Abstract
Activation of the NF-κB pathway requires the formation of Met1-linked ‘linear’ ubiquitin chains on NEMO, which is catalysed by the Linear Ubiquitin Chain Assembly Complex (LUBAC) E3 consisting of HOIP, HOIL-1L and Sharpin. Here, we show that both LUBAC catalytic activity and LUBAC specificity for linear ubiquitin chain formation are embedded within the RING-IBR-RING (RBR) ubiquitin ligase subunit HOIP. Linear ubiquitin chain formation by HOIP proceeds via a two-step mechanism involving both RING and HECT E3-type activities. RING1-IBR catalyses the transfer of ubiquitin from the E2 onto RING2, to transiently form a HECT-like covalent thioester intermediate. Next, the ubiquitin is transferred from HOIP onto the N-terminus of a target ubiquitin. This transfer is facilitated by a unique region in the C-terminus of HOIP that we termed ‘Linear ubiquitin chain Determining Domain’ (LDD), which may coordinate the acceptor ubiquitin. Consistent with this mechanism, the RING2-LDD region was found to be important for NF-κB activation in cellular assays. These data show how HOIP combines a general RBR ubiquitin ligase mechanism with unique, LDD-dependent specificity for producing linear ubiquitin chains.
Keywords: E3 ligase, HHARI, Parkin, RNF31, TRIAD
Introduction
Ubiquitin conjugation is a highly versatile system for conferring post-translational modifications, since this 76-amino acid protein can make a variety of chains that signal to different downstream effectors. The ubiquitins in these chains are usually linked between the ubiquitin C-terminus of the donor ubiquitin and any of the seven lysines in the acceptor ubiquitin, but the donor ubiquitin can also link to the amino group in the N-terminal methionine of the acceptor ubiquitin, leading to the formation of linear ubiquitin chains.
Linear ubiquitin chains are assembled by the Linear Ubiquitin Chain Assembly Complex (LUBAC), which plays a critical role in the activation of the NF-κB pathway that is involved in various functions, including cell survival and inflammation. NF-κB activation can be induced by, for example, cytokines or DNA damage, which lead to LUBAC-mediated ubiquitination of NEMO with linear ubiquitin chains (Tokunaga et al, 2009; Niu et al, 2011). This linear ubiquitination of NEMO causes IKKβ phosphorylation and activation. Subsequently, IκBα is degraded and free NF-κB translocates to the nucleus to activate the transcription of target genes (Kirisako et al, 2006; Haas et al, 2009; Iwai and Tokunaga, 2009; Tokunaga et al, 2009).
LUBAC consists of at least three different proteins, HOIP (RNF31), HOIL-1L (RBCK1) and Sharpin (Kirisako et al, 2006; Gerlach et al, 2011; Ikeda et al, 2011; Tokunaga et al, 2011). HOIP and HOIL-1L belong to the RING-in-between-RING (RBR) class of E3 ligases. However, only the RBR domain of HOIP and not HOIL-1L is required for linear ubiquitin chain formation by LUBAC and subsequent IKKβ phosphorylation (Kirisako et al, 2006; Hostager et al, 2010). Nevertheless, a combination of HOIP with either HOIL-1L or Sharpin is the minimal requirement for linear ubiquitin chain catalysis (Kirisako et al, 2006; Gerlach et al, 2011).
The RBR class of E3 ligases, also known as the TRIAD class (two RING fingers and DRIL (double RING linked)), was first described in 1999 (Morett and Bork, 1999; van der Reijden et al, 1999). The structures of the separate RING domains and the in-between RING (IBR) have been solved (PDB entry 1WIM, report to be published; Capili et al, 2004; Beasley et al, 2007); however, it remains unclear how the RBR forms a functional unit. RING1 has a classical RING fold, which is typically used for E2–E3 interactions (Zheng et al, 2000; Deshaies and Joazeiro, 2009). Also, RING2 interacts with different E2s in yeast-two-hybrid studies and the cysteine and histidine distribution of RING2 suggests that it forms a RING domain (Hristova et al, 2009; Markson et al, 2009; Marteijn et al, 2009; van Wijk et al, 2009). However, even though Zn2+ stoichiometry analysis indicates that all RING domains in Parkin coordinate two zinc ions (Hristova et al, 2009), the solution structure of HHARI RING2 does not have a classical RING-fold and coordinates only one zinc ion per monomer. Furthermore, the HHARI RING2 domain was recently shown to form a thioester adduct with ubiquitin (HHARI∼ubiquitin) on a free cysteine as an intermediate step in the ubiquitin transfer (Wenzel et al, 2011), similar to that found in HECT domains. Although the thioester adduct could not be visualized on the RBR-protein Parkin in the same study, mechanistic analysis indicated that both RBR proteins include a HECT-like step in the ubiquitin transfer.
An intact RBR domain is necessary for efficient E3-ligase functioning, however Parkin IBR-RING2 can mediate the formation of ubiquitin linkages in the absence of RING1 (Matsuda et al, 2006; Chew et al, 2011). In addition to the interaction of both RING domains with E2 enzymes, the RBR of Parkin also interacts non-covalently with ubiquitin during chain formation (Chaugule et al, 2011).
The specificity for ubiquitin chain types is regulated completely at the level of the E3 ligase in HECT domains (Wang and Pickart, 2005; Kim and Huibregtse, 2009). In contrast, with RING domain E3 ligases the E2 enzymes contribute to the chain types that are formed. Some E2s directly mediate the formation of specific ubiquitin chains via the non-covalent binding of an acceptor ubiquitin, positioning a particular lysine residue to attack the thioester bond between the E2 and the donor ubiquitin (Petroski and Deshaies, 2005; Eddins et al, 2006; Wickliffe et al, 2011). A single RING E3 can recruit several of these E2s and makes different chains dependent on the E2 specificity (Christensen et al, 2007; Kim et al, 2007). Occasionally, the chain type that is being formed by a combination of a RING E3 and a less specific E2, such as Ube2D3, is determined by the specific E2–E3 combination (Wu-Baer et al, 2003; Nishikawa et al, 2004).
So far, LUBAC is the only E3 ligase complex that is known to promote linear ubiquitin chain formation. Although it contains RING domains, its ubiquitin chain forming specificity overrides that of the collaborating E2 enzymes. Thus even highly specific E2s that are known to catalyse the formation of very specific chain types, such as Ube2K that forms K48 linked chains (Chen et al, 1991), will form linear ubiquitin chains in the presence of LUBAC (Kirisako et al, 2006). Therefore, chain type specificity is thought to be embedded within the LUBAC E3, but it is unclear how this specificity is organized.
We performed an in-vitro analysis of HOIP ubiquitin chain assembly activity to investigate the mechanism underlying linear ubiquitin chain formation by LUBAC. We show that a truncated form of HOIP is active in in-vitro linear chain formation in the absence of HOIL-1L and Sharpin. The catalytic activity and specificity for linear ubiquitin chain assembly of LUBAC is completely embedded within HOIP RING2 and a newly identified Linear ubiquitin chain Determining Domain (LDD) in the C-terminus of HOIP. Furthermore, we show that the ubiquitin thioester is first transferred from the E2 onto HOIP and is subsequently linked to a target ubiquitin that is docked on the LDD. This study strengthens the knowledge on the general mechanism for RBR-mediated ubiquitin chain formation and provides novel mechanistic insights in linear ubiquitin chain assembly by HOIP.
Results
Linear ubiquitin chain formation specificity is embedded within HOIP
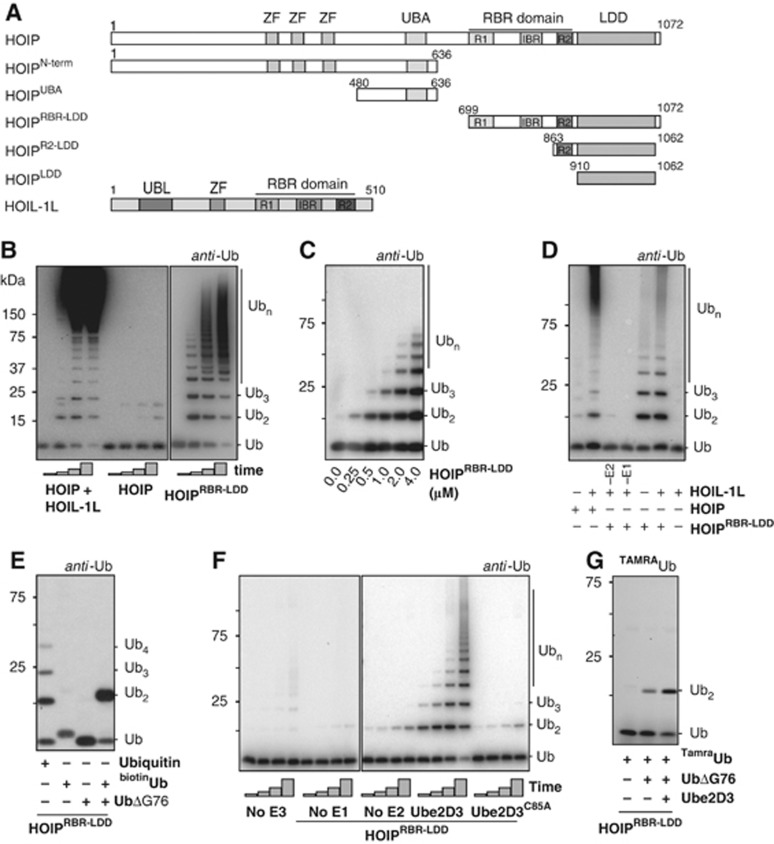
To study linear chain formation, we expressed full-length human HOIL-1L, full-length HOIP and a series of HOIP deletion constructs in E. coli. We used synthetic genes that are optimized for bacterial expression (Figure 1A) and used the purified proteins for in-vitro reactions, analysing free ubiquitin chain formation. As expected, full-length HOIP alone was not active in forming ubiquitin chains, but in the presence of HOIL-1L robust chain formation was observed (Figure 1B).
Figure 1.
HOIP mediates linear ubiquitin chain formation. (A) HOIP and HOIL-1L constructs used in this study. Ubiquitin-Like domain (UBL), Npl4 Zinc Finger (ZF), Ubiquitin-Associated domain (UBA), Linear chain Determining Domain (LDD) and a RING-in-Between-RING domain (RBR) consisting of two RING domains (R1 and R2) and an in-between RING domain (IBR). The domain borders of the UBL, ZF, UBA and RBR domains are drawn to scale according to Uniprot definitions ( www.uniprot.org). (B) Ubiquitin chain formation with Ube2L3 in combination with 2 μM of the different E3s after 0, 10, 20 and 40 min. Standard reaction conditions are described in Materials and methods. Reactions were performed without NaCl. (C) Increasing amounts of ubiquitin chain formation with increasing concentrations of HOIPRBR-LDD. Reactions were performed in the presence of Ube2D3 and were stopped after 1 h. (D) HOIPRBR-LDD cannot be further activated by HOIL-1L in a 1-h reaction with Ube2L3. Reactions were performed without NaCl. (E) Ubiquitin chain formation by HOIPRBR-LDD with N-terminally blocked ubiquitin (biotinubiquitin) and C-terminally truncated ubiquitin (UbiquitinΔGly76). Reactions were stopped after 90 min at 32°C. (F) HOIPRBR-LDD activity with and without Ube2D3 and the Ube2D3 active site mutant (Ube2D3C85A). Reactions were stopped after 15, 30, 60 and 120 min. (G) Di-ubiquitin linkage formation with TAMRAubiquitin in the presence and absence of ubiquitinΔGly76 and Ube2D3 after 2 h.
Since previous published data were derived from assays performed in the absence of sodium chloride (Kirisako et al, 2006; Gerlach et al, 2011) under conditions that are far from physiological (∼150 mM NaCl), we tested the influence of NaCl and pH on the in-vitro reactions. In the absence of salt, the reactions were more active and it was easier to visualize detailed chains (Supplementary Figure S1A and B), but the overall pattern of the bands on gel remained the same. Furthermore, the proteins were only active in conditions above pH 7 and raising the pH up to 9.5 caused a minor extra activation of the reactions (Supplementary Figure S1C). We mainly used reaction conditions with 150 mM NaCl at pH 8; however, conditions without NaCl are used in some of our experiments as a tool to improve visualization of the activity of the LUBAC proteins.
Next, we used an N-terminally truncated form of HOIP, which includes only the RBR domain and the C-terminal region that we have named Linear ubiquitin chain Determining Domain (HOIPRBR-LDD, Figure 1A). The sequence of the LDD is not conserved between RBR proteins, and Psi-BLAST searches and a Phyre threading analysis on this region reveal that it is exclusive to HOIP. Nevertheless, between HOIP orthologues the LDD is highly conserved (Supplementary Figure S1D), which suggests that the LDD functions specifically in the context of the upstream RBR domain in HOIP. When we tested HOIPRBR-LDD for in-vitro activity we found, surprisingly, that this construct does not require HOIL-1L and Sharpin for in-vitro activity (Figure 1B and C). HOIPRBR-LDD does not contain the UBA domain that is needed for the interaction with HOIL-1L and Sharpin (Kirisako et al, 2006; Tokunaga et al, 2009, 2011), explaining why the activity of HOIPRBR-LDD is hardly increased by the addition of HOIL-1L in the reactions (Figure 1D).
As HOIL-1L and Sharpin have been shown to be important for HOIP activity, we wondered whether the short RBR-LDD construct of HOIP retained the specificity for making N-terminally linked ubiquitin chains. We tested chain formation, using either Ube2D3 (UbcH5c) or Ube2L3 (UbcH7) as E2 enzymes. In both cases, HOIPRBR-LDD forms ubiquitin chains with lysine-less ubiquitin (K0) and mutated ubiquitins that contain either a single lysine or a lysine point mutation (Supplementary Figure S1E). In addition, when the ubiquitin N-terminus is blocked with a His tag, a biotin or a TAMRA-label, the ubiquitin chain formation is eliminated (Figure 1E; Supplementary Figure S1F and G), indicating that the accessibility of the N-terminus is critical for this reaction. Combinations of any of the N-terminally blocked ubiquitins with ubiquitinΔGly76, which can only function as an acceptor ubiquitin, produces solely di-ubiquitins (Figure 1E; Supplementary Figure S1F and G), confirming that a free ubiquitin N-terminus is essential for ubiquitin chain formation by HOIP. Consequently, the RBR-LDD in the C-terminus of HOIP is sufficient for the linear ubiquitin chain formation specificity of the LUBAC E3 and does not require the presence of other LUBAC subunits.
Since HOIL-1L and Sharpin are essential for full-length HOIP activity, but not for the HOIPRBR-LDD, it seems that the catalytic centre is not available for catalysis in the full-length protein. The UBL domains of either HOIL-1L or Sharpin have to bind to the UBA domain of HOIP, which lies N-terminally of the catalytic RBR-LDD, to activate the proteins in the NF-κB pathway (Sieber et al, 2012; Yagi et al, 2012). This could suggest some level of auto-inhibition within HOIP, similar to that seen in the RBR-protein Parkin, where the N-terminal UBL is binding to the C-terminal ubiquitin binding domain to block the catalytic centre (Chaugule et al, 2011). Therefore, we tested if the N-terminus of HOIP can inhibit HOIPRBR-LDD in trans. Full-length HOIP, HOIPN-term or HOIPUBA was added to the reaction with HOIPRBR-LDD, but the constructs did not inhibit the HOIPRBR-LDD-mediated chain formation (Supplementary Figure S1H). Apparently, the covalent linkage of the N-terminal domains to the RBR is required for the inhibition, either by increasing the local concentration or by arranging some position-specific conformational change that can be released by the Sharpin or HOIL-1L interaction. Consequently, the exact mechanism by which the catalytic domain is kept in an inactive state in full-length HOIP remains to be resolved.
The active LUBAC E3 mediates the specific formation of linear ubiquitin chains in cooperation with many different E2 enzymes that are normally highly specific in the formation of different types of ubiquitin chains (Kirisako et al, 2006). This ability to override the E2 specificity is retained in HOIPRBR-LDD. It specifically catalyses the formation of linear ubiquitin chains in the presence of the E2s Ube2D3, which can mediate the formation of many different types of lysine-linked ubiquitin chains (Kim et al, 2007), and Ube2L3, which targets to cysteines (Wenzel et al, 2011), indicating that the E2s are important to deliver the activated ubiquitin to the complex, but do not contribute to the chain type specificity.
HOIP has E2-independent linear chain forming activity
Interestingly, we observed very weak chain formation activity with HOIPRBR-LDD even with an inactive Ube2D3 mutant (C85A) (Figure 1F). Therefore, we analysed the HOIPRBR-LDD activity in the absence of E2 enzymes and still observed HOIP-dependent activity (Figure 1F), confirming that the E3 does not require an E2 for activity. However, in the absence of the E1 no activity is observed (Figure 1F). The chains formed in the E2-independent reaction are exclusively linear ubiquitin chains (Figure 1G). A similar E2-independent activity was recently described for the RBR-protein Parkin (Chew et al, 2011), indicating that this may be a general feature of RBR proteins. Nevertheless, E2-independent activity is unlikely to reflect a physiological activity, since the reaction is much more efficient in the presence of an E2∼Ub thioester. However, these data emphasize that linear chain specificity does not rely on E2s, but is completely embedded within HOIP.
HOIP RING1 and IBR are involved in E2-mediated activity
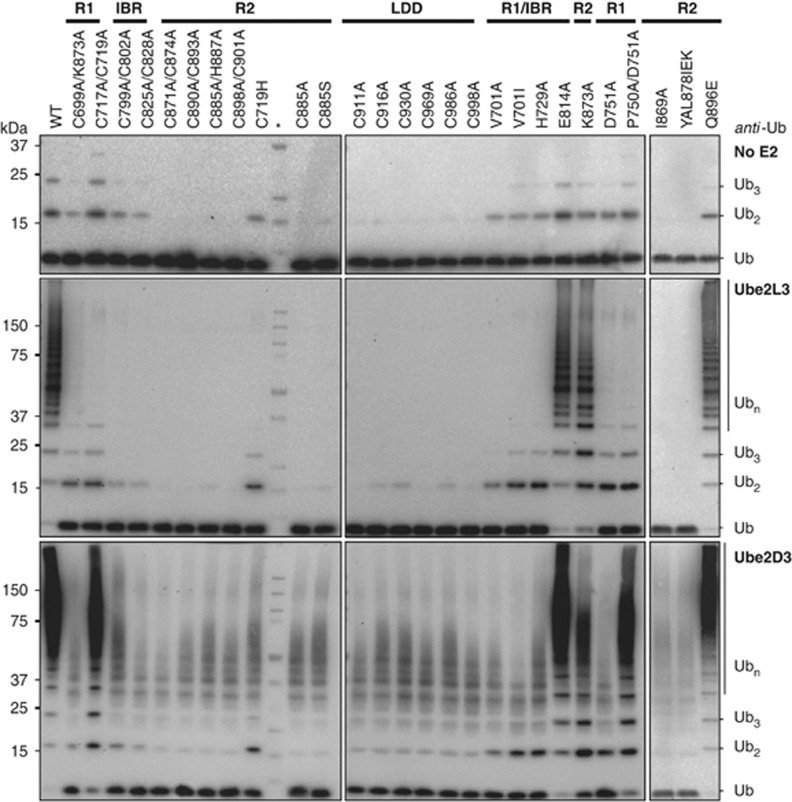
We next examined how HOIP promotes linear ubiquitin chain formation. To address this point, we made a series of point mutations and deletion constructs to unravel the contributions of the various domains within HOIPRBR-LDD (Supplementary Figure S2). The activities of all point mutants that are used in this study are shown in Figure 2 and are summarized in Supplementary Figure S2. The effect of the mutations in the different domains of HOIP will be discussed throughout this article.
Figure 2.
All domains in HOIPRBR-LDD are involved in ubiquitin chain formation. Ubiquitin chain formation with HOIPRBR-LDD mutants in the presence and absence of the E2 Ube2L3 or Ube2D3. Reactions were stopped after 6 h. The molecular weight marker is indicated by the asterisk (*). The solid bars indicate the location (RING1 (R1), IBR, RING2 (R2), LDD) of the mutations within HOIPRBR-LDD.
First, the importance of RING1 and the IBR were analysed. RING and IBR domains coordinate two zinc ions via eight Cys/His residues, whereby each zinc ion is coordinated by four Cys/His residues. Cysteine mutations in RING1 that disrupt the coordination of the zinc ions caused reduced E2-dependent activity with both Ube2D3 and Ube2L3 (Figure 2). Also HOIPRBR-LDD V701A, which was designed to interfere directly with the E2–E3 interaction but not to disrupt the RING-fold (Brzovic et al, 2003), inhibited the ubiquitin chain formation. Interestingly, the C717, 719A mutant solely disrupted Ube2L3-dependent activity and not Ube2D3-mediated chain formation, revealing a difference in the binding interface between HOIP and different E2s. Nevertheless, the complete set of mutants reveals that RING1 is essential for the activity with both E2s. The E2-independent activity of HOIPRBR-LDD was not affected by the RING1 mutations, indicating a classical RING-type role for RING1 where the RING domain catalyses the transfer from the E2 onto a target site. The IBR cysteine mutants also influenced the E2-dependent ubiquitin chain assembly, but not E2-independent activity (Figure 2). Therefore, both RING1 and the IBR are important for E2-mediated ubiquitin chain formation by HOIP.
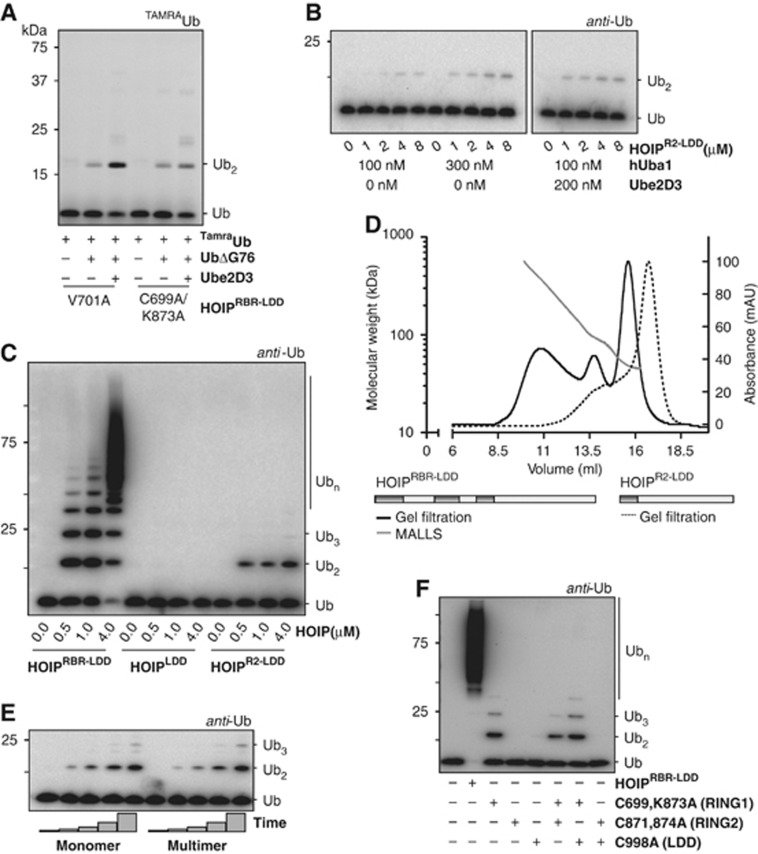
HOIPR2-LDD forms the minimal domain for linear ubiquitin chain formation
The linear ubiquitin chain assembly specificity of HOIP is preserved in HOIP RING1 mutants (Figure 3A) and RING1/IBR mutations do not affect the E2-independent activity (Figure 2), indicating that these domains are not used in the actual linkage formation between two ubiquitins. Accordingly, when the RING1 and IBR domains are deleted (HOIPR2-LDD, Figure 1A), the ability to form ubiquitin linkages in an E2-independent manner is retained (Figure 3B). HOIPR2-LDD cannot be further activated by Ube2D3 (Figure 3B; Supplementary Figure S3), showing the importance for RING1-IBR in E2-dependent activity. Thus, a completely intact HOIPRBR-LDD is needed for efficient ubiquitin chain formation that is facilitated by the E2, but the intrinsic ubiquitin chain assembly activity is located more C-terminally in the RING2 and the LDD.
Figure 3.
HOIPR2-LDD is the catalytic centre for in cis linear ubiquitin chain assembly. (A) Di-ubiquitin formation with HOIPRBR-LDD RING1 mutants with TAMRAubiquitin in the presence and absence of ubiquitinΔGly76 and Ube2D3. Reactions were stopped after 60 min (B) HOIPR2-LDD-mediated ubiquitin chain assembly after 2 h cannot be activated by Ube2D3. (C) Activity of different HOIP constructs after 1 h in the presence of Ube2D3 reveals that HOIPLDD is not active. (D) S200 10/300 gel filtration elution profile of HOIPRBR-LDD (black line) and HOIPR2-LDD (dotted line). Maximum absorbance has been set at 100 mAU. The according calculated molar mass from the MALLS for HOIPRBR-LDD (grey dots) is plotted against the elution volume. Three peaks are identified, correlating to HOIPRBR-LDD monomers (42.9 kDa), dimers (85.8 kDa) and multimers (>85.8 kDa). (E) Ubiquitin chain formation by HOIPRBR-LDD monomers and multimers. Monomers and multimers were taken from corresponding single fractions from the middle of the different peaks of the gel filtration profile and used immediately (without concentrating the samples) in ubiquitin chain formation reactions. HOIPRBR-LDD was assayed at 0.5 μM in the presence of Ube2D3. Time points were taken after 0, 10, 20, 40 and 60 min. (F) Combinations of HOIP mutants do not rescue ubiquitin chain formation activity with Ube2D3 (assay performed in absence of NaCl).
We aimed at mapping the regions in HOIPR2-LDD that are essential for ubiquitin chain catalysis. The importance of RING2 was explored by comparing the activity of HOIPR2-LDD and a construct that lacks all of the RBR domain (HOIPLDD) (Figure 1A). Although HOIPR2-LDD can still form E2-independent di-ubiquitin linkages, HOIPLDD is catalytically inactive even at high concentrations (Figure 3C). In addition, single cysteine-to-alanine mutants of HOIPRBR-LDD RING2 are catalytically inactive (Figure 2). Therefore, RING2 is essential for ubiquitin chain assembly. Next, the relevance of the LDD in ubiquitin chain formation was investigated. We were unable to express constructs of HOIP that lack the LDD and all LDD mutants are catalytically inactive (Figure 2). Nevertheless, the LDD alone is not sufficient for catalysis. Consequently, the integrity of both RING2 and the LDD is needed for linear ubiquitin chain assembly by HOIP.
HOIP mediates ubiquitin chain formation in cis
The presence of multiple copies of HOIP within LUBAC (Kirisako et al, 2006) suggests that HOIP might assemble ubiquitin chains in trans. Therefore, we next examined if the ubiquitin chain formation reaction is catalysed by single HOIP molecules (in cis) or by the cooperation of multiple copies of HOIP (in trans). The gel filtration profile and multi-angle laser light scattering (MALLS) of HOIPRBR-LDD show that the protein is purified as a mixture of monomers, dimers and multimers (Figure 3D). Nevertheless, the different fractions of the gel filtration profile show equal activity in free ubiquitin chain formation assays (Figure 3E), implying that the multimerization of HOIPRBR-LDD is not a requirement for activity.
To confirm these data, we combined inactive HOIPRBR-LDD mutants in chain formation assays to test whether they would collaborate to rescue the ability to form ubiquitin linkages. RING1 mutants are affected in E2-dependent chain formation and LDD mutants cannot support the formation of the isopeptide bond between two ubiquitins. Consequently, a combination of a RING1 mutant and an LDD mutant is expected to be effective in chain formation if the reaction occurs in trans. The combination of a RING1 and an LDD mutant did not lead to effective ubiquitin chain assembly, showing that the mutants do not complement each other (Figure 3F). Furthermore, a combination of a RING1 mutant and a RING2 mutant, or a RING2 mutant and an LDD mutant did not result in chain formation (Figure 3F), suggesting again that HOIPRBR-LDD proteins act individually and do not collaborate in ubiquitin chain formation. Finally, HOIPR2-LDD is purified mainly as a monomer (Figure 3D) and is still active in E2-independent chain formation. Therefore, we conclude that multimerization of HOIP is not a requirement for activity and linear ubiquitin chain formation is catalysed within single HOIP molecules.
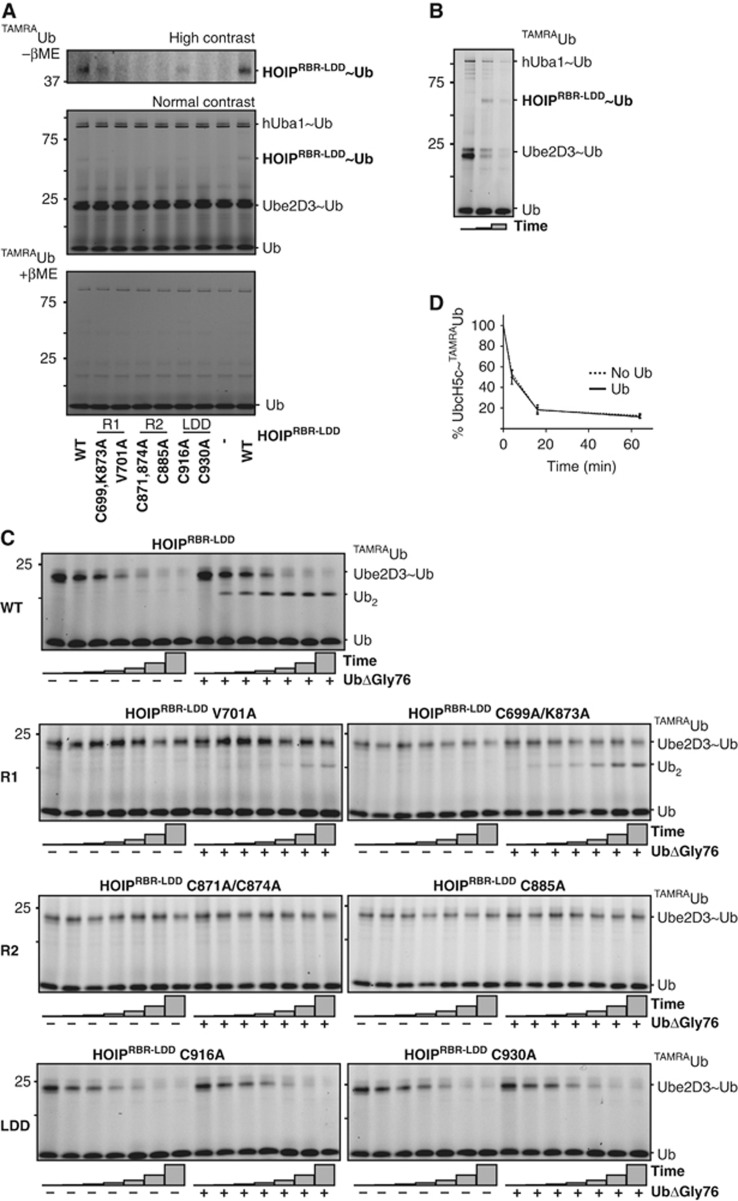
HOIP forms a reversible covalent intermediate with ubiquitin
In light of the HECT-like character of RING2 in other RBR proteins (Wenzel et al, 2011), we tested whether HOIP could make a covalent thioester intermediate. We used single-cycle turnover assays, with pre-charged E2∼TAMRAubiquitin thioester and were able to trap an E3-ubiquitin intermediate with HOIPRBR-LDD. A covalent E3∼TAMRAubiquitin complex could be visualized on non-reducing gels using anti-HOIP western blotting or, more clearly, by the TAMRA signal of ubiquitin (Figure 4A and B; Supplementary Figure S4A, Ube2D3; Supplementary Figure S4B, Ube2L3). The HOIPRBR-LDD∼ubiquitin intermediate could be disrupted by the addition of reducing loading buffer, which illustrates that HOIPRBR-LDD forms a reversible covalent bond with ubiquitin in cooperation with both Ube2D3 and Ube2L3.
Figure 4.
E2∼ubiquitin discharge and chain formation are two separate events. (A) Formation of a reversible covalent intermediate between HOIP and TAMRAubiquitin with different HOIPRBR-LDD mutants. Ube2D3 was used as the E2 enzyme. The TAMRA signal is visualized on a reduced gel and at two contrast levels on a non-reduced gel. (B) Single-cycle turnover assay monitoring Ube2D3∼TAMRAubiquitin discharge and HOIP∼TAMRAubiquitin formation after 0, 4 and 16 min at 37°C. (C) Single-cycle turnover assays showing Ube2D3∼TAMRAubiquitin discharge by HOIP mutants (left half of each gel), and di-ubiquitin formation upon the addition ubiquitinΔGly76 (right half of each gel). Discharge reactions were stopped after 0, 2, 4, 8, 16, 32 and 64 min. RING1 (R1), RING2 (R2). (D) Ube2D3∼TAMRAubiquitin discharge rates in the presence and absence of an acceptor ubiquitin. Standard error for the Ube2D3∼TAMRAubiquitin was calculated over three independent experiments.
Here, we show for a second RBR protein the presence of an E3∼ubiquitin thioester bond. The covalent HOIP∼ubiquitin is transient, as indicated by the low signals, however the bond could be detected in the RING1 mutants as well as in the C916A LDD mutant (Figure 4A; Supplementary Figure S4B). The RING2 mutants were completely impaired in forming this intermediate (Figure 4A; Supplementary Figure S4B). RING2 has been suggested in the literature as the actual site for the E3∼ubiquitin thioester in RBR proteins, although visualization of the E3∼thioester has only been successful for HHARI (Wenzel et al, 2011). We could not assign the thioester forming cysteine, since several cysteines in RING2 are impaired in thioester formation. The HOIP Cys885 that aligns with the thioester-forming Cys357 in HHARI, could not form an oxyester HOIP∼ubiquitin intermediate, when mutated to serine (Supplementary Figure S4C). However, this could be due to detection limits of the assay, since the reaction is less favourable. Unlike the LDD, RING2 is conserved between RBR proteins (Supplementary Figure S4D) and it is essential for E2∼ubiquitin discharge and E3∼ubiquitin formation. Therefore, it seems likely that RING2 provides the actual site on which the E3∼ubiquitin is formed.
HOIP-mediated ubiquitin transfer from the E2 onto a target is a two-step mechanism
To understand how the different domains within HOIPRBR-LDD contribute to the assembly of ubiquitin chains, we monitored the in vitro E2∼ubiquitin discharge and di-ubiquitin formation in single-cycle turnover assays with TAMRAubiquitin and the selected purified HOIPRBR-LDD mutants. The amino-terminus of TAMRAubiquitin is not available for linear ubiquitin chain formation and can only be linked to a ubiquitin with a free N-terminus by HOIPRBR-LDD. This feature allowed us to uncouple the discharge of ubiquitin from the E2 active site cysteine (in the absence of ubiquitinΔGly76) and the formation of the isopeptide bond between the N- and C-terminus of two ubiquitins (in the presence of ubiquitinΔGly76).
HOIPRBR-LDD completely discharged TAMRAubiquitin from the E2 over time in the single-cycle turnover assays and formed di-ubiquitins when ubiquitinΔGly76 was added to the reaction (Figure 4C; Supplementary Figure S4E). The E2∼ubiquitin discharge is less efficient when RING1 mutants are used in the reaction and also the amount of di-ubiquitin that is formed declines (Figure 4C; Supplementary Figure S4E). This confirms the role of RING1 in E2-mediated activity. Nevertheless, the E2-independent activity of the RING1 mutants is hardly affected (Figure 2), showing that RING1 is less important for the E2-independent driven activity and the ubiquitin linkage formation.
The discharge of the ubiquitin from the E2 on HOIP and the linkage of the ubiquitin to a target ubiquitin by RING2-LDD were uncoupled in the single-cycle turnover assays. Although LDD mutants do not have any ubiquitin linkage formation activity in ubiquitin chain formation reactions, they are capable of efficiently discharging ubiquitin from the E2 (Figure 4C; Supplementary Figure S4E). This indicates that the LDD is not involved in the destabilization of the E2∼ubiquitin thioester, but is critical for ubiquitin chain assembly. Apparently, the trans-thiolation of the ubiquitin from the E2 onto HOIP is independent of ubiquitin chain assembly. Accordingly, the E2∼ubiquitin discharge efficiency is not dependent on the presence of an acceptor ubiquitin to which the donor ubiquitin can be transferred (Figure 4D). Interestingly, RING2 mutants are impaired in both the E2∼ubiquitin discharge and the ubiquitin linkage formation (Figure 4C; Supplementary Figure S4E), suggesting that RING2 is required for both steps of the ubiquitin transfer. This central role for RING2 in the transfer of the ubiquitin further supports its role as acceptor site for the E3∼ubiquitin intermediate. Consequently, efficient ubiquitin chain formation is initiated by the E2-dependent delivery of ubiquitin to HOIP RING2 and is completed by subsequent LDD-mediated ubiquitin chain assembly.
HOIP LDD catalyses the final step of the ubiquitin transfer
The binding of ubiquitin to ubiquitin docking sites in E3 ligases is suggested to play a role in ubiquitin chain formation specificity by bringing a specific ubiquitin lysine residue in close proximity of the ubiquitin thioester bond (Deshaies and Joazeiro, 2009). HOIP has several known ubiquitin interaction motifs (UBA and the NFZs), but since these are not present in HOIPRBR-LDD they cannot explain the linear ubiquitin chain formation. We found that the LDD is important for linking ubiquitins together, but not for E2∼ubiquitin discharge and HOIP∼ubiquitin intermediate formation. Therefore, we wondered whether the LDD could function as a ubiquitin docking site.
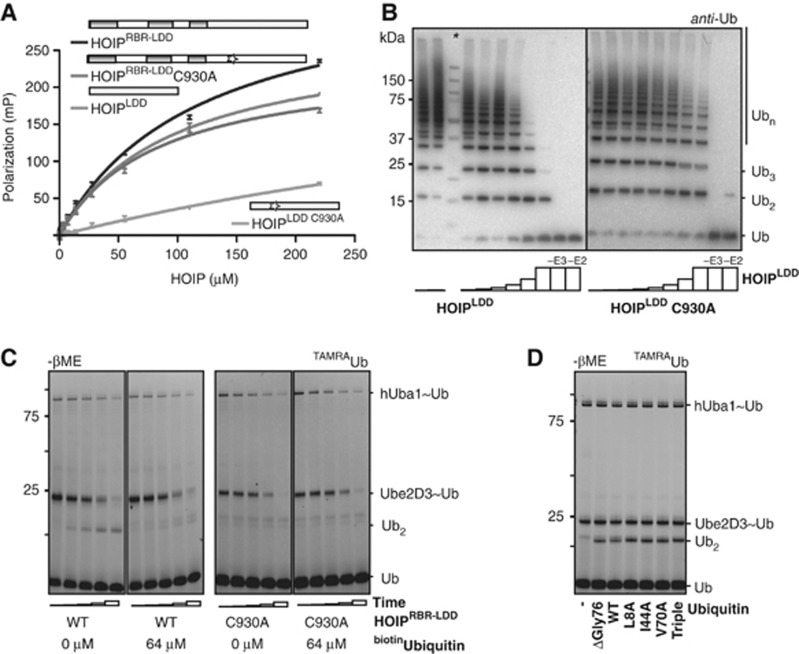
We measured the affinity of ubiquitin for HOIPRBR-LDD and HOIPLDD by fluorescent polarization (FP) with TAMRA-labelled ubiquitin (TAMRAubiquitin and ubiquitinTAMRA). Both HOIPRBR-LDD and HOIPLDD interacted with ubiquitinTAMRA with an affinity of ∼100 μM, and do not bind to the free TAMRA dye, showing that the LDD does interact with ubiquitin (Figure 5A; Supplementary Figure S5A and B). Then, we analysed the effect of the C930A mutant of the LDD, which is impaired in ubiquitin chain formation, on the affinity for ubiquitin in the FP assays. Unexpectedly, in the context of the full HOIPRBR-LDD, the C930A mutation did not affect affinity for ubiquitinTAMRA. In contrast, in the LDD alone, the HOIPLDD C930A has a greatly reduced affinity for ubiquitinTAMRA (Figure 5A; Supplementary Figure S5A). The loss of activity of the HOIPRBR-LDD C930A was not caused by unfolding of the protein as was shown by the gel filtration profile (Supplementary Figure S2D). Therefore, the loss of binding of the LDD C930A mutant indicates interference with ubiquitin binding. In the longer construct, the mutation is possibly not strong enough to disrupt the complete interaction and just interferes with the proper ubiquitin orientation for chain formation or a second ubiquitin interaction site may be present elsewhere outside the LDD.
Figure 5.
HOIP LDD interacts with the acceptor ubiquitin. (A) FP assay of ubiquitinTAMRA binding to HOIP, showing increase in FP as a function of [HOIP], HOIPRBR-LDD KD=118±8 μM, HOIPRBR-LDD C930A KD=83±9.2 μM, HOIPLDD KD=97±7 μM, HOIPLDD C930A KD=734±395 μM. Standard deviations were calculated over three repeats. (B) HOIPLDD and HOIPLDD C930A inhibition on ubiquitin chain formation by HOIPRBR-LDD in a concentration series of 0, 1, 2, 4, 8, 16, 32 and 64 μM. Control reactions at the highest concentration of HOIPLDD do not contain either HOIPRBR-LDD (−E3) or Ube2D3 (−E2). The molecular weight marker is indicated by the asterisk (*). (C) Single-cycle turnover assays in the presence and absence of the acceptor ubiquitin-competitor: biotinubiquitin. The TAMRA signal visualizes di-ubiquitin formation by HOIPRBR-LDD and Ube2D3∼ubiquitin discharge by HOIPRBR-LDD C930A after 0, 2, 4, 8 and 16 min. (D) Di-ubiquitin formation between TAMRAubiquitin and different ubiquitin mutants visualized by the TAMRA signal on a non-reduced gel. T=10 min. UbiquitinTriple=L8A, I44A, V70A triple mutant.
The interaction between the LDD and ubiquitin was verified in in-vitro ubiquitin chain formation assays. First, HOIPLDD was titrated into the ubiquitin chain reaction with HOIPRBR-LDD. The increasing amounts of HOIPLDD inhibited HOIPRBR-LDD-mediated ubiquitin chain formation, presumably by competing away the freely available ubiquitin (Figure 5B). Importantly, addition of the HOIPLDD C930A to linear ubiquitin chain formation assays did not inhibit HOIPRBR-LDD-mediated ubiquitin chain formation (Figure 5B). The loss of inhibition by the LDD C930A mutant indicates that this site is indeed important for ubiquitin interaction.
We then tested if the interaction between the acceptor ubiquitin and HOIP is needed for di-ubiquitin formation in a single-cycle turnover assay. Ube2D3 was loaded with TAMRAubiquitin, after which the HOIPRBR-LDD-dependent discharge of the E2∼TAMRAubiquitin, and the linkage of TAMRAubiquitin to wild-type ubiquitin was monitored. To test if the acceptor ubiquitin interacts with HOIP during the di-ubiquitin formation, a biotinubiquitin, which cannot act as an acceptor, was added during the discharge reaction to compete with the wild-type ubiquitin (Figure 5C). Under these conditions, the E2∼TAMRAubiquitin discharge and HOIP∼ubiquitin intermediate formation were unaffected by the presence of biotinubiquitin, showing that the transfer of the donor ubiquitin from the E2 onto HOIP was not affected (Figure 5C; Supplementary Figure S5C). In contrast, the di-ubiquitin formation was inhibited by the biotinubiquitin, suggesting that the N-terminally blocked ubiquitin competes with the wild-type ubiquitin for binding to HOIP in the final step of the ubiquitin transfer.
These results are in line with the fact that the LDD mutants are impaired in ubiquitin–ubiquitin linkage formation but not in E2∼ubiquitin discharge (Figure 4C; Supplementary Figure S4E), showing that the LDD does not interact with the donor ubiquitin, but rather with the acceptor ubiquitin. Interestingly, the LDD/ubiquitin interaction does not require the ubiquitin hydrophobic patch, which is used by many ubiquitin interaction motifs (Dikic et al, 2009), since point mutants of the hydrophobic patch still accept TAMRAubiquitin via HOIPRBR-LDD (Figure 5D; Supplementary Figure S5D). Consequently, the interaction between the LDD and ubiquitin is specific to HOIP, which is in line with the unique linear ubiquitin specificity that is evoked by HOIP.
HOIP RING2 and LDD are crucial for NF-κB activation in cells
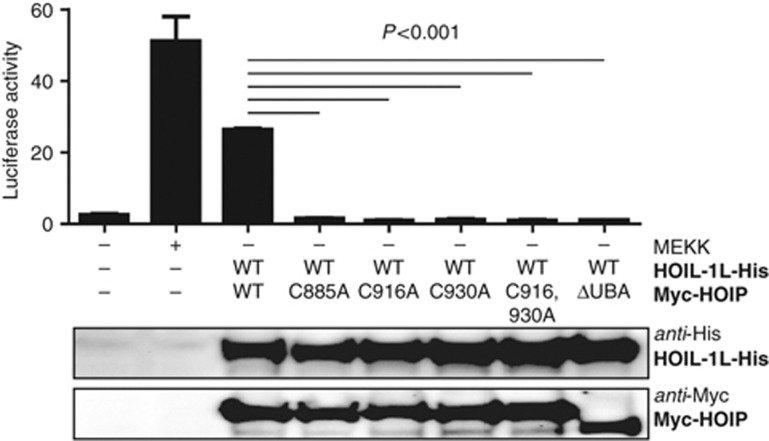
Both RING2 and the LDD of HOIP are essential for linear ubiquitin chain formation in vitro. To determine the biological relevance of this finding, we tested wild-type HOIP and the RING2 C885A, LDD C916A, LDD C930A, single and LDD C916,930A double mutants in HEK293FT cells. First, we verified the expression levels of the mutants and their capacity to interact with HOIL-1L in pull-down assays. All of the mutants retained the capacity to bind to HOIL-1L (Supplementary Figure S6A), showing that the mutations did not cause major deficiencies in the folding of the full-length protein. Next, we tested the activity of the mutants in NF-κB luciferase reporter assays. In line with published data (Tokunaga et al, 2009), the combined expression of HOIP and HOIL-1L resulted in NF-κB activation to levels approaching those when using MEK-kinase 1 (Figure 6; Nemoto et al, 1998). However, the combined expression of HOIL-1L with any of the HOIP cysteine mutants did not activate NF-κB. As expected, the observed NF-κB signalling activity was dependent on the presence of the HOIL-1L interaction domain, the ubiquitin-associated domain (UBA domain) in HOIP, as well as on the co-expression with HOIL-1L, since neither wild-type nor HOIP mutants were active without it (Supplementary Figure S6B). These data show that RING2 and the LDD are essential for linear ubiquitin chain formation and LUBAC-mediated NF-κB activation in cells.
Figure 6.
HOIP RING2 and LDD are essential for NF-κB pathway activation. Dual Luciferase reporter assay for NF-κB activation. Full-length HOIP wild-type or HOIP cysteine mutants were co-expressed together with HOIL-1L (see lower panel) and a luciferase reporter construct, containing five NF-κB binding sites. A luciferase renilla construct was used as transfection control. Firefly luciferase values were normalized to renilla luciferase values. Normalized luciferase activity of 5 × NF-κB reporter vector (upper panel) is shown as mean±s.e.m. (*P<0.001, Student’s t-test, representative experiment of n=4).
Discussion
NF-κB activation is an important signal during immune and DNA-damage responses. Upon stimulation, the formation of linear ubiquitin chains on NEMO forms an essential early event in the activation of the pathway. These linear ubiquitin chains are assembled by LUBAC, which contains the two RBR E3 ligases HOIP and HOIL-1L. It is poorly understood how RBR proteins form functional units to mediate the assembly of ubiquitin chains or how linear ubiquitin chain specificity is determined by the LUBAC E3.
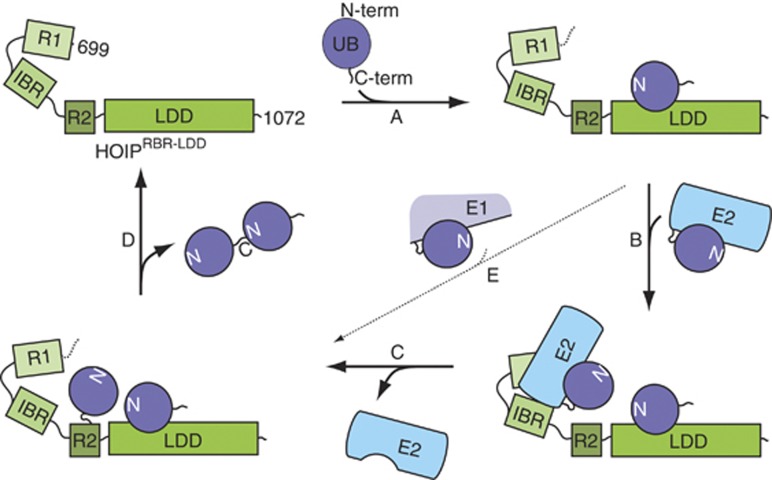
Our results give a detailed insight in the molecular mechanism by which linear ubiquitin chains are formed by HOIP (Figure 7). We show that removing the N-terminal 698 residues of HOIP unmasks the full linear ubiquitin chain formation activity and bypasses the need for HOIL-1L and Sharpin. Within this active region of HOIP, the first two domains (RING1-IBR) are needed for the catalysis of the E2-mediated delivery of ubiquitin to the E3, but the mechanism that directs the linkage to the N-terminus of a target ubiquitin is embedded within HOIP RING2-LDD. The relevance of this activity in the context of the full-length protein within LUBAC was verified by experiments in HEK293FT cells, showing that single point mutations in either RING2 or the LDD impair NF-κB signalling.
Figure 7.
Model for HOIPRBR-LDD-mediated ubiquitination. Linear ubiquitin chain assembly requires both (A) the binding and correct orientation of an acceptor ubiquitin by the LDD and (B) the recruitment of an E2∼ubiquitin to RING1-IBR. The ubiquitin is transferred from the E2 onto the acceptor ubiquitin in two independent steps. (C) First, the ubiquitin thioester is transferred from the E2 onto RING2 and (D) second, the ubiquitin is covalently linked to the N-terminus of the acceptor ubiquitin that is oriented by the LDD. (E) The E2-dependent activity can be bypassed by a less pronounced E2-independent activity.
The transfer of ubiquitin from the E2 onto an acceptor ubiquitin is mediated by HOIP in a two-step mechanism. First, the ubiquitin thioester is transferred from the E2 onto HOIP, most likely on RING2, to form a reversible covalent intermediate (Figure 7C). This step can be catalysed by the RING1-IBR mediated interaction with an E2∼Ub thioester. Second, the ubiquitin is transferred from HOIP onto the N-terminus of the target ubiquitin to form an isopeptide bond. This uncoupling of the E2 catalysed step from the transfer step to the acceptor ubiquitin explains why E2 enzymes do not affect the chain type specificity of HOIP.
The LDD is essential for the specific transfer of the donor ubiquitin from HOIP onto the acceptor ubiquitin (Figure 7). We have shown that the interaction between the LDD and the acceptor ubiquitin is important during this process, suggesting that it functions as a ubiquitin docking domain for the acceptor ubiquitin. The need for a C-terminal ubiquitin interaction domain within HOIP is likely to reflect a general feature for ubiquitin chain catalysis of RBR proteins, since Parkin also contains a recently identified ubiquitin interaction domain, which is located just before RING2 of the RBR domain, that is used in ubiquitin chain formation (Chaugule et al, 2011).
Among RBRs, the RING2-LDD uniquely promotes linear ubiquitin chain formation. This selectivity for the amino-terminus is exquisite since the ubiquitin N-terminus and K63 are located close to each other, indicating that precise positioning of the acceptor ubiquitin by the LDD is very important. It seems plausible that RING2-LDD provides additional contributions to selective targeting, possibly by preferring the chemical constellation of the ubiquitin amino-terminus over a lysine amino group.
Materials and methods
Construction of plasmids
Codon optimized cDNA for E. coli expression of HOIP and HOIL-1L was obtained from Genscript. The cDNA was subcloned into pGEX-6P-1 vectors (GE Healthcare) with an N-terminal GST tag for E. coli expression. HOIPRBR-LDD, HOIPR2-LDD and HOIPLDD were cloned into a pETNKI-His-3C-LIC-amp vector for E. coli expression (Luna-Vargas et al, 2011). Mammalian expression constructs pcDNA3.1-HOIL-1L-His, pcDNA3.1-Myc-HOIP and pcDNA3.1-Myc-HOIP-ΔUBA563–616 were kindly provided by Dr K Iwai (Osaka University, Japan). UbiquitinΔGly76, Ubiquitin single and triple point mutations and point mutations in HOIP were introduced by using the QuikChange Mutagenesis Kit from Stratagene (La Jolla, CA, USA). The luciferase NF-κB reporter construct, pNF-κB-Luc, and the positive control pFC-MEKK were obtained from Agilent Technologies. Renilla luciferase vector, pRL-null, was obtained from Promega.
General, proteins and antibodies
Ubiquitin, hUba1, Ube2D3 and Ube2L3 were expressed and purified as described previously (Pickart and Raasi, 2005; Buchwald et al, 2006; Marteijn et al, 2009; El Oualid et al, 2010). TAMRAUbiquitin, ubiquitinTAMRA, His6ubiquitin and biotinubiquitin (inhibition assay) were generously provided by Remco Merkx, Dharjath Hameed and Huib Ovaa (El Oualid et al, 2010). BiotinUbiquitin (di-ubiquitin formation assays) and ubiquitin lysine mutants were obtained from Boston Biochem.
Protein expression and purification
Full-length HOIP and HOIL-1L were expressed in E. coli Bl21 (DE3) pLysS cells by induction with 0.8 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) and 0.2 mM ZnSO4 overnight at 18°C. Cells were resuspended in 20 mM Tris/HCl pH 8.0, 100 mM NaCl, 5 mM β-mercapto-ethanol (βME) and Complete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). Cells were lysed by a high-pressure EmulsiFlex-C5 device (Avestin, Mannheim, Germany). Initial purification was achieved by binding the proteins to glutathione beads, washing the beads with buffer supplemented with 0.5 M NaCl and elution in buffer containing 50 mM GSH. The GST tag was cleaved by incubation with 3C protease, followed by gel filtration (Superose6) online with a GST column in 20 mM Hepes/HCl pH 8, 100 mM NaCl, 1 μM ZnCl2 and 5 mM βME.
HOIPRBR-LDD (699–1072) was produced in E. coli Bl21 (DE3) pLysS cells. Expression was induced by the addition of 0.4 mM IPTG and 10 μM ZnCl2 at an OD600 of 0.8 in LB medium supplemented with 50 μg/ml carbenicillin and chloramphenicol. Expressions were further cultivated overnight at 16°C. The cells were lysed in 50 mM Tris/HCl pH 8, 150 mM NaCl, 2 mM imidazole, 1 μM ZnCl2, 5 mM βME in the presence of complete EDTA-free protease inhibitor cocktail (Roche), DNAse and 1 mM MgCl2 by a high-pressure EmulsiFlex-C5 device (Avestin). Cleared lysate was incubated with Talon beads. The protein was eluted from the beads in buffer containing 200 mM imidazole and was subsequently loaded on a Resource-Q column. The His tag was cleaved in solution with 3C protease. Further purification was achieved by a Heparin column followed by size-exclusion chromatography (Superdex 200) in buffer containing 25 mM Hepes/HCl pH 7.0, 150 mM NaCl, 1 μM ZnCl2 and 5 mM βME.
HOIPRBR-LDD point mutants, HOIPR2-LDD and HOIPLDD were expressed and purified as described for HOIPRBR-LDD, excluding the cleavage of the His tag and size-exclusion chromatography. For comparison with the HOIPRBR-LDD point mutants, wild-type HOIPRBR-LDD was prepared following the same protocol.
In-vitro ubiquitin chain formation
In-vitro ubiquitin chain formation reactions were performed in standard conditions, unless specified otherwise. Standard conditions for ubiquitin chain formation were 100 nM hUba1, 600 nM of the indicated E2, 1 μM E3, 15 μM ubiquitin and 10 mM ATP in buffer containing 20 mM Hepes/HCl pH 8, 150 mM NaCl, 10 mM MgCl2, 0.5 mM DTT. Reactions were performed at 37°C and stopped by the addition of protein loading buffer containing βME. Samples were separated on 4–12% Nu-PAGE gels (Invitrogen) in MES buffer and analysed by western blot using ubiquitin antibody (P4D1, Santa Cruz biotechnology) and HRP conjugated anti-Mouse antibody (Bio-Rad, Hercules, CA, USA).
Single-cycle turnover assays
Single-cycle turnover assays were performed in the same buffer conditions as described for the ubiquitin chain formation. TAMRAubiquitin (500 nM) was loaded onto E2- (600 nM) in an ATP (1 mM)-dependent manner via hUba1 (100 nM) in 120 μl final reaction volume for 20 min at 37°C. The charging reaction was terminated by depleting the ATP with 2U apyrase. After 5 min incubation at room temperature, the sample was divided into smaller aliquots to compare the effects of the addition of HOIPRBR-LDD (1 μM) and ubiquitinΔGly76 (500 nM). BiotinUbiquitin was added simultaneously with HOIPRBR-LDD and wtUbiquitin in the acceptor ubiquitin competition assays. Reactions were performed at 37°C and stopped by the addition of non-reducing loading buffer on ice. Samples were analysed on 4–12% NU-PAGE gels (Invitrogen) in MES buffer and the TAMRA signal was visualized on a ChemiDoc XRS (Bio-Rad). Band quantification of Ube2D3∼TAMRAubiquitin was done with the ImageJ program ( http://imagej.nih.gov/ij). Loading differences were accounted for by measuring the total amount of TAMRA signal per lane. Percentages normalized for the total amount of Ube2D3∼TAMRAubiquitin at T=0. Western blot analysis was performed using anti-HOIP (ab85294, Abcam) and HRP-conjugated anti-Rabbit (Bio-Rad) antibodies.
Covalent HOIP∼ubiquitin intermediate formation
E2∼ubiquitin was prepared as described for the single-cycle turnover assays in buffer containing 20 mM Hepes pH 8.5 and 5 mM βME. After the addition of Apyrase, HOIP (2 μM) was added to the mixture. Reactions were performed for 5 min on ice. The TAMRA signal was visualized on a ChemiDoc XRS (Bio-Rad) and HOIPRBR-LDD was visualized on non-reducing western blots with anti-HOIP antibody (ab85294, Abcam). Sample loading buffer was supplemented with 1 M Urea to partially unfold the proteins.
Fluorescence polarization assays
The fluorescence anisotropy of the C-terminal TAMRA-labelled ubiquitin (1 nM) in binding buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM βME and 1 g/l chicken ovalbumin) was measured on a Perkin-Elmer EnVision 2010 Multilabel Reader. The binding was measured in 75 μl reactions. Serial 1:1 dilutions, starting at 220 μM HOIP, were performed in three repeats. Reactions were incubated for 20 min at 4°C before the measurements. The samples were excited at 531 nm and the emission was measured at 579 nm, with correction for both the buffer background and G-factor of the instrument. The assays were performed in ‘non-binding surface flat bottom’ black 96-well plates (Corning) at room temperature. The resulting binding isotherms (anisotropy versus HOIP concentration) were fit to a 1:1 non-linear binding model (Y=Bmax × X/(Kd+X)). All experimental data were processed using Ms Excel and Prism 4.03 (GraphPad Software, Inc.).
Multi-angle laser light scattering
MALLS experiments were performed on a Mini-Dawn light scattering detector (Wyatt Technology) in line with a Superdex S200 10/30 column at 4°C in buffer containing 25 mM Hepes/HCl pH 7, 150 mM NaCl, 1 μM ZnCl2 and 5 mM βME. Refractive index and light scattering detectors were calibrated against toluene and BSA. Data were analysed using the Astra software.
Cell culture and transient transfection
HEK293FT cells were cultured in Dulbecco modified Eagle medium (DMEM; GIBCO) supplemented with 10% non-heat-inactivated fetal calf serum (GIBCO), 1% penicillin/streptomycin (MP Biomedical), 1% non-essential amino acids (GIBCO) and 1% L-glutamine (MP Biomedical). Cells were cultured in 24-well plates at 37°C supplied with 5% CO2.
For transient expression, 400 ng plasmid DNA (pcDNA3.1-HOIL-1L-His, pcDNA3.1-Myc-HOIP, pcDNA3.1-Myc-HOIP-mutants, pFC-MEKK) was used. Empty vector pcDNA3.1 was used to compensate for differences in DNA amounts. Furthermore, 400 ng of luciferase NF-κB reporter construct and 200 ng of Renilla luciferase vector were added to the transfection medium. In total, 2 μg of DNA was transfected in each condition. Transfection was performed at 60% confluence with lipofectamine 2000 (Invitrogen). Each condition was experimentally tested in triplicate.
NF-κB transactivation assay
As readout for NF-κB activation we performed a Dual luciferase™ reporter assay (Promega). Forty-eight hours after transfection, cells were washed with PBS and lysed in 100 μl of passive lysis buffer (Promega) for 1 h. Luciferase assays were performed according to the protocol provided by the manufacturer (Promega). Luciferase signals were measured on the Lumat LB 9507 (EG&G Berthold). Western blot analysis was performed to confirm protein expression. Proteins of total lysates generated as described above were separated on 10% polyacrylamide gels and transferred onto polyvinylidenefluoride (PDVF) membranes (Bio-Rad). PVDF membranes were probed with anti-Myc (Santa Cruz) and anti-His (Abcam) primary antibodies, followed by probing with HRP-conjugated secondary antibodies. Antibody signal was visualized by chemiluminescence using the Bio-Rad ChemiDox XRS+.
Supplementary Material
Acknowledgments
We thank Rick Hibbert for contributing to the cloning and purification of ubiquitin mutants, and suggesting the basis for the biotinubiquitin inhibition experiment. Francesca Mattiroli for contributing to the E2 purifications. Dr K Iwai for mammalian expression constructs of HOIP and HOIL-1L, Remco Merkx for TAMRA-labelled ubiquitin and Dharjath Hameed for biotinubiquitin. Members of the Sixma laboratory for discussions on the project. Rick Hibbert, Francesca Mattiroli, Danny Sahtoe, Flora Groothuizen and Fred van Leeuwen for critical reading of the manuscript. These studies were funded by an ERC advanced grant and EU Spine2C FP6 grant.
Author contributions: JJS designed and performed the in-vitro experiments and wrote the manuscript. WJvD contributed to protein expression, purification and western blots. DM and SMN designed and performed the cell culture-based experiments. BAvdR designed and supervised the cell culture-based experiments. TKS designed and supervised experiments and wrote the manuscript. All authors critically read the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Beasley SA, Hristova VA, Shaw GS (2007) Structure of the Parkin in-between-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson’s disease. Proc Natl Acad Sci USA 104: 3095–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D 3rd, Fukuda M, Ohta T, Klevit R (2003) Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA 100: 5646–5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK (2006) Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25: 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capili AD, Edghill EL, Wu K, Borden KL (2004) Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J Mol Biol 340: 1117–1129 [DOI] [PubMed] [Google Scholar]
- Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H (2011) Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J 30: 2853–2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Niles EG, Pickart CM (1991) Isolation of a cDNA encoding a mammalian multiubiquitinating enzyme (E225K) and overexpression of the functional enzyme in Escherichia coli. J Biol Chem 266: 15698–15704 [PubMed] [Google Scholar]
- Chew KC, Matsuda N, Saisho K, Lim GG, Chai C, Tan HM, Tanaka K, Lim KL (2011) Parkin mediates apparent E2-independent monoubiquitination in vitro and contains an intrinsic activity that catalyzes polyubiquitination. PLoS One 6: e19720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol 14: 941–948 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C (2006) Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol 13: 915–920 [DOI] [PubMed] [Google Scholar]
- El Oualid F, Merkx R, Ekkebus R, Hameed DS, Smit JJ, de Jong A, Hilkmann H, Sixma TK, Ovaa H (2010) Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew Chem Int Ed Engl 49: 10149–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596 [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36: 831–844 [DOI] [PubMed] [Google Scholar]
- Hostager BS, Fox DK, Whitten D, Wilkerson CG, Eipper BA, Francone VP, Rothman PB, Colgan JD (2010) HOIL-1L interacting protein (HOIP) as an NF-kappaB regulating component of the CD40 signaling complex. PLoS ONE 5: e11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova VA, Beasley SA, Rylett RJ, Shaw GS (2009) Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J Biol Chem 284: 14978–14986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K, Tokunaga F (2009) Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep 10: 706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HC, Huibregtse JM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 29: 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas MP, Christodoulou E, Alfieri A, van Dijk WJ, Stadnik M, Hibbert RG, Sahtoe DD, Clerici M, Marco VD, Littler D, Celie PH, Sixma TK, Perrakis A (2011) Enabling high-throughput ligation-independent cloning and protein expression for the family of ubiquitin specific proteases. J Struct Biol 175: 11. [DOI] [PubMed] [Google Scholar]
- Markson G, Kiel C, Hyde R, Brown S, Charalabous P, Bremm A, Semple J, Woodsmith J, Duley S, Salehi-Ashtiani K, Vidal M, Komander D, Serrano L, Lehner P, Sanderson CM (2009) Analysis of the human E2 ubiquitin conjugating enzyme protein interaction network. Genome Res 19: 1905–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, van der Meer LT, Smit JJ, Noordermeer SM, Wissink W, Jansen P, Swarts HG, Hibbert RG, de Witte T, Sixma TK, Jansen JH, van der Reijden BA (2009) The ubiquitin ligase Triad1 inhibits myelopoiesis through UbcH7 and Ubc13 interacting domains. Leukemia 23: 1480–1489 [DOI] [PubMed] [Google Scholar]
- Matsuda N, Kitami T, Suzuki T, Mizuno Y, Hattori N, Tanaka K (2006) Diverse effects of pathogenic mutations of Parkin that catalyze multiple monoubiquitylation in vitro. J Biol Chem 281: 3204–3209 [DOI] [PubMed] [Google Scholar]
- Morett E, Bork P (1999) A novel transactivation domain in parkin. Trends Biochem Sci 24: 229–231 [DOI] [PubMed] [Google Scholar]
- Nemoto S, DiDonato JA, Lin A (1998) Coordinate regulation of IkappaB kinases by mitogen-activated protein kinase kinase kinase 1 and NF-kappaB-inducing kinase. Mol Cell Biol 18: 7336–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa H, Ooka S, Sato K, Arima K, Okamoto J, Klevit RE, Fukuda M, Ohta T (2004) Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J Biol Chem 279: 3916–3924 [DOI] [PubMed] [Google Scholar]
- Niu J, Shi Y, Iwai K, Wu ZH (2011) LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J 30: 3741–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123: 1107–1120 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Raasi S (2005) Controlled synthesis of polyubiquitin chains. Methods Enzymol 399: 21–36 [DOI] [PubMed] [Google Scholar]
- Sieber S, Lange N, Kollmorgen G, Erhardt A, Quaas A, Gontarewicz A, Sass G, Tiegs G, Kreienkamp HJ (2012) Sharpin contributes to TNFalpha dependent NFkappaB activation and anti-apoptotic signalling in hepatocytes. PLoS ONE 7: e29993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K (2011) SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471: 633–636 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- van der Reijden BA, Erpelinck-Verschueren CA, Lowenberg B, Jansen JH (1999) TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci 8: 1557–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, de Vries SJ, Kemmeren P, Huang A, Boelens R, Bonvin AM, Timmers HT (2009) A comprehensive framework of E2-RING E3 interactions of the human ubiquitin-proteasome system. Mol Syst Biol 5: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Pickart CM (2005) Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J 24: 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M (2011) The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Baer F, Lagrazon K, Yuan W, Baer R (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem 278: 34743–34746 [DOI] [PubMed] [Google Scholar]
- Yagi H, Ishimoto K, Hiromoto T, Fujita H, Mizushima T, Uekusa Y, Yagi-Utsumi M, Kurimoto E, Noda M, Uchiyama S, Tokunaga F, Iwai K, Kato K (2012) A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex. EMBO Rep 13: 462–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102: 533–539 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.