Abstract
The regulation of Ubiquitin (Ub) conjugates generated by the complex network of proteins that promote the mammalian DNA double-strand break (DSB) response is not fully understood. We show here that the Ub protease POH1/rpn11/PSMD14 resident in the 19S proteasome regulatory particle is required for processing poly-Ub formed in the DSB response. Proteasome activity is required to restrict tudor domain-dependent 53BP1 accumulation at sites of DNA damage. This occurs both through antagonism of RNF8/RNF168-mediated lysine 63-linked poly-Ub and through the promotion of JMJD2A retention on chromatin. Consistent with this role POH1 acts in opposition to RNF8/RNF168 to modulate end-joining DNA repair. Additionally, POH1 acts independently of 53BP1 in homologous recombination repair to promote RAD51 loading. Accordingly, POH1-deficient cells are sensitive to DNA damaging agents. These data demonstrate that proteasomal POH1 is a key de-ubiquitinating enzyme that regulates ubiquitin conjugates generated in response to damage and that several aspects of the DSB response are regulated by the proteasome.
Keywords: 53BP1, double-strand break repair, DUB, proteasome, ubiqutiin
Introduction
Double-stranded breaks (DSBs) have the potential to cause cell death or transformation and multicellular organisms have evolved a complex network of factors to repair these lesions. There are two main mechanisms of repair, non-homologous end-joining (NHEJ), which occurs throughout the cell cycle and homologous recombination (HR), which uses the homologous sister chromatid as a template and occurs in late S phase and G2. The assembly of repair proteins to sites of DSBs is coordinated by post-translational modifications, in particular by ubiquitin conjugation.
The p53 binding protein (53BP1) acts to promote NHEJ in several contexts, inhibits HR, and is part of a mechanism that shields lesions arising in replication stress through mitosis and G1 (FitzGerald et al, 2009; Harrigan et al, 2011; Lukas et al, 2011). 53BP1 specifically binds to methylated histones, particularly H4K20me2, which may be increased at sites of DNA damage (Pei et al, 2011). In addition, Ub conjugation events promoted by ligases RNF8 and RNF168, which are recruited to sites of DNA repair through ATM-mediated phosphorylation events, are thought to contribute to exposure of the mark following damage (Panier and Durocher, 2009). Two, possibly overlapping, mechanisms are recognised. In the first, factors that bind H4K20me2 are removed from chromatin to allow 53BP1 binding. This appears to be through K48-poly-Ub generated chiefly by RNF8. Modification of the tudor-domain containing demethylases, JMJD2A/B, is associated with their clearance from chromatin and subsequent degradation (Mallette et al, 2012). Extraction of K48-poly-Ub by the valosin-containing protein (VCP/p97) is partially required for the assembly of 53BP1 and may involve the removal of the H4K20me2 binding protein, L3MBTL1 (Acs et al, 2011; Lok et al, 2011; Meerang et al, 2011). The requirement for K48-poly-Ub is not certain however as a mutant RNF8 unable to form these linkages nevertheless is able to promote the response to DSBs (Lok et al, 2011).
The second mechanism involves RNF8/RNF168 activity with the E2 conjugating enzyme Ubc13 to mediate K63-poly-Ub (Kolas et al, 2007; Zhao et al, 2007; Bekker-Jensen et al, 2010). These chains are conjugated to histones H2A and H2AX, and potentially other proteins local to the DSB, and their generation promotes the recruitment of 53BP1 through an unknown mechanism presumed to function at the level of histone remodelling or relaxation (Huen et al, 2007; Kolas et al, 2007; Mailand et al, 2007; Wang and Elledge, 2007; Zhao et al, 2007; Doil et al, 2009; Stewart et al, 2009; Bekker-Jensen et al, 2010).
The Receptor Associated Protein 80 (RAP80) complex (also called the BRCA1-A complex) is independently assembled at locations adjacent to 53BP1 at the DSB by the direct K63-poly-Ub binding properties of RAP80 (Kim et al, 2007; Mailand et al, 2007; Sobhian et al, 2007; Wang and Elledge, 2007; Wang et al, 2007, 2009; Yan et al, 2007; Sato et al, 2009; Shao et al, 2009; Mok and Henderson, 2010; Coleman and Greenberg, 2011; Hu et al, 2011; Chapman et al, 2012). RAP80 and the associated protein BRCC36 (BRCA1/BRCA2-Containing Complex 36) are associated with inhibition of HR shortly after induction of DSBs (Coleman and Greenberg, 2011; Hu et al, 2011). This complex also contains the Breast Cancer Associated Gene 1 (BRCA1), which is implicated in the response to DNA cross-links and in promoting DNA resection in the presence of 53BP1 (Bouwman et al, 2010; Bunting et al, 2010, 2012). BRCA1 also generates poly-Ub chains at DSBs although whether this is relevant to genome integrity remains unclear (Reid et al, 2008; Drost et al, 2011; Shakya et al, 2011; Zhu et al, 2011).
The accumulation of RAD51, the recombinase required for DSB repair by HR, also requires RNF8, and is related to the extraction activity of p97/VCP but is independent of RNF168-generated K63-Ub chains (Stewart et al, 2007; Meerang et al, 2011; Sy et al, 2011). The post-replication repair Ub ligase RAD18 may also contribute by interacting both with RAD51C and with K63-poly-Ub or K48-poly-Ub generated by RNF8 (Huang et al, 2009).
Modification with Ub can be reversed by Ub proteases called de-ubiquitinating enzymes (DUBs) and some DUB activities can modulate DNA damage signalling and repair by editing Ub-conjugates: BRCC36 specifically hydrolyses Ub-K63 polymers and regulates 53BP1 accumulation; overexpression of the H2A DUB, USP3, abolishes RAP80 and 53BP1 localisation; and USP16 is required for local transcriptional restoration after recovery from a DSB (Joo et al, 2007; Doil et al, 2009; Shao et al, 2009; Shanbhag et al, 2010). Given the complexity and degree of involvement of poly-Ub in DNA repair it is highly likely that further DUB activities form part of the DSB response.
Three DUBs are associated with the proteasome (Finley, 2009), and the proteasome itself has been implicated in the eukaryotic response to DSBs. In yeast, its subunits are required to maintain chromosomal stability and promote DNA repair (Spataro et al, 1997; Krogan et al, 2004; Ben-Aroya et al, 2010). Mammalian cells treated with proteasome core inhibitors exhibit persistent MDC1 damage foci, poor recruitment of BRCA1 and 53BP1 to DSBs, reduced gene conversion in HR and increased sensitivity to DNA damaging agents (Dantuma et al, 2006; Gudmundsdottir et al, 2007; Jacquemont and Taniguchi, 2007; Mailand et al, 2007; Murakawa et al, 2007; Shi et al, 2008; Meerang et al, 2011). Depletion of 20S and 19S subunits has also been shown to reduce RAD51 foci formation (Jacquemont and Taniguchi, 2007).
A more direct role for the proteasome in the mammalian DSB response has been suggested by the localisation of the 20S into repair foci, the interaction of the HR protein BRCA2 (Breast Cancer Associated gene 2) with the 19S proteasomal activator, and the identification of a non-essential 19S subunit, Deleted in Split-Hand/Split-Foot 1 (DSS1), as a protein required for HR repair (Ustrell et al, 2002; Gudmundsdottir et al, 2004; Blickwedehl et al, 2007, 2008; Gudmundsdottir et al, 2007; Liu et al, 2010; Holloman, 2011; Levy-Barda et al, 2011). In addition many repair proteins, like the majority of cellular proteins, are turned over by proteasomal degradation.
Here, we tested the hypothesis that further DUBs are integral to the mammalian DSB response. In a screen for DUBs that regulate poly-Ub clearance we identified POH1, the intrinsic DUB of the 19S proteasome lid. We show that POH1 promotes the correct coordination of the cellular response to DSBs. It is required to restrict two aspects of 53BP1 accumulation and consequently influences NHEJ. Further, it promotes HR repair independently of 53BP1 through the promotion of RAD51 loading. Our data confirm the notion that further DUBs form part of the DSB response and reveal that several portions of the response require proteasomal involvement.
Results
Identification of POH1 in regulation of DSB-associated Ub conjugates
To examine the possibility that further mammalian DUBs are part of the regulation of Ub conjugates at sites of DNA repair, we screened pools of small interfering RNAs (siRNAs) to identify DUBs whose depletion resulted in a reduced ability to clear Ub conjugates after release from Hydroxyurea (HU). This identified a pool directed against POH1 (Figure 1A, see Supplementary Figure 1A for Ub conjugates measure after release from HU, Supplementary Figure 1B for validation with single siRNA duplexes and Supplementary Figure 1C for the influence of POH1 siRNA on cell-cycle transit).
Figure 1.
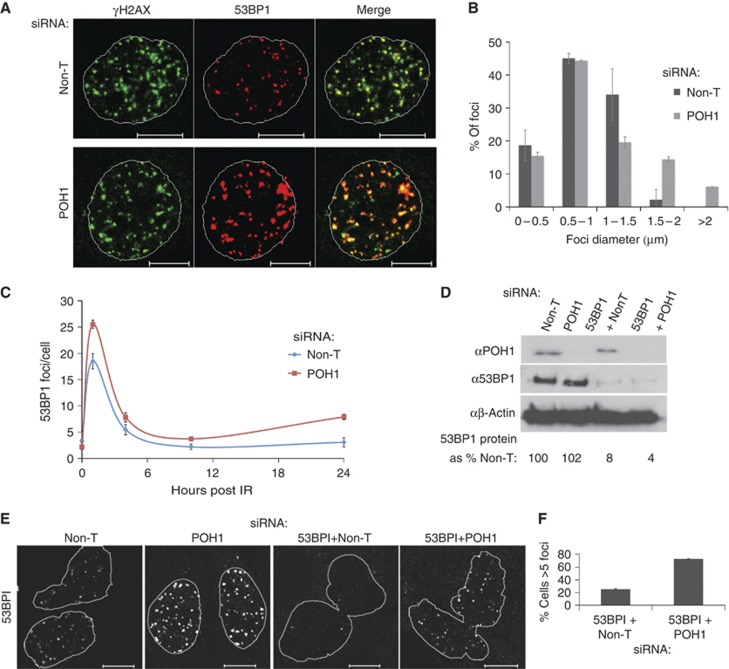
Identification of POH1 in the regulation of DSB-associated Ub conjugates. (A) Identification of POH1 in a screen of DUBs. Scatter plot of averaged Z-scores of FK2 luminescence readings from duplicate library screens of siRNA pools targeting the 103 known or predicted mammalian DUBs. Serum-starved HeLa cells were plated onto individual pools of siRNA, and after 24 h placed into complete media with 3 mM HU for 16 h, then released into complete media (without HU) and fixed after a further 16 h. (Under these conditions control cells show clearance of conjugates see Supplementary Figure 1A). Z-score indicates the number of standard deviations from the population mean. Positive score indicates pools exhibiting increased FK2-Ub and negative those with decreased FK2-Ub. 95 and 99% confidence levels are shown. (B) POH1 is required to reduce conjugates produced following HU treatment. HeLa cells were transfected with non-targeting (Non-T) or POH1 siRNAs and then treated with 3 mM HU or untreated for a further 8 h. Conjugated Ub levels were detected with monoclonal anti-Ub conjugate antibody (FK2) and measured upon addition of chemiluminescent substrate, relative to non-specific HRP antibody control. Relative luminescence units (RLUs) of mean FK2 Ub conjugates are expressed (three replicates/treatment). (C) Ub conjugate foci are larger in POH1-depleted cells. U20S transfected with Non-T or POH1 siRNAs exposed to 2 Gy irradiation, and fixed 1 h later. Cells were incubated with FK2 (anti-Ub conjugates) and anti-γH2AX antibodies. The white line shows the outline of the DNA stained by Hoechst. (D) Quantification of Ub conjugate foci diameter. U20S transfected with Non-T or POH1 siRNAs and treated as above. After imagining by confocal microscopy the diameter of each foci was measured (n=100 foci/treatment). (E) Exogenous POH1 incorporates into the 19S particle. 293T cells were transfected with siRNA-resistant forms of Flag-POH1 and mutant Flag-POH1 (JAMMM). Flag containing complexes were immunoprecipitated (IP) and analysed by immunoblotting with antibody to PSMD4, a subunit of the 19S base and anti-flag. (F) siRNA-resistant POH1, but not JAMM-mutant POH1, can reduce IR-induced Ub conjugate foci in POH1-depleted cells. Representative images of U20S cells treated with Non-T or POH1-D siRNA and transfected with siRNA-resistant Flag-POH1 or siRNA-resistant mutant Flag-POH1 (JAMMM) before exposure to 5 Gy irradiation. Cells were fixed 1 h post IR and immunostained with FK2 and anti-flag antibodies. White arrows indicate POH1-expressing cells. All scale bars throughout are 10 μm. (G) Quantification of mean fluorescence intensity of nuclear FK2-Ub foci. U20S cells transfected with Non-T and POH1 (D) siRNA and co-transfected with siRNA-resistant POH1 and POH1-JAMMM. The fluorescence intensity of nuclear FK2-Ub foci was measured using Zeiss confocal software (0–250 arbitrary intensity units) 30 cells for each condition (600 foci total).
POH1 is active in the context of the 19S (Patterson-Fortin et al, 2010) and is required for ubiquitin-dependent protein degradation (Yao and Cohen, 2002; Cooper et al, 2009). We compared the impact of POH1 depletion on untreated and HU-treated cells. Depletion of POH1 increased Ub conjugates in the absence of treatment and exposure of POH1-depleted cells to HU increased Ub conjugate levels further (Figure 1B), indicating that in addition constitutive ubiquitin processing POH1 is also required to reduce conjugates generated on genotoxic stress. We next examined DNA-damage foci induced by irradiation (IR) and found that depletion of POH1 increased the size and intensity of Ub conjugates detected by the FK2 antibody (Figure 1C and D). These data suggest that Ub conjugates induced by DSBs may be processed through the 19S.
POH1 protease is a zinc-dependent metalloprotease in the JAB1/MPN/MOV34 (JAMM) family (reviewed in Nijman et al, 2005, illustrated in Supplementary Figure 1D and E). To examine any requirement for its catalytic activity in the processing of DSB-associated Ub conjugates, we expressed siRNA-resistant forms of the DUB; either wild-type or mutated within its JAMM motif (H113→A and H115→A, referred to throughout as JAMMM). Both forms of exogenous POH1 co-precipitated the 19S component PSMD4 from cells, indicating incorporation of the exogenous protein into the 19S particle (Figure 1E). siRNA-resistant POH1 was able to restore the accumulation of Ub conjugates within IR-induced nuclear foci to levels seen in control siRNA-treated cells, whereas the JAMMM mutant could not (Figure 1F and G). Thus, the DUB activity of POH1 is required to restrain Ub conjugate levels at foci that occur in response to DNA damage.
POH1 restricts 53BP1 accumulation
Ub conjugates are central in directing 53BP1 recruitment to DSBs and to DNA lesions sequestered in G1. We examined untreated and irradiated cells depleted for POH1 for 53BP1 foci. Accumulations were larger following POH1 depletion both in irradiated cells (Figure 2A and B) and in naive cells (Supplementary Figure 2A). Co-staining with antibody to γH2AX revealed that 53BP1 accumulation extended beyond the region labelled for γH2AX (Figure 2A) and 53BP1 occupied a larger volume of the nucleoplasm (Supplementary Figure 2A ‘projections’). Cyclin A-positive and -negative cells also exhibited larger 53BP1 foci when depleted for POH1, suggesting that its influence on 53BP1 occurs in G1, S and G2 phases of the cell cycle (Supplementary Figure 2B and C). Cells expressing the catalytic mutant enzyme exhibited larger foci of 53BP1 than those expressing WT protein suggesting a dominant-negative impact of integration of the mutant protein into the 19S (Supplementary Figure 2D) and indicating a requirement for POH1 DUB activity in restricting 53BP1 assemblies.
Figure 2.
POH restricts 53BP1 accumulation at sites of DSBs. (A) 53BP1 foci are larger in POH1-depleted cells. U20S transfected with Non-T or POH1 siRNAs exposed to 2 Gy irradiation, and fixed 1 h later. Cells were incubated with anti-53BP1 and anti-γH2AX antibodies. The white line shows the outline of the DNA stained by Hoechst. (B) Quantification of 53BP1 foci diameter. U20S transfected with Non-T or POH1 siRNAs and treated as above. After imaging by confocal microscopy, the diameter of each foci was measured. The graph illustrates proportion of foci in bins of increasing diameter (n=100 foci/treatment, 2 repeats). (C) Clearance of 53BP1 foci in POH1 siRNA and Non-T-treated cells. Quantification of 53BP1 foci per cell in U20S treated with Non-T or POH1 siRNAs before exposure to 2 Gy irradiation and fixed at various times in recovery, before staining with anti-53BP1 and imaging by confocal microscopy (3 replicates of 50 cells/time point). (D) Depletion of POH1 does not increase 53BP1 protein levels. U20S transfected with Non-T, POH1, 53BP1 siRNA or co-transfected 53BP1 and POH1 siRNA were lysed and immunoblotted with anti-53BP1, anti-POH1 or anti-β-actin antibodies. The % of 53BP1 knockdown is based on quantification using Image J. (E) Depletion of POH1 restores 53BP1 foci in cells with low level of 53BP1 protein. U20S transfected with Non-T, POH1 or 53BP1 siRNAs as above, exposed to 2 Gy irradiation and fixed 1 h later before incubation with anti-53BP1 antibody. The white line shows the outline of the DNA stained by Hoechst. (F) Quantification of cells with 53BP1 foci. U20S transfected with 53BP1 and Non-T siRNA or 53BP1 and POH1 siRNA together, exposed to 2 Gy irradiation and fixed 1 h later before incubation with anti-53BP1 antibody. Cells were scored for the presence or absence of 53BP1 foci (>5 foci/cell) (100 cells/condition, 2 repeats).
More 53BP1 foci were evident on POH1 depletion following exposure to IR than in cells treated with non-targeting siRNA possibly as a result of the enlargement of previously cryptic accumulations. Enlarged foci were nevertheless cleared after IR and reduced 4–10 h post IR, similar to control cells (Figure 2C). Thus, POH1 acts to antagonise acute 53BP1 accumulation, but is not required in its clearance.
We considered that an explanation for the enlarged 53BP1 accumulations may be reduced degradation of the protein. We observed that expression of POH1-JAMMM increased 53BP1 protein levels slightly (Supplementary Figure 2E). However, depletion of POH1 did not increase 53BP1 levels and the half-life of 53BP1 was not significantly affected (Supplementary Figure 2F). 53BP1 is highly expressed in cells, so to test the possibility of increased 53BP1 levels relevant to accumulation we examined the impact of POH1 depletion in cells expressing low levels of 53BP1 protein (Figure 2D). In 53BP1 siRNA-treated cells, few 53BP1 foci were formed on exposure to IR (Figure 2E). Following co-depletion with POH1, 53BP1 protein levels remained low, yet strikingly, the ability of 53BP1 to form DNA-damage induced foci was not lost (Figure 2D–F). These data show that POH1 acts as a powerful antagonist to 53BP1 assembly at sites of DNA damage and excludes increased 53BP1 protein levels as the main level of regulation.
Proteasomal activity restricts 53BP1 spreading
The majority of the 19S is found associated with the 20S core (in the 26S) (Geng and Tansey, 2012). Cell and in vitro experiments have shown that core degradation and 19S de-ubiquitination are linked so that disruption of the core results in inhibition of POH1 DUB activity (Verma et al, 2002). To address whether the 20S is functionally linked to 53BP1 accumulation, we examined cells in which proteasome function was impaired either by depletion of the proteasomal core factor, PSMA6, or by MG132 treatment complemented with exogenous Ub. The introduction of exogenous Ub is necessary to overcome the cellular starvation of free Ub caused by proteasomal inhibition (Supplementary Figure 3A and B). Both conditions resulted in enlarged 53BP1 accumulations (Supplementary Figure 3C and D). These data indicate that the 20S core is functionally linked to the restriction of 53BP1 accumulation and that the 19S regulates 53BP1 in the context of the 26S proteasome.
53BP1 tandem tudor domain is required for enlarged foci
To understand whether increased 53BP1 assemblies are formed through direct interaction with methylated histones or through another mechanism we generated the 53BP1 mutation, D1521→R, which prevents tudor-domain binding to methylated histones (Huen et al, 2007). Exogenous WT 53BP1 formed enlarged foci in POH1-depleted cells but D1521R-53BP1 formed very few foci in control or in POH1-depleted cells (Supplementary Figure 4A–C). In the cells in which the mutant did accumulate into foci these were not enlarged on POH1 depletion (Supplementary Figure 4D). Thus, POH1 is likely to be regulating the canonical pathway of 53BP1 recruitment and not an alternative pathway.
RNF8/RNF168 and POH1 play opposing roles in 53BP1 recruitment
RNF8 or RNF168 Ub ligases are required to promote 53BP1 foci formation. However, low expression of these ligases retains the ability to promote 53BP1 accumulations if either JMJD2A/B or the K63-specific DUB, BRCC36 is also co-depleted. These factors are antagonistic to 53BP1 accumulation, JMJD2 proteins compete for chromatin marks bound by 53BP1 while BRCC36 hydrolyses K63 chains that promote 53BP1 recruitment (Shao et al, 2009; Mallette et al, 2012). We tested the relationship between RNF8/168 and POH1 and found that co-depletion of POH1 with either ligase allowed 53BP1 foci formation (Figure 3A–C). Further exogenous POH1-JAMMM partially restored 53BP1 foci in RNF8-depleted cells (Supplementary Figure 5). These data demonstrate opposing roles for RNF8/168 and the POH1 DUB in 53BP1 recruitment.
Figure 3.
RNF8/RNF168 and POH1 play opposing roles in 53BP1 accumulation. (A) Depletion of POH1 restores 53BP1 foci in cells depleted of RNF8 or RNF168. U20S transfected with Non-T, RNF8 or RNF168 siRNAs or co-transfected with RNF8/RNF168 siRNAs with POH1 siRNA and exposed to 2 Gy irradiation and fixed 1 h later before incubation with anti-53BP1 antibody. The white line shows the outline of the DNA stained by Hoechst. (B) Protein levels in POH1 and RNF8/168 siRNA-treated cells. U20S transfected with Non-T, POH1, RNF8, RNF168 siRNA or a combination with POH1 siRNA, lysed, immunoblotted with anti-53BP1, anti-RNF8 (left panel) or anti-RNF168 (right panel), anti-POH1 and anti-β-actin antibodies. (C) Quantification of cells with 53BP1 foci. U20S transfected with Non-T, RNF8 or RNF168 siRNA or siRNA to RNF8 and RNF168 and POH1 together, scored for the presence or absence of 53BP1 foci (>5 foci/cell) (100 cells/condition, 2 repeats).
POH1 DUB activity is associated with maintenance of JMJD2A on chromatin
The tudor domains of JMJD2A/B bind H4K20me2 with higher affinity than the 53BP1 tudor domain (Mallette et al, 2012). To assess whether chromatin mark availability is altered in POH1-depleted cells, we tested the ability of JMJD2A to compete with 53BP1 accumulation. In control cells, JMJD2A expression inhibited 53BP1 foci formation, whereas in POH1-depleted cells 53BP1 foci formed, albeit smaller (Figure 4A). Expression of the JMJD2A tudor domain mutant (D939→R) had no impact on 53BP1 confirming the activity of JMJD2A is through its ability to interact with methylated chromatin. Since 53BP1 accumulation is partially resistant to competition by JMJD2A in POH1 depleted cells, these data are consistent with an increased chromatin mark presence/availability.
Figure 4.
POH1 influences JMJD2A chromatin occupancy. (A) POH1-depleted cells are partially resistant to JMJD2A overexpression. U20S transfected with Non-T or POH1 siRNA for 24 h, before transfection with WT Flag-JMJD2A or JMJD2A with the mutation D939→R (DR) for a further 48 h. Cells were then exposed to 2 Gy irradiation and fixed 1 h later before incubation with anti-53BP1 and anti-Flag antibodies and imaged by confocal microscopy (left—expression of WT-JMJD2A only is shown). Red is Flag-JMJD2A, green is 53BP1, blue is DNA stained by Hoechst. Flag-JMJD2A expressing cells are illustrated with a white arrow. The graph (right) shows quantification of the presence of 53BP1 foci (>5 foci/cell) in Flag-JMJD2A expressing cells. (Cells were scored for 100 cells/condition, 2 repeats.) (B) POH1 promotes the maintenance of JMJD2A on chromatin. U20S treated with Non-T control or POH1 siRNAs before being left untreated or exposed to 2 Gy irradiation and the 200 mM NaCl-resistant chromatin fraction prepared after 1 h recovery. Chromatin (nuclear insoluble) was resuspended in SDS–PAGE buffer and boiled before immunoblotting with antibodies for JMJD2A (top panel), Histone-3, lysine 9 tri-methylation, H3K9me3 (middle panel) or Histone-3, H3, (third panel) as a loading control. In the bottom panel, the nuclear soluble extract was blotted for JMJD2A. (C) POH1 DUB activity promotes the maintenance of JMJD2A on chromatin. U20S transfected with Flag-POH1 wild type or Flag-POH1-JAMMM for 24 h then treated with Non-T control or POH1 siRNAs for a further 72 h and the insoluble chromatin fraction prepared. Chromatin was resuspended in SDS–PAGE buffer and boiled before immunoblotting with antibodies as for (B). In the bottom panel, the material in the nuclear extract removed on 200 mM NaCl was blotted for JMJD2A. (D) Lysine 63 but not lysine 48 of Ub is required to promote 53BP1 recruitment in cells treated with proteasome inhibitor. Quantification of 53BP1 foci in MG132-treated U20S cells transfected with Myc-LacZ (myc) or myc-tagged forms of Ub; wild-type (WT), lysine 63- to -arginine mutant (K63R), or lysine 48- to -arginine mutant (K48R). All cells were treated with 5 μM MG132 for 2 h before irradiation (5 Gy) and fixed after 1 h recovery then immunostained with antibodies to 53BP1 and to myc. 53BP1 foci were counted in myc-positive cells (n=100 cells/condition).
We considered that the resistance may relate to JMJD2A itself and assessed chromatin-associated JMJD2A. While IR induced the loss of JMJD2A from chromatin in cells treated with control siRNA, JMJD2A was reduced from chromatin with or without damage in POH1-depleted cells (Figure 4B). JMJD2A is a lysine demethylase that catalyses the removal of di- and tri-methylated H3K9 and H3K36 (Whetstine et al, 2006) and consistent with its reduced chromatin presence H3K9me3 was increased on POH1 depletion (Figure 4B). We introduced siRNA-resistant WT and JAMMM POH1 into cells and transfected POH1 siRNA and found that the expression of WT but not mutant POH1 correlated with the presence of JMJD2A on chromatin (Figure 4C), indicating that the DUB activity of POH1 is required to maintain JMJD2A on chromatin.
Constitutive loss of JMJD2A from chromatin potentially circumvents the need for induced K48-poly-Ub mediated JMJD2 eviction on DNA damage. K48-poly-Ub chains are short-lived at sites of DSBs (Meerang et al, 2011; Feng and Chen, 2012) and we have been unable to detect them using linkage-specific antibodies. To assess the requirement for Ub linkages in 53BP1 accumulation, we instead compared cells treated with proteasome inhibitor and complemented with exogenous Ub or Ub mutants. This showed that WT Ub, K48→R-Ub, but not K63→R-Ub was capable of restoring 53BP1 foci in MG132-treated cells (Figure 4D). Thus, K63 linkages but not K48 linkages are required to promote 53BP1 accumulation under conditions of proteasome dysfunction.
POH1 regulates K63-linked Ub in the DNA damage response
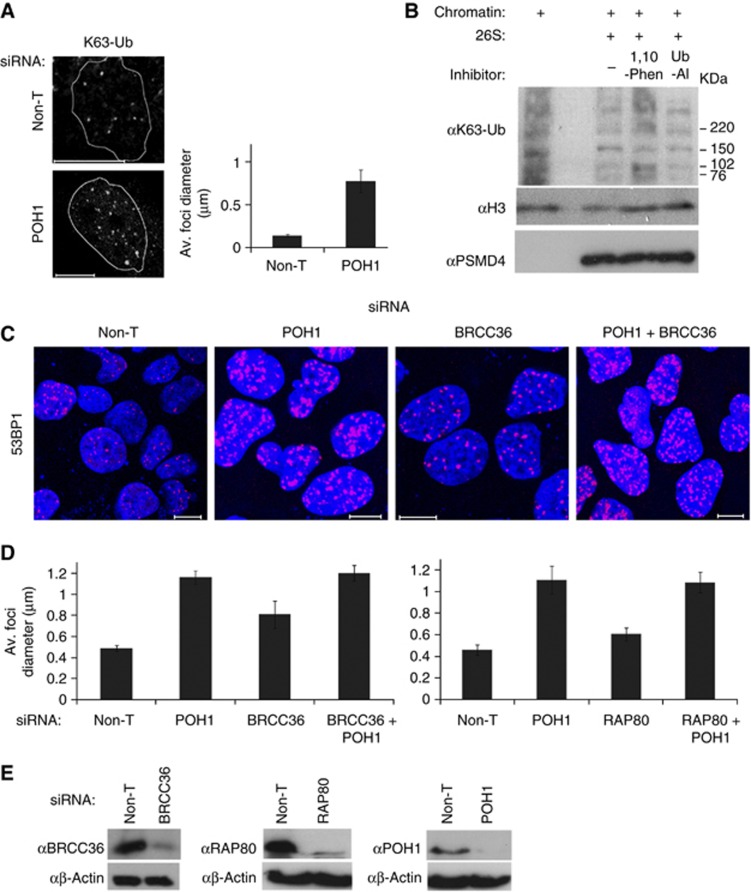
We next assessed whether K63-linked Ub accumulation to sites of DNA damage may be regulated by POH1. Using a chain-specific antibody to ubiquitin linked through K63 we found that POH1-depleted cells exhibited enlarged K63-Ub foci after damage (Figure 5A), suggesting that POH1 may act to process this conjugate. POH1 cleaves poly-Ub ‘en bloc’ near the site of substrate conjugation prior to degradation but is also able to hydrolyse K63-Ub isopeptide linkages (Yao and Cohen, 2002; Cooper et al, 2009). To examine whether POH1 can have a direct impact on K63-Ub chromatin modification, we purified chromatin from POH1-depleted cells and incubated this with purified proteasomes and inhibitors either of cysteine proteases, Ub-aldehyde (Ub-Al), or metalloproteases, using the zinc-chelating agent 1,10-o-phenanthroline that abrogates the activity of POH1 in the proteasome (Cooper et al, 2009). 1,10-o-phenanthroline, but not Ub-Al inhibited the reduction of high molecular weight K63-linked Ub in chromatin preparations (Figure 5B). These data suggest that the proteasomal metalloprotease, POH1, is active in reducing these conjugates.
Figure 5.
POH1 regulates K63-linked Ub in the DNA damage response. (A) POH1-depleted cells exhibit increased K63-Ub accumulation on DNA damage. U20S treated with Non-T control or POH1 siRNAs, then exposed to 2 Gy irradiation and 1 h recovery before staining with anti-K63-Ub antibody, imaged by confocal microscopy (left) and measured. In the graph (right), the average foci diameter is shown (>130 foci/condition). (B) Proteasomal metalloprotease activity is required to process high molecular weight K63-Ub on chromatin from POH1-depleted cells. U20S treated with POH1 siRNAs before exposure to 2 Gy irradiation and allowed 1 h to recover before cells were harvested and 200 mM NaCl-resistant chromatin fraction prepared. These preparations were resuspended in proteasome buffer containing protease inhibitors, and ATP (first lane), plus 1 μg purified 26S proteasome (second lane), with 4 mM 1,10-o-phenanthroline (1,10-Phen, third lane) or 0.5 μM Ub-aldehyde (fourth lane) and incubated for 2 h before resolution on an SDS–PAGE gel and immunoblotted using an antibody specific for K63-linked Ub chains, K63-Ub (top panel), and Histone-3 H3, (middle panel) and 19S component PSMD4 (bottom panel) as loading controls. (C) Comparison of BRCC36, POH1 and co-depletion of BRCC36 and POH1 on 53BP1 foci. U20S treated with Non-T, BRCC36, POH1 or both BRCC36 and POH1 siRNAs, for 72 h exposed to 5 Gy irradiation, and following 1 h recovery stained with anti-53BP1 antibody then imaged by confocal microscopy. (D) Comparison of depletions of BRCC36 or RAP80 with POH1 on 53BP1 foci size. U20S treated with Non-T, BRCC36, RAP80, POH1 or co-transfected with BRCC36 and POH1 siRNAs, or RAP80 and POH1 for 72 h and exposed to 5 Gy irradiation, and following 1 h recovery stained with anti-53BP1 antibody and foci measured from confocal images (n=>50/foci per treatment). (E) Knockdown of RAP80, BRCC36 and POH1 protein levels. U20S treated with the siRNAs for 72 h. Lysates immunoblotted with antibodies against BRCC36, RAP80, POH1 and β-actin.
Relationship between POH1 and BRCC36
POH1 is the second JAMM-type Ub protease found to regulate Ub conjugates at sites of DSBs. The JAMM protease BRCC36 is a component of the BRCA1-A complex and a K63 linkage-specific DUB that also acts to restrain Ub conjugate generation at DSBs and antagonises 53BP1 accumulation (Sobhian et al, 2007; Feng et al, 2009; Shao et al, 2009; Wang et al, 2009; Patterson-Fortin et al, 2010). We assessed the relationship between the 19S and the BRCA1-A complex in regulating 53BP1 by combining RAP80 or BRCC36 and POH1 depletions. Combinations of depletions did not increase 53BP1 foci size over that seen in cells depleted for POH1 alone (Figure 5C–E), indicating that the RAP80 complex and 19S are in the same pathway consistent with the role of POH1 in restraining K63-linked Ub. Notably, the requirement for POH1 in the restriction of 53BP1 accumulations was greater than RAP80 or BRCC36 indicating a larger, or additional, role related to 53BP1 assemblies. This feature is consistent with the function of POH1 in promoting JMJD2A retention. Taken together, our data show that POH1 is required to constrain two aspects of chromatin associated with 53BP1 accumulation, K63-Ub modification and JMJD2A eviction, to restrict 53BP1 accumulation.
No evidence of an influence on BRCA1/RAP80
RAP80 and BRCA1 are also recruited to chromatin at sites of DNA damage via K63-linked Ub. However, BRCA1 foci were not restored in RNF8-depleted cells by co-depletion of POH1 (Supplementary Figure 6A). POH1 siRNA had little impact on BRCA1 foci sizes nor influenced RAP80 accumulation (Figure 6B and C). Further, while transfection with an siRNA against the 19S subunit PSMD7/rpn8 resulted in enlarged 53BP1 foci it also had no influence on BRCA1 accumulation (Supplementary Figure 6D). Thus while the proteasome acts to restrict 53BP1 accumulation, there is no evidence of an impact on BRCA1.
Figure 6.
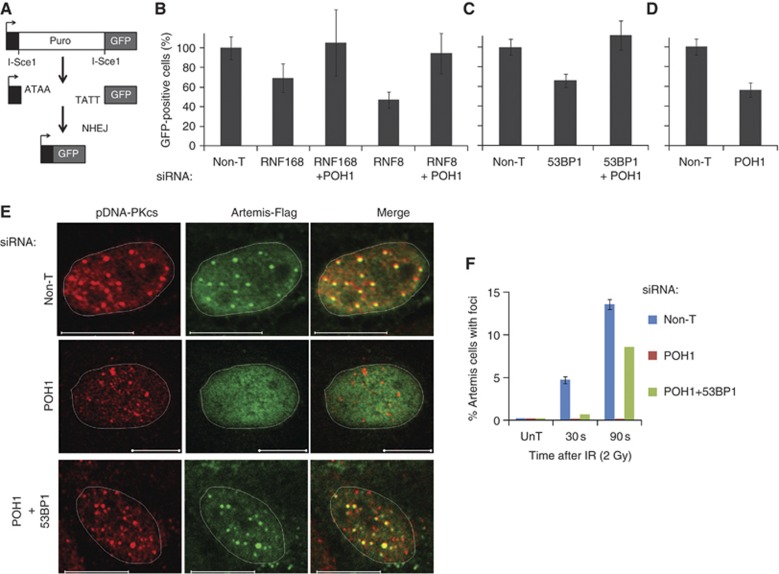
POH1 regulates end-joining DNA repair. (A) Cartoon of the EJ5-GFP construct. This was integrated into HeLa cells for the non-homologous end-joining (NHEJ) assay. Following transient RFP (transfection control) and I-SceI expression, RFP and GFP+ cells are analysed by FACS, repair by NHEJ restores the GFP expression cassette (drawn after Bennardo et al, 2008). (B–D) POH1 regulates NHEJ. HeLa cells bearing EJ5-GFP were treated with the siRNA sequences shown and the NHEJ assay undertaken. Each experiment was performed in triplicate, the average of two experiments, six technical repeats is shown. GFP+ cells are expressed as a % of Non-T cells within each experiment. (E, F) POH1 inhibits Artemis accumulation that can be partially rescued by 53BP1 co-depletion. (E) U2OS transfected with Artemis-Flag before transfection with Non-T, POH1 or POH1 and 53BP1 siRNA were exposed to 2 Gy irradiation (taking 50 s) and incubated for a further 30 or 90 s before sucrose extraction on ice for 2 min and fixation in 4% PFA. Cells at 90 s are shown stained with anti-DNA-PKcs (pS2056) and anti-flag. (F) Quantification of cells with Artemis foci. Cells exhibiting pDNA-PKcs foci and positive for Flag-Artemis were scored for the presence of Artemis foci (n=>100 cells/condition).
POH1 regulates end-joining DSB repair
53BP1 acts to promote the repair of DSBs by NHEJ in several contexts, including in simple euchromatic environments (Xie et al, 2007; Difilippantonio et al, 2008; Dimitrova et al, 2008; Bothmer et al, 2010; Noon et al, 2010; Coleman and Greenberg, 2011). We first examined the influence of POH1 on end-joining DNA repair using an integrated I-SceI-based assay (Figure 6A) in ligase-depleted contexts. RNF168 or RNF8 depletion each reduced NHEJ, whereas co-depletion of either ligase with POH1 did not (Figure 6B), suggesting that the influence of POH1 on 53BP1 recruitment maybe reflected in DNA repair. To test this directly, we reduced 53BP1 expression itself with siRNA treatment and examined the impact of co-depletion with POH1. Strikingly, this also allowed end-joining (Figure 6C) again correlating the influence of POH1 on 53BP1 accumulation with NHEJ (Figure 2D and E). These data suggest that the influence of POH1 on 53BP1 accumulation, rather than on chromatin or other factors, is able to regulate end-joining DNA repair. Depletion of POH1 alone reduced end-joining (Figure 6D), and since this is not the case when both 53BP1 and POH1 are depleted (i.e., in cells where 53BP1 foci are less exaggerated; Figure 2E), these data imply that excessive 53BP1 accumulation is inhibitory to repair detected on this substrate.
To examine how this might be mediated, we examined the phosphorylation of the NHEJ factor DNA-dependent protein kinase catalytic subunit (DNA-PKcs) at Ser2056 and the localisation of the NHEJ factor Artemis. Both proteins rapidly recruit to sites of DNA damage (Uematsu et al, 2007; Miller et al, 2011). Indeed seconds after IR a proportion of Flag-Artemis expressing cells exhibited foci that colocalised with pDNA-PKcs (Figure 6E). However in cells depleted for POH1, pDNA-PKcs was evident but Artemis did not form foci over the time-frame studied. Consistent with the impact on end-joining repair depletion of both 53BP1 and POH1 resulted in a partial restoration of Artemis foci with DNA-PK (Figure 6E and F). Together, these data establish that POH1 regulates end-joining DNA repair through 53BP1 and suggest that excessive 53BP1 alters the assembly of NHEJ factors.
The 19S enrichment at sites of DNA damage requires Ub conjugation and processing
The proteasome is associated with transcription and chromatin remodelling and is enriched on damaged chromatin (Blickwedehl et al, 2007; Ben-Aroya et al, 2010; Chou et al, 2010; Levy-Barda et al, 2011). We confirmed enrichment of the endogenous 19S to sites of DSBs using antibodies against 19S subunits (Supplementary Figure 7A and B). To examine whether Ub conjugation associated with the DSB response is required to increase the local concentrations of the 19S, we depleted several pathway components. siRNA to RNF8, Ubc13, Ub (UBA52) and to a lesser extent BRCA1, each reduced colocalisation of the 19S subunit PSMC5 with γH2AX (Figure 7A; Supplementary Figure 7C). Further RNF8 and Ubc13 depletion prevented enrichment of chromatin adjacent to the DSB site in chromatin immunoprecipitation (ChIP) experiments (Figure 7B and C). Since reduction of DNA-repair E3 Ub ligases, the Ub conjugating enzyme associated with K63-Ub production, Ubc13 and Ub itself resulted in poor enrichment at damaged sites we suggest that Ub conjugation at sites of DSBs is central in promoting accumulation at sites of DNA damage.
Figure 7.
19S enrichment at sites of DSBs requires Ub conjugation and processing. (A) 19S enrichment at sites of DNA damage is promoted by repair E3 ligases. U20S treated with Non-T, RNF8 or BRCA1 siRNAs before exposure to 5 Gy irradiation and staining with the antibodies to γH2AX and the 19S subunit PSMC5. (B) 19S enrichment to chromatin around DSB break site requires RNF8. HeLa expressing oestrogen receptor-IPpo-I (HeLa-IPpo-I) treated with Non-T or RNF8 siRNA before treatment with 4-hydroxytamoxifen (4-OHT) to induce endonuclease translocation into the nucleus. Chromatin immunoprecipitation (ChIP) was performed with the indicated antibodies and PCR adjacent to an IPpo-I cut site amplified. The mean fold increase in PCR product between induced and uninduced cells is shown (three replicates/antibody). (C) 19S enrichment to chromatin around DSB break site requires Ubc13. HeLa-IPpo-I treated with Non-T or Ubc13 siRNA and subject to ChIP as above (three replicates). (D) POH1-JAMMM colocalises poorly with γH2AX on irradiation. U20S transfected with expression constructs for RFP-POH1 and RFP-POH1-JAMMM for 48 h before exposure to 5 Gy irradiation, triton extraction and staining with the antibodies to γH2AX. (E) POH1, but not POH1-JAMMM, enriches chromatin near DNA damage sites. HeLa-IPpo-I I transfected with Flag-POH1 or Flag-POH1-JAMMM for 24 h, treated ±4-OHT for 16 h. ChIP was performed with anti-Flag antibodies and the mean fold increase in enrichment in PCR product between induced and uninduced cells is shown (three replicates/condition). Inset shows cells similarly transfected with Flag-POH1 and analysed by SDS–PAGE and immunoblot for exogenous POH1 and β-actin loading control.
We examined whether the catalytic mutant of POH1 can also enrich at sites of DNA damage. Immunofluorescence and ChIP analysis revealed greater association of WT POH1 than POH1-JAMMM (Figure 7D and E), suggesting that DUB activity is required for localisation to DSB sites.
To understand the mechanism of poor POH1-JAMMM localisation, we addressed what role loss of DUB activity has on Ub conjugate interaction of the 19S. Both mutant and wild-type Flag-POH1 co-purified high molecular weight Ub, indicating interaction of both WT and mutant 19S complexes with Ub conjugates (Supplementary Figure 8A). However, POH1-JAMMM particles had a reduced capacity to release or process these conjugates (Supplementary Figure 8A). When WT and POH1-JAMMM particles were purified from cells expressing Ub bearing only lysine 63 following IR, wild-type, but not JAMMM particles co-purified the mutant Ub, suggesting that JAMMM particles have a reduced ability to interact with Ub conjugates formed on IR (Supplementary Figure 8B). A model in which conjugate trapping by the catalytically dead 19S particle inhibits further interactions is consistent with these observations.
POH1 promotes HR independently of 53BP1
Proteasome components have previously been implicated in RAD51 foci formation (Jacquemont and Taniguchi, 2007) and we confirmed the requirement for POH1 in promoting RAD51 foci formation following IR and camptothecin treatment (Supplementary Figure 9A and B). Using an integrated I-SceI-based gene conversion substrate (Supplementary Figure 9C) we found that POH1 catalytic activity is required for normal levels of HR repair (Figure 8A). Importantly, the expression of RAD51 was not lost and we found that POH1-depleted cells exhibited a normal cell-cycle distribution (Supplementary Figure 9D and E) so that poor HR in these cells cannot be explained by altered cell cycle or an absence of RAD51.
Figure 8.
POH1 promotes homologous recombination and RAD51 loading. (A) POH1 activity is required for homologous recombination repair of double-strand breaks. HeLa cells bearing an integrated gene conversion substrate (described in Supplementary Figure 9C) treated with Non-T or POH1 (D) siRNA and transfected with expression constructs for siRNA-resistant Flag-POH1 versions indicated, RFP and Sce-I for a further 24 h. Each experiment was performed in triplicate, the average of two experiments is shown. (B) 53BP1 or BRCC36 depletion does not improve HR in POH1-depleted cells. Gene conversion recombination assay of cells treated with Non-T, POH1, 53BP1, BRCC36 siRNA or combinations of POH1 and 53BP1 or BRCC36 siRNA sequences. (Mean of three replicates shown.) (C) DSS1 foci require POH1. U20S treated with Non-T, or POH1 siRNAs for 72 h before exposure to 2 Gy irradiation and staining with antibodies to DSS1 and imaged by confocal microscopy (left). In the graph (right), the average foci diameter of 53BP1 foci in RFP-Ub positive cells is shown (50 foci/condition). (D) DSS1 antibody enriches chromatin near DNA damage sites in control but not in POH1-depleted cells. HeLa-IPpo-I cells treated with Non-T or POH1 siRNA before treatment with 4-OHT and ChIP performed with anti-DSS1 antibody. The mean fold enrichment in PCR product between uninduced and induced is shown (mean of three replicates/antibody). (E) RAD51 foci in POH1-depleted cells can be rescued by DSS1 expression. U20S transfected with POH1 siRNA for 24 h, before transfection with myc-DSS1 for a further 48 h. Cells were exposed to 2 Gy irradiation and fixed 4 h later before incubation with anti-RAD51 antibody and anti-myc antibody and imaged by confocal microscopy. Red is myc-DSS1, white is RAD51, white line outlines DNA stained. (F) Poor HR repair in POH1-depleted cells can be rescued by DSS1 expression. HeLa cells bearing an integrated gene conversion substrate were treated with Non-T control or POH1 siRNAs for 24 h before transfection with myc-DSS1 for a further 24 h before transfection with Sce-I for a further 24 h. GFP indicates conversion. The % GFP cells are expressed relative to those in Non-T control cells (mean of three replicates).
K63-poly-Ub generated at sites of DSBs recruits factors inhibitory to resection in HR: RAP80, BRCC36 and 53BP1 (Bunting et al, 2010; Coleman and Greenberg, 2011; Hu et al, 2011). We speculated that POH1 may promote HR by restricting the assembly of these proteins. To test this notion, we examined proteins involved in resection, the BRCA1-CtBP-interacting protein (CtIP) and the ssDNA binding protein RPA (replication protein A). Both were recruited into foci to the same degree in POH1-depleted and control-treated cells following camptothecin treatment, which causes DNA damage primarily in S phase (Supplementary Figure 10A). In G2-phase cells following IR slightly reduced numbers of RPA foci were evident, in particular fewer smaller foci (Supplementary Figure 10B) suggesting DNA resection may be affected in G2 cells. To test whether these observations have a functional impact, we examined depletion of BRCC36 and 53BP1 in cells with POH1 depletion on HR repair. As expected, depletion of BRCC36 or 53BP1 alone increased HR as measured by gene conversion, however, when combined with POH1 siRNA neither improved HR (Figure 8B). Consequently, POH1 is likely to be required in another aspect of HR instead of, or in addition to, DNA-end resection, and in contrast to the relationship in NHEJ, its influence on HR is primarily not through 53BP1.
POH1 promotes the enrichment of DSS1 at sites of DSBs
We examined the recruitment of some of the proteins that act to mediate RAD51 loading. The accumulation of BRCA2 and its interacting protein PALB2 was unaffected by POH1 depletion (Supplementary Figure 11A). We also examined the small acidic protein DSS1, a component of the 19S proteasome, BRCA2 co-factor and component of various other complexes (Wilmes et al, 2008). DSS1 failed to accumulate into foci in cells treated with POH1 siRNA and anti-DSS1 antibody did not enrich for DNA around break sites when cells were depleted for POH1 (Figure 8C and D). These data indicate that a population of DSS1 is enriched at damage sites with the 19S. Further, we found that catalytically active, but not mutant, POH1 interacted with BRCA2, and BRCA1, following HU treatment consistent with a role promoting HR (Supplementary Figure 11B). We have been unable to detect endogenous DSS1 by immunoblot. Although POH1 depletion had little influence on the low-level expression of exogenous DSS1 (Supplementary Figure 11C), we cannot discount that its expression may be lost following depletion of POH1. Taken together, these data suggest an association of the 19S proteasome with factors that promote RAD51 loading. However as many proteins, including BRCA proteins are also degraded via the proteasome (Choudhury et al, 2004; Schoenfeld et al, 2004), which may contribute to our data, we also wished to consolidate this notion through another means. We therefore tested the impact of exogenous DSS1 expression on HR in POH1-depleted cells. Overexpression of DSS1 restored both RAD51 foci and HR repair in these cells (Figure 8E and F), confirming that POH1 promotes RAD51 accumulation/loading and consistent with the observation that DSS1 recruitment, or expression, is limited on POH1 depletion.
POH1 is required for cell viability in response to DNA damage
Our data predict that POH1 is required for cell survival following exposure to agents that cause DSBs. Since complete POH1 depletion reduces cell viability (Gallery et al, 2007; Byrne et al, 2010), we first identified a dose of POH1 siRNA that reduced POH1 expression but allowed cell survival for 2 weeks. Exposure of partially depleted cells to IR, cisplatin or HU, revealed that POH1 is required for resistance to each agent, consistent with its ability to promote cellular responses to DNA damage (Figure 9A–D).
Figure 9.
POH1 is required for cellular resistance to DNA damaging agents. (A) Partial depletion of POH1. Immunoblot of HeLa lysates transfected with Non-T and POH1 (F) siRNA at 2.5 nM, lysed and examined for POH1 expression and β-actin loading control. (B–D) POH1 is required for resistance to DNA damaging agents. HeLa transfected with 2.5 nM POH1 siRNA and Non-T siRNA, irradiated, treated with 0.5–50 μM cisplatin for 1 h, or 2–10 mM HU for 8 h. Cells were diluted, plated in triplicate and incubated at 37°C for 2 weeks after which colonies were counted.
Discussion
Ub conjugation is an important and complex modification in the DSB response and significant roles for additional DUBs were expected. We have shown that a major regulator of the DNA-damage poly-Ub cascade is the proteasome, and within it the JAMM-type protease POH1.
POH1 has a global influence on cellular Ub conjugates. Here, we have demonstrated that both its constitutive activity and its specific role in restricting Ub conjugates at sites of DNA damage are important in promoting the normal response to DSBs.
POH1 is important in the two main pathways employed to repair DSBs. In NHEJ, it acts to restrain 53BP1 accumulation, through countering both K63-Ub modification and JMJD2A chromatin eviction, while in HR repair it acts independently of its influence on 53BP1 and instead promotes RAD51 loading.
53BP1 plays a significant role in the preservation of genomic integrity and the mechanisms that promote its accumulation at DSB sites following DNA damage include the removal of H4K20me2 binding proteins by ubiquitination, and possibly degradation, to allow the exposure of H4K20me2 for 53BP1 binding (Acs et al, 2011; Mallette et al, 2012). Of the known H4K20me2 binding proteins removed in this way JMJD2A has the highest affinity (Mallette et al, 2012). Knockdown of POH1 results in constitutive loss of JMJD2A from chromatin, increased H3K9me2 and 53BP1 foci were partially resistant exogenous JMJD2A expression. Our data support the notion that eviction and degradation of JMJD2A occur as two separate steps, since POH1 depletion inhibits degradation, but does not prevent JMJ2DA eviction. We cannot rule out that other H4K20me2 binding proteins are also removed, and we anticipate that chromatin occupancy of the related demethylase JMJD2B is also promoted by POH1. The mechanism of promotion of JMJD2A chromatin occupancy is not known and will be an important area of future study. It is notable that JMJD2A is modified in the absence of DNA damage by ubiquitin ligases FbxL4 and FBX022 (Tan et al, 2011; Van Rechem et al, 2011) and that depletion of FBX022 reduced H3K9me3 suggesting increased chromatin occupancy of the JMJD2 without Ub modification (Tan et al, 2011). We speculate that POH1 may function in equilibrium with non-DNA repair associated ligases to regulate the degree of Ub modification and thus eviction of JMJD2A. Other mechanisms such as increased expression or reduced degradation of a factor that promotes eviction are also possible.
Our data show that POH1 and the 19S are enriched on damaged chromatin by the activity of ubiquitin conjugation components where it acts to counter K63-Ub accumulation in the same pathway as the K63-Ub chain-specific DUB BRCC36. We suggest that POH1 DUB processes K63-linked Ub on chromatin, which in turn restricts 53BP1 spread.
POH1 regulation of 53BP1 and end-joining DNA repair acts in opposition to RNF8/RNF168. Ligase activity would appear to be needed to overcome POH1 promotion of chromatin occupation by JMJD2A and K63-Ub processing to allow 53BP1 recruitment (Figure 10A and B).
Figure 10.
Models of POH1 function in the DSB response. (A) Model of POH1-mediated restriction of 53BP1. POH1 in the 19S (accompanied by the 20S, not shown) is required to maintain JMJD2A on chromatin. This is through its DUB/degradation activity. JMJD2 proteins retain interaction with H4K20me2 and compete with 53BP1 for the chromatin mark restricting 53BP1 spread. RNF8/168 (RNF8 only shown) generates ubiquitin conjugates of K48 linkages. These modify JMJD2 proteins and L3MBTL1 (not shown) and promote their removal from chromatin. The ligases also modify histones with K63-linked ubiquitin. The combined activity of the removal of chromatin mark binding proteins and K63-poly-Ub generation promotes the accumulation of 53BP1 to the mark. POH1 also counters the K63-poly-Ub on chromatin to further restrict 53BP1 accumulation. (B) Chromatin in the absence of POH1 activity. H4K20me2 marks are exposed, and H3K9me3 is increased (not shown). RNF8/RNF168 ligase activity modifies chromatin with K63-poly-ubiquitin over a larger expanse and 53BP1 accumulation is exaggerated. (C) Proposal for POH1-mediated promotion of RAD51 loading. The 19S interacts with BRCA1 and BRCA2 at sites of DNA damage and POH1 promotes DSS1 enrichment and the loading of RAD51.
Somewhat surprisingly our data point to the possibility that excessive 53BP1 is repressive to end-joining, at least on the substrate measured here. One possibility is the excessive spreading of 53BP1, and subsequent increased mobility of the broken regions (Dimitrova et al, 2008; Bothmer et al, 2011) may encourage end-joining to inappropriate junctions increasing translocations and resulting in a reduced end-joining product for relatively near broken ends, such as those in the substrate used to measure repair in this report. Alternatively, or additionally, it is possible that the protective role of 53BP1 in preventing unrepaired ends from degradation (Liu et al, 2012) has spread to overprotect the DNA ends and prevent some NHEJ factor accessibility. Our data suggest that Ku and DNA-PK are correctly associated but that Artemis is not, indicating that the accumulation or kinetics of end-joining factors at sites of DSBs can be affected by excessive 53BP1. Artemis is required for end processing in a subset of substrates such as those with overhangs and hairpins (Ma et al, 2002), whether simpler lesions are also affected has not been assessed.
The contribution of POH1 to the regulation of two elements of 53BP1 recruitment is insufficient to explain why the spreading of K63-Ub chains is not reflected in increased assembly of the RAP80/BRCA1-A complex which also has K63-poly-Ub binding properties. One possibility is that K63-linked Ub is not the only determinant for RAP80/BRCA1-A accumulation and recently binding to SUMO as well as ubiquitin has been reported as part of the RAP80 recruitment (Hu et al, 2012). Further, it seems likely that different populations of K63-linked Ub may be required to target 53BP1 and RAP80/BRCA1 to different specific territories at the DSB (Mok and Henderson, 2010; Chapman et al, 2012). Further support for two separate recruitment pathways is the report of a lysine mutant of MDC1 that inhibits RAP80 but not 53BP1 accumulation (Strauss and Goldberg, 2011; Strauss et al, 2011).
The 19S and RAP80/BRCA1-A complexes are intriguingly similar (as previously discussed by Wang et al, 2009) but why the cell uses two similar complexes in the processing of RNF8/RNF168-dependent K63-poly-Ub is not clear. It may be a reflection of the discrete populations of K63-poly-Ub at damaged chromatin, the need to modulate the different functions of the recruited proteins perhaps in relation to the cell cycle or chromatin states, or the need to dampen this modification, and its consequences, effectively.
In addition to the regulation of 53BP1 and end-joining repair, we have shown that POH1 promotes HR repair. Despite the antagonistic influence of POH1 on K63-poly-Ub it appears that increased HR-repressive factors recruited by this modification are largely not responsible, or not wholly responsible, for the defect in HR repair in POH1-depleted cells. Instead our findings point to a role for POH1 later in the pathway, associated with RAD51 loading. BRCA1 is partially required for recruitment of the 19S and recently the SUMO-targeted E3 ubiquitin ligase RNF4 has been demonstrated to accumulate at sites of DNA damage and to be partially required for recruitment of the proteasome (Galanty et al, 2012; Yin et al, 2012) it is tempting to speculate that ligases retain proteasome association by the sequential production of ubiquitin conjugates in the damage response.
Our data indicate that RAD51 loading is defective in POH1-depleted cells since expression of the small nucleotide-like protein DSS1 is capable of restoring repair. DSS1 acts as a co-factor with BRCA2 to promote RAD51 loading on RPA-ssDNA (Liu et al, 2010) but may also have BRCA2-independent activities in HR repair (Krogan et al, 2004). Our data point to a requirement for POH1 in the recruitment or expression of DSS1 and in the interaction with BRCA1 and BRCA2. We propose that the simplest model is that 19S enrichment promotes DSS1 recruitment to DSBs to augment RAD51 loading onto ssDNA (Figure 10C). We do not rule out the potential for an additional defect in RAD51 loading in these cells.
Since proteasome particles can carry the 19S activator at both ends of the 20S barrel, or be mixed (hybrid), with the other end occupied by other activators, a complex and nuanced role of the proteasome at DSBs can be envisaged. In a recent report, a fraction of the proteasomal activator PA28γ that drives degradation independently of Ub was found located to sites of DSBs. PA28γ was required for a proportion of NHEJ, but in contrast to POH1 in the 19S, PA28γ acted to restrict RAD51 localisation to damaged sites and reduce HR (Levy-Barda et al, 2011). Thus, different roles for different activators, or the balance of activators, on the proteasome core in the DSB response is possible. Together with our findings, these allude to the possibility of cross-talk between Ub-mediated degradation pathways and the DSB response, with implications for conditions associated with impaired proteasome function, such as cardiac and neurodegenerative diseases (Lehman, 2009; Yu and Kem, 2010).
In conclusion, our data reveal novel roles for the 19S proteasome in the DNA damage response, in particular showing it has a restrictive influence on 53BP1 assembly. The findings presented suggest that the proteasome has the capacity to tune DNA repair responses by balancing the activity of ubiquitin ligases.
Materials and methods
A full list of antibodies can be found in Supplementary Tables 1 and 2.
Immunoprecipitations
Anti-flag immunoprecipitations performed with M2-affinity gel (Sigma), and anti-GFP with GFP-trap (Chromotech) according to manufacturer’s instructions.
Chromatin preparations
These were done according to the method described in Mallette et al (2012).
Cell culture
HeLa, U20S and 293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% Fetal Calf Serum, Penicillin, Streptomycin and 1 × L-glutamine at 37°C, 5% CO2.
siRNA experiments and screen conditions
Cells were transfected with a final concentration of 100 nM siRNA, harvested 48–72 h after transfection. Knockdown with the DUB siGENOME RTF library (Thermo Fisher) was achieved using the reverse transfection method specified by Dharmacon (Thermo Fisher). HeLa cells were fixed (4% PFA followed by 1% Triton X-100) 16 h later (a total of 64 h after plating) and stained with monoclonal FK2-HRP conjugate (Biomol, Exeter, UK) and washed before detection with SuperSignal Chemiluminescent ELISA substrate (Pierce). A full list of siRNA sequences can be found in Supplementary Table 3.
Constructs and transfections
pCI-neo.POH1 was a gift from Dr Ribeiro’s laboratory (Institute of Parasitology, McGill University, Quebec). pCBASceI (J Stark, Beckman Research Institute) was used to express I-SceI to induce damage in HeLa DR-GFP3 cells. pBABE HA-ER-I-PpoI was a gift of Michael B Kastan, (St Jude Children’s Research Hospital, Memphis, TN, USA). GFP-DSS1 and myc-DSS1 were purchased from Origene (Rockville, MD, USA), HA-DSS1 was a gift of Christopher Lord (Institute of Cancer Research, London, UK), GFP-BRCA1 and Ub constructs have been previously described (Morris and Solomon, 2004). A full list of primers used for mutagenesis used can be found in Supplementary Table 4. Cells were transfected using FuGene 6 Transfection reagent from Hoffmann-La Roche, (Basel, Switzerland) and fixed 48–72 h later.
Colony assays
Seventy-two hours after siRNA transfection, cells were either treated with 0.5–50 μM cisplatin for 1 h 1–10 mM HU for 8 h or irradiated at 1–8 Gy using a gamma cell 1000 Elite irradiator (Caesium-137 source). Cells were diluted, plated in triplicate and incubated at 37°C for 2 weeks, with media changed every 3–4 days. Colonies were fixed with 100% methanol, stained with crystal violet (0.05% in distilled water) and counted.
Immunofluorescence microscopy
Cells were plated onto glass coverslips and fixed for staining with 4% PFA for 10 min followed by 5 min permeabilisation with 1% Triton X-100 or as otherwise described. All scale bars are 10 μm. Resolution of DSS1, proteasome subunits, RPA and CtIP-GFP was achieved with prior extraction in sucrose/triton or NP-40 buffer, respectively. (Sucrose/triton extraction buffer: 0.02–0.025% Triton in 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes (pH 6.8), 1 mM EGTA, and protease inhibitors. NP-40 buffer: 0.01–0.02% NP-40 in 20 mM HEPES pH 7.5, 20 mM NaCl, 5 mM MgCl2 1 mM DTT and protease inhibitors). Fixed cells were blocked and antibodies diluted in 10% FCS in PBS. All antibodies were incubated at room temperature for 1 h. Following antibody staining, cells were additionally stained with Hoechst to allow visualisation of the nuclei. Cells were examined using a Zeiss LSM 510 confocal microscope with three lasers giving excitation lines at 633, 543 and 488 nm. Data from channels were collected sequentially using the appropriate band-pass filters. Data were collected with eight-fold averaging at a resolution of 1024 × 1024 pixels, using an optical slice of between 0.5 and 1 mm using a × 63 objective within the Zeiss Axioplan-II microscope.
Quantification of foci
All images within an experiment were captured at identical exposures selected so as to avoid saturation at any individual focus. Intra-nuclear foci were either counted by hand from confocal images or using an automated approach. Counting by hand used LSM image browser software (Zeiss, Germany), a ‘focus’ was measured from where its intensity exceeded 100 arbitrary intensity units, all were measured across the horizontal plane. Automated foci counting was performed using Image J (NIMH, MD, USA) built-in macro functions. Cell nuclei and foci were separated from the background using the auto-threshold algorithm and adjacent objects were divided using the watershed routine. The signal-to-noise ratio for each focus was calculated and foci with SNR <2.0 were discarded. Automated counting was calibrated on previous foci counted manually. (As a comparison, average RPA foci/nucleus of cells treated with camptothecin were 116.7±4.9 (s.e.) in manual counting and 114.15±9.5 (s.e.) for automated counting of the same 13 nuclei).
ChIP assays
ChIP assays were performed as previously described (Berkovich et al, 2008), in which oestrogen receptor-fused IPpo-I endonuclease is translocated rapidly to the nucleus on 4-hydroxytamoxifen (4-OHT) addition. PCR amplification of the Chromosome 1 site adjacent to the IPpo-I endonuclease target site PCR was analysed using SYBR Green reaction in 96-well plates on ABI-7900 real-time PCR machine.
Fluorescence-activated cell sorting
For cell-cycle analysis, cells were fixed with 70% ethanol, incubated for 4 h with RNase A (250 μg/ml), resuspended in propidium iodide (10 μg/ml) and analysed by FlowJo or WinMDI software was used to reveal the percentage of cells in each phase of the cell cycle.
HR and end-joining assays
HeLa DR-GFP3 and EJ-7 cells were transfected with stated siRNAs and/or expression constructs, followed 24 h later by co-transfection with pCBASceI-I and red fluorescent protein (RFP) to induce damage and control for transfection efficiency, respectively. Cells were incubated for a further 48 h before being fixed with 4% PFA. Green fluorescent protein (GFP) indicates gene conversion/NHEJ depending on the assay. In fluorescence-activated cell sorting (FACS) analysis, the red cells were gated and the number of green fluorescent cells counted as a measure of repair (those cells able to repair the damage caused by expressed I-SceI). Assays were performed in triplicate. Assays combining siRNA and protein expression were either gated on using the RFP tag of the transfected protein or using RFP co-transfection as a surrogate marker for transfection efficiency and transcriptional competence.
Proteasome assays
Immunopurification of Flag complexes was achieved as described above. Purified 26S proteasome was purchased from Biomol. Complexes or chromatin was then incubated at 37°C, in proteasome buffer (50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 40 mM NaCl, 1 mM Zinc Sulphate, 2 mM ATP, 10% glycerol, 2 mg/ml BSA and 1 mM DTT) in the presence of protease inhibitors and where stated 1 μM Ub-Al for the times indicated. Unbound material was removed by subsequent washing with 10 ml of buffer before re-precipitation and suspension in SDS–PAGE loading buffer.
Supplementary Material
Acknowledgments
Thanks to Grant Stewart and Ellen Solomon for discussions and Penny Jeggo for sharing unpublished results. Thanks to Jeremy Stark (Beckman Research Institute) for DR-GFP3 EJ5-GFP and pCBASce-1 plasmids, Michael Kastan (St Jude Children’s Research Hospital, Memphis, Tennessee, USA) for HA-ER-I-PpoI retrovirus plasmids, Andrew Turnell (University of Birmingham) for PSMC5 antibody, Paula Ribeiro (Institute of Parasitology, McGill University, Quebec) for pCI-neo.POH1, Christopher Lord (Institute of Cancer Research, London) for HA-DSS1. Steve Jackson (University of Cambridge, UK) for Flag-Artemis, Roger Grand (University of Birmingham, UK) GFP-CtIP cells and Stéphane Richard (McGill University Canada) for Flag-JMJD2A constructs. This work was supported by the Breast Cancer Campaign: LRB (2006NovPHD13), JJ (2010MayPR01) JRM and JB (2006MaySF06), HS (2010NovPhD02), Cancer Research UK, LRB, RMD (C8820/A9494 and C8820/A15265), the Medical Research Council (6900577) DW and Breakthrough Breast Cancer, VS.
Author contributions: LRB performed the screen, validation and functional experiments, RMD performed repair assays, immunoblot experiments and cell-cycle analysis. JJ performed immunoprecipitation and immunoblot experiments, VS performed knockdown and foci quantification experiments, AG performed ChIP and repair experiments and DW performed cell-cycle profile analysis, HS performed MG132 rescue experiments. JB made ubiquitin mutants and FF wrote the automated foci counting algorithm and performed analysis. RMD, AG and LRB performed data analysis. All authors reviewed and commented on the manuscript. JRM designed the study, performed experiments and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat Struct Mol Biol 18: 1345–1350 [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Bartek J, Lukas J, Mailand N (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12: 80–86sup pp 1–12 [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S, Agmon N, Yuen K, Kwok T, McManus K, Kupiec M, Hieter P (2010) Proteasome nuclear activity affects chromosome stability by controlling the turnover of Mms22, a protein important for DNA repair. PLoS Genet 6: e1000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark JM (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4: e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovich E, Monnat RJ Jr, Kastan MB (2008) Assessment of protein dynamics and DNA repair following generation of DNA double-strand breaks at defined genomic sites. Nat Protoc 3: 915–922 [DOI] [PubMed] [Google Scholar]
- Blickwedehl J, Agarwal M, Seong C, Pandita RK, Melendy T, Sung P, Pandita TK, Bangia N (2008) Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc Natl Acad Sci USA 105: 16165–16170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blickwedehl J, McEvoy S, Wong I, Kousis P, Clements J, Elliott R, Cresswell P, Liang P, Bangia N (2007) Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat Res 167: 663–674 [DOI] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Di Virgilio M, Bunting SF, Klein IA, Feldhahn N, Barlow J, Chen HT, Bosque D, Callen E, Nussenzweig A, Nussenzweig MC (2011) Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 42: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC (2010) 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. J Exp Med 207: 855–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, Hiddingh S, Thanasoula M, Kulkarni A, Yang Q, Haffty BG, Tommiska J, Blomqvist C, Drapkin R, Adams DJ, Nevanlinna H, Bartek J, Tarsounas M, Ganesan S, Jonkers J (2010) 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 17: 688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Kozak ML, Kim JM, Wong N, Lopez-Contreras AJ, Ludwig T, Baer R, Faryabi RB, Malhowski A, Chen HT, Fernandez-Capetillo O, D’Andrea A, Nussenzweig A (2012) BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol Cell 46: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, Xu X, Deng CX, Finkel T, Nussenzweig M, Stark JM, Nussenzweig A (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A, McLaren RP, Mason P, Chai L, Dufault MR, Huang Y, Liang B, Gans JD, Zhang M, Carter K, Gladysheva TB, Teicher BA, Biemann HP, Booker M, Goldberg MA, Klinger KW, Lillie J, Madden SL, Jiang Y (2010) Knockdown of human deubiquitinase PSMD14 induces cell cycle arrest and senescence. Exp Cell Res 316: 258–271 [DOI] [PubMed] [Google Scholar]
- Chapman JR, Sossick AJ, Boulton SJ, Jackson SP (2012) BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci advance online publication, 2 May 2012 doi:; DOI: 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA 107: 18475–18480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury AD, Xu H, Baer R (2004) Ubiquitination and proteasomal degradation of the BRCA1 tumor suppressor is regulated during cell cycle progression. J Biol Chem 279: 33909–33918 [DOI] [PubMed] [Google Scholar]
- Coleman KA, Greenberg RA (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem 286: 13669–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE (2009) K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J 28: 621–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Groothuis TA, Salomons FA, Neefjes J (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J Cell Biol 173: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, Sleckman BP, Nussenzweig A (2008) 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 456: 529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456: 524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, Lukas J, Lukas C (2009) RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136: 435–446 [DOI] [PubMed] [Google Scholar]
- Drost R, Bouwman P, Rottenberg S, Boon U, Schut E, Klarenbeek S, Klijn C, van der Heijden I, van der Gulden H, Wientjens E, Pieterse M, Catteau A, Green P, Solomon E, Morris JR, Jonkers J (2011) BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell 20: 797–809 [DOI] [PubMed] [Google Scholar]
- Feng L, Chen J (2012) The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 19: 201–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Huang J, Chen J (2009) MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev 23: 719–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF (2009) 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans 37: 897–904 [DOI] [PubMed] [Google Scholar]
- Galanty Y, Belotserkovskaya R, Coates J, Jackson SP (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 26: 1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallery M, Blank JL, Lin Y, Gutierrez JA, Pulido JC, Rappoli D, Badola S, Rolfe M, Macbeth KJ (2007) The JAMM motif of human deubiquitinase Poh1 is essential for cell viability. Mol Cancer Ther 6: 262–268 [DOI] [PubMed] [Google Scholar]
- Geng F, Tansey WP (2012) Similar temporal and spatial recruitment of native 19S and 20S proteasome subunits to transcriptionally active chromatin. Proc Natl Acad Sci USA 109: 6060–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Ashworth A (2007) The proteasome is involved in determining differential utilization of double-strand break repair pathways. Oncogene 26: 7601–7606 [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir K, Lord CJ, Witt E, Tutt AN, Ashworth A (2004) DSS1 is required for RAD51 focus formation and genomic stability in mammalian cells. EMBO Rep 5: 989–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP (2011) Replication stress induces 53BP1-containing OPT domains in G1 cells. J Cell Biol 193: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloman WK (2011) Unraveling the mechanism of BRCA2 in homologous recombination. Nat Struct Mol Biol 18: 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Paul A, Wang B (2012) Rap80 recruitment to DNA double strand breaks requires binding to both sumo- and ubiquitin-conjugates. J Biol Chem 287: 25510–25519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM (2011) RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 25: 685–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, Chen J (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol 11: 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131: 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont C, Taniguchi T (2007) Proteasome function is required for DNA damage response and fanconi anemia pathway activation. Cancer Res 67: 7395–7405 [DOI] [PubMed] [Google Scholar]
- Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, Chang C, Wang H (2007) Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature 449: 1068–1072 [DOI] [PubMed] [Google Scholar]
- Kim H, Chen J, Yu X (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205 [DOI] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318: 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, Datta N, Cagney G, Buratowski S, Emili A, Greenblatt JF (2004) Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell 16: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Lehman NL (2009) The ubiquitin proteasome system in neuropathology. Acta Neuropathol 118: 329–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Barda A, Lerenthal Y, Davis AJ, Chung YM, Essers J, Shao Z, van Vliet N, Chen DJ, Hu MC, Kanaar R, Ziv Y, Shiloh Y (2011) Involvement of the nuclear proteasome activator PA28gamma in the cellular response to DNA double-strand breaks. Cell Cycle 10: 4300–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang W, Dubois RL, Yamamoto K, Wolner Z, Zha S (2012) Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc Natl Acad Sci USA 109: 3903–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lok GT, Sy SM, Dong SS, Ching YP, Tsao SW, Thomson TM, Huen MS (2011) Differential regulation of RNF8-mediated Lys48- and Lys63-based poly-ubiquitylation. Nucleic Acids Res 40: 196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J (2011) 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 13: 243–253 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131: 887–900 [DOI] [PubMed] [Google Scholar]
- Mallette FA, Mattiroli F, Cui G, Young LC, MJ Hendzel, Mer G, Sixma TK, Richard S (2012) RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J 31: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M, Ritz D, Paliwal S, Garajova Z, Bosshard M, Mailand N, Janscak P, Hubscher U, Meyer H, Ramadan K (2011) The ubiquitin-selective segregase VCP/p97 orchestrates the response to DNA double-strand breaks. Nat Cell Biol 13: 1376–1382 [DOI] [PubMed] [Google Scholar]
- Miller L, Foradori CD, Lalmansingh AS, Sharma D, Handa RJ, Uht RM (2011) Histone deacetylase 1 (HDAC1) participates in the down-regulation of corticotropin releasing hormone gene (crh) expression. Physiol Behav 104: 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok MT, Henderson BR (2010) A comparison of BRCA1 nuclear localization with 14 DNA damage response proteins and domains: identification of specific differences between BRCA1 and 53BP1 at DNA damage-induced foci. Cell Signal 22: 47–56 [DOI] [PubMed] [Google Scholar]
- Morris JR, Solomon E (2004) BRCA1: BARD1 induces the formation of conjugated ubiquitin structures, dependent on K6 of ubiquitin, in cells during DNA replication and repair. Hum Mol Genet 13: 807–817 [DOI] [PubMed] [Google Scholar]
- Murakawa Y, Sonoda E, Barber LJ, Zeng W, Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, Boulton SJ, Takeda S (2007) Inhibitors of the proteasome suppress homologous DNA recombination in mammalian cells. Cancer Res 67: 8536–8543 [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol 12: 177–184 [DOI] [PubMed] [Google Scholar]
- Panier S, Durocher D (2009) Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 8: 436–443 [DOI] [PubMed] [Google Scholar]
- Patterson-Fortin J, Shao G, Bretscher H, Messick TE, Greenberg RA (2010) Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J Biol Chem 285: 30971–30981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470: 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T (2008) E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proc Natl Acad Sci USA 105: 20876–20881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA (2004) BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol 24: 7444–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, deRooij DG, Hirsch S, Ravi K, Hicks JB, Szabolcs M, Jasin M, Baer R, Ludwig T (2011) BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334: 525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao G, Lilli DR, Patterson-Fortin J, Coleman KA, Morrissey DE, Greenberg RA (2009) The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc Natl Acad Sci USA 106: 3166–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Ma Z, Willers H, Akhtar K, Scott SP, Zhang J, Powell S (2008) Disassembly of MDC1 foci is controlled by ubiquitin-proteasome-dependent degradation. J Biol Chem 283: 31608–31616 [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316: 1198–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spataro V, Toda T, Craig R, Seeger M, Dubiel W, Harris AL, Norbury C (1997) Resistance to diverse drugs and ultraviolet light conferred by overexpression of a novel human 26 S proteasome subunit. J Biol Chem 272: 30470–30475 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, Nakada S, Ylanko J, Olivarius S, Mendez M, Oldreive C, Wildenhain J, Tagliaferro A, Pelletier L, Taubenheim N, Durandy A, Byrd PJ, Stankovic T, Taylor AM, Durocher D (2009) The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136: 420–434 [DOI] [PubMed] [Google Scholar]
- Stewart GS, Stankovic T, Byrd PJ, Wechsler T, Miller ES, Huissoon A, Drayson MT, West SC, Elledge SJ, Taylor AM (2007) RIDDLE immunodeficiency syndrome is linked to defects in 53BP1-mediated DNA damage signaling. Proc Natl Acad Sci USA 104: 16910–16915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss C, Goldberg M (2011) Recruitment of proteins to DNA double-strand breaks: MDC1 directly recruits RAP80. Cell Cycle 10: 2850–2857 [DOI] [PubMed] [Google Scholar]
- Strauss C, Halevy T, Macarov M, Argaman L, Goldberg M (2011) MDC1 is ubiquitylated on its tandem BRCT domain and directly binds RAP80 in a UBC13-dependent manner. DNA Repair (Amst) 10: 806–814 [DOI] [PubMed] [Google Scholar]
- Sy SM, Jiang J, Dong SS, Lok GT, Wu J, Cai H, Yeung ES, Huang J, Chen J, Deng Y, Huen MS (2011) Critical roles of ring finger protein RNF8 in replication stress responses. J Biol Chem 286: 22355–22361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MK, Lim HJ, Harper JW (2011) SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol Cell Biol 31: 3687–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ (2007) Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol 177: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustrell V, Hoffman L, Pratt G, Rechsteiner M (2002) PA200, a nuclear proteasome activator involved in DNA repair. EMBO J 21: 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rechem C, Black JC, Abbas T, Allen A, Rinehart CA, Yuan GC, Dutta A, Whetstine JR (2011) The SKP1-Cul1-F-box and leucine-rich repeat protein 4 (SCF-FbxL4) ubiquitin ligase regulates lysine demethylase 4A (KDM4A)/Jumonji domain-containing 2A (JMJD2A) protein. J Biol Chem 286: 30462–30470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ (2002) Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615 [DOI] [PubMed] [Google Scholar]
- Wang B, Elledge SJ (2007) Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 104: 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Hurov K, Hofmann K, Elledge SJ (2009) NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev 23: 729–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y (2006) Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481 [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bergkessel M, Bandyopadhyay S, Shales M, Braberg H, Cagney G, Collins SR, Whitworth GB, Kress TL, Weissman JS, Ideker T, Guthrie C, Krogan NJ (2008) A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol Cell 32: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R (2007) Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 28: 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM (2007) The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res 67: 6647–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE (2002) A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407 [DOI] [PubMed] [Google Scholar]
- Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT (2012) SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev 26: 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Kem DC (2010) Proteasome inhibition during myocardial infarction. Cardiovasc Res 85: 312–320 [DOI] [PubMed] [Google Scholar]
- Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S (2007) A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol Cell 25: 663–675 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, Gage FH, Verma IM (2011) BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature 477: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.