Abstract
The “methionine cycle” plays a critical role in the regulation of concentrations of (S)-adenosylmethionine (AdoMet), the major biological methyl donor. We set out to study sequence variation in genes encoding the enzyme that synthesizes AdoMet in liver, methionine adenosyltransferase 1A (MAT1A) and the major hepatic AdoMet using enzyme, glycine N-methyltransferase (GNMT), as well as functional implications of that variation. We resequenced MAT1A and GNMT using DNA from 288 subjects of three ethnicities, followed by functional genomic and genotype-phenotype correlation studies performed with 268 hepatic biopsy samples. We identified 44 and 42 polymorphisms in MAT1A and GNMT, respectively. Quantitative Western blot analyses for the human liver samples showed large individual variation in MAT1A and GNMT protein expression. Genotype-phenotype correlation identified two genotyped single-nucleotide polymorphisms (SNPs), reference SNP (rs) 9471976 (corrected p = 3.9 × 10−10) and rs11752813 (corrected p = 1.8 × 10−5), and 42 imputed SNPs surrounding GNMT that were significantly associated with hepatic GNMT protein levels (corrected p values < 0.01). Reporter gene studies showed that variant alleles for both genotyped SNPs resulted in decreased transcriptional activity. Correlation analyses among hepatic protein levels for methionine cycle enzymes showed significant correlations between GNMT and MAT1A (p = 1.5 × 10−3) and between GNMT and betaine homocysteine methyltransferase (p = 1.6 × 10−7). Our discovery of SNPs that are highly associated with hepatic GNMT protein expression as well as the “coordinate regulation” of methionine cycle enzyme protein levels provide novel insight into the regulation of this important human liver biochemical pathway.
Introduction
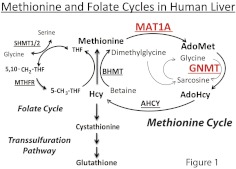
(S)-Adenosylmethionine (AdoMet), the methyl donor for most biological methylation reactions (Cantoni, 1951a,b), is synthesized from methionine and ATP by methionine adenosyltransferase (MAT) (Fontecave et al., 2004). It is then used as a methyl donor for reactions catalyzed by methyltransferase enzymes to produce methylated compounds and (S)-adenosylhomocysteine (AdoHcy) (Clarke et al., 2003). AdoHcy is subsequently converted to homocysteine, which can be either remethylated to form methionine, completing the “methionine cycle,” or converted to cysteine and GSH by the transsulfuration pathway (Fig. 1). The methionine cycle and “folate cycle” shown in Fig. 1 have been implicated in the pathophysiology of diseases as diverse as cancer (Weinstein et al., 2006; Kasperzyk et al., 2009; Maruti et al., 2009; Stevens et al., 2010), cardiovascular disease (Arnesen et al., 1995; Frosst et al., 1995; Nygard et al., 1995; Clarke et al., 2003), and psychiatric illness (Smythies et al., 1997; Main et al., 2010).
Fig. 1.
Methionine and folate cycles in the human liver. AHCY, adenosylhomocysteine hydrolase; MTHFR, methylenetetrahydrofolate reductase; Hcy, homocysteine.
The human liver expresses several unique methionine cycle enzymes (Fig. 1). In particular, MAT2A encodes the enzyme that catalyzes AdoMet biosynthesis in nonhepatic tissues and in fetal liver, but MAT1A is expressed only in adult liver and encodes the enzyme that catalyzes AdoMet synthesis in the liver (Gil et al., 1996; Mato et al., 1997). Once formed, AdoMet is used by methyltransferase enzymes to generate methylated compounds, with AdoHcy as a reaction product. The tetrameric enzyme glycine N-methyltransferase (GNMT; E.C. 2.1.1.20) is highly expressed in liver (1–3% of total soluble protein) and poorly expressed in other tissues except the prostate and pancreas (Kerr, 1972; Heady and Kerr, 1973). GNMT catalyzes the methylation of glycine to form sarcosine and AdoHcy (Blumenstein and Williams, 1960). The biological role of sarcosine is not well understood, but a metabolomic study identified sarcosine as a metabolic biomarker for the progression of prostate cancer (Sreekumar et al., 2009). There is also evidence that the reaction catalyzed by GNMT is a major factor regulating hepatic AdoMet concentrations (Mudd and Poole, 1975; Mudd et al., 1980; Balaghi et al., 1993). In addition to MAT1A and GNMT, betaine homocysteine methyltransferase (BHMT) is another methionine cycle enzyme that is expressed primarily in the liver (Li et al., 2008). BHMT catalyzes the hepatic remethylation of homocysteine to form methionine by transferring a methyl group from betaine to homocysteine (Skiba et al., 1982) (Fig. 1).
Rare mutations in MAT1A have been associated with persistent hypermethioninemia without elevation in either circulating homocysteine or tyrosine concentrations (Chamberlin et al., 2000). GNMT mutations have also been linked to hypermethioninemia (Mudd et al., 2001; Luka et al., 2002; Augoustides-Savvopoulou et al., 2003). These reports raise the possibility that common DNA sequence variation in the MAT1A and GNMT genes might modulate hepatic AdoMet concentrations and, as a result, hepatic methylation (Fig. 1). In the present study, we set out to systematically resequence both genes, followed by functional genomic studies and hepatic genotype-phenotype correlation analyses in an attempt to identify functional variants in MAT1A and GNMT that might contribute to variation in the regulation of AdoMet metabolism as well as methionine and folate cycle function.
Materials and Methods
DNA Samples and Gene Resequencing.
DNA samples for gene resequencing were obtained from the Coriell Cell Repository (Camden, NJ). In particular, Human Variation Panel samples from 96 European American (EA), 96 African American (AA), and 96 Han Chinese American (HCA) subjects (sample sets HD100CAU, HD100AA, and HD100CHI, respectively) were used in the resequencing studies. These samples had been collected, anonymized, and deposited by the National Institute of General Medical Sciences. All subjects had provided written informed consent for the use of their DNA for research purposes. Our studies were reviewed and approved by the Mayo Clinic Institutional Review Board. Details of the DNA sequencing methods have been described previously (Ji et al., 2007). All MAT1A exons, intron-exon splice junctions, and ∼1 kilobase (kb) of both 5′ and 3′ flanking regions (FRs) were amplified using the polymerase chain reaction (PCR), and the amplicons were sequenced on both strands in the Mayo Clinic Molecular Biology Core Facility using dye terminator sequencing chemistry. Because GNMT is much smaller than MAT1A, the entire GNMT gene as well as approximately 1 kb of its 5′ and 3′ FRs were sequenced. Accession numbers for the reference sequences used were NM_000429.2. for MAT1A and NM_018960.4 for GNMT. Sequence chromatograms were analyzed using Mutation Surveyor (SoftGenetics, LLC, State College, PA). Sequences of primers used to perform the PCR amplifications and gene resequencing studies are listed in Supplemental Table 1.
Functional Characterization of MAT1A Nonsynonymous Single-Nucleotide Polymorphisms.
Mammalian expression constructs were created for wild-type (WT) MAT1A by subcloning the open reading frame of MAT1A from the Origene clone SC119881 (Origene, Rockville, MD) into pcDNA3.1D/V5-His-TOPO (Invitrogen, Carlsbad, CA) in frame with the V5-His tag, and site-directed mutagenesis was used to create variant allozyme expression constructs. These expression constructs were used to transfect COS-1 cells to obtain recombinant MAT1A allozymes for use in quantitative Western blot analyses and for the assay of allozyme enzyme activity. Bacterial expression constructs were also created for MAT1A WT and variant allozymes and were expressed in BL21 Escherichia coli. This bacterially expressed MAT1A was used to perform substrate kinetic experiments, as described previously (Wang et al., 2003).
Structural analysis of MAT1A allozymes was performed by using the 2.1-Å resolution crystal structure of human MAT1A bound to AdoMet (Protein Data Bank accession code 2OBV). Visualization and analysis of the MAT1A structures and the computational “mutation” of E238K was performed using the graphics program Crystallographic Object-Oriented Toolkit (Coot) (Purcell et al., 2007). Additional details regarding the MAT1A structural analysis are described in the supplemental data.
MAT1A and GNMT Western Blot Analyses of Human Hepatic Biopsy Samples.
Two hundred sixty-eight human adult liver surgical biopsy samples were obtained from the Mayo Clinic (Rochester, MN). These samples were from EA women undergoing medically indicated surgery. Tissue samples from only one sex were used to eliminate the possibility of confusion as a result of sex-dependent differences in enzyme protein expression. Characteristics of the patients from whom the biopsies were obtained have been described previously (Feng et al., 2009; Zhang et al., 2009; Nordgren et al., 2011). Use of these anonymized surgical biopsy samples was reviewed and approved by the Mayo Clinic Institutional Review Board. Cytosol extracted from the hepatic tissue was stored at −80°C before use.
Quantitative Western blot analyses were performed for both MAT1A and GNMT using the 268 liver cytosol preparations. A rabbit polyclonal antibody generated by Cocalico Biologicals, Inc. (Reamstown, PA), against MAT1A amino acids 208 to 228 and a commercial mouse GNMT polyclonal antibody (Sigma-Aldrich, St. Louis, MO) were used to perform these studies. Purified His-tagged protein standards for MAT1A (50 ng) as well as a pooled sample of hepatic high-speed supernatant as a standard for GNMT were loaded on each gel and were stained for either MAT1A or GNMT protein. Levels of endogenous β-actin were also assayed for each gel and were used as a loading control. Specifically, each of the 268 liver cytosol samples was loaded in triplicate, and values for MAT1A and GNMT immunoreactive protein for the triplicate samples were calculated on the basis of the relative intensity of protein bands on the gel compared with an appropriate protein standard assayed on the same gel.
Tag Single-Nucleotide Polymorphism Selection and Genotyping of Hepatic Biopsy DNA.
Genotype data from a variety of sources were used to select tag single-nucleotide polymorphisms (SNPs) for genes encoding proteins in the methionine and folate cycles (see Fig. 1), including MAT1A and GNMT, for use in genotyping DNA samples from the same 268 human liver biopsy samples that we had phenotyped for level of hepatic MAT1A and GNMT protein. Specifically, a total of 768 tag SNPs for methionine and folate cycle genes, including 42 for MAT1A and 5 for GNMT, were selected using SNP genotype data from 168 unrelated HapMap white subjects (http://www.hapmap.org, data release 27/phase II + III) as well as data from our resequencing studies for these genes (Shield et al., 2004; Martin et al., 2006; Feng et al., 2009; Nordgren et al., 2011) and SNP data obtained by using Illumina 550K and 510S genome-wide BeadChips (Illumina, Inc., San Diego, CA) and Coriell Institute-generated Affymetrix 6.0 genome-wide SNP genotypes for DNA from the 96 EA subjects included in the Coriell Institute Human Variation Panel (Camden, NJ), the same cell lines from which we obtained the DNA for our gene resequencing experiments. The SNPs were selected to tag across the genes and to include approximately 20 kb of flanking sequence, with r2 ≥ 0.8 and a minor allele frequency of ≥0.025. LDselect (Carlson et al., 2004) was used to perform tagging, and genotyping of DNA from the 268 hepatic biopsy samples was performed in the Mayo Genotyping Shared Resource using Illumina GoldenGate technology (Illumina, Inc.). Of the 42 MAT1A tag SNPs, 4 were excluded because of call rates of <94%, leaving 38 MAT1A SNPs and 5 GNMT SNPs for use in the genotype-phenotype correlation analysis.
Genotype-Phenotype Correlation Analysis and 1000 Genomes Project Imputation.
In addition to SNPs genotyped using the DNA samples, genotypes of untyped SNPs across both MAT1A and GNMT were imputed using 1000 Genomes Project and HapMap (phase II, release 22) data as reference sets. Tag SNP genotypes in the 268 hepatic DNA samples were the genetic background upon which imputation was performed. The software package MaCH 1.0 (Li et al., 2006) was used to perform imputation. To increase the likelihood of detecting functionally important variants at a distance from the genes, variants within 200 kb on either side of the two genes were included in the imputation. Imputation quality estimates were determined by masking 10% of the genotypes at random and imputing the masked genotypes to compare actual and imputed masked genotypes. Estimated allelic dosage values for the imputed genotypes were then used, in addition to the genotyped SNPs, to perform the association analyses. Associations between MAT1A or GNMT protein levels and MAT1A or GNMT genotypes for both imputed and typed SNPs were performed by using Spearman's rank correlations and were tested versus zero using a Wald test. Genotype-phenotype correlations for MAT1A SNPs or GNMT SNPs with log2-transformed protein levels for the two genes were calculated using PLINK (Purcell et al., 2007). Pairwise linkage disequilibrium (LD) was determined by using SNAP (Johnson et al., 2008). Mapping of transcription factors for SNPs with low p values for the association analysis was performed by using the Encyclopedia of DNA Elements (ENCODE) data on the University of California Santa Cruz genome browser website (http://genome.ucsc.edu/) (Boyle et al., 2011).
GNMT Reporter Gene and Quantitative Reverse Transcriptase-PCR Assays.
Reporter gene assays were used to functionally characterize SNPs with low p values for the genotype-phenotype correlation analyses. Specifically, DNA sequences [200 to 300 base pairs (bp)] harboring the SNP loci selected for study were cloned into a pGL3-promoter luciferase reporter vector that contained an SV40 promoter upstream of the luciferase gene (Promega, Madison, WI). One microgram of each reporter gene construct was then cotransfected into the human hepatocellular carcinoma HepG2 cell and LNCaP human prostate cancer cell lines (American Type Culture Collection, Manassas, VA), with 20 ng of the pRL-TK Renilla luciferase vector as a control for transfection efficiency, followed by dual-luciferase assay performed 24 h after transfection (Promega). Two independent transfection studies were performed for each reporter gene construct, with triplicate independent transfections for each construct in each experiment. Values for relative activity were expressed as a percentage of the pGL3-promoter activity for vectors without an insert. Comparisons were made between pGL3 reporter gene constructs containing WT and variant nucleotides at the SNP loci. DNA samples used to amplify SNP regions were selected from the Coriell Institute Human Variation Panel DNA samples because the genotypes of these samples were known on the basis of both our gene resequencing studies and the genome-wide association studies genotyping data available for these samples. Sequences of primers that were used to amplify genomic regions containing the SNP selected for study during the reporter gene assay are listed in Supplemental Table 2.
Finally, quantitative reverse transcriptase-PCR (qRT-PCR) was used to determine the level of GNMT expression in a series of cell lines. Specifically, total RNA was isolated from LNCaP, HEK293T, PC-3, DU145, and HepG2 cells as well as frozen liver tissues by using Quick-RNA Mini Prep kit (Zymo Research, Irvine, CA). Real-time PCR was performed with the power SYBR Green RNA-to-CT 1-step Kit (Applied Biosystems, Carlsbad, CA) using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control gene. The PCR conditions were 1 cycle of 30 min at 48°C, 10 min at 95°C, 40 cycles of 15 s at 95°C, 1 min at 60°C, followed by 1 cycle of 15 s at 95°C, 1 min at 60°C, and 15 s at 95°C. Primers for performing qRT-PCR for GNMT and GAPDH were purchased from QIAGEN (Valencia, CA; QT00026285 for GNMT and QT01192646 for GAPDH).
Results
MAT1A and GNMT Resequencing.
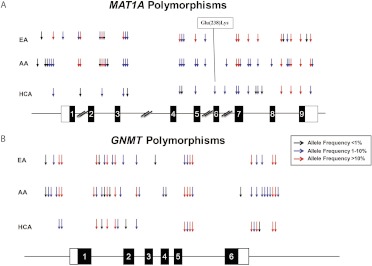
Sanger sequencing was used to resequence the exons, splice junctions, and ∼1000 bp of both the 5′ and the 3′ FRs of MAT1A as well as the entire GNMT gene, including ∼1000 bp of 5′ and 3′ FR, using 288 DNA samples, 96 each from AA, EA, and HCA subjects (Tables 1 and 2; Fig. 2). MAT1A resequencing identified 44 polymorphisms, 27 in EA, 36 in AA, and 20 in HCA, including one nonsynonymous (ns) SNP (G712>A, E238K). This SNP was present, heterozygous, in DNA from three HCA subjects. Eleven of these 44 polymorphisms were novel, i.e., they had not been deposited in dbSNP or assigned a reference SNP (rs) identifier number. GNMT resequencing identified 42 polymorphisms, 22 in EA, 32 in AA, and 19 in HCA samples. Unlike the situation with MAT1A, we did not identify any nsSNPs during GNMT resequencing. Twenty seven of the 42 GNMT polymorphisms were novel. All polymorphisms observed during our MAT1A and GNMT resequencing were in Hardy-Weinberg equilibrium (p > 0.05).
TABLE 1.
Human MAT1A genetic polymorphisms
Exons and untranslated regions (UTRs) are numbered relative to the A (nucleotide 1) in the ATG translation initiation codon in exon 1. Negative numbers were assigned to positions 5′ to that location, and positive numbers were assigned to positions 3′. Nucleotides located within introns are numbered on the basis of their distance from the nearest splice junction, with distances from 3′ splice junctions assigned positive numbers and distances from 5′ splice junctions assigned negative numbers. Exon sequences are bold. Polymorphisms identified previously are noted by reference SNP (rs) number.
| Polymorphism Location | Nucleotide Sequence Change | Amino Acid Change | MAF |
rs No. | NCBI36/hg18 (dbSNP 130) | ||
|---|---|---|---|---|---|---|---|
| AA | EA | HCA | |||||
| 5′ FR (−1015) | G > A | 0.000 | 0.005 | 0.000 | 82040174 | ||
| 5′ FR (−823) | A > T | 0.005 | 0.000 | 0.000 | 82039982 | ||
| 5′ FR (−806) | G > C | 0.005 | 0.000 | 0.000 | 82039965 | ||
| 5′ FR (−686) | C > A | 0.016 | 0.000 | 0.000 | 82039845 | ||
| 5′ FR (−625) | C > T | 0.016 | 0.000 | 0.000 | 82039784 | ||
| 5′ FR (−424) | T > C | 0.026 | 0.147 | 0.026 | rs17677908 | 82039583 | |
| 5′ UTR (−153) | C > T | 0.000 | 0.042 | 0.000 | rs11595587 | 82039312 | |
| Intron 1 (111) | G > A | 0.115 | 0.073 | 0.000 | rs3862534 | 82038958 | |
| Intron 1 (−9) | C > G | 0.469 | 0.188 | 0.005 | rs10887721 | 82035334 | |
| Intron 2 (14) | G > A | 0.005 | 0.031 | 0.000 | 82035234 | ||
| Intron 2 (199) | Deletion of TAAT | 0.245 | 0.281 | 0.042 | rs71482773 | 82035046 | |
| Intron 2 (−306) | A > C | 0.146 | 0.005 | 0.000 | rs28539197 | 82034080 | |
| Intron 2 (−211) | C > T | 0.026 | 0.000 | 0.000 | 82033985 | ||
| Intron 2 (−162) | G > A | 0.005 | 0.000 | 0.000 | 82033936 | ||
| Intron 3 (96) | T > C | 0.068 | 0.276 | 0.057 | rs2236569 | 82033556 | |
| Intron 3 (−104) | G > C | 0.052 | 0.068 | 0.000 | 82030632 | ||
| Intron 3 (−54) | T > C | 0.026 | 0.099 | 0.005 | rs71481597 | 82030582 | |
| Intron 4 (90) | C > T | 0.068 | 0.286 | 0.021 | rs2282367 | 82030326 | |
| Intron 4 (274) | G > A | 0.026 | 0.099 | 0.005 | rs71481596 | 82030142 | |
| Intron 4 (313) | C > T | 0.052 | 0.073 | 0.000 | rs41284066 | 82030103 | |
| Exon 5 (426) | C > T | 0.068 | 0.286 | 0.021 | rs1143694 | 82030032 | |
| Exon 5 (429) | C > T | 0.010 | 0.000 | 0.000 | 82030029 | ||
| Intron 5 (−47) | G > A | 0.052 | 0.073 | 0.000 | 82026377 | ||
| Exon 6 (712) | G > A | E238K | 0.000 | 0.000 | 0.016 | 82026168 | |
| Intron 6 (−95) | C > T | 0.000 | 0.000 | 0.016 | 82025030 | ||
| Intron 6 (−85) | G > A | 0.021 | 0.000 | 0.000 | 82025020 | ||
| Exon 7 (870) | G > A | 0.120 | 0.297 | 0.021 | rs10788546 | 82024834 | |
| Exon 7 (882) | C > T | 0.120 | 0.297 | 0.021 | rs10887711 | 82024822 | |
| Exon 7 (885) | A > T | 0.010 | 0.000 | 0.000 | rs17851642 | 82024819 | |
| Intron 7 (44) | C > T | 0.026 | 0.151 | 0.026 | rs55855057 | 82024709 | |
| Intron 7 (68) | Deletion of A | 0.010 | 0.000 | 0.000 | 82024685 | ||
| Intron 7 (75) | G > A | 0.000 | 0.000 | 0.005 | 82024678 | ||
| Intron 7 (98) | C > T | 0.074 | 0.276 | 0.021 | rs10788545 | 82024655 | |
| Intron 7 (274) | C > T | 0.000 | 0.000 | 0.005 | 82024479 | ||
| Exon 8 (1005) | C > T | 0.016 | 0.000 | 0.000 | rs61734474 | 82024336 | |
| Intron 8 (14) | T > C | 0.260 | 0.469 | 0.443 | rs2994388 | 82024242 | |
| Intron 8 (15) | G > A | 0.000 | 0.005 | 0.000 | 82024241 | ||
| Intron 8 (114) | G > A | 0.021 | 0.021 | 0.000 | rs72809554 | 82024142 | |
| Intron 8 (336) | G > A | 0.026 | 0.000 | 0.000 | 82023920 | ||
| Intron 8 (−44) | C > T | 0.135 | 0.223 | 0.531 | rs4933327 | 82023663 | |
| Exon 9 (1131) | T > C | 0.260 | 0.468 | 0.438 | rs2993763 | 82023574 | |
| 3′ UTR (1207) | C > A | 0.000 | 0.005 | 0.000 | 82023498 | ||
| 3′ UTR (1255) | C > T | 0.245 | 0.234 | 0.026 | rs7087728 | 82023450 | |
| 3′ UTR (1261) | G > T | 0.026 | 0.000 | 0.000 | 82023444 | ||
TABLE 2.
Human GNMT genetic polymorphisms
Exons and untranslated regions (UTRs) are numbered relative to the A (nucleotide 1) in the ATG translation initiation codon located in exon 1. Negative numbers were assigned to positions 5′ to that location, and positive numbers were assigned to positions 3′. Nucleotides located within introns (intervening sequences) are numbered on the basis of their distance from the nearest splice junction, with distances from 3′ splice junctions assigned positive numbers, and distances from 5′ splice junctions assigned negative numbers. Exon sequences are bold. Polymorphisms identified previously are noted by rs number.
| Polymorphism Location | Nucleotide Sequence Change | Amino Acid Change | MAF |
rs No. | NCBI36/hg18 (dbSNP 130) | ||
|---|---|---|---|---|---|---|---|
| AA | EA | HCA | |||||
| 43035794 | |||||||
| 5′ FR (−690) | C > T | 0.005 | 0.005 | 0.000 | 43035829 | ||
| 5′ FR (−655) | C > T | 0.010 | 0.000 | 0.000 | 43035899 | ||
| 5′ FR (−585) | T > C | 0.021 | 0.000 | 0.000 | 43035937 | ||
| 5′ FR (−547) | C > T | 0.000 | 0.010 | 0.000 | 43035948 | ||
| 5′ FR (−536) | Deletion of GTTACCGT | 0.016 | 0.000 | 0.000 | 43035995 | ||
| 5′ FR (−489) | C > G | 0.443 | 0.297 | 0.057 | rs11752813 | 43036439 | |
| 5′ FR (−45) | C > T | 0.542 | 0.313 | 0.068 | rs10948059 | 43036730 | |
| Intron 1 (41) | G > A | 0.156 | 0.000 | 0.000 | rs5031030 | 43036736 | |
| Intron 1 (47) | G > T | 0.099 | 0.401 | 0.628 | rs2296805 | 43036832 | |
| Intron 1 (143) | C > A | 0.000 | 0.005 | 0.000 | 43036925 | ||
| Intron 1 (236) | Deletion of AC | 0.000 | 0.000 | 0.005 | 43037339 | ||
| Intron 1 (650) | G > A | 0.089 | 0.000 | 0.000 | rs58057801 | 43037414 | |
| Intron 1 (725) | C > T | 0.005 | 0.016 | 0.000 | 43037511 | ||
| Intron 1 (−417) | C > T | 0.242 | 0.422 | 0.758 | 43037638 | ||
| Intron 1 (−290) | C > G | 0.005 | 0.000 | 0.000 | 43037806 | ||
| Intron 1 (−122) | G > A | 0.000 | 0.021 | 0.000 | 43037817 | ||
| Intron 1 (−111) | A > G | 0.266 | 0.422 | 0.745 | rs7760250 | 43037821 | |
| Intron 1 (−107) | C > T | 0.000 | 0.000 | 0.053 | 43037867 | ||
| Intron 1 (−61) | C > T | 0.016 | 0.000 | 0.000 | 43037873 | ||
| Intron 1 (−55) | C > G | 0.000 | 0.010 | 0.000 | 43038039 | ||
| Exon 2 (318) | C > T | 0.000 | 0.000 | 0.005 | 43038287 | ||
| Intron 2 (232) | A > G | 0.188 | 0.052 | 0.053 | rs3800292 | 43038437 | |
| Intron 2 (−41) | A > G | 0.052 | 0.000 | 0.000 | 43038746 | ||
| Intron 3 (−42) | C > A | 0.000 | 0.005 | 0.000 | 43038855 | ||
| Exon 4 (519) | G > A | 0.027 | 0.000 | 0.000 | 43038924 | ||
| Exon 4 (588) | C > T | 0.005 | 0.000 | 0.000 | 43039187 | ||
| Intron 5 (22) | Deletion of G | 0.218 | 0.083 | 0.122 | 43039193 | ||
| Intron 5 (28) | C > T | 0.158 | 0.039 | 0.025 | rs4987174 | 43039202 | |
| Intron 5 (37) | G > A | 0.495 | 0.522 | 0.131 | rs4987173 | 43039239 | |
| Intron 5 (74) | G > C | 0.066 | 0.394 | 0.635 | rs2296804 | 43039505 | |
| 3′ UTR (968) | G > A | 0.006 | 0.000 | 0.000 | 43039614 | ||
| 3′ FR (1070) | Deletion of TTAT | 0.089 | 0.395 | 0.621 | rs5875822 | 43039708 | |
| 3′ FR (1174) | C > G | 0.000 | 0.000 | 0.010 | 43039732 | ||
| 3′ FR (1193) | G > A | 0.042 | 0.026 | 0.146 | rs736158 | 43039757 | |
| 3′ FR (1223) | G > A | 0.000 | 0.000 | 0.005 | 43039839 | ||
| 3′ FR (1305) | G > A | 0.026 | 0.042 | 0.000 | rs1051218 | 43039871 | |
| 3′ FR (1337) | Insertion of T | 0.026 | 0.000 | 0.000 | 43040026 | ||
| 3′ FR (1492) | C > A | 0.016 | 0.000 | 0.000 | 43040058 | ||
| 3′ FR (1524) | G > A | 0.026 | 0.000 | 0.000 | 43040178 | ||
| 3′ FR (1664) | G > T | 0.458 | 0.489 | 0.120 | rs1129187 | 43040180 | |
| 3′ FR (1646) | T > C | 0.271 | 0.416 | 0.760 | rs1129186 | 43040220 | |
| 3′ FR (1686) | A > G | 0.026 | 0.000 | 0.000 | 43035794 | ||
Fig. 2.
Human MAT1A and GNMT polymorphisms observed during gene resequencing. The figure shows a schematic representation of the human (A) MAT1A and (B) GNMT gene structures, with arrows indicating the locations of polymorphisms observed during the resequencing studies. Black rectangles represent exons encoding the opening reading frame, and open rectangles represent portions of exons encoding untranslated region sequences. The colors of arrows indicate minor allele frequencies.
MAT1A Allozyme Functional Characterization.
Our gene resequencing effort identified one MAT1A nsSNP (G712>A, E238K) with a minor allele frequency of 0.016 in DNA from HCA subjects. Mammalian and bacterial expression constructs were created for both WT and the Lys238 variant to generate recombinant MAT1A allozymes as well as bacterially purified protein that could both be assayed for MAT1A allozyme activity and used to perform substrate kinetic experiments. Alterations in amino acid sequence as a result of genetic polymorphisms can have functional consequences either because of changes in protein quantity or altered substrate kinetics. However, the MAT1A Lys238 variant allozyme did not differ significantly from the WT protein in terms of either (see the MAT1A allozyme apparent Km values, enzyme activities, and relative protein quantities listed in Table 3). In addition, structural analysis of the human MAT1A allozymes showed that the WT Glu238 residue was surface-exposed with a side chain exposed to solvent. Substitution of Glu238 with the hydrophilic lysine residue present in the variant enzyme could be easily accommodated by WT protein folding. Finally, E238K is distant from both the dimer and tetramer interfaces and, therefore, would be unlikely to interfere with the formation of either oligomeric form of the protein. These structural predictions all supported our observations of a lack of functional implications for this variant allozymes (see structural analysis in the supplemental data).
TABLE 3.
MAT1A allozyme functional genomics
Values are mean ± S.E.M. for three independent determinations. The variant allozyme values did not differ significantly (p > 0.05) from those for the WT allozyme for either apparent Km values or enzyme activity (measured using bacterially expressed protein) or relative protein quantity after expression in COS-1 cells.
| MAT1A Allozyme |
Km |
Allozyme Activity | Protein Quantity | |
|---|---|---|---|---|
| Methionine | ATP | |||
| μM | % of WT | % of WT | ||
| WT | 315 ± 35 | 938 ± 85 | 100 ± 2 | 100 ± 3 |
| Lys238 | 327 ± 54 | 894 ± 76 | 102 ± 3 | 104 ± 4 |
Hepatic Genotype-Phenotype Correlation Analysis.
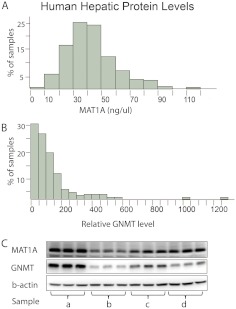
To determine whether DNA sequence variation in the MAT1A and GNMT genes might play a role in variation in the expression of these proteins in the human hepatic tissue where both enzymes are predominantly expressed, we assayed levels of MAT1A and GNMT protein expression in 268 adult human liver biopsy samples by performing quantitative Western blot analyses. Both enzymes showed large individual variation, with a 100- and 1000-fold variation in MAT1A and GNMT protein expression, respectively (Fig. 3). The data in Fig. 3 show a Gaussian frequency distribution for levels of MAT1A protein (Fig. 3A) but a skewed distribution of GNMT protein levels, with many samples that displayed low levels and two “outlier” points with very high levels of protein (Fig. 3B). The figure also shows representative Western blots for both enzymes (Fig. 3C).
Fig. 3.
Human hepatic liver MAT1A and GNMT protein levels. A, MAT1A protein-level frequency distribution. B, GNMT protein-level frequency distribution. C, representative Western blot analysis gel with a, b, c, and d representing individual samples assayed in triplicate.
As the next step in our analysis, we genotyped 42 tag SNPs for MAT1A and 5 tag SNPs for GNMT using DNA samples from the same 268 subjects from whom the liver biopsy samples used to perform the Western blot analyses had been obtained. Overall call rate of these 42 SNPs was more than 98.0%, but call rates were < 94% for 4 SNPs, so those SNPs were excluded from the subsequent analysis. In addition, using the genotyped SNPs as a scaffold, we performed imputation using 1000 Genomes Project data across both genes out to 200 kb from both the 5′ and 3′ ends of the genes (1000 Genomes Project Consortium, 2010). Imputation identified an additional 150 MAT1A and 61 additional GNMT SNPs with MaCH “Rsq” values (an estimate of the squared correlation between imputed and true genotypes) of more than 0.3. Only SNPs for EA subjects were imputed because the liver biopsy samples had been obtained entirely from EA subjects.
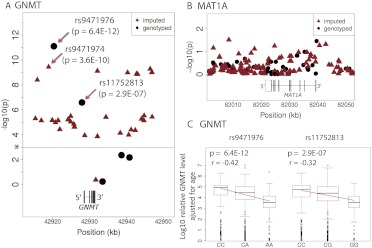
Hepatic GNMT protein levels were significantly correlated with age (r = −0.17, p = 0.006), but that was not the case for MAT1A (r = 0.06, p = 0.30). Therefore, the GNMT protein levels used in the association analysis were adjusted for age. Two of the genotyped GNMT tag SNPs, rs9471976 and rs11752813, were significantly associated with GNMT protein levels, with p values of 6.4 × 10−12 and 2.88 × 10−7, (p = 3.9 × 10−10 and p = 2.5 × 10−7 after correction for multiple comparisons), respectively (Fig. 4A). The correlation of GNMT protein expression with genotype for these two SNPs is shown graphically in Fig. 4C. Similar analyses performed with imputed GNMT SNPs identified 38 additional markers within 200 kb on either side of the gene that were significantly associated with GNMT protein levels (p < 10−4), with rs9471974 being the most significant imputed SNP (p = 3.6 × 10−10). No MAT1A SNPs were significantly associated with MAT1A protein levels (minimum p = 0.03, p = 1.5 after correction for multiple comparisons; Fig. 4B). A list of all SNPs with p values <10−4 for association with GNMT protein levels is summarized in Supplemental Table 3.
Fig. 4.
Genotype-phenotype correlations for human hepatic MAT1A and GNMT. Negative log-transformed p values for single SNP associations with human liver (A) GNMT protein levels and (B) MAT1A protein levels using both genotyped SNPs and SNPs imputed by using 1000 Genomes Project data. Figure 4, A and B, shows SNPs within 20 kb of the 3′ and 5′ ends of the genes and includes only imputed SNPs with imputation quality score Rsq values >0.3. Black circles represent genotyped tag SNPs, and red triangles represent imputed SNPs. *, the threshold for significance after Bonferroni correction for GNMT. The structures of both genes are also shown schematically. C, the relationship between SNP genotypes for the GNMT rs9471976 and rs11752813 SNPs and hepatic GNMT protein is shown. Spearman's rank correlation coefficients (r) and p values are also shown.
GNMT Reporter Gene Studies.
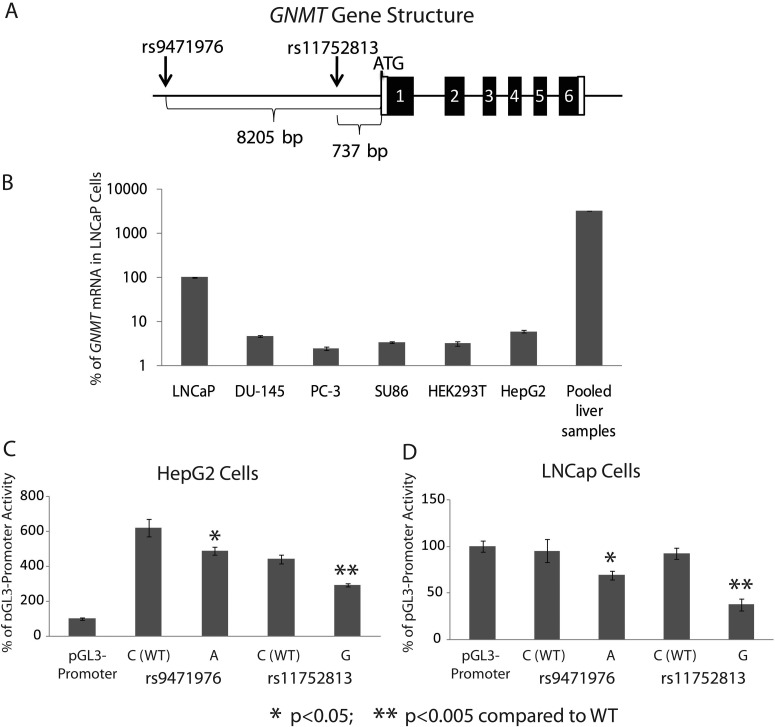
GNMT resequencing had not identified any polymorphisms that altered the encoded amino acid sequence. Therefore, our functional genomic experiments for GNMT focused on SNPs that were significantly associated with hepatic GNMT protein levels. Specifically, reporter gene constructs were created for the two genotyped SNPs with the lowest p values for association with GNMT protein levels (rs9471976 and rs11752813). The locations of these SNPs relative to the site of GNMT transcription initiation are shown in Fig. 5A. As a first step, we next surveyed a series of cell lines in an attempt to determine whether they might express GNMT. In particular, we performed qRT-PCR with mRNA preparations from HepG2, HEK293T, SU86, PC-3, DU-145, and LNCaP cell lines as well as pooled human liver sample preparations. As shown in Fig. 5B, compared with the HepG2 cell preparations, only LNCaP showed a relatively high level of GNMT mRNA expression. Please note that the y-axis is a logarithmic scale. Therefore, we used LNCaP and HepG2 cells in our reporter gene studies because LNCap is highly expressed GNMT and because of HepG2 cells' origin from hepatic tissue—the major organ known to highly express GNMT. Results from dual-luciferase assays performed with HepG2 cells showed that DNA sequences around both rs9471976 and rs11752813 could enhance pGL3-promoter activity by up to 6-fold (Fig. 5C). There were statistically significant differences in terms of their ability to drive transcription between WT and variant allele sequences for both SNPs in HepG2 and LNCap cells (Fig. 5, C and D). In both cases, the effect of variant SNP genotypes was consistent with the liver genotype-phenotype association results, i.e., the G allele in rs11752813 and the A allele in rs9471976 were both associated with lower levels of hepatic GNMT protein. However, because these two SNPs, rs9471976 and rs11752813, had been chosen for genotyping by using a SNP tagging strategy, they may not represent the functional SNPs but rather may be in LD with other variants that have functional significance.
Fig. 5.
GNMT qRT-PCR in a series of cell lines as well as dual luciferase reporter gene assays for the GNMT rs9471976 and rs11752813 SNPs performed with HepG2 and LNCaP cells. A, schematic representation of the GNMT gene with the locations of rs9471976 and rs11752813. B, GNMT mRNA measured by qRT-PCR in a series of cell lines expressed as a percentage of the value for LNCaP cells. C, luciferase reporter gene assays for HepG2 cells. D, luciferase reporter assays for LNCaP cells. Each bar for the reporter gene studies represents relative luciferase activity reported as a percentage of the pGL3-promoter construct activity. Values represent mean ± S.E.M. for six independent transfections.
Coordinate Regulation of the Hepatic Folate-Methionine Cycles.
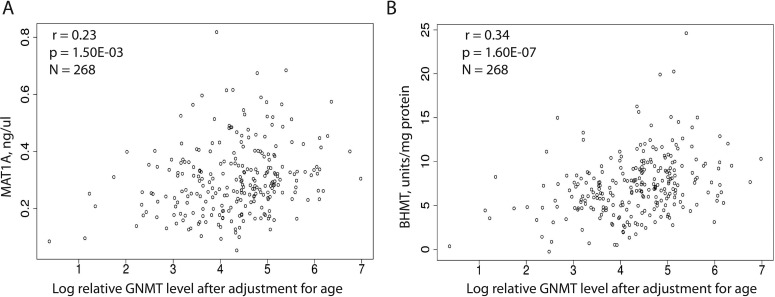
Finally, in an attempt to begin to understand the overall regulation of the methionine and folate cycles in human liver, we performed association analyses among levels of hepatic protein for other enzymes in these pathways that had also been assayed in the same 268 liver samples used to perform the present study. Specifically, we correlated BHMT and serine hydroxymethyltransferase 1 (SHMT1) protein levels as well as the level of enzyme activity for another important methyltransferase enzyme, catechol O-methyltransferase (COMT) (Feng et al., 2009; Zhang et al., 2009; Nordgren et al., 2011) with levels of MAT1A and GNMT protein expression (Table 4). Like MAT1A and GNMT, BHMT is predominantly expressed in hepatic tissue (Li et al., 2008). Because there was a significant correlation between age and COMT activity (p = 0.02), and because GNMT protein level was also correlated with age (p = 0.006) in these samples, data for the genotype-phenotype association analyses described earlier and for the hepatic methionine and folate cycle correlation analysis described here were adjusted for the age of the subject from whom the liver biopsy has been obtained for GNMT protein and COMT activity levels. In the 268 liver cytosol preparations studied, there was a significant correlation between GNMT and BHMT protein levels (r = 0.34, p = 1.60 × 10−7 after correction for multiple comparisons) and between GNMT and MAT1A protein levels (r = 0.23, corrected p = 1.50 × 10−3) (see Fig. 6). Level of activity for another AdoMet-dependent methyltransferase, COMT, was also associated with expression level of protein for these same two enzymes, but less significantly (p = 0.03 and 0.04, respectively). These results raise the possibility of coordinate regulation of methionine cycle enzyme protein levels in human hepatic tissue. Our association analysis of GNMT SNPs with levels of hepatic protein expression also showed that the rs9471976 and rs11752813 GNMT SNPs were significantly associated with SHMT1 protein level (p = 0.007 and 0.02, respectively).
TABLE 4.
Spearman correlations and p values for correlations of levels of protein expression measured in 268 hepatic biopsy samples
The three enzymes that are expressed primarily in liver, GNMT, MAT1A, and BHMT, are bolded. The p values listed in the table have been corrected for multiple comparisons.
| rs Value p value | COMT*a | BHMT | SHMT1 | MAT1A | GNMTa |
|---|---|---|---|---|---|
| COMT*a | 0.03 | 0.03 | 0.04 | 0.02 | |
| BHMT | 0.18 | 0.18 | 0.89 | 1.60E-07 | |
| SHMT1 | 0.18 | 0.15 | 1.00 | 0.17 | |
| MAT1A | −0.13 | 0.10 | 0.00 | 1.50E-03 | |
| GNMTa | 0.19 | 0.34 | 0.15 | 0.23 |
, enzyme activity was used to perform the correlation analysis for COMT.
Values for relative protein expression or enzyme activity were adjusted for age.
Fig. 6.
Correlations between levels of MAT1A and BHMT protein in adult human liver biopsy samples plotted against log GNMT protein levels in the same samples adjusted for age. A, GNMT versus MAT1A. B, GNMT versus BHMT. Spearman's correlation coefficients, r, as well as p values for associations after correction for multiple comparisons are shown. In both panels, the level of log GNMT protein has been corrected for patient age.
Discussion
AdoMet is the major methyl donor for biological methylation reactions, including those involved in DNA and histone methylation as well as the methyl conjugation of hormones, neurotransmitters, and drugs (Mato et al., 1997). In the adult human liver, AdoMet is synthesized from methionine and ATP by a reaction catalyzed by MAT1A (Kluijtmans et al., 2003), and a major process in its hepatic utilization is the GNMT-catalyzed formation of sarcosine and AdoHcy (Kluijtmans et al., 2003). These reactions represent critical steps in the hepatic methionine cycle (see Fig. 1). Because of the importance of methylation for a variety of cellular functions, intercellular AdoMet concentrations and the AdoMet/AdoHcy ratio have been reported to contribute to variation in biological processes ranging from the epigenetic regulation of gene expression to biogenic amine neurotransmitter biosynthesis and metabolism (Reynolds et al., 1984; Carney et al., 1987; Ulrey et al., 2005). In addition, AdoMet concentrations are a major factor determining levels of “downstream” metabolites in the methionine cycle such as AdoHcy, homocysteine, and GSH. For example, elevated plasma homocysteine levels have been associated with increased risk for cardiovascular disease, and individual variation in homocysteine concentrations is thought to be influenced by genetic factors. However, to date, the C667T variant (rs1801133) in the methylene tetrahydrofolate reductase gene is one of only a small number of common genetic polymorphisms that are known to be associated with variation in circulating homocysteine concentrations (Frosst et al., 1995). In an attempt to begin to dissect the possible role of genetic variation in genes encoding enzymes in the methionine cycle on a variety of cellular processes, in the present study we set out to determine common genetic variation in genes encoding the hepatic AdoMet synthesizing and degrading enzymes, MAT1A and GNMT. To do that, we took a systematic approach that began with gene resequencing, followed by imputation using 1000 Genomes Project data, functional genomic experiments, and genotype-phenotype correlation analyses performed with human hepatic biopsy samples. The gene resequencing studies identified common DNA sequence variation across all nine MAT1A exons, exon-intron splice junctions, and across the entire GNMT gene, as well as ∼1 kb of 5′ and 3′ FRs for both genes (Fig. 2; Supplemental Tables 1 and 2). Resequencing of 288 DNA samples identified one nsSNP in MAT1A, but this variant did not appear to have functional consequences (Table 3). Using 1000 Genomes Project data that were released soon after we completed our gene resequencing studies, we were able to extend our ability to scan these genes for functional markers out to 200 kb on either side of both genes.
Genotype-phenotype association analyses were then performed using 268 adult human surgical hepatic biopsy samples because the liver is the organ in which both MAT1A and GNMT are predominantly expressed. We observed two GNMT genotyped SNPs, rs9471976 and rs11752813, with p values for association with level of hepatic GNMT protein expression of 6.4 × 10−12 and 2.9 × 10−7, respectively, and 38 imputed SNPs within ±200 kb surrounding GNMT that were significantly associated with hepatic GNMT protein levels (p < 10−4). Many of these SNPs were in LD and were located in a region within 10 kb 5′ to GNMT, suggesting a possible role for this region in the regulation of GNMT transcription. Results of reporter gene studies performed with both HepG2 and LNCaP cells suggested that sequences around the rs9471976 and rs11752813 SNPs could increase transcription up to 6-fold, and there were significant differences in reporter gene activity between the WT and variant alleles (Fig. 5). In contrast, none of the 42 genotyped or 150 imputed MAT1A SNPs were highly associated with hepatic MAT1A protein levels (p > 0.03) (see Fig. 4B). These observations indicate that genetic regulation of GNMT expression in the human liver might be influenced by SNPs in a region at the 5′ end of the GNMT gene. Whether these polymorphisms influence concentrations of hepatic AdoMet or other methionine cycle metabolites, e.g., homocysteine, will have to be determined in the course of future experiments.
Finally, we examined the possibility of the coordinate regulation of hepatic methionine and folate cycle protein expression by determining correlations among protein expression or the activities of enzymes in this pathway in the 268 liver biopsy samples that we had used to perform the Western blot analyses (Table 4). Among the six proteins included in that analysis, GNMT, MAT1A, and BHMT are expressed predominantly in adult human liver, whereas COMT and SHMT1 are ubiquitously expressed. In a previous analysis of methionine and folate cycle mRNA expression in the Human Variation Panel lymphoblastoid cell lines that were used as a source of DNA for our gene resequencing studies (Hebbring et al., 2012), we observed strong correlations among mRNA expression levels for SHMT1, SHMT2, COMT, MAT2A, and MAT2B (Hebbring et al., 2012). Unfortunately, MAT1A, GNMT, and BHMT are not expressed in lymphoblastoid cells. The results of these two complementary association analyses for hepatic tissue and lymphoblastoid cells both raise the possibility of the “coordinate regulation” of the expression of genes encoding methionine and folate cycle enzymes. In summary, the results of the present study, when joined with those of previous reports, indicate the existence of the coordinate regulation of methionine cycle enzymes, with possible functional consequences for methylation in the human liver.
Supplementary Material
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grants R01-GM28157, U19-GM61388 (The Pharmacogenomics Research Network), R21-GM86689]; the National Institutes of Health National Cancer Institute [Grant R01-CA132780]; the National Institutes of Health National Center for Research Resources [Grant KL2-RR024151] (KL2 Mentored Career Development Award); a Gerstner Family Mayo Career Development Award in Individualized Medicine; and a PhRMA Foundation Center of Excellence Award in Clinical Pharmacology.
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
 The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- AdoMet
- (S)-adenosylmethionine
- MAT
- methionine adenosyltransferase
- AdoHcy
- (S)-adenosylhomocysteine
- GNMT
- glycine N-methyltransferase
- BHMT
- betaine homocysteine methyltransferase
- EA
- European American
- AA
- African American
- HCA
- Han Chinese American
- SNP
- single-nucleotide polymorphism
- rs
- reference SNP
- kb
- kilobase
- FR
- flanking region
- PCR
- polymerase chain reaction
- WT
- wild type
- LD
- linkage disequilibrium
- bp
- base pair
- qRT-PCR
- quantitative reverse transcriptase-PCR
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ns
- nonsynonymous
- SHMT1
- serine hydroxymethyltransferase 1
- COMT
- catechol-O-methyltransferase.
Authorship Contributions
Participated in research design: Ji, Nordgren, Hebbring, Peng, Yee, and Weinshilboum.
Conducted experiments: Ji, Nordgren, Y. Chai, Hebbring, Peng, Pelleymounter, Moon, X. Chai, Zhang, and Wieben.
Performed data analysis: Ji, Nordgren, Hebbring, Jenkins, Abo, Fridley, Yee, and Weinshilboum.
Wrote or contributed to the writing of the manuscript: Ji, Nordgren, Abo, Peng, Yee, and Weinshilboum.
References
- The 1000 Genomes Project Consortium (2010) A map of human genome variation from population-scale sequencing. Nature 467:1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnesen E, Refsum H, Bønaa KH, Ueland PM, Førde OH, Nordrehaug JE. (1995) Serum total homocysteine and coronary heart disease. Int J Epidemiol 24:704–709 [DOI] [PubMed] [Google Scholar]
- Augoustides-Savvopoulou P, Luka Z, Karyda S, Stabler SP, Allen RH, Patsiaoura K, Wagner C, Mudd SH. (2003) Glycine N -methyltransferase deficiency: a new patient with a novel mutation. J Inherit Metab Dis 26:745–759 [DOI] [PubMed] [Google Scholar]
- Balaghi M, Horne DW, Wagner C. (1993) Hepatic one-carbon metabolism in early folate deficiency in rats. Biochem J 291 (Pt 1):145–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstein J, Williams GR. (1960) The enzymatic N-methylation of glycine. Biochem Biophys Res Comm 3:259–263 [Google Scholar]
- Boyle AP, Song L, Lee BK, London D, Keefe D, Birney E, Iyer VR, Crawford GE, Furey TS. (2011) High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res 21:456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni GL. (1951a) Activation of methionine for transmethylation. J Biol Chem 189:745–754 [PubMed] [Google Scholar]
- Cantoni GL. (1951b) Methylation of nicotinamide with soluble enzyme system from rat liver. J Biol Chem 189:203–216 [PubMed] [Google Scholar]
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. (2004) Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 74:106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney MW, Toone BK, Reynolds EH. (1987) S-adenosylmethionine and affective disorder. Am J Med 83:104–106 [DOI] [PubMed] [Google Scholar]
- Chamberlin ME, Ubagai T, Mudd SH, Thomas J, Pao VY, Nguyen TK, Levy HL, Greene C, Freehauf C, Chou JY. (2000) Methionine adenosyltransferase I/III deficiency: novel mutations and clinical variations. Am J Hum Genet 66:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Lewington S, Landray M. (2003) Homocysteine, renal function, and risk of cardiovascular disease. Kidney Int Suppl:S131–S133 [DOI] [PubMed] [Google Scholar]
- Feng Q, Keshtgarpour M, Pelleymounter LL, Moon I, Kalari KR, Eckloff BW, Wieben ED, Weinshilboum RM. (2009) Human S-adenosylhomocysteine hydrolase: common gene sequence variation and functional genomic characterization. J Neurochem 110:1806–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M, Atta M, Mulliez E. (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29:243–249 [DOI] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP. (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10:111–113 [DOI] [PubMed] [Google Scholar]
- Gil B, Casado M, Pajares MA, Boscá L, Mato JM, Martín-Sanz P, Alvarez L. (1996) Differential expression pattern of S-adenosylmethionine synthetase isoenzymes during rat liver development. Hepatology 24:876–881 [DOI] [PubMed] [Google Scholar]
- Heady JE, Kerr SJ. (1973) Purification and characterization of glycine N-methyltransferase. J Biol Chem 248:69–72 [PubMed] [Google Scholar]
- Hebbring SJ, Chai Y, Ji Y, Abo RP, Jenkins GD, Fridley B, Zhang J, Eckloff BW, Wieben ED, Weinshilboum RM. (2012) Serine hydroxymethyltransferase 1 and 2: gene sequence variation and functional genomic characterization. J Neurochem 120:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Moon I, Zlatkovic J, Salavaggione OE, Thomae BA, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. (2007) Human hydroxysteroid sulfotransferase SULT2B1 pharmacogenomics: gene sequence variation and functional genomics. J Pharmacol Exp Ther 322:529–540 [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. (2008) SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 24:2938–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasperzyk JL, Fall K, Mucci LA, Håkansson N, Wolk A, Johansson JE, Andersson SO, Andrén O. (2009) One-carbon metabolism-related nutrients and prostate cancer survival. Am J Clin Nutr 90:561–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr SJ. (1972) Competing methyltransferase systems. J Biol Chem 247:4248–4252 [PubMed] [Google Scholar]
- Kluijtmans LA, Young IS, Boreham CA, Murray L, McMaster D, McNulty H, Strain JJ, McPartlin J, Scott JM, Whitehead AS. (2003) Genetic and nutritional factors contributing to hyperhomocysteinemia in young adults. Blood 101:2483–2488 [DOI] [PubMed] [Google Scholar]
- Li F, Feng Q, Lee C, Wang S, Pelleymounter LL, Moon I, Eckloff BW, Wieben ED, Schaid DJ, Yee V, et al. (2008) Human betaine-homocysteine methyltransferase (BHMT) and BHMT2: common gene sequence variation and functional characterization. Mol Genet Metab 94:326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Ding J, Abecasis GR. (2006) Mach 1.0: rapid haplotype reconstruction and missing genotype inference. 2006 ASHG Annual Meeting; 2006 Oct 9–13; New Orleans, LA American Society of Human Genetics [Google Scholar]
- Luka Z, Cerone R, Phillips JA, 3rd, Mudd HS, Wagner C. (2002) Mutations in human glycine N-methyltransferase give insights into its role in methionine metabolism. Hum Genet 110:68–74 [DOI] [PubMed] [Google Scholar]
- Main PA, Angley MT, Thomas P, O'Doherty CE, Fenech M. (2010) Folate and methionine metabolism in autism: a systematic review. Am J Clin Nutr 91:1598–1620 [DOI] [PubMed] [Google Scholar]
- Martin YN, Salavaggione OE, Eckloff BW, Wieben ED, Schaid DJ, Weinshilboum RM. (2006) Human methylenetetrahydrofolate reductase pharmacogenomics: gene resequencing and functional genomics. Pharmacogenet Genomics 16:265–277 [DOI] [PubMed] [Google Scholar]
- Maruti SS, Ulrich CM, White E. (2009) Folate and one-carbon metabolism nutrients from supplements and diet in relation to breast cancer risk. Am J Clin Nutr 89:624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato JM, Alvarez L, Ortiz P, Pajares MA. (1997) S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther 73:265–280 [DOI] [PubMed] [Google Scholar]
- Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti G, Caruso U, Lorini R, Watkins D, Matiaszuk N, Rosenblatt DS, et al. (2001) Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia. J Inherit Metab Dis 24:448–464 [DOI] [PubMed] [Google Scholar]
- Mudd SH, Ebert MH, Scriver CR. (1980) Labile methyl group balances in the human: the role of sarcosine. Metabolism 29:707–720 [DOI] [PubMed] [Google Scholar]
- Mudd SH, Poole JR. (1975) Labile methyl balances for normal humans on various dietary regimens. Metabolism 24:721–735 [DOI] [PubMed] [Google Scholar]
- Nordgren KK, Peng Y, Pelleymounter LL, Moon I, Abo R, Feng Q, Eckloff B, Yee VC, Wieben E, Weinshilboum RM. (2011) Methionine adenosyltransferase 2A/2B and methylation: gene sequence variation and functional genomics. Drug Metab Dispos 39:2135–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nygård O, Vollset SE, Refsum H, Stensvold I, Tverdal A, Nordrehaug JE, Ueland M, Kvåle G. (1995) Total plasma homocysteine and cardiovascular risk profile. The Hordaland Homocysteine Study. JAMA 274:1526–1533 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds EH, Carney MW, Toone BK. (1984) Methylation and mood. Lancet 2:196–198 [DOI] [PubMed] [Google Scholar]
- Shield AJ, Thomae BA, Eckloff BW, Wieben ED, Weinshilboum RM. (2004) Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatry 9:151–160 [DOI] [PubMed] [Google Scholar]
- Skiba WE, Taylor MP, Wells MS, Mangum JH, Awad WM., Jr (1982) Human hepatic methionine biosynthesis. Purification and characterization of betaine:homocysteine S-methyltransferase. J Biol Chem 257:14944–14948 [PubMed] [Google Scholar]
- Smythies JR, Gottfries CG, Regland B. (1997) Disturbances of one-carbon metabolism in neuropsychiatric disorders: a review. Biol Psychiatry 41:230–233 [DOI] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457:910–914 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Stevens VL, McCullough ML, Sun J, Gapstur SM. (2010) Folate and other one-carbon metabolism-related nutrients and risk of postmenopausal breast cancer in the Cancer Prevention Study II Nutrition Cohort. Am J Clin Nutr 91:1708–1715 [DOI] [PubMed] [Google Scholar]
- Ulrey CL, Liu L, Andrews LG, Tollefsbol TO. (2005) The impact of metabolism on DNA methylation. Hum Mol Genet 14:R139–R147 [DOI] [PubMed] [Google Scholar]
- Wang SH, Kuo SC, Chen SC. (2003) High-performance liquid chromatography determination of methionine adenosyltransferase activity using catechol-O-methyltransferase-coupled fluorometric detection. Anal Biochem 319:13–20 [DOI] [PubMed] [Google Scholar]
- Weinstein SJ, Stolzenberg-Solomon R, Pietinen P, Taylor PR, Virtamo J, Albanes D. (2006) Dietary factors of one-carbon metabolism and prostate cancer risk. Am J Clin Nutr 84:929–935 [DOI] [PubMed] [Google Scholar]
- Zhang J, Ji Y, Moon I, Pelleymounter LL, Ezequel Salavaggione O, Wu Y, Jenkins GD, Batzler AJ, Schaid DJ, Weinshilboum RM. (2009) Catechol O-methyltransferase pharmacogenomics: human liver genotype-phenotype correlation and proximal promoter studies. Pharmacogenet Genomics 19:577–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.