated to explain the concentration data obtained after 15 mg/kg RES, 5 mg/kg R3S, and 3.5 mg/kg R3G i.a. administration. An open three-compartment model with elimination from the central compartment (model 1, Fig. 3A) was found to characterize well the concentration-time profiles for RES after its i.a. bolus administration. The predicted and observed RES plasma concentrations from model 1 are shown in Fig. 3B. An open two-compartment model (model 2, Fig. 3C) best described the observed R3S concentration after R3S 5 mg/kg i.a. bolus administration (Fig. 3D). An open three-compartment model with a delay compartment (model 3, Fig. 3E) and elimination from central compartment was used to characterize the concentration-time profile of R3G after R3G 3.5 mg/kg i.a. bolus administration (Fig. 3F). A delay compartment was included to better describe the data (Davis et al., 2000), because a secondary peak was observed in R3G concentration that might be due to enterohepatic cycling. The delay site is characterized by two parameters that are estimated from the data: the delay time and the number of delay compartments. Mass entering the delay site passes through each of the delay compartments. The delay time was fixed as 180 min based on visual examination of data and 10 delay compartments were used. Table 2 shows the compartmental pharmacokinetic parameter estimates of RES, R3S, and R3G. Run tests were performed for models 1, 2 and 3 and resulted in large p values (p = 0.26, 0.25, and 0.26, respectively), indicative of a lack of run of signs. The mean clearance estimate of RES and preformed R3G and R3S from models 1, 2, and 3 were comparable with the noncompartmental clearance estimates, respectively (Tables 1 and 2).
Fig. 3.
Compartmental modeling of RES, R3G, and R3S disposition. A, three-compartment PK model 1 describing the disposition of RES after administration of RES (15 mg/kg i.a.). Vc, volume of the central compartment; k, first-order rate constants for RES disposition. B, observed average RES concentration (data points) and PK model 1 predicted (solid line) RES concentration-time profiles after RES administration, and plot of weighted residuals versus predicted RES concentration (inset). C, two-compartment PK model 2 describing the disposition of R3S after administration of R3S (5 mg/kg i.a.). Vc,R3S, volume of the central compartment; k, first-order rate constants for R3S disposition. D, observed average R3S concentration (data points) and PK model 2 predicted (solid line) R3S concentration-time profiles after R3S administration, and plot of weighted residuals versus predicted R3S concentration (inset). E, enterohepatic cycling PK model 3 describing the disposition of R3G after administration of R3G (3.5 mg/kg i.a.). Vc,R3G, volume of the central compartment; k, first-order rate constants for R3G disposition. F, observed average R3G concentration (data points) and PK model 3 predicted (solid line) R3G concentration-time profiles after R3G administration, and plot of weighted residuals versus predicted R3G concentration (inset). [Note: The notation used throughout is k(from, to). In SAAM II software, the rate constants have a different notation k(to, from).]
TABLE 2.
Compartmental pharmacokinetic parameters of RES, R3S, and R3G administered as parent compound using model 1, 2 and 3, respectively
| Parameters | RES, Model 1 | R3S, Model 2 | R3G, Model 3 |
|---|---|---|---|
| Estimate, CV% | |||
| Vc, RES (l/kg) | 1.77 (27.29) | N/A | N/A |
| Cl, RES (ml · min−1 · kg−1) | 104.54 (3.97) | N/A | N/A |
| k1,0 (min−1) | 0.06 (25.66) | N/A | N/A |
| k1,2 (min−1) | 0.23 (23.74) | N/A | N/A |
| k2,1 (min−1) | 0.06 (14.74) | N/A | N/A |
| k1,3 (min−1) | 0.09 (24.73) | N/A | N/A |
| k3,1 (min−1) | 0.004 (11.84) | N/A | N/A |
| Vc, R3S (l/kg) | N/A | 0.69 (11.43) | N/A |
| Cl, R3S (ml · min−1 · kg−1) | N/A | 63.11 (3.56) | N/A |
| k1,0 R3S (min−1) | N/A | 0.09 (9.37) | N/A |
| k1,2 R3S (min−1) | N/A | 0.15 (7.17) | N/A |
| k2,1 R3S (min−1) | N/A | 0.012 (7.45) | N/A |
| Vc, R3G (l/kg) | N/A | N/A | 0.23 (10.95) |
| Cl, R3G (ml · min−1 · kg−1) | N/A | N/A | 10.63 (6.81) |
| k1,0 R3G (min−1) | N/A | N/A | 0.05 (11.86) |
| k1,2 R3G (min−1) | N/A | N/A | 0.04 (15.53) |
| k2,1 R3G (min−1) | N/A | N/A | 0.03 (14.97) |
| k1,3 R3G (min−1) | N/A | N/A | 0.07 (9.06) |
| k3,1 R3G (min−1) | N/A | N/A | 0.002 (11.27) |
N/A, not applicable.
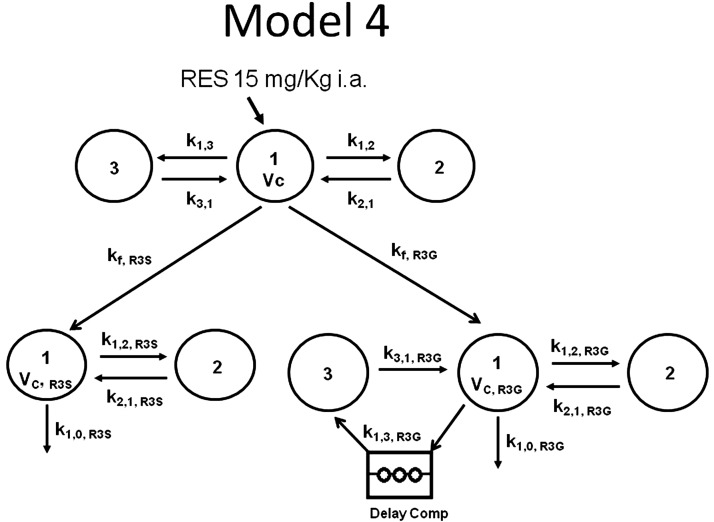
Simulation of in vivo-formed metabolites using model 4.
A great deal of time was invested in trying to fit comprehensive models to RES and its in vivo-formed metabolites simultaneously, but the models did not converge [possibly due to the large number of parameters (n = 17) in relation to the data collected]. Hence, simulations were performed instead of model-fitting. The parameters obtained from models 1, 2, and 3 were fixed in model 4 and then the in vivo-formed R3S and R3G after RES administration (Fig. 4) predicted under two conditions. In the first condition, the elimination clearance of the preformed metabolites was assumed to be equal to that of the in vivo-formed metabolites. For this process, all of the parameters in model 4 were fixed using parameters from models 1, 2, and 3. Next, this result was used to predict formation rate constants kf,R3S and kf,R3G (see Fig. 4). Rate constants kfR3S and kf,R3G were converted to clearance parameters by multiplying with central volume of distribution (Vc). Simulations were performed but resulted in poor fit of the predicted versus observed plasma concentrations of RES (Fig. 5A), R3S (Fig. 5B), and R3G (Fig. 5C).
Fig. 4.
PK model 4 describing the disposition of in vivo-formed metabolite R3S and R3G after RES (15 mg/kg i.a.) administration. Individual models for RES, R3S, and R3G are as described in Fig. 3. Vc, volumes of central compartments; k, first-order disposition rate constants; kf, first-order formation rate constants for RES metabolites. [Note: The notation used throughout is k(from, to). In SAAM II software, the rate constants have a different notation k(to, from).]
Fig. 5.
Observed and PK model 4 simulated concentration-time profiles of RES (A), R3S (B), and R3G (C) after RES administration, assuming elimination clearance of preformed metabolites to be equal to in vivo-formed metabolites.
In the second condition, the elimination clearance of preformed metabolites was assumed to be dissimilar to in vivo-formed metabolites. For this process, all of the parameters in model 4 were fixed using parameters from models 1, 2, and 3 except for the elimination rate constants of R3S (k1,0, R3S) and R3G (k1,0, R3G). The parameters kf,R3S (0.032 min−1), kf,R3G (0.030 min−1), k1,0, R3S (0.453 min−1), and k1,0, R3G (0.295 min−1) were predicted from the model (Fig. 4). Next the simulation was performed by fixing kf,R3S, kf,R3G, k1,0, R3S, and k1,0, R3G providing a good fit of observed versus model predicted RES, R3S, and R3G plasma concentrations after RES 15 mg/kg i.a. administration as shown in Fig. 6, A, B, and C, respectively. The second condition also provided a more realistic estimate of the fraction of RES metabolized to R3S and R3G to be 52 and 48%, respectively. Parameters were estimated under the assumption that there was no elimination of RES other than R3S and R3G. Elimination clearances of in vivo-formed R3S (313.08 ml · min−1 · kg−1) and R3G (67.86 ml · min−1 · kg−1) (Table 3) predicted under the second condition were found to be higher than the elimination clearances of preformed R3S (76.29 ml · min−1 · kg−1) and R3G (13.78 ml · min−1 · kg−1; Table 1).
Fig. 6.
Observed and PK model 4 simulated concentration-time profiles of RES (A), R3S (B), and R3G (C) after RES administration, assuming elimination clearance of preformed metabolites to be not equal to in vivo-formed metabolites.
TABLE 3.
Comparison of elimination or systemic clearance of in vivo-formed metabolites and preformed metabolites
Discussion
RES pharmacokinetics has been reported previously (Marier et al., 2002; Boocock et al., 2007; Brown et al., 2010; Patel et al., 2010). RES is extensively metabolized into its sulfated and glucuronidated metabolites in mammals (Goldberg et al., 2003; Walle et al., 2004). This study characterized the kinetics of the major metabolites of RES, R3S, and R3G, by administering the preformed metabolites. This is the first report of the metabolite kinetics of R3S and R3G after the administration of these preformed metabolites and their quantification against synthetic standards. The utility of the present work lies in development of models to explain the complex kinetics of highly conjugated substrates such as polyphenols. Furthermore, models such as those presented here will be further developed in future studies to evaluate 1) kinetics of conjugated metabolites that might be active and 2) potential interactions between polyphenols and coadministered drugs, e.g., enzyme induction or inhibition.
Similar to rats (Juan et al., 2010), RES was extensively metabolized to R3G in mice in the present study. Both RES and R3G underwent enterohepatic recirculation. The systemic clearance of RES observed in the present study (118.77 ml · min−1 · kg−1; Table 1) was comparable to that reported in rats [195 ml · min−1 · kg−1 183 and ml · min−1 · kg−1 reported by Marier et al., 2002 and Kapetanovic et al., 2011]. The high clearance is likely due to the rapid metabolism of RES. It is interesting to note that the value for total body clearance (118.77 ml · min−1 · kg−1; Table 1) greatly exceeds hepatic blood flow rate (90 ml · min−1 · kg−1) in mice (Davies and Morris, 1993). With the hepatic extraction ratio of RES assumed to be 100%, the maximum possible hepatic clearance is calculated to be 90 ml · min−1 · kg−1. Extrahepatic clearance (28.77 ml · min−1 · kg−1) was calculated as the difference between total body clearance (118.77 ml · min−1 · kg−1 for 15 mg/kg i.a. dose) and hepatic clearance (90 ml · min−1 · kg−1), which represents 24.22% of total body clearance. This result indicates the possibility of extrahepatic metabolism playing a role in the clearance of RES. A clear secondary peak was observed when preformed R3G was administered (Fig. 2C), which can be attributed to the enterohepatic circulation of R3G. Enterohepatic circulation of R3G has also been shown earlier (Marier et al., 2002). It is interesting to note that no RES was observed in plasma after administration of preformed R3S or preformed R3G.
Preformed major metabolites of RES (R3S and R3G) were administered to delineate the metabolite kinetics and to determine the fraction of RES converted to respective metabolites. The present data (Table 1) and literature reports clearly indicate much greater conversion of RES to R3S and R3G (Yu et al., 2002; Wenzel and Somoza, 2005) than predicted by eqs. 1 and 2, which assume similar kinetics of preformed versus in vivo-formed metabolite (Table 1). Because these formation clearances are calculated based on preformed metabolite data, the difference in preformed metabolite kinetics compared with in vivo-formed metabolite kinetics might be responsible for underprediction of metabolite formation clearances. This underprediction prompted the use of a modeling approach to delineate the kinetics of in vivo-formed metabolites.
The average plasma concentration-time profile of RES after 15 mg/kg RES administration was explained by a three-compartment model (Fig. 3A). Similar models have been used to explain disposition of drugs undergoing enterohepatic cycling (Hasselström and Säwe, 1993). RES was modeled to be distributed from a central compartment into two peripheral compartments, with elimination from the central compartment. It has been shown earlier (Colburn, 1982) that enterohepatic circulation increases the apparent volume of distribution. Therefore, a very high volume of distribution of RES can be partially attributed to tissue binding and partially to enterohepatic circulation of RES.
R3S plasma profile was explained by a two-compartment model with elimination from the central compartment (Fig. 3C). Although enterohepatic cycling of R3S is also a possibility, a two-compartment model was found to explain well the plasma profile of R3S after R3S (preformed) administration. An enterohepatic circulation model described the disposition of R3G after R3G (preformed) administration (Fig. 3E). Similar models have been developed for morphine 3-glucuronide (Ouellet and Pollack, 1995), morphine (Dahlström and Paalzow, 1978), phenolphthalein (Colburn et al., 1979), and isoflavones (Moon et al., 2006). These models used either a series of cycling compartments linked by first-order rate constants or a single compartment with a lag time to account for the delay observed in the appearance of a secondary peak in plasma. A similar approach was used in the present model by using a delay compartment that comprised 10 compartments with a single rate constant (k1,3, R3G) and a fixed delay time of 3 h. Thus, in Fig. 3E, compartment 1 depicts the blood as well as quickly equilibrating tissues. Compartment 2 depicts more slowly equilibrating tissues and compartment 3 can depict the intestinal compartment. The rate constant k1,3, R3G represents several different processes including biliary transport of R3G, transit of R3G through the gastrointestinal lumen, and possible hydrolysis of glucuronides into RES. The rate constant k3,1, R3G may denote absorption of re-formed RES and its subsequent glucuronidation into R3G or absorption of R3G from the lower intestine. A lag time of 3 h is included between biliary transport and absorption to account for transit from the liver to the site of deglucuronidation, and subsequent metabolism and reabsorption. The clearance estimates obtained after compartmental analysis of pooled data were found to be comparable with those estimated with individual data by noncompartmental methods (Tables 1 and 2).
Models 1, 2, and 3 were combined to form a comprehensive model 4 to predict the in vivo-formed R3S and R3G after RES administration (Fig. 4). Simulation using the assumption that elimination clearances of preformed and in vivo-formed metabolites are the same, led to a poor overlap of the observed and predicted in vivo-formed R3S and R3G (Fig. 5). Simulation using the assumption that elimination clearances of preformed and in vivo-formed metabolites are different, led to a much improved prediction (Fig. 6). The second approach also gave a more realistic formation ratio of R3S and R3G as 52 and 48%, respectively. This value was comparable with the formation ratio of in vivo-formed R3S (46%) and R3G (54%) predicted by Colom et al. (2011). With the second approach, the elimination clearance of R3S and R3G used for the simulation was higher than the preformed metabolites' elimination clearances. It has been suggested that phase II metabolites such as glucuronides and sulfates are more hydrophilic and preformed metabolites may experience difficulty penetrating into an eliminating organ, and hence the extent of its elimination may be less than that of in vivo-generated metabolites, whose entry into the eliminating organ is in the form of a more lipophilic precursor (Pang et al., 1984; Pang, 1985). The formation and elimination clearances of the formed and preformed metabolites are markedly different. Differences in metabolite kinetics of preformed and in vivo-formed metabolites were clearly visible in the present study.
It has been observed by Pang and coworkers (Pang et al., 2008; Pang, 2009) that although preformed metabolite administration might not be able to provide a complete correlation of the formed metabolite time course, the accompanying information can be incorporated to build a comprehensive PK model. This can improve the predictions of in vivo-formed metabolites. In the present work, model 4 was useful for purposes of predicting the disposition of metabolites as well as RES exposure. This and similar models can be further developed and improved to predict events such as interactions with xenobiotics that lead to enzyme induction or inhibition.
As a first study of RES metabolite PK, the present study did not include data collection such as urine, feces, or bile. This is an obvious limitation of the study, because additional data would provide a more detailed picture of RES disposition. In addition, single-dose plasma data collected here did not aid in discerning elimination from the central versus peripheral compartment (Berezhkovskiy, 2004; Yates and Arundel, 2008). Thus, a criticism of the models presented is the assumption of elimination solely from the central compartment. If peripheral elimination were to play a role in the elimination of RES or its metabolites (e.g., metabolism in tissues kinetically different from the central compartment), the steady-state volume of distribution estimates might be predicted inaccurately with the present models.
In summary, the kinetics of R3S and R3G were studied for the first time by administering the preformed metabolites. PK models were developed to adequately explain the kinetics of RES and its two major metabolites, R3S and R3G. Preformed and in vivo-formed R3S and R3G kinetics were compared, and a marked difference was observed between the preformed and in vivo-formed metabolite kinetics. Due to observed kinetic differences between in vivo-formed metabolites and preformed metabolites, safety and toxicity studies conducted with preformed metabolites are useful only when there is a similarity in the kinetics of preformed and in vivo-formed metabolites, or when sufficient tissue exposure of preformed metabolites is ensured. However, achieving high tissue exposure can be especially difficult for very hydrophilic metabolites.
Supplementary Material
Acknowledgments
We are grateful for support supplied by Temple University School of Pharmacy.
This work was supported in part by the National Institutes of Health National Cancer Institute [Grants R03-CA133943, R03-CA159389].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
- MIST
- drug metabolites in safety testing
- PK
- pharmacokinetics
- R3S
- trans-resveratrol-3-sulfate
- R3G
- trans-resveratrol-3-O-glucuronide
- RES
- trans-3,5,4′-trihydroxystilbene
- R4′G
- trans-resveratrol-4′-O-glucuronide
- R4′S
- trans-resveratrol-4′-sulfate
- i.a.
- intra-arterial administration
- AUC
- area under the plasma concentration-time curve
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- APAP
- acetaminophen
- IS
- internal standard
- LOQ
- limit of quantitation
- MRT
- mean residence time
- Cl
- clearance
- CV
- coefficient of variation.
Authorship Contributions
Participated in research design: Sharan and Nagar.
Conducted experiments: Sharan.
Contributed new reagents or analytic tools: Sharan, Iwuchukwu, and Canney.
Performed data analysis: Sharan, Zimmerman, and Nagar.
Wrote or contributed to the writing of the manuscript: Sharan, Zimmerman, and Nagar.
References
- Akaike H. (1974) A new look at the statistical model identification. IEEE Trans Automat Contr AC-19:716–723 [Google Scholar]
- Baillie TA, Cayen MN, Fouda H, Gerson RJ, Green JD, Grossman SJ, Klunk LJ, LeBlanc B, Perkins DG, Shipley LA. (2002) Drug metabolites in safety testing. Toxicol Appl Pharmacol 182:188–196 [DOI] [PubMed] [Google Scholar]
- Berezhkovskiy LM. (2004) Volume of distribution at steady state for a linear pharmacokinetic system with peripheral elimination. J Pharm Sci 93:1628–1640 [DOI] [PubMed] [Google Scholar]
- Boocock DJ, Faust GE, Patel KR, Schinas AM, Brown VA, Ducharme MP, Booth TD, Crowell JA, Perloff M, Gescher AJ, et al. (2007) Phase I dose escalation pharmacokinetic study in healthy volunteers of resveratrol, a potential cancer chemopreventive agent. Cancer Epidemiol Biomarkers Prev 16:1246–1252 [DOI] [PubMed] [Google Scholar]
- Brown VA, Patel KR, Viskaduraki M, Crowell JA, Perloff M, Booth TD, Vasilinin G, Sen A, Schinas AM, Piccirilli G, et al. (2010) Repeat dose study of the cancer chemopreventive agent resveratrol in healthy volunteers: safety, pharmacokinetics, and effect on the insulin-like growth factor axis. Cancer Res 70:9003–9011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colburn WA. (1982) Pharmacokinetic and biopharmaceutic parameters during enterohepatic circulation of drugs. J Pharm Sci 71:131–133 [DOI] [PubMed] [Google Scholar]
- Colburn WA, Hirom PC, Parker RJ, Milburn P. (1979) A pharmacokinetic model for enterohepatic recirculation in the rat: phenolphthalein, a model drug. Drug Metab Dispos 7:100–102 [PubMed] [Google Scholar]
- Colom H, Alfaras I, Maijó M, Juan ME, Planas JM. (2011) Population pharmacokinetic modeling of trans-resveratrol and its glucuronide and sulfate conjugates after oral and intravenous administration in rats. Pharm Res 28:1606–1621 [DOI] [PubMed] [Google Scholar]
- Dahlström BE, Paalzow LK. (1978) Pharmacokinetic interpretation of the enterohepatic recirculation and first-pass elimination of morphine in the rat. J Pharmacokinet Biopharm 6:505–519 [DOI] [PubMed] [Google Scholar]
- Davies B, Morris T. (1993) Physiological parameters in laboratory animals and humans. Pharm Res 10:1093–1095 [DOI] [PubMed] [Google Scholar]
- Davis TM, Daly F, Walsh JP, Ilett KF, Beilby JP, Dusci LJ, Barrett PH. (2000) Pharmacokinetics and pharmacodynamics of gliclazide in Caucasians and Australian Aborigines with type 2 diabetes. Br J Clin Pharmacol 49:223–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick CB, Obach RS. (2010) Metabolites in safety testing: “MIST” for the clinical pharmacologist. Clin Pharmacol Ther 87:345–350 [DOI] [PubMed] [Google Scholar]
- Gabrielsson J, Weiner D. (2006) Pharmacokinetic and Pharmacodynamic Data Analysis: Concepts and Applications, 4th ed, Swedish Pharmaceutical Press, Stockholm [Google Scholar]
- Goldberg DM, Yan J, Soleas GJ. (2003) Absorption of three wine-related polyphenols in three different matrices by healthy subjects. Clin Biochem 36:79–87 [DOI] [PubMed] [Google Scholar]
- Hasselström J, Säwe J. (1993) Morphine pharmacokinetics and metabolism in humans. Enterohepatic cycling and relative contribution of metabolites to active opioid concentrations. Clin Pharmacokinet 24:344–354 [DOI] [PubMed] [Google Scholar]
- Hastings KL, El-Hage J, Jacobs A, Leighton J, Morse D, Osterberg RE. (2003) Drug metabolites in safety testing. Toxicol Appl Pharmacol 190:91–92 [DOI] [PubMed] [Google Scholar]
- Hoshino J, Park EJ, Kondratyuk TP, Marler L, Pezzuto JM, van Breemen RB, Mo S, Li Y, Cushman M. (2010) Selective synthesis and biological evaluation of sulfate-conjugated resveratrol metabolites. J Med Chem 53:5033–5043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwuchukwu OF, Sharan S, Canney DJ, Nagar S. (2012) Analytical method development for synthesized conjugated metabolites of trans-resveratrol, and application to pharmacokinetic studies. J Pharm Biomed Anal 63:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan ME, Maijó M, Planas JM. (2010) Quantification of trans-resveratrol and its metabolites in rat plasma and tissues by HPLC. J Pharm Biomed Anal 51:391–398 [DOI] [PubMed] [Google Scholar]
- Kapetanovic IM, Muzzio M, Huang Z, Thompson TN, McCormick DL. (2011) Pharmacokinetics, oral bioavailability, and metabolic profile of resveratrol and its dimethylether analog, pterostilbene, in rats. Cancer Chemother Pharmacol 68:593–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marier JF, Vachon P, Gritsas A, Zhang J, Moreau JP, Ducharme MP. (2002) Metabolism and disposition of resveratrol in rats: extent of absorption, glucuronidation, and enterohepatic recirculation evidenced by a linked-rat model. J Pharmacol Exp Ther 302:369–373 [DOI] [PubMed] [Google Scholar]
- Meng X, Maliakal P, Lu H, Lee MJ, Yang CS. (2004) Urinary and plasma levels of resveratrol and quercetin in humans, mice, and rats after ingestion of pure compounds and grape juice. J Agric Food Chem 52:935–942 [DOI] [PubMed] [Google Scholar]
- Moon YJ, Sagawa K, Frederick K, Zhang S, Morris ME. (2006) Pharmacokinetics and bioavailability of the isoflavone biochanin A in rats. AAPS J 8:E433–E442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso T, Iwaki M, Tanino T, Ikeda K, Paku T, Horibe Y, Suzuki H. (1998) Pharmacokinetics of aniracetam and its metabolites in rats. J Pharm Sci 87:594–598 [DOI] [PubMed] [Google Scholar]
- Ouellet DM, Pollack GM. (1995) Biliary excretion and enterohepatic recirculation of morphine-3-glucuronide in rats. Drug Metab Dispos 23:478–484 [PubMed] [Google Scholar]
- Pang KS. (1985) A review of metabolite kinetics. J Pharmacokinet Biopharm 13:633–662 [DOI] [PubMed] [Google Scholar]
- Pang KS. (2009) Safety testing of metabolites: expectations and outcomes. Chem Biol Interact 179:45–59 [DOI] [PubMed] [Google Scholar]
- Pang KS, Cherry WF, Terrell JA, Ulm EH. (1984) Disposition of enalapril and its diacid metabolite, enalaprilat, in a perfused rat liver preparation. Presence of a diffusional barrier for enalaprilat into hepatocytes. Drug Metab Dispos 12:309–313 [PubMed] [Google Scholar]
- Pang KS, Kwan KC. (1983) A commentary: methods and assumptions in the kinetic estimation of metabolite formation. Drug Metab Dispos 11:79–84 [PubMed] [Google Scholar]
- Pang KS, Morris ME, Sun H. (2008) Formed and preformed metabolites: facts and comparisons. J Pharm Pharmacol 60:1247–1275 [DOI] [PubMed] [Google Scholar]
- Patel KR, Brown VA, Jones DJ, Britton RG, Hemingway D, Miller AS, West KP, Booth TD, Perloff M, Crowell JA, et al. (2010) Clinical pharmacology of resveratrol and its metabolites in colorectal cancer patients. Cancer Res 70:7392–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson PG, Wienkers LC. (2009) Handbook of Drug Metabolism. Informa Healthcare USA, New York [Google Scholar]
- Prueksaritanont T, Lin JH, Baillie TA. (2006) Complicating factors in safety testing of drug metabolites: kinetic differences between generated and preformed metabolites. Toxicol Appl Pharmacol 217:143–152 [DOI] [PubMed] [Google Scholar]
- Smith DA, Obach RS. (2005) Seeing through the mist: abundance versus percentage. Commentary on metabolites in safety testing. Drug Metab Dispos 33:1409–1417 [DOI] [PubMed] [Google Scholar]
- Walle T, Hsieh F, DeLegge MH, Oatis JE, Jr, Walle UK. (2004) High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab Dispos 32:1377–1382 [DOI] [PubMed] [Google Scholar]
- Wenzel E, Somoza V. (2005) Metabolism and bioavailability of trans-resveratrol. Mol Nutr Food Res 49:472–481 [DOI] [PubMed] [Google Scholar]
- Yates JW, Arundel PA. (2008) On the volume of distribution at steady state and its relationship with two-compartmental models. J Pharm Sci 97:111–122 [DOI] [PubMed] [Google Scholar]
- Yu C, Shin YG, Chow A, Li Y, Kosmeder JW, Lee YS, Hirschelman WH, Pezzuto JM, Mehta RG, van Breemen RB. (2002) Human, rat, and mouse metabolism of resveratrol. Pharm Res 19:1907–1914 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.