Abstract
The Nrd1-Nab3-Sen1 (NNS) complex pathway is responsible for transcription termination of cryptic unstable transcripts and sn/snoRNAs. The NNS complex recognizes short motifs on the nascent RNA, but the presence of these sequences alone is not sufficient to define a functional terminator. We generated a homogeneous set of several hundreds of artificial, NNS-dependent terminators with an in vivo selection approach. Analysis of these terminators revealed novel and extended sequence determinants for transcription termination and NNS complex binding as well as supermotifs that are critical for termination. Biochemical and structural data revealed that affinity and specificity of RNA recognition by Nab3p relies on induced fit recognition implicating an α-helical extension of the RNA recognition motif. Interestingly, the same motifs can be recognized by the NNS or the mRNA termination complex depending on their position relative to the start of transcription, suggesting that they function as general transcriptional insulators to prevent interference between the non-coding and the coding yeast transcriptomes.
Keywords: cryptic unstable transcripts, hidden transcription, Nrd1p-Nab3p-Sen1p complex, transcriptional insulators, transcription termination
Introduction
The concept of pervasive and/or hidden transcription has emerged in the last few years from studies revealing that the transcribed fraction of eukaryotic genomes is considerably higher than expected from early annotations based on RNA steady-state abundance and phylogenetic conservation (Johnson et al, 2005; Jacquier, 2009). Whether pervasive transcription predominantly pollutes the expression of meaningful genetic information or increases the regulatory potential of the cell remains matter of debate. What is clear, however, is that it has to be controlled to maintain the stability and intelligibility of the coding transcriptome. RNA degradation and termination of transcription are crucial in this respect.
In Saccharomyces cerevisiae, termination of RNA polymerase II (RNAPII) transcription occurs via two major pathways (Kuehner et al, 2011). Transcription termination of mRNA coding genes depends on a multi-subunit complex, composed by the Cleavage and Polyadenylation Factor and the Cleavage Factors IA and IB (hereafter referred to as the CPF complex). The CPF complex is recruited to the nascent RNA when the latter contains signals that are recognized by RNA binding subunits, among which, Hrp1p and Rna15p. Termination occurs concomitantly or shortly after cleavage of the nascent transcript and polyadenylation by the poly(A) polymerase Pap1p, which is required for subsequent export to the cytoplasm and translation (Mandel et al, 2008; Kuehner et al, 2011; Millevoi and Vagner, 2011).
The second pathway plays a central role in the control of pervasive transcription as well as in the biogenesis of sn- and snoRNAs (Steinmetz et al, 2001; Thiebaut et al, 2006; Arigo et al, 2006b; Gudipati et al, 2008). It is dependent on an essential protein complex constituted by the RNA-binding proteins Nrd1p and Nab3p and the putative helicase Sen1p (hereafter referred to as the NNS complex). The targets of the NNS complex include transcription units producing short 200–600 nt unstable RNAs dubbed CUTs (Cryptic Unstable Transcripts) (Wyers et al, 2005; Thiebaut et al, 2006; Arigo et al, 2006b). Contrary to the CPF pathway, termination by the NNS pathway is coupled to degradation of the transcript produced or trimming of the precursor in case of sn- and snoRNAs. The RNAs are polyadenylated by the TRAMP complex, containing a different poly(A) polymerase encoded by the TRF4 gene (LaCava et al, 2005; Vanacova et al, 2005; Wyers et al, 2005; Egecioglu et al, 2006), which stimulates degradation by the nuclear exosome, a complex with exo- and endonuclease activities borne by its catalytic subunits, Rrp6p and Dis3p (Lebreton and Seraphin, 2008; Schmid and Jensen, 2008; Chlebowski et al, 2011). One important specificity of the NNS pathway is that it functions almost exclusively within a window of <1000, bp after transcription initiation (Jenks and Reines, 2005; Steinmetz et al, 2006; Kopcewicz et al, 2007; Gudipati et al, 2008). This is thought to relate to the preferential interaction of Nrd1p with the RNAPII carboxy terminal domain (CTD) phosphorylated on serine 5 of its heptapeptide repeats (Ser5-P), which predominates early in transcription (Vasiljeva et al, 2008; Mayer et al, 2011; Tietjen et al, 2011).
CUTs constitute the largest share of hidden transcription in yeast, and are produced by at least as many transcription units as mRNA coding genes (Wyers et al, 2005; Davis and Ares, 2006; Houalla et al, 2006; Neil et al, 2009; Xu et al, 2009). These RNAs are widespread, generally originating from bidirectional promoters associated with mRNA coding genes. They are often found in intergenic regions and in several cases they overlap mRNA coding genes, either in the sense or in antisense orientation. Termination by the NNS complex most often prevents full transcriptional overlap, which would be disruptive. At the same time, overlapping non-coding transcription has been clearly involved in the regulation of gene expression, exemplified by the nucleotide biogenesis and glycolysis pathways (Kuehner and Brow, 2008; Thiebaut et al, 2008; Neil et al, 2009). Thus, the NNS pathway plays a pivotal role in shaping the balance between regulation and protection of the coding transcriptome.
Understanding the sequence motifs that encode termination signals is a prerequisite to decrypt the mechanism of NNS-dependent termination pathway and its impact in controlling pervasive transcription. Transcription termination by the NNS pathway critically requires the interaction of Nrd1p and Nab3p with the nascent transcript containing GUAA/G and UCUU tetranucleotides, respectively (Carroll et al, 2004, 2007; Steinmetz et al, 2006; Hobor et al, 2011; Lunde et al, 2011). Although the importance of these motifs has been clearly established in several studies with model termination substrates, their presence is not sufficient to univocally define terminators. The abundance of these motifs is highly variable among the characterized terminators, ranging from one to more than ten (Thiebaut et al, 2006; Arigo et al, 2006a; Kuehner and Brow, 2008), strongly suggesting that additional sequences and/or a particular arrangement of motifs are required for defining bona fide NNS-dependent terminators.
Because a large number of natural cryptic transcripts are known, most of which are NNS-dependent, termination signals could theoretically be extracted from these sequences. However, the existence of strong sequence biases complicates the statistical analysis. For instance, it is not trivial to define a robust background model, that is, a set of neutral elements relative to which the test set can be judged to contain over- or under-represented words. Also, the co-existence of multiple, positive or negative selective pressures complicate the recognition of specific signals in the natural genomic environment. For instance, GUAA and GUAG are indeed underrepresented in coding regions, but whether this is due to the presence of stop codons (underlined) or to their role in NNS-dependent termination remains undetermined. Similarly, intergenic regions are enriched in termination motifs, but also in regulatory signals for the initiation of transcription.
The strategy we undertook in the present study allows circumventing both limitations. We adopted an in vivo SELEX strategy using an original genetic system and selected short terminators of uniform length among a pool of random sequences. This has provided a large winning set of sequences selected exclusively on their ability to induce termination, mostly by the NNS pathway. Importantly, this strategy also provided a very robust background model in the set of non-selected sequences, allowing a very reliable statistical evaluation of the overrepresentation of sequence motifs in terminators. We identified and validated extended binding sites for Nrd1p and Nab3p as well as novel, AU-rich motifs that are also bound by the complex. NMR titration experiments revealed that Nab3p recognizes its extended site via an induced fit mechanism and allowed identifying a novel region of the protein that critically contributes to the specificity of the interaction. Importantly, we show that the overall affinity of the NNS complex for the nascent RNA is not always limiting for termination. Rather, the arrangement of sites and their association in supermotifs is critical for function. Finally, we demonstrate that the same sequence motifs can be recognized by either the NNS or the CPF complex, depending merely on the distance from the transcriptional start site. Thus, both pathways have adapted to recognize largely overlapping signals, in spite of the different protein composition and fate of the transcripts produced. These results have important implications for the mechanism of NNS complex termination and its function in the control of pervasive transcription.
Results
In vivo selection of artificial CUTs from a naive pool of sequences
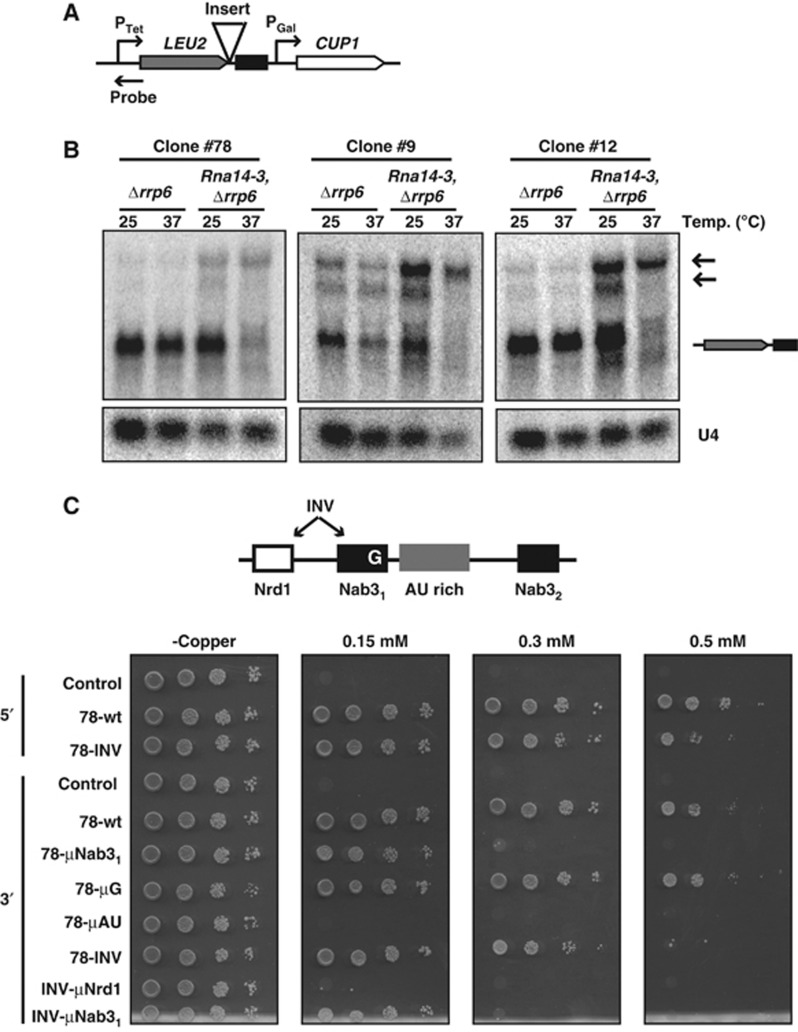
To isolate the elements dictating NNS-dependent termination from overlapping signals that might be present in natural CUTs, we devised a strategy to select for terminators from a naïve pool of sequences. We constructed a reporter system containing two strong promoters in tandem, PTet and PGal (Supplementary Figure 1A). The two promoters are separated by a test sequence and PGal drives expression of CUP1, which confers copper-resistant growth to yeast. Transcription from the upstream PTet promoter interferes with transcription from PGal, generating copper-sensitive yeasts unless the test sequence contains a terminator, which allows expression of CUP1 and copper-resistant growth (Supplementary Figure 1B–D). A pool of naïve sequences containing a random region of 120 nt obtained by chemical synthesis was introduced in the reporter system by recombination and subjected to two rounds of selection on copper-containing medium (see Materials and methods; Figure 1A and Supplementary Figure 2A).
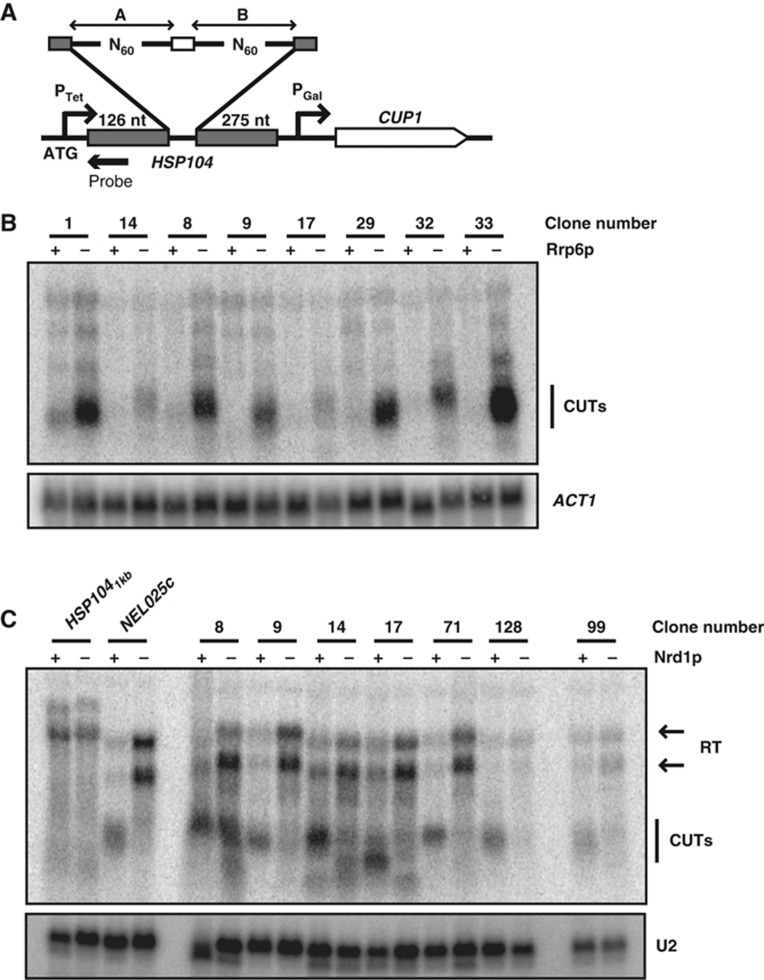
Figure 1.
In vivo selection of transcriptional terminators. (A) Scheme of the reporter construct used for the selection. The structure of the random sequences is shown on the top. A white box indicates a short central constant region, introduced for cloning purposes. The position of the probe used for the northern analyses in (B) and (C) is marked by an arrowhead. (B) Northern blot analysis of selected clones in a wild-type or Δrrp6 strain as indicated. Short unstable transcripts resulting from transcription termination at the inserted sequences are labelled CUTs. ACT1 mRNA is used as a loading control. (C) Northern blot analysis of selected clones to assess NNS complex dependency. Analysis was performed in a PGal-NRD1, Δrrp6 strain, grown either on galactose (‘+’ lanes) or on glucose for 6 h (‘−’ lanes) to deplete Nrd1p. Small unstable transcripts derived from termination at the selected region (CUTs) are observed in the presence of Nrd1p. HSP1041 kb contains the first 1 kb of HSP104 coding sequences as a negative control for termination. As a positive control, we used sequences from the NEL025c CUT that induces NNS complex-dependent termination. Transcriptional read-through (RT) at the selected terminators (or the control) produces transcripts that terminate at the downstream CUP1 terminator or a cryptic terminator in the GAL1 promoter (marked by two arrowheads). The U2 RNA is used as a loading control. Figure source data can be found with the Supplementary data.
Sequencing of roughly 130 inserts from the selected pool allowed defining two classes of potential terminators. Approximately 70% of the selected sequences were enriched in the previously identified Nrd1p- and Nab3p-binding sites, suggesting that these are NNS-dependent terminators (Supplementary Figure 2B). The other class of sequences contained a different set of motifs that will be described elsewhere (Colin et al, in preparation). We will hereafter only refer to the sequences belonging to the first class.
Northern blot analyses largely validated the predominant occurrence of NNS complex-dependent termination in the selected clones. We observed expression of the short and functional CUP1 transcript in all the constructs analysed but not in a negative control containing a copper-sensitive clone (data not shown). Short transcripts driven by PTet and terminating upstream of PGal were specifically observed in the selected clones. These RNAs were generally strongly stabilized in the absence of the nuclear exosome subunit Rrp6p (Figure 1B and data not shown), confirming the production of unstable transcripts, which is characteristic of the NNS pathway. Importantly, termination of these clones was affected by metabolic depletion of Nrd1p (Figure 1C) leading to the appearance of long read-through transcripts.
Taken together, these results indicate that the pool of selected sequences is strongly enriched for artificial, fully functional CUT-like NNS-dependent terminators.
Statistical analyses of artificial CUTs lead to the identification of putative new motifs involved in NNS-dependent termination
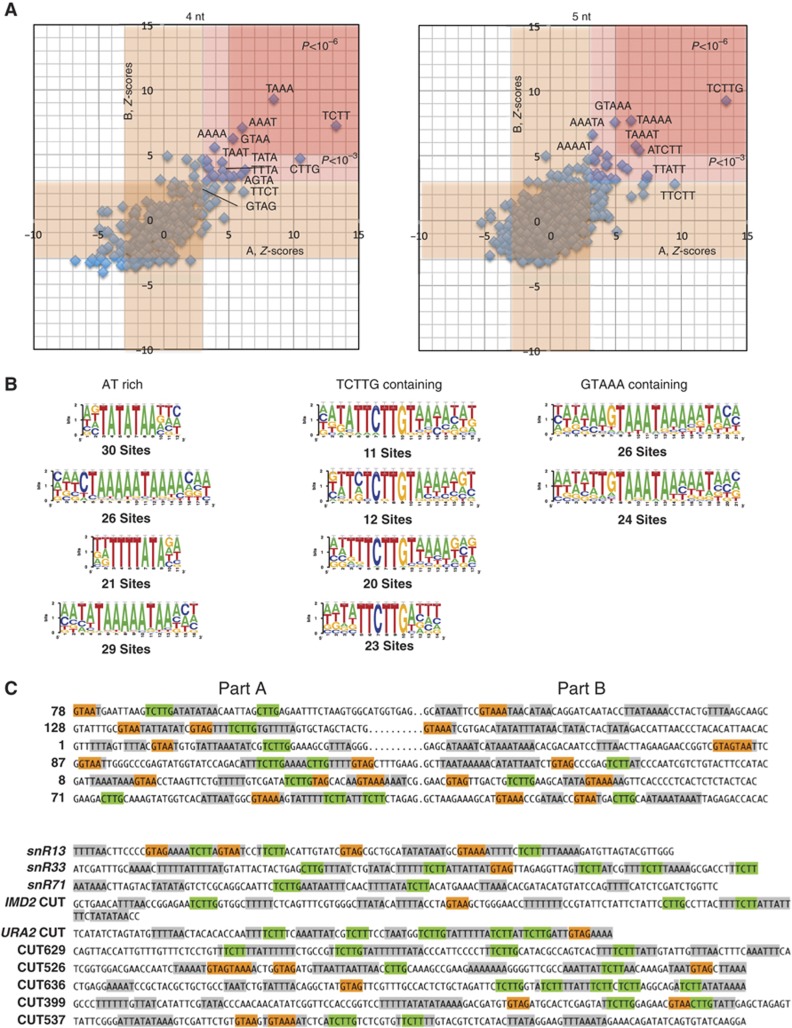
We bulk sequenced the inserts with a paired ends protocol as described in Materials and methods. The presence of overrepresented motifs was statistically evaluated relative to the robust background model provided by the pool of non-selected sequences using RSAT (van Helden, 2003). After filtering, ∼700 selected clones were analysed against a neutral set of roughly 8000 non-selected sequences. Since the starting pool of random sequences contains two regions with a different sequence bias (Figure 1A, regions A and B; see Materials and methods) we evaluated the statistical significance of overabundant motifs of four and five nucleotides separately in the two regions. The selection nicely converged towards a common set of motifs found in the two regions. As expected, the known sites (UCUU, GUAA and GUAG) recognized by Nrd1p and Nab3p (Carroll et al, 2004) were all significantly overrepresented (Figure 2A; Supplementary Figure 3), although GUAG was less prominent, possibly suggesting a minor role for this motif. The highly significant abundance of the UCUU-overlapping tetranucleotide CUUG in both sets suggested that the two motifs are part of an extended motif (UCUUG), which was confirmed by pentanucleotide analysis (P<3 × E−14). This extended motif is by far the most prominent feature detected in the set of artificial terminators but also in natural CUTs (P=7.5 × E−44) and is markedly excluded in protein coding regions (P=1 × E−46, see Materials and methods). No strong preference was observed for the nucleotide preceding UCUU, while GUAA sites followed by an A and preceded by an A or a U were significantly enriched over other GUAA-containing pentanucleotides (Supplementary Figure 3), suggesting that (A/U)GUAAA represents an extended binding site for Nrd1p.
Figure 2.
Statistical analysis of the motifs overrepresented in the selected terminators. (A) Dispersion plot of the Z-values for all possible tetra- and pentanucleotides in the two regions (A, x axis and B, y axis) of the selected sequences. Z-values are calculated by RSAT relative to the observed frequency distribution of tetra- and pentanucleotides in the non-selected sequences, determined separately for the A and B regions due to the different sequence bias in the starting pool. Two red zones indicate P-values associated to the Z-scores that are respectively lower than 10−3 (light red) or 10−6 (darker red). Exact values for the most represented oligonucleotides are indicated in Supplementary Figure 3. A light brown region indicates a Z-score range of 3 around the centre of the distribution. The sequence of the oligonucleotides with the highest Z-scores is indicated on the graphic. (B) Sequence logos of extended motifs determined by pattern assembly. RSAT uses the most frequent overlapping oligonucleotides to generate larger motifs. Logos are determined based on the occurrence of these supermotifs in the sequences analysed. The number of sequences used to generate the logos is indicated below each logo. Supermotifs were classed according to the presence or absence of Nrd1p- or Nab3p-binding sites or AU-rich motifs. (C) Examples of sequences containing supermotifs. Previously identified Nrd1p- and Nab3p-binding sites as well as their new extended versions are indicated in orange and green, respectively, while the AU-rich motifs are shown in grey. The first set of sequences (numbered) represents clones that have been validated by northern analyses. The second set of sequences includes natural known NNS-dependent terminators, including well-characterized snoRNAs (SNR13, SNR33 and SNR71) and CUTs (IMD2 and URA2 CUTs) and other CUTs identified in a previous genome-wide analysis (Xu et al, 2009).
Surprisingly, AU-rich motifs were strongly overrepresented in parts A and B of artificial CUTs to the same extent as Nrd1p and Nab3p canonical or extended sites (Figure 2A; Supplementary Figure 3). For instance, the sequences UAAA and AAAU were generally enriched to a higher extent (P<E−8) than the bona fide Nrd1p-binding sites GUAA and GUAG. This finding strongly suggests that AU-rich motifs are major determinants for termination by the NNS pathway. To assess the possible functional association of these motifs, we performed a pattern assembly analysis using RSAT (Figure 2B and C). The program assembles overlapping motifs that are statistically overrepresented to obtain larger consensus regions and generates logos based on occurrences in the set of sequences analysed (van Helden, 2003). Besides extended AU-rich regions, we observed a significant association between the UCUUG or the GUAA sequences and AU motifs, suggesting that proximity between these sites is functionally relevant within larger termination supermotifs. Importantly, such motifs are frequently observed in natural sequences including the well-characterized SNR13 and SNR33 terminators or the IMD2 and URA2 CUTs (Figure 2C).
Overall, these analyses point to the sequences UCUUG and A/UGUAAA as the main Nrd1p–Nab3p binding sites. They also strongly point to the existence of supermotifs as major determinants for NNS-dependent termination.
Mutational analysis of an artificial CUT confirms the role of the new motifs in termination by the NNS pathway
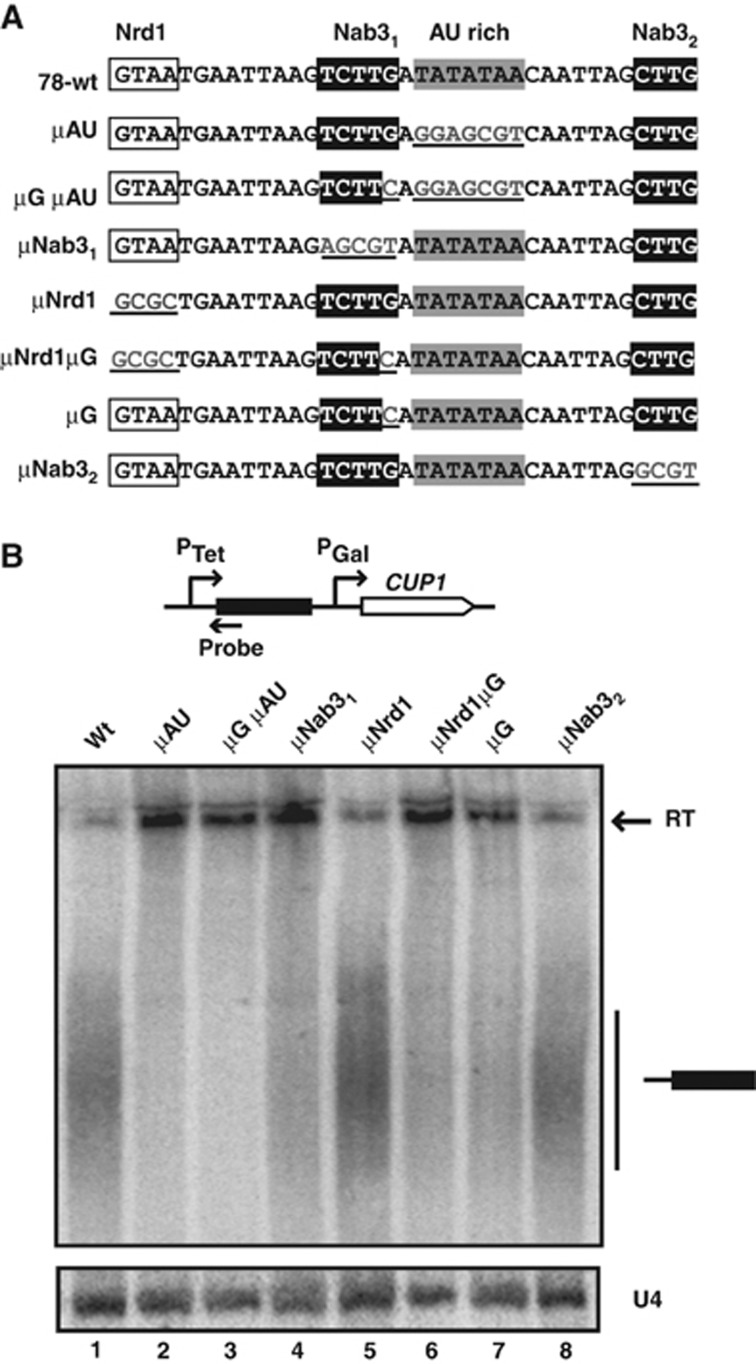
The analyses presented above suggested that specificity for transcription termination by the NNS complex might rely on longer and more complex arrangements of sites. We therefore undertook a mutational analysis of one of our artificial CUTs (clone #78) containing some of the selected motifs clustered in a short region that is necessary and sufficient for NNS-dependent termination (Supplementary Figure 4). This region contains a Nrd1p-binding site (GUAA), two variants of the Nab3p-binding site (UCUUG, Nab31 and CUUG, Nab32) and an AU-rich motif (UAUAUAA) (Figure 3A). Importantly, the AU-rich motif is located immediately downstream of the Nab31-binding site, defining a putative supermotif.
Figure 3.
Mutational analysis of the sequence motifs in clone 78. (A) Position of the motifs and sequence of the mutations introduced. (B) Northern blot analysis to detect termination induced by the different constructs. Transcripts were separated by 6% PAGE for better resolution of the short species. Read-through transcripts (RT) are indicated by a black arrow. Note that the two read-through species shown in Figure 1 are not resolved in this experimental set-up. The small RNA U4 was used as a loading control. Figure source data can be found with the Supplementary data.
Interestingly, among the three binding sites for the NNS complex, only the extended Nab31 site, UCUUG, was required for termination as assessed by copper resistance assays and northern blot analysis of mutated constructs (Figure 3B; Supplementary Figure 5, compare constructs μNrd1, μNab31 and μNab32). Importantly, the G nucleotide extending the Nab31 site was essential in this context as its mutation alone (μG) led to a termination defect similar to that observed for μNab31 (Figure 3B, compare lanes 1, 4 and 7). Finally, mutation of the AU-rich motif alone (μAU) dramatically impaired termination (Figure 3B, compare lanes 1 and 2), indicating that this sequence plays an essential role in NNS-dependent termination. The double mutants tested did not worsen the termination defects observed in single mutants (Figure 3B, lanes 3 and 6), even when the latter were both defective (i.e., μGμAU), likely indicating a threshold effect. Together, these results indicate that the G extending the Nab31 site and the AU-rich motif are important elements for NNS-dependent termination. Also, they indicate that the binding sites for the NNS complex are not all functionally equivalent.
The arrangement of sequence motifs dictates the efficiency and specificity of a terminator
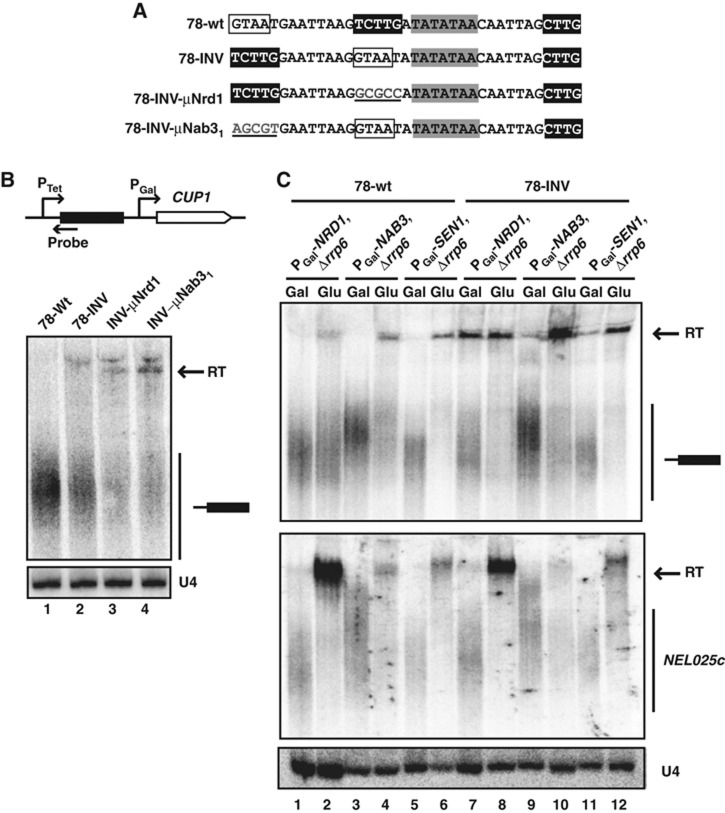
The latter results might suggest that the impact of a given motif on termination is context dependent and that, in the appropriate configuration, robust termination can be induced by a few ‘strong’ elements. Because only the Nab31 site is indispensable for termination, and this site is associated to the AU-rich motif in one supermotif (Figure 2), we set out to investigate whether this proximity defines a termination-proficient context. To this end, we first inverted the positions of the Nrd1 and the Nab31 sites in construct 78-INV (Figure 4A) and asked whether the acquired proximity with the AU-rich sequence makes the Nrd1 site functionally important for termination. Note that in this novel configuration the two sites constitute a GUAA-AU, supermotif (Figure 2). The efficiency of termination in construct 78-INV was only slightly diminished relative to construct 78 (Figure 4B, lanes 1 and 2). Strikingly, however, when the Nrd1-binding site was mutated in this position (78-INV-μNrd1), termination was impaired (Figure 4B, lanes 2 and 3, Supplementary Figure 5), indicating that this site becomes significant when associated to the AU-rich motif. Consistent with this notion, mutation of the AU-rich motif in this context completely abolishes termination (Supplementary Figure 5). Interestingly, the Nab31 site set apart from the AU-rich region retained some functionality, as it was still necessary for efficient termination (Figure 4B, lanes 2 and 4).
Figure 4.
Impact of the arrangement of termination motifs on termination. (A) Sequence of the terminator variants analysed. (B) Northern blot analysis of RNAs derived from the constructs indicated. Read-through transcripts (RT) are indicated by a black arrow. (C) Northern blot analysis of constructs 78-wt and 78-INV upon metabolic depletion of the different components of the NNS complex. Cells were grown on glucose for 6 h (to deplete Nrd1p) or 14 h (to deplete Nab3p and Sen1p). Detection of the transcripts expressed from PTet is shown on the top. Expression of the NEL025c CUT (bottom) was monitored to verify the correct depletion of the proteins. The small RNA U4 was used as a loading control.
The above results predict that substrates with different functional sites should be differentially sensitive to impairment of either Nrd1p or Nab3p function. For instance, clone 78 (that does not contain functional Nrd1 sites) is expected to be less dependent on Nrd1p while 78-INV (containing Nrd1 and Nab3 functional sites) should be dependent on both proteins. Indeed, metabolic depletion of Nrd1p impaired recognition of the 78-INV terminator or the endogenous NEL025c as a control but only affected to a limited extent termination of clone 78 (Figure 4C, lanes 1–2 and 7–8). Nrd1p depletion was effective as very low to undetectable levels of protein were observed after 2 h of metabolic depletion and longer depletion times did not further affect termination (Supplementary Figure 6 and data not shown). Conversely, termination induced by both 78 and 78-INV sequences was markedly dependent on Nab3p (Figure 4C, lanes 3, 4, 9 and 10). Finally, metabolic depletion of Sen1p impinged similarly on termination of both constructs, consistent with the notion that this factor is required for termination but not for the recognition of termination signals (Figure 4C, lanes 5, 6, 11 and 12).
These findings confirm the importance of the supermotifs containing canonical NNS complex binding sites associated with AU-rich regions. This underscores the notion that the arrangement of termination motifs, and not their mere presence on the nascent transcript, determines the ‘strength’ and specificity of NNS-dependent terminators.
The AU-rich motif is recognized by the NNS complex in vitro
The prominent enrichment of AU-rich motifs in artificial CUTs (Figure 2), together with our mutational analysis (Figure 3) defines these elements as novel termination signals for the NNS pathway. The CPF-CF components Hrp1p and Rna15p recognize similar sequences (Kessler et al, 1997; Valentini et al, 1999; Gross and Moore, 2001) and have been involved, directly or indirectly, in termination of snoRNA (Fatica et al, 2000) and CUTs (Kuehner and Brow, 2008). It was important to assess whether in vivo these two proteins recognize the AU-rich element in the context of the NNS pathway. Therefore, we performed northern blot analysis of RNAs derived from clone 78 expressed in PGal-HRP1, Δrrp6 or rna15-3, Δrrp6 strain. As shown in Supplementary Figure 7, no significant effect on termination was observed upon metabolic depletion of Hrp1p or temperature inactivation of Rna15p, while termination of the SUA7 gene was clearly impaired in the same conditions, indicating that inactivation of both proteins was effective. A similar experiment performed with a PGal-PAB1, Δrrp6 also showed that depletion of Pab1p, a poly(A) binding protein, did not impinge on the recognition of the terminator (data not shown). Therefore, the impact of AU-rich elements in NNS complex-dependent termination does not relate to a possible role of Hrp1p, Rna15p or Pab1p in this pathway.
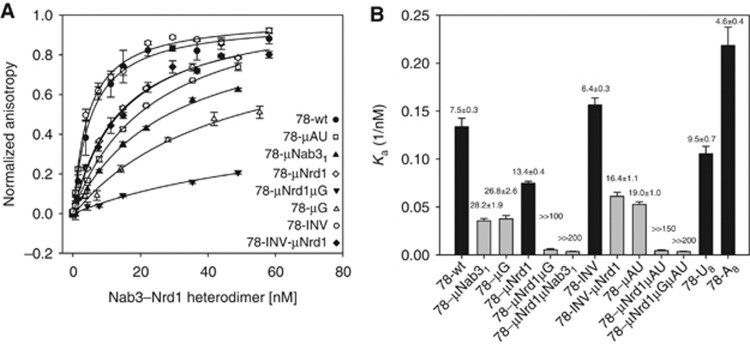
To assess whether the NNS complex recognizes this element directly, we performed fluorescence anisotropy (FA) binding assays using recombinant Nrd1p–Nab3p heterodimer (Carroll et al, 2007) and synthetic RNA versions of clone 78 or its mutant derivatives (Figure 5A and B). As expected, mutation of the Nrd1 and Nab31 sites significantly decreased the affinity of the NNS complex for the RNA. Importantly, mutation of the AU-rich motif to a randomly chosen sequence (clone μAU) or an (AC)4 motif (data not shown) affected binding similarly to mutation of the Nrd1 site, indicating that this sequence is an important determinant of the interaction, at least in vitro. Replacing the AU-rich motif in clone 78 with a stretch of As (78-A8) or Us (78-U8) restored both termination and wild-type interaction with the complex relative to the μAU construct (Supplementary Figure 8; Figure 5).
Figure 5.
Fluorescence anisotropy assessment of Nrd1p–Nab3p heterodimer binding to wild-type and mutant terminators. (A) Binding isotherms for equilibrium binding of Nrd1p–Nab3p heterodimer to various mutants of clone 78, monitored by FA. In all, 10 nM fluorescently labelled RNA was titrated by Nrd1p–Nab3p heterodimer, the data were fitted using a single-site binding model, the data were normalized for visualization purposes. (B) Summary of association constants (Ka) derived from FA affinity measurements and termination proficiency for the wild-type and mutant constructs. The ionic strength and pH of the binding buffer were the same for all the measurements. The Ka values are derived from the equilibrium dissociation constants (Kd, indicated on top of every histogram, nM) calculated from the best fit to the data using a single-site binding isotherm (Heyduk and Lee, 1990). Black bars: termination proficient; grey bars: termination-deficient sequences. Each data point represents triplicate assays (error bars represent standard deviation).
Combining several mutations strongly decreased binding, indicating that the different motifs all contribute independently to the interaction. However, the decrease in affinity upon mutation of the different sites correlates only to a limited extent with the efficiency of termination. For instance mutations μNrd1 and μAU affect similarly binding to Nrd1p–Nab3p (Figure 5), which does not reflect the radically different behaviour of these sequences in termination assays (Figure 3A and B; Supplementary Figure 5). Also, sequences 78-μNrd1 and 78-INV-μNrd1 bound the complex with virtually identical affinity (Figure 5A and B), yet only 78-μNrd1 is able to terminate transcription (Figures 3B and 4B).
These results indicate that the NNS complex recognizes directly the AU-rich termination motif. They also strongly suggest that above a given threshold, the overall affinity of the complex for the nascent RNA is not the limiting factor for termination.
Interaction with the 3′ guanine extension of the extended Nab3p-binding site remodels the surface of Nab3p RNA-recognition motif
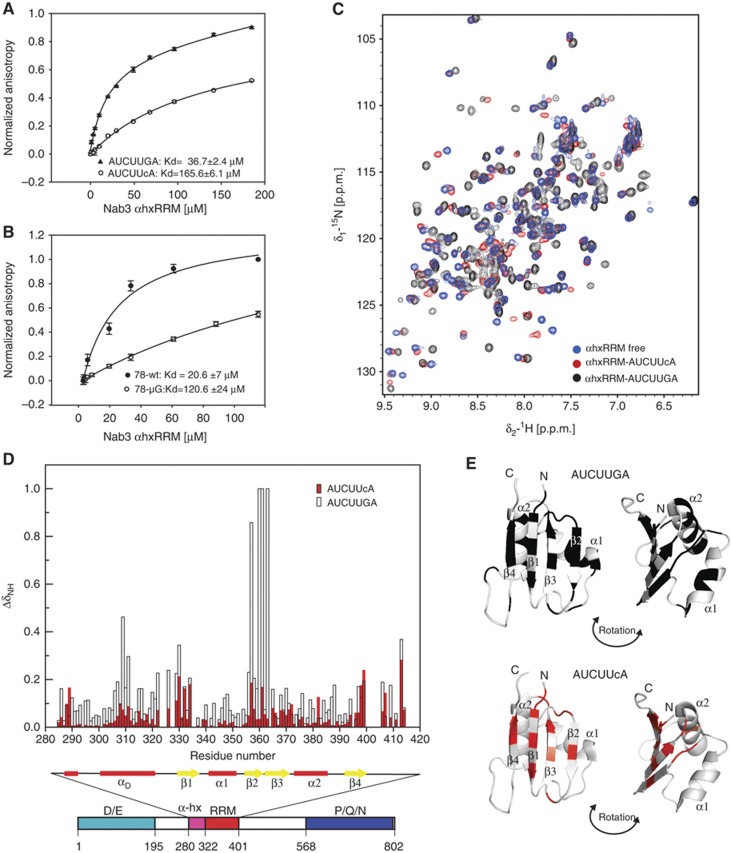
The strong impact on termination of the 3′-end guanine extension of the Nab3-binding site (Figure 3) prompted us to evaluate its contribution to NNS complex binding. Interestingly, mutation of the G alone (78-μG) caused the same decrease in affinity as the μNab31 mutation (Figure 5), suggesting that this nucleotide plays a critical role in RNA recognition by Nab3p. However, this is surprising in the light of the previously determined structure of the RNA-recognition motif (RRM) of Nab3 in complex with 5′-UCUU-3′ RNA (Hobor et al, 2011; Lunde et al, 2011), which revealed that the Nab3 RRM (321–415) specifically recognizes only the first three nucleotides of the UCUU substrate. Therefore, we undertook a more detailed structural analysis of the recognition of this nucleotide by Nab3p employing NMR spectroscopy.
The Nab3 RRM (321–415) construct previously used in structural studies showed virtually the same binding affinity to UCUU and UCUUG (data not shown) and displayed the same 1H-15N HSQC spectra of Nab3p RRM when bound to UCUU or UCUUG (Supplementary Figure 9). This indicates that the last G of UCUUG is not recognized by this Nab3p domain, yet it contrasts with the FA measurements obtained using the Nrd1-Nab3 heterodimer reported above (Figure 5). Since the latter experiments were performed with a larger Nab3p fragment, new constructs were designed with N- and C-terminal extensions to the RRM. We found that a construct containing a 40 amino-acids N-terminal extension of the RRM, encompassing a long α-helix (amino acids 283–415, referred to as αhxRRM throughout the text) binds to UCUUG with significantly higher affinity (37±2 μM versus 170±8 μM) than the original RRM construct (amino acids 321–415, data not shown). The UCUUc mutant displayed a significant drop in the affinity, confirming the importance of 3′-end guanine of the Nab3-binding site for recognition by αhxRRM (Figure 6A). Akin to binding experiments with short RNA substrates, identical results were obtained in the context of clone 78 (Figure 6A and B). Importantly, NMR titration experiments showed a very specific interaction between UCUUG and αhxRRM as evidenced by large perturbations of chemical shifts in the 1H-15N HSQC spectra of αhxRRM (Figure 6C and D). The largest perturbations occur in the distal N-terminal α-helix (αD) and the β2-strand of the αhxRRM (Figure 6D). Mapping these perturbations on the previously determined structure of the Nab3 RRM (Hobor et al, 2011) show that the binding of UCUUG provides additional changes at the β2-strand and α-helices when compared to the binding of UCUUC (Figure 6E). This strongly indicates that the recognition of the G-containing Nab3 termination site involves an induced fit mechanism, in which αD helix and the flanking regions are rearranged on the canonical RRM upon the RNA binding.
Figure 6.
Specific recognition of the 3′-terminal guanine extension by an extended Nab3p RNA recognition domain. Equilibrium binding of αhxRRM with: (A) AUCUUGA and AUCUUcA and (B) clone 78, wild-type and μG mutant, monitored by FA measurements (the data were normalized for visualization purposes). Values correspond to the average of three independent experiments (error bars represent standard deviation). (C) 1H-15N HSQC NMR spectra of Nab3 αhxRRM alone (in blue) and in the presence of 1 eq of 5′-AUCUUGA-3′ (in black) or 1 eq of 5′-AUCUUcA-3′ (in red) measured on 600 MHz spectrometer at 303 K. The conformation of αhxRRM undergoes significant changes upon binding to AUCUUGA as reflected in the chemical shift changes when titrated with AUCUUGA RNA. The chemical changes are less significant upon binding to AUCUUcA. (D) Quantification of chemical shift perturbations of Nab3 αhxRRM upon binding to AUCUUGA (black) or AUCUUcA (red) RNA. The combined chemical shift perturbations (([wHNΔδHN]2+[wNΔδN]2)1/2), where wHN=1 and wN=0.154 are weight factors of the nucleus, are plotted versus the residue numbers and schematic figure of secondary structure elements. Significant changes occur in the β1 and β2 strands and in the distal α-helical element (αD). (E) Chemical shift changes (Δδ) upon RNA binding mapped to the surface of the Nab3 RRM structure (Hobor et al, 2011). The upper part shows Δδ of αhxRRM with AUCUUGA RNA (black), and the lower part shows Δδ of αhxRRM with AUCUUCA RNA (red). Only Δδ>0.075 are displayed.
These results provide a mechanistic explanation for the importance of the 3′-end guanine extension of the Nab3-binding site. They also allow defining an additional motif in the Nab3p RRM that mediates a conformational change occurring upon the specific recognition of the conserved G of the Nab3-binding site.
Overlapping recognition of termination signals by the NNS- and the CPF-dependent pathways
It has been previously shown that natural CUTs can be used as terminators by the CPF-dependent pathway when mislocalized towards the 3′end of longer transcriptional units (Kopcewicz et al, 2007; Gudipati et al, 2008; Kuehner and Brow, 2008). This either suggests that the two termination pathways recognize identical or overlapping signals, or that the natural NNS terminators analysed in the previous studies are ‘polluted’ by independent CPF/CF non-overlapping termination signals (e.g., derived from other transcription units). Artificial NNS-dependent terminators provide a unique opportunity to distinguish between these possibilities. Clone 78 is particularly informative in this respect since it is insensitive to mutation or depletion of critical CPF/CF components (Supplementary Figure 7). This terminator is therefore unlikely to contain signals selected by virtue of a CPF-dependent selective pressure. Therefore, we set out to test whether termination signals selected based on NNS dependency could be used by the CPF pathway when moved away from the transcriptional start site. For this purpose, we modified the reporter system by inserting a 1.1-kb ORF (LEU2) between the PTet and PGal promoter and cloned the sequence of clone 78 and two additional artificial, NNS-dependent terminators immediately downstream of this ORF (Figure 7A). These terminators that are NNS dependent in a promoter proximal position also induced termination when localized at the 3′-end of the 1.1 kb LEU2 gene (Figure 7A and data not shown). However, in this position stable RNAs were produced and termination was no longer dependent on the NNS pathway (Figure 7A; Supplementary Figure 10). Rather, termination was impaired by heat inactivation of the thermosensitive mutant Rna14-3p, which is an essential component of the CPF complex (Figure 7A). These results indicate that signals selected for NNS-dependent termination can be also recognized by the CPF complex in the appropriate context. Importantly, mutation of most signals that impaired NNS-dependent termination also markedly affected termination induced by the CPF pathway, indicating considerable overlap for substrate recognition by the two pathways. Interestingly, mutation of the G extending the Nab31 site was ineffective in a promoter distal location (Figure 7C, 78-μG 3′), further supporting the notion that this particular nucleotide is a major determinant of the specific recognition of termination substrates by the NNS complex. Together, these results strongly suggest that the two major yeast termination pathways recognize very similar motifs in their substrates. Recognition of these motifs is most likely operated by different factors depending on the distance from the transcriptional start site.
Figure 7.
The NNS and the CPF pathways have largely overlapping sequence determinants. (A) A scheme of the reporter construct containing the 1.1-kb LEU2 gene inserted between the PTet and the PGal promoters. Artificial CUT sequences were cloned by recombination downstream of LEU2 as indicated. A stuffer fragment from the HSP104 gene (486 nt) is present after the terminators insertion point as polymerases committed to CPF-dependent termination are known to transcribe downstream of the terminator. (B) Northern blot analysis of RNAs derived from clone 78 and two additional artificial terminators cloned downstream of LEU2. These constructs were expressed in an rna14-3 strain to assess the dependency on the CPF pathway. A Δrrp6 strain was employed to stabilize read-through transcripts generated by mutation of Rna14p. (C) Copper growth assay of cells expressing the constructs containing the artificial terminators cloned downstream of LEU2 as indicated (3′). A scheme indicates the positions of the relevant motifs. Sequences are reported in Figures 3A and 4A. For comparison, clones 78 and 78-INV inserted in a 5′ position (inducing termination by the NNS pathway) are also included in the test set as indicated.
Discussion
The emerging concept that transcription occurs pervasively irrespective of canonical gene borders (Jacquier, 2009) raises the important question of how the cell ‘protects’ regulatory regions from invasive polymerases. ‘Wild’ polymerases have to be controlled because potentially disruptive for the expression of neighbouring genes and genome stability. Transcription termination operated by the NNS complex plays a role of utmost importance in this respect.
This work has been inspired by the consideration that the sequence motifs recognized by the NNS complex are seemingly insufficient to provide the required specificity and efficiency to the process. From our very sensitive in vivo SELEX approach we revealed extended Nrd1p- and Nab3p-binding sites together with novel, AU-rich motifs. We show that transcription termination only to a limited extent correlates with the affinity of NNS complex binding and that the arrangement of motifs is a critical parameter. Importantly, Nab3p binds its extended site by an induced fit mechanism whereby a novel module distinct from the canonical RRM specifically recognizes a strongly conserved G in the Nab3 site. Finally, we show that the same sequence motifs selected for NNS-dependent termination can also be recognized by the CPF pathway indicating that the two termination pathways have adapted to recognize largely overlapping signals in spite of the very different factors involved and the different fate of the RNAs produced.
In vivo selection of artificial terminators
Our in vivo SELEX strategy overcomes many of the limitations inherent to the search for termination motifs in natural CUTs. First, we could use a robust neutral model by estimating background words frequencies in a non-selected pool of >8000 sequences. Second, sequences were specifically selected for their ability to induce transcription termination, thus limiting alternative selective pressures generating motifs that might pollute the statistical analyses. From our analysis of the winning pool we can roughly estimate the ‘evolutionary cost’ to generate a functional, NNS-dependent terminator. This could be a difficult task for the disperse nature of the information present in these terminators. However, we also selected another class of terminators that contains a single motif of 8 nt that is necessary and sufficient for termination (Colin et al, in preparation). Because the two classes of terminators have approximately equal abundance, the informational content of NNS-dependent terminators is equivalent to ‘fixing’ 8 contiguous nucleotides, that is, roughly 16 bits of information (Schneider et al, 1986). Since NNS terminators contain split motifs, this is consistent with the notion that more than two sites of four nucleotides are required to induce termination.
Statistical analysis of overrepresented motifs indicates that the GUAG sequence, a known Nrd1p-binding site, was enriched to a lesser extent relative to the other binding sites of the complex, indicating that this particular motif is less important for binding or termination than GUAA (Nrd1p binding) or UCUU (Nab3p binding). We also found that these tetranucleotides are actually part of extended binding sites, U/AGUAAA and UCUUG, respectively, considerably increasing the informational content of individual termination motifs. These extended sites are similar but not identical to the consensus derived from in vivo crosslinking data (respectively UGUAG and GNUUCUGU for Nrd1p and Nab3; Creamer et al, 2011). The differences might pertain to crosslinking biases (which tend to favour Us) or to the different neutral model used to evaluate the statistical significance as discussed above. The presence of a G extending the Nab3 site was first noticed in experiments detecting RNAs associated with the NNS complex (Hogan et al, 2008) and confirmed in more recent crosslinking approaches (Creamer et al, 2011; Wlotzka et al, 2011). We show here that mutation of the G extending the Nab3 site is highly disruptive both for termination and for NNS complex binding. Importantly, we provide a mechanistic explanation for this effect by showing that this nucleotide is specifically recognized by long N-terminal α-helical extension of the Nab3p RRM and that binding provokes changes in the structure of the protein. These structural changes impact the strength and the geometry of the interaction and might explain why CUUG is strongly preferred as a Nab3p crosslinking site relative to UCUU (Wlotzka et al, 2011). It is certainly possible that the structural perturbations induced by RNA binding, besides affecting the strength and specificity of the interaction, are transmitted to other domains of the protein, allosterically altering the function of the complex in termination.
AU-rich motifs are prominent NNS complex-dependent termination signals
Remarkably, we found that AU-rich motifs are strongly overrepresented and functionally important in our set of synthetic terminators to similar or higher levels than ‘canonical’ Nrd1p-binding sites. We provide important clues on the mechanistic impact of this motif in termination. We show that it cannot be ascribed to recognition by Hrp1p or Rna15p, two proteins involved in the CPF pathway and known to bind AU-rich motifs (Kessler et al, 1997; Valentini et al, 1999; Gross and Moore, 2001; Mandel et al, 2008; Supplementary Figure 7). Rather, this motif contributes to NNS complex binding to a higher extent than a GUAA Nrd1p-binding site (Figure 5) and can be substituted, both for binding and for termination, by stretches of As or Us. This strongly suggests that recognition of AU-rich termination signals depends on the NNS complex, although we cannot exclude that another factor also recognizes these motifs concomitantly or sequentially in vivo.
These findings are relevant to the function of natural terminators. One of the CUTs generated by upstream transcription initiation at the IMD2 locus is paradigmatic in this respect. This strong NNS pathway terminator (starting at position −67 relative to the IMD2 AUG) (Kuehner and Brow, 2008) contains only one UCUU motif that is necessary but not sufficient for termination. However, it also contains an upstream CUUG motif and a downstream, prominent, AU-rich region, whose sequence is identical to that selected in clone 78. Importantly, random mutations that affect termination were identified within these motifs (Kuehner and Brow, 2008 and our unpublished results), which was previously unexplained and can now be rationalized in the light of our results.
The high AT richness of intergenic regions (roughly 66%) is thought to contribute to the establishment of nucleosome-free regions (NFRs), from where transcription generally originates. We suggest that the AT richness of NFRs also has an additional role in providing a favourable background for termination signals that could be generated at a low evolutionary cost. Thus, the same sequence background would favour the generation of transcription and protect it from interference due to ‘invading’ polymerases. The unexplained occurrence of 3′ NFRs between convergent genes (Neil et al, 2009; Xu et al, 2009) could even be due to a secondary effect of the AT richness of termination signals.
Significance of termination supermotifs
Our analysis of synthetic terminators reveals the statistically significant presence of supermotifs containing canonical NNS complex binding sites associated to AU-rich motifs and we show by mutational analysis that this association is functionally important. It is possible that the close proximity of the AU-rich motif favours cooperative binding of the complex to the RNA as previously suggested for closely positioned Nrd1 and Nab3 sites (Carroll et al, 2007). However, our experiments also indicate that the overall affinity of the complex for the terminator is not necessarily limiting for termination. Indeed, constructs 78-μNrd1 and 78-INV-μNrd1 bind the complex with very similar affinity (Figure 5), yet only the latter (in which the association Nrd1 site-AU-rich motif is lost) cannot induce transcription termination. This indicates that it is the local environment determined by a critical arrangement of motifs that favours termination, rather than the global load of NNS complex on the nascent RNA. ‘Hot spots’ of termination could therefore be determined by the local high affinity recognition of the supermotif, possibly in association with additional factors.
It is also possible that termination requires an additional or alternative read-out of the AU-rich motif than simple NNS complex binding, possibly by the elongating polymerase. For instance, it is possible that AT-rich regions induce RNAPII pausing and that the close juxtaposition of a pausing element with protein binding on the nascent RNA elicits termination. This would be reminiscent of the mechanism of intrinsic termination in bacteria whereby a T-stretch inducing the pause is immediately preceded by a GC-rich hairpin. Base pairing within the stem disrupts critical polymerase–nucleic acid contacts in the elongation complex and induces termination (Peters et al, 2011). Binding of the NNS complex might be analogous to hairpin formation in sequestering the sequence immediately upstream of the polymerase.
Termination supermotifs might be essential for short CUTs, as for the IMD2 and URA2 CUTs (Figure 2), and the small size of our artificial CUTs might have favoured the prominent selection of these motifs. We suggest that, in the absence of termination supermotifs, multiple, less efficient termination event occur independently, the combination of which is required to fully prevent polymerase read through. This might explain the requirement for multiple NNS complex binding sites previously demonstrated for NEL025c and autoregulation of the NRD1 gene (Thiebaut et al, 2006; Arigo et al, 2006a).
Overlapping termination signals are recognized by the CPF and NRD1 pathway
Because most of our artificial terminators led to the production of Rrp6-sensitive transcripts and were sensitive to the NNS pathway, it is unlikely that CPF-dependent terminators are significantly present in our winning set. We suggest that these results from the predominance of the position effect that strongly favours NNS complex-dependent termination proximally to the transcription start site (Jenks and Reines, 2005; Steinmetz et al, 2006; Kopcewicz et al, 2007; Gudipati et al, 2008). However, in spite of a selective pressure favouring NNS complex-dependent termination (as verified for clone 78), these terminators also contain CPF signals as they are recognized by the CPF complex when re-localized at >1 kb from the transcription start site (Figure 7). Natural NNS-dependent terminators can be recognized by the CPF complex when mislocalized (Jenks and Reines, 2005; Steinmetz et al, 2006; Kopcewicz et al, 2007; Gudipati et al, 2008). However, whether these natural sequences contain independent and non-overlapping signals directing termination by either pathway is unclear, and even suggested in the case of the SNR13 terminator (Steinmetz et al, 2006). Strikingly, we show that termination signals for the two pathways largely overlap. Indeed, not only the AU-rich region is required for CPF-dependent termination (which could have been expected) (Gross and Moore, 2001; Proudfoot, 2011) but also the integrity of the Nrd1 and Nab3 sites. The only exception is mutation of the G residue mediating Nab3p-specific contacts, further underscoring the functional importance of this residue. This finding is remarkable in the light of the different machineries involved in the two pathways and the different fate of the RNAs produced. It is theoretically possible that a common ‘recognition module’ exists that is shared by the two termination complexes. However, this is not supported by experimental evidence since, with the exception of a few pcf11 alleles, mutations in factors belonging to one given pathway do not affect significantly the other (Kim et al, 2006). We suggest that the two termination pathways have adapted independently to recognize highly similar signals, possibly converging on sequences that alter the processivity of the polymerase or favour its propensity to terminate. The fact that termination signals are bi-functional has important functional implications for the control of pervasive transcription. They would constitute efficient transcriptional insulators to halt both polymerases initiating in the immediate vicinity, for example, producing 5′ or 3′ ORF-overlapping CUTs, as well as polymerases deriving from remote initiation events, for example, reading through termination signals of neighbouring ORFs. Use of the same signals for both termination mechanisms would be highly economical in terms of evolutionary cost for a compact genome as that of S. cerevisiae.
Materials and methods
Construction of yeast strains, standard molecular biology analyses and proteins purification procedures are reported in Supplementary methods. Yeast strains, plasmids and oligonucleotides used in this work are listed in Supplementary Tables 1–3.
Generation of the pool of random sequences and in vivo selection
The library of random sequences for the in vivo selection was generated from two chemically synthesized oligonucleotides containing a 60-nt variable region flanked by two constant regions of 13 nt (5′-end) and 20 nt (3′-end) (oligonucleotides DL1698 and DL1665, Supplementary Table 3). In all, 500 pmol of each oligonucleotide was annealed over their 20 nt complementary 3′-ends and filled in using the klenow fragment of E. coli polymerase I. After purification, the mixture was PCR amplified with primers DL1702 and DL1666 (Supplementary Table 3) that anneal to the 5′ constant regions of DL1698 and DL1665 and extend the homology region for subsequent cloning of the random pool by recombination. The final pool contains two regions of random sequence (A and B, Figure 1A) separated by a constant segment of 20 nt. Transformation of a Δcup1 yeast strain with the pool yielded roughly 105 colonies. These were directly replica plated on galactose medium containing 0.3 mM copper for selection based on expression of the CUP1 gene under control of the PGal promoter on the reporter. Copper-resistant clones were pooled, the inserts were amplified by PCR with oligonucleotides directed against the constant regions and cloned by recombination in pDL367 for a second round of selection. This selection strategy effectively minimized the emergence of false positives due to rearrangements of the vector, generally leading to loss of the doxycycline repressible promoter. Roughly 100 of the copper-resistant clones were sequenced manually before large scale sequencing. Fifteen clones were subjected to northern blot analysis to verify the occurrence of termination between Ptet and Pgal.
Deep sequencing and statistical analyses
Inserts from the winning or the starting pools were amplified by PCR using primers containing a two nucleotides barcode. The primers included adaptors for flow cell amplification and annealing sites for sequencing primers. Paired ends sequencing was performed on an Illumina GAIIx platform. Sequences of parts A and B were coupled with a home-made algorithm. Roughly 30% of sequences of the winning pool were excluded from the analysis because containing NNS complex-independent terminators (Colin et al, in preparation). The remaining roughly 700 distinct sequences were submitted to subsequent statistical analyses with RSAT (van Helden, 2003). Motifs of four and five nucleotides that are significantly overabundant and constitute potential termination signals were identified by comparing frequencies observed in the winning pool relative to frequencies observed in the starting pool (8000 sequences). Since the two oligonucleotides used to generate parts A and B of the starting pool are on the opposite strand, the nucleotide bias due to the chemical synthesis is different in the two regions as assessed from sequencing (part A: 22.7% (A); 19.4% (C); 29.2% (G) and 28.7% (T); part B: 31.3% (A); 26.8% (C); 18.6% (G) and 23.1% (T)). Since this strongly influences the frequency of each motif in the starting pool, we adopted a separate background model for parts A and B of the pool. P-values were calculated by RSAT with a correction for overlapping occurrences. Extended motifs were identified from overrepresented hexanucleotides using the pattern assembly and convert-matrix tools of RSAT. Pattern assembly aligns overlapping overrepresented motifs to generate larger elements that can be converted to matrices. Logos are generated by RSAT based on these matrices. P-values for the overrepresentation of TCTTG in natural CUTs (Gudipati et al, submitted) and underrepresentation in ORFs have been calculated estimating expected frequencies from input sequences (CUTs or ORFs) with a Markov chain model of order 2.
FA assays
The equilibrium binding of Nab3 RRM, Nab3 αhxRRM and the Nab3191–565–Nrd11–548 heterodimer to their specific substrates was analysed by FA. The RNA oligonucleotides were 5′ fluorescein labelled. The labelled RNA oligonucleotides were purchased from Sigma-Aldrich, the lyophilized samples were dissolved in water. The measurements were conducted on a FluoroMax-4 spectrofluorometer (Horiba Jobin-Yvon, USA). The instrument was equipped with a thermostatted cell holder with a Neslab RTE7 water bath (Thermo Scientific, USA). The whole system was operated using FluorEssence software (version 2.5.3.0, Horiba Jobin-Yvon). The fluorescein fluorophore was excited at 488 nm and its emission was collected at 520 nm. The width of both excitation and emission monochromatic slits was 14 nm for the 1 nM substrates, and 8 nm for the 10 nM substrates, the integration time was set to 3 s in both cases. All experiments were carried out at 25°C in a stirred 1.9 ml quartz cuvette. A fixed delay of 30 s was set between each aliquot addition and start of the measurement to allow the reaction to reach equilibrium. This delay was sufficient, as no further change in anisotropy was observed. Every data point is an average of three measurements.
The data were analysed in SigmaPlot 11 software (Systat Software, USA). The experimental isotherms were fit to a single-site binding model according to Heyduk and Lee (1990) using non-linear least squares regression or with single-site saturation binding model. The data were normalized for visualization purposes.
NMR analyses
All NMR spectra of 2.5 mM uniformly 15N-labelled Nab3 ahxRRM and RRM in 50 mM sodium phosphate buffer (pH 8.0), 300 mM NaCl, 10 mM β-mercaptoethanol (90% H2O/10% D2O) were recorded on Bruker AVANCE 600 spectrometer equipped with a cryoprobe at a sample temperature of 30°C. All 1H-15N HSQC spectra were acquired with 8 scans, 1024 points in 1H and 256 increments in 15N dimension, processed standardly with TopSpin (Bruker BioSpin) and analysed in Sparky (Goddard, TD and Kneller, DG, SPARKY 3, University of California, San Francisco, USA). Spectra for NMR titration were measured on a 15N isotopically enriched αhxRRM and unlabelled AUCUUG/CA, and on a 15N isotopically enriched RRM and unlabelled UCUU or UCUUG. In all titrations, RNA was added stepwise (in 4–6 steps) in small volumes into protein solutions. For each addition of the RNAs, the 1H-15N HSQC spectra were acquired. In the course of the titrations, the resonances moved from their initial positions, which correspond to the free form, in a stepwise directional manner until they reached their final positions, which correspond to the fully bound state. These data indicate that, in all complexes, the proteins are in fast exchange between their free and bound forms relative to the NMR time scale.
Supplementary Material
Acknowledgments
We would like to thank F Lacroute for help with yeast genetics, D Gautheret and T Barthel for help in analysing deep sequencing data and T Villa for critical reading of the manuscript. This work was supported by the Danish National Research Foundation (DL), the ANR (DL, ANR-08-Blan-0038-01), the CNRS (DL), the project ‘CEITEC—Central European Institute of Technology’ (RS, CZ.1.05/1.1.00/02.0068) from European Regional Development Fund, Czech Science Foundation (RS, P305/12/G034, P305/10/1490). OP and RKG received fellowships from EMBO and Région Ile de France, respectively. FH is in receipt of the Brno City Municipality Scholarship for Talented PhD Students. This research was carried out within the scope of the Associated European Laboratory LEA ‘Laboratory of Nuclear RNA Metabolism’. This work has benefited from the facilities and expertise of the high throughput sequencing platform of IMAGIF (Centre de Recherche de Gif— www.imagif.cnrs.fr).
Author contributions: OP designed and performed experiments and wrote the paper. FH designed and performed experiments. JB performed experiments. KK performed experiments and analysed the data. YD-C analysed the data. RKG performed experiments. RS designed experiments, analysed the data and wrote the paper. DL designed experiments, analysed the data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arigo JT, Carroll KL, Ames JM, Corden JL (2006a) Regulation of yeast NRD1 expression by premature transcription termination. Mol Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL (2006b) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL (2007) Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Pradhan DA, Granek JA, Clarke ND, Corden JL (2004) Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol Cell Biol 24: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A, Tomecki R, Lopez ME, Seraphin B, Dziembowski A (2011) Catalytic properties of the eukaryotic exosome. Adv Exp Med Biol 702: 63–78 [DOI] [PubMed] [Google Scholar]
- Creamer TJ, Darby MM, Jamonnak N, Schaughency P, Hao H, Wheelan SJ, Corden JL (2011) Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway: Nrd1, Nab3, and Sen1. PLoS Genet 7: e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M Jr (2006) Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 103: 3262–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Henras AK, Chanfreau GF (2006) Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA 12: 26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Morlando M, Bozzoni I (2000) Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J 19: 6218–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moore CL (2001) Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol Cell Biol 21: 8045–8055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Villa T, Boulay J, Libri D (2008) Phosphorylation of RNA polymerase CTD dictates transcription termination choice. Nat Struct Mol Biol 15: 786–794 [DOI] [PubMed] [Google Scholar]
- Heyduk T, Lee JC (1990) Application of fluorescence energy transfer and polarization to monitor Escherichia coli cAMP receptor protein and lac promoter interaction. Proc Natl Acad Sci USA 87: 1744–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobor F, Pergoli R, Kubicek K, Hrossova D, Bacikova V, Zimmermann M, Pasulka J, Hofr C, Vanacova S, Stefl R (2011) Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J Biol Chem 286: 3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO (2008) Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol 6: e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houalla R, Devaux F, Fatica A, Kufel J, Barrass D, Torchet C, Tollervey D (2006) Microarray detection of novel nuclear RNA substrates for the exosome. Yeast 23: 439–454 [DOI] [PubMed] [Google Scholar]
- Jacquier A (2009) The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet 10: 833–844 [DOI] [PubMed] [Google Scholar]
- Jenks MH, Reines D (2005) Dissection of the molecular basis of mycophenolate resistance in Saccharomyces cerevisiae. Yeast 22: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Johnson JM, Edwards S, Shoemaker D, Schadt EE (2005) Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21: 93–102 [DOI] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, Zhao J, Gross S, Silver PA, Moore CL (1997) Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3′-end formation in yeast. Genes Dev 11: 2545–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Vasiljeva L, Rando OJ, Zhelkovsky A, Moore C, Buratowski S (2006) Distinct pathways for snoRNA and mRNA termination. Mol Cell 24: 723–734 [DOI] [PubMed] [Google Scholar]
- Kopcewicz KA, O'Rourke TW, Reines D (2007) Metabolic regulation of IMD2 transcription and an unusual DNA element that generates short transcripts. Mol Cell Biol 27: 2821–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Brow DA (2008) Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol Cell 31: 201–211 [DOI] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C (2011) Unravelling the means to an end: RNA polymerase II transcription termination. Nat Rev Mol Cell Biol 12: 283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lebreton A, Seraphin B (2008) Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim Biophys Acta 1779: 558–565 [DOI] [PubMed] [Google Scholar]
- Lunde BM, Horner M, Meinhart A (2011) Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM. Nucleic Acids Res 39: 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L (2008) Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65: 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Soding J, Cramer P (2011) Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S (2011) Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A (2009) Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042 [DOI] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R (2011) Bacterial transcription terminators: the RNA 3′-end chronicles. J Mol Biol 412: 793–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ (2011) Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH (2008) The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci 33: 501–510 [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stormo GD, Gold L, Ehrenfeucht A (1986) Information content of binding sites on nucleotide sequences. J Mol Biol 188: 415–431 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Ng SB, Cloute JP, Brow DA (2006) cis- and trans-Acting determinants of transcription termination by yeast RNA polymerase II. Mol Cell Biol 26: 2688–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M, Colin J, Neil H, Jacquier A, Seraphin B, Lacroute F, Libri D (2008) Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol Cell 31: 671–682 [DOI] [PubMed] [Google Scholar]
- Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D (2006) Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ (2011) Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol 17: 1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentini SR, Weiss VH, Silver PA (1999) Arginine methylation and binding of Hrp1p to the efficiency element for mRNA 3′-end formation. RNA 5: 272–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J (2003) Regulatory sequence analysis tools. Nucleic Acids Res 31: 3593–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A (2008) The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlotzka W, Kudla G, Granneman S, Tollervey D (2011) The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. EMBO J 30: 1790–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, Libri D, Jacquier A (2005) Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM (2009) Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.