Abstract
Acute myeloid leukemia (AML) is a disease characterized by uncontrolled proliferation of clonal neoplastic hematopoietic precursor cells. This leads to the disruption of normal hematopoiesis and bone marrow failure. Major breakthroughs in the past have contributed to our understanding of the genetic failures and the changed biology in AML cells that underlie the initiation and progression of the disease. It is now recognized that not only genetic but also epigenetic alterations are similarly important in this process. Since these alterations do not change the DNA sequences and are pharmacologically reversible, they have been regarded as optimal targets for what is now known as epigenetic therapy. In this review, we will discuss our current understanding of normal epigenetic processes, outline our knowledge of epigenetic alterations in AML, and discuss how this information is being used to improve current therapy of this disease.
Acute myeloid leukemia (AML) is characterized by uncontrolled proliferation of clonal neoplastic hematopoietic precursor cells, leading to disruption of normal hematopoiesis and bone marrow failure. Molecular and cytogenetic analyses have been used to define genetic defects that contribute to the initiation and maintenance of this disease. Recurrent chromosomal rearrangements can be detected in more than 50% of AML cases and have been considered one of the most important prognostic factors for the prediction of clinical outcome.1 However, approximately 45% of AML cases have been shown to harbor normal karyotypes, although in the majority of these cases, one or more defects can be detected at the molecular level.2–7 The resulting molecular aberrations in these patients have been used to predict clinical outcome and/or, as in the case of internal tandem duplication of FLT3, the mutant proteins have been used as therapeutic targets.
In contrast to the aforementioned genetic aberrations that lead to irreversible structural DNA changes, epigenetic alterations result in a loss of gene function but do not modify the DNA coding sequence and can be reversed pharmacologically. In this review, we will outline briefly our current understanding of epigenetic modifications necessary for normal cell functions and describe efforts to unravel alterations that have occurred in these epigenetic patterns and contributed to myeloid leukemogenesis. Furthermore, we will highlight current clinical approaches that use this emerging information to develop novel epigenetic strategies.
EPIGENETICS, THE INTERPLAY OF DNA METHYLATION, CHROMATIN, AND NON-CODING RNAs
Once thought to be a simple package for DNA sequences, chromatin is now appreciated as an exceedingly important and complex aspect of normal cell biology. Several decades of research have revealed that modifications of DNA and chromatin proteins play a central role in virtually all functions associated with DNA, including transcriptional control, chromosome stability, DNA repair, and DNA replication. A complex interplay between DNA and chromatin modifications encodes a layer of information that acts in addition to the genetic code. Once established, this “epigenetic” information is preserved during cellular replication and, in certain cases, can even be passed through the germ line and inherited by subsequent generations.8 Epigenetic information has been shown to be facilitated by three primary factors: DNA methylation, histone modifications, and non-coding RNA.
DNA Methylation
The most well-studied epigenetic mediator is DNA methylation. In mammals, a family of DNA methyltransferase enzymes (DNMTs) catalyzes the addition of a methyl group to the 5’ position of the cytosine ring specifically in CpG dinucleotide sequences. This is the only abundant modification known to occur in mammalian DNA (~70% of CpGs; ~1%–2% of total DNA). The biological importance of DNA methylation is highlighted by the fact that targeted deletion of all DNMT enzymes known to possess methyltransferase activity results in lethality in mice. Deletion of Dnmt1, the principal DNMT enzyme that primarily functions to copy DNA methylation to newly synthesized DNA during replication (maintenance methylation), results in embryonic lethality.9 Deletion of Dnmt3a and Dnmt3b, DMNTs that have been shown to be involved in both maintenance and de novo methylation activities, also produces a lethal phenotype.10 In humans, mutations in DNMT3B cause ICF syndrome, a disease that is marked by chromomsomal instability, developmental defects, and immunodeficiency.11 Mutations in the MECP2 gene that encodes a protein that binds specifically to methylated DNA cause Rett syndrome, which is associated with several neurological defects.12 In contrast, DNA demethylation is thought to occur passively by failing to maintain methylation during DNA synthesis, although active demethylation also has been observed in specific biological contexts, especially during development,13 thereby suggesting the possibility that this activity is mediated by enzymatic processes.14,15
Although several non-mutually exclusive roles for DNA methylation have been described, including chromosome stability16 and genome compartmentalization,17 the vast majority of the data available strongly support a role in transcriptional regulation. The genomic distribution of CpG dinucleotides and their subsequent methylation is remarkably non-random; relative to the genome at large, CpGs are highly enriched in short sequences called CpG islands (CGIs) commonly found within or very near the 5' regions of genes. CpGs within CGIs are strongly resistant to methylation in normal somatic tissues, whereas most non-5′, non-CGI CpGs are methylated. The reverse may occur in cancer cells. A multitude of studies have demonstrated an inverse correlation between methylation of CGIs and gene expression. DNA hypomethylation is generally permissive for transcription, whereas DNA hypermethylation is associated with gene repression. Indeed, pharmacologically induced DNA demethylation can result in re-expression of hypermethylated genes. Most evidence supports that DNA methylation exerts its repressive effects either by directly blocking transcription factor binding or via a host of DNA binding proteins that specifically interact with methylated DNA. For example, DNA methylation blocks the binding of CTCF, a key protein that functions to insulate actively transcribed domains from neighboring heterchromatin.18 Proteins that share a conserved methyl-CpG-binding domain motif, MBD1-4 and MeCP2, along with Kaiso make up a family of proteins that selectively bind to methylated DNA and recruit additional factors with repressive properties.19
Histone Modifications
The four core histone proteins (H2A, H2B, H3, and H4) along with 147 bp of DNA comprise the nucleosome, the repeated structural unit of chromatin. The N-terminal histone tails protrude from the core assembly and are subject to a vast array of covalent modifications, including acetylation, methylation, ubiquitination, adenosine triphosphate (ADP)-ribosylation, and sumolation of lysines (K), methylation of arginine (R) residues, and phosphorylation of serines and threonines (for review see Kouzarides20). Together, these modifications are thought to comprise the “histone code,” a complex series of modifications that, when integrated, signal for either active euchromatic or silent heterochromatic states. Histone modifications function by, with the exception of methylation, reversing the net electrical charge of the histone tails thus reducing the affinity of inter-nucleosome and/or DNA-histone interactions. Acetylation of histones H3 and H4 by protein complexes that retain histone acetyltransferase (HAT) activity are a hallmark of active transcription. This activity is opposed by histone deacetylases (HDACs) that remove acetyl groups, promoting the formation of heterochromatin. Histone modifications also function by enhancing or decreasing the affinity for co-activator or co-repressor complexes. For example, acetylation promotes the association of several transcriptional activator complexes through the specific interaction of bromo domain-containing proteins. Histone methylation also functions in this manner and can signal for either eu- or heterochromatin though the interaction of several proteins that contain specific chromo-like and PHD domains. Methylation of H3K9 and/or H3K27 by various histone methyltransferases (HMTs), including Suv39h1, Suv39h2, EHMT1, EHMT2/G9a, SETBD1/ESET, and Ezh2 promote the association of heterochromatin protein 1 (HP1), a major component of heterochromatin, and other repressive polycomb group proteins. Methylation of H3K4 signals for euchromatin by recruiting ATP-dependent chromatin remodeling complexes and other factors that activate transcription. Importantly, protein complexes that are recruited to chromatin commonly have enzymatic activities that in addition modify other histone tail residues to promote further the intended chromatin state. Crosstalk between histone modifications forms multiple layers of information that provide stability to the euchromatic or heterochromatic state.
Non-Coding RNAs
Small non–(protein)-coding RNAs have been shown to influence gene expression through a variety of mechanisms in many organisms. RNA-mediated gene repression is mainly achieved through post-transcriptional gene silencing via the expression of an RNA molecule that is complementary (in the anti-sense orientation) to a target transcript. Homology-dependent transcript degradation is mediated by the Argonaut family of proteins that initially process and stabilize anti-sense RNAs, and later facilitate transcript cleavage or transcriptional interference.21 MicroRNAs (miRNAs) are small RNA molecules (19–23 nucleotides in length) that target specific 3' untranslated regions (UTRs) of genes and cause reduced protein production in a gene-specific manner. Although RNA-mediated post-translational gene silencing is not strictly defined as epigenetic, miRNAs are likely to play an important role in the regulation of genes with important epigenetic functions. In a recent study, DNMT3A and DNMT3B were found to be modulated by miR-29, a miRNA gene that is downregulated in lung cancer.22 Decreased miR-29 expression results in elevated DNMT3A and -3B, which contributes to aberrant hypermethylation of genes with tumor-suppressor functions.
Non-coding RNAs also have been shown to influence gene expression by inducing changes in chromatin conformation.23 While these processes have been well described in plants and some lower organisms, less is known about this function in mammals. Interestingly, X-chromosome inactivation in mammals is dependent on the coating of the repressed chromosome with RNA from the XIST gene in mammals, preceding changes in DNA methylation and histone modifications.24 As the activity of DNMTs and histone modifying enzymes do not appear to be contingent on the underlying sequence, RNA-based mediation of epigenetic phenomena would assist to explain the locus-specific nature of DNA methylation and histone modifications.
DNA Methylation and Histones: An Integrated View of Epigenetic Transcriptional Regulation
Recent findings have highlighted a strong interdependence between DNA methylation and histone modifications. A first line of evidence came from the discovery that DNMT1 interacts with HDAC125 and HDAC2,26 demonstrating a functional relationship between histone acetylation and DNA methylation. Experimental induction of DNA demethylation leads to histone acetylation, and demethylation can be caused by the HDAC inhibitor, trichostatin A.27 the histone methyltransferase that methylates H3K9, Suv39h, associates with DNMT128 and the H3K27 methylase, Ezh2, associates with all three catalytically active DNMTs.29 Furthermore, several ATP-dependent chromatin remodeling complexes that interact with chromatin in a histone modification-dependent manner influence DNA methylation, including Suv39h,30 ATRX,31 and Lsh.32 Lsh associates with Dnmt3a and Dnmt3b, demonstrating how this functional relationship might occur.33 A major question that remains to be answered is which of these epigenetic marks drives the establishment and maintenance of chromatin states. Two recent studies found that H3K4 methylation blocks the developmental acquisition of DNA methylation, indicative of a predominant role of histones in determining chromatin state and acquisition of other epigenetic marks.34,35 Other evidence shows that upon promoter activation, DNA demethylation precedes H3K4 methylation.36 As DNA methylation is the only epigenetic modification to be covalently associated with the DNA itself, CpG methylation status is in the unique position to provide long-term stability of gene expression states. Histone sliding and eviction occurs frequently in transcribed regions.37 DNA methylation may enable a lasting “chromatin memory” that endures the prolonged cellular lifespan of mammalian cells and persists through activities such as transcription and mitosis.
Tissue-Specificity and Development of Genome-Wide Epigenetic Patterns
The importance of epigenetic information is underscored by the fact that genome-wide patterns are unique between virtually all healthy cell types and tissues from within an individual.38,39 Commonly these patterns are altered during disease.40 Normal cellular differentiation and tissue development coincide with developmental acquisition of genome-wide patterns of DNA methylation. It is theorized that epigenetic factors are involved in the control of cellular totipotency, differentiation, and lineage selection. In support of this hypothesis, embryonic stem cells have distinct patterns of histone modifications that simultaneously signal for active and repressive chromatin, a precursor state to the choice of cell fate.41 Following cell lineage selection, tissue-specific hypermethylation of 5′ regions of genes generally correlates with gene repression in the particular tissue.39,42 However, as only a small proportion of genes demonstrate tissue specificity in DNA methylation patterns, the roles of other epigenetic factors in this process await evaluation on a genome-wide scale. Interestingly, DNA methylation differences between tissues are vastly more common in non-5′ regions.38 These changes may be involved in the regulation of non-coding RNAs or modulation of regional chromatin structure via interactions with the nuclear matrix. In support of this, MeCP2 binds CpGs in AT-rich sequence contexts43 and associates with matrix proteins.44
Observations showing that malignant cells maintain aberrant epigenetic marks, particularly DNA methylation, compared to their normal counterparts has dominated the field of epigenetics. Global hypomethylation is commonly observed in cancerous cells with, paradoxically, locus-specific hypermethylation.45 Mechanisms of tumorigenesis based on hypermethylation and silencing of tumor-suppressor genes have been well-described; whereas the role of global hypomethylation is less clear as very few loci outside of CGIs, which are normally unmethylated, have been investigated. Most studies have invoked a strategy of investigating the DNA methylation status of candidate loci, based on their biological function and/or involvement in other types of cancer. Non-biased, genome-wide approaches have determined that epigenetic changes in cancer occur at relatively high frequency and display tumor type-specific patterns.46 It is now appreciated that epigenetic changes in cancer are generally more abundant in comparison to genetic changes.47 As genome-wide scanning approaches become more comprehensive, critical information about the patterns of epigenetic aberrations in cancer will become clearer.
EPIGENETIC ALTERATIONS IN AML
While major efforts have led to a better understanding of molecular defects encoded in the DNA sequences of malignant AML cells, our understanding of how epigenetic alterations contribute to myeloid leukemogenesis remains to be fully elucidated. Aberrant epigenetics in AML were first reported in 1987 with the description of altered DNA methylation in the 5′ regulatory region of the calcitonin gene in primary leukemia samples.48 Subsequently, Pfeifer et al reported that overall levels of 5-methylcytosine do not significantly change in newly diagnosed acute leukemias49 while they decrease at relapse, suggesting that global loss of methylation (hypomethylation) is not part of the initiating steps of leukemogeneis, but rather it constitute an additional “hit” leading to therapy resistance.49 Follow-up studies confirmed global hypomethylation of the leukemia genome and identified repetitive sequences as the targets for DNA hypomethylation.50
Subsequent studies in malignant blasts focused mainly on hypermethylation of genes that are unmethylated and expressed in normal non-malignant cells. It became clear that hypermethylation correlates with gene silencing and similar to a genetic mutation, inactivate gene transcription. The list of genes hypermethylated in AMLs includes tumor-suppressor genes with well-established functions in cell cycle control, apoptosis, or DNA repair.51–59
In a first attempt to estimate the extent of hypermethylation in AML, Melki et al used bisulfite genomic sequencing to study hypermethylation in the promoter regions of eight cancer-related genes in 20 AML patients.60 Nineteen of the 20 patients showed methylation in at least one gene, and 15 had aberrant methylation in two or more promoters. Based on this study, the authors concluded that AML might be characterized by a general deregulation of CpG island methylation. In a similar study analyzing the DNA methylation status of 14 gene promoter–associated CpG islands in AML,61 the authors described concordance of DNA methylation events, and the existence of a methylator phenotype was suggested. With the development of a novel genome scan for aberrant DNA methylation, an assay called restriction landmark genomic scanning (RLGS), it became possible to evaluate the DNA methylation status of thousands of gene promoters within a single assay. Surprising results out of these studies from genome scans in AML cells were that the levels of global promoter methylation could reach up to 8.3% of all CGIs.62 Based on an estimated 29,000 CGIs in the human genome,63 this number would translate into more than 2,000 aberrantly methylated CGIs or the silencing of over 2000 genes. Interestingly, the patterns of DNA methylation were non-random, which suggests an underlying mechanism that leads to specific methylation of target genes or a selection process leading to the enrichment of cells with specific subset genes that have been inactivated by DNA methylation. Furthermore, comparing DNA methylation profiles from different tumor types to those patterns observed in AML revealed that each tumor type has a characteristic pattern and identified tumor type-specific DNA methylation events,46 again supporting the argument that DNA methylation events could develop into potential diagnostic and prognostic biomarkers.
Currently it is unclear what the mechanisms are that lead to aberrant DNA methylation in a leukemia genome. Several reports have described the active recruitment of DNMTs to target sites by onco-fusion proteins.64,65 A well-characterized example is provided by Di Croce et al. for the onco-fusion protein PML-RAR, generated by the translocation of chromosome 15 (location of the PML gene) and chromosome 17 (location of the retinoic receptor alpha) in acute promyelocytic leukemia.64 In this study it was shown that PML-RAR onco-fusion protein has the ability to recruit DNA methyltransferases to target promoters containing a retinoic acid response element. This recruitment leads to DNA methylation of target promoters (eg, RARβ2) and subsequent gene silencing. This epigenetic silencing involves additional recruitment of polycomb repressive complex 266 and indirect interaction through an HDAC3-mediated complex with MBD1.67 However, this mechanism alone does not explain the complexity of epigenetic silencing seen in primary leukemic cells since it was found that PML-RAR translocations are rarely associated with RARβ2 promoter methylation.68 A similar mechanism has been proposed also for t(8;21) AML, where the AML1-ETO fusion protein was shown to recruit DNMT1, in addition to HDACs, on the promoter regions of target genes.64,65 Recently, other studies have provided evidence for epigenetic control of the gene expression patterns of precursor or stem cells.69 A large proportion of aberrantly methylated genes show repressive histone H3K27 methylation in addition to an active mark H3K4 methylation in embryonic stem cells.70–72 Another possibility would be a random targeting of promoter sequences followed by a selection process supporting the outgrowth or survival of cells containing a specific DNA methylation pattern in addition to other genetic aberrations.73
The importance of chromatin changes associate with alterations in DNA methylation patterns in leukemia is exemplified by alterations including the mixed lineage leukemia (MLL) gene. MLL is a frequent partner for recurrent translocations in acute leukemias, possesses a DNA binding domain, and is a H3K4 methyltransferase. In normal cells, MLL positively regulates gene expression of several genes, including HOX genes, whereas the fusion proteins including the MLL portion have lost the H3K4 methyltransferase activity, resulting in deregulation of target genes and transformation into leukemic stem cells.74 In contrast, the partial duplication of MLL-PTD, occurring mostly in AML with a normal karyotype,75 unlike the MLL chimeric fusion proteins, results in an enlongated protein that retains all functional domains of the MLL wild-type protein, including the C-terminal SET domain that confers histone methyltransferase activity.76 Nevertheless, the presence of this chimeric protein results in aberrant expression of the HOX gene and silencing of the concurrently present MLL wild-type allele via epigenetic alterations.77
EPIGENETIC THERAPIES IN AML
Pharmacologic reversal of aberrant epigenetic changes that silence genes important in hematopoiesis can restore normal bone marrow function and lead to clinical disease response in AML. We review here clinical results with three different epigenetic-targeted therapeutic approaches: (1) hypomethylation with azanucleosides, (2) histone deacetylation with HDAC inhibitors, and (3) dual targeting of aberrant methylation and deacetylation with combinations of these agents.
Clinical Studies With Hypomethylating Agents
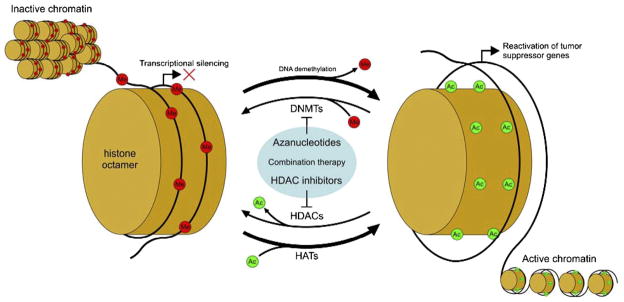
Although there are no drugs in this category specifically approved for AML, currently there are two hypomethylating agents approved by the US Food and Drug Administration (FDA) for treatment of myelodysplastic syndrome (MDS): 5-azacytidine (Vidaza, AZA; Pharmion Corp) and 5-aza-2'-deoxycytidine (decitabine, Dacogen; MGI Pharma/SuperGen). Both of these azanucleosides have activity in myeloid neoplasms, although the optimal doses and schedules of these agents in AML are still under investigation. Introduced decades ago as cytotoxic agents, these compounds seem to act via demethylation/differentiation rather than cytotoxicity, when used at doses far lower than the maximum tolerated dose (MTD). These compounds exert their hypomethylating activity by competing with the endogenous pool of deoxynucleosides for incorporation into newly synthesized DNA. Once incorporated into DNA, azanucleosides covalently bind and sequester DNMTs, leading to loss of promoter hypermethylation and re-expression of silenced genes (Figure 1).
Figure 1.
Mechanism of reactivation of silenced genes in cancer using azanucleosides and HDAC (histone deacetylase) inhibitors. Aberrant silencing of tumor-suppressor genes is maintained by factors that control chromatin states. DNMT (DNA methyltransferase) and HDAC enzymes cooperatively work to methylate DNA and deacetylate histones, respectively. Treatment with azanucleosides reduces DNMT activity and thus promotes net DNA demethylation (occurring during DNA replication or by active processes), while HDAC inhibitors promote a net gain of histone acetylation. In combination, these two therapies work synergistically to promote and maintain active chromatin states. HAT, histone acetyltransferase.
AZA was approved for use in MDS patients based on a Cancer and Leukemia Group B (CALGB) trial.78,79 Treatment with low-dose AZA (75 mg/m2 day subcutaneously for 7 days each month) resulted in a significantly better response, longer median time to AML progression, and decreased probability of leukemic transformation compared with best supportive care. The AZA arm had a complete remission (CR) rate of only 7%, but the majority of patients had evidence of clinical benefit and improved quality of life. Interestingly, most responders showed clinical effects beginning in the third or fourth month, suggesting that repetitive exposure of malignant cells to intermittent, low-dose AZA may affect only a fraction of cells during each cycle and that clinical response may thus not be apparent initially. Preliminary results of a new phase III, multicenter, randomized study of AZA versus conventional care regimens (several permitted) in higher risk MDS patients demonstrated a significant survival advantage for patients receiving AZA for a median of nine cycles at the same dose as in the CALGB study. Two-year overall survival (OS) was significantly higher for the AZA group (51% v 26%, P <.0001) with a median OS also superior for the AZA group (24.4 months v 15 months). There was no increase in early mortality detected between in the AZA group versus best supportive care.80 Low-dose AZA as a single-agent also has been explored in AML; a CR rate of 13% was reported at the annual meeting of the American Society of Hematology (ASH) in 2007.81
Decitabine also was recently approved by the FDA for the treatment of MDS on the basis of favorable clinical response for poor-risk patients compared with best supportive care. The schedule of decitabine administered was 15 mg/m2 intravenously over 3 hours, every 8 hours, for nine doses, every 6 weeks with a significantly higher response rate for decitabine (17%, including a 9% CR rate) versus best supportive care (P <.001).82 However, even lower doses of decitabine have shown promising results, with a remarkable 39% CR rate in a single-center report when administered at 20 mg/m2 intravenously daily for 5 days, every 28 days.83
Single-agent studies with decitabine in AML also have shown activity. In a dose-finding study of 5–20 mg/m2/d over 1 hour for 5 days a week × 2 weeks (10 doses) in a range of hematologic malignancies (n = 50, including 44 with AML/MDS), the response rate of for 15 mg/m2/d was 11/17 (65%), including six CRs.84 Among the patients with AML, 14% (5/37) had a CR. Dose-dependent global demethylation of DNA by decitabine at 5–20 mg/m2/d was associated with a clinical response in AML patients.85 Recently, Cashen et al reported a phase II study of single-agent decitabine in older, untreated AML patients who were unable to receive conventional induction therapy.86 Decitabine was given in doses of 20 mg/m2 over 1 hour, for 5 consecutive days every 4 weeks, resulting in a 26% response rate (seven of 27 patients). At The Ohio State University, a more intensive, 10-day schedule of inductions with decitabine, ie, 20 mg/m2 over 1 hour, for 10 consecutive days every 4 weeks is currently being tested, based on promising clinical and pharmacodynamic results from a phase I study (discussed later in this review). Further studies with decitabine in a novel role, maintenance therapy for AML in first CR after completion of intensive induction and consolidation, are also being pursued by different groups.
Clinical Studies With HDAC Inhibitors
The potential for therapy targeting aberrant HDAC activity in AML was demonstrated a decade ago.87 Several groups have demonstrated that the fusion protein PML-RAR aberrantly recruits HDACs, thereby inducing chromatin hypoacetylation and gene silencing. Warrell et al initially reported that in a patient with refractory acute promyelocytic leukemia (APL) and clinically resistant to all-trans retinoic acid (ATRA), PB administered intravenously twice daily in combination with ATRA led to eventual complete morphologic, cytogenetic, and molecular remission along with a time-dependent increase in histone acetylation.87,88 A platform for further development of PB as a regulator of transcription via HDAC inhibition was subsequently established by Gore et al. This group demonstrated feasibility and achievement of pharmacologically relevant PB plasma levels with a prolonged infusion of PB in patients with MDS and AML, although few hematologic responses were observed.89
Since these initial studies, clinical responses with HDAC inhibitors as single agents in subsets of AML other than t(15;17) have been noted, but overall results have been disappointing. AML activity of depsipeptide in several phase I/II single-agent studies was limited to blast count reductions.90 In other clinical trials, oral administration of suberoylanilide hydroxamic acid (SAHA) has been investigated in a phase I study of patients with MDS or refractory leukemia.91 Of 34 AML/MDS patients on the study, only two achieved CR and two incomplete CR (CRi, failure to recover normal neutrophils and/or platelet counts without evidence of leukemia). Several groups have studied valproic acid with or without ATRA in AML92 with rare clinical responses. Gojo et al recently reported results of a phase I study of MS-275 administered orally (weekly, duration of treatment varied) to 38 AML/MDS patients.93 Although no objective responses were observed, correlative studies confirmed histone H3/H4 acetylation in most patients by week 2 or 3 of therapy.
Combination of Hypomethylating Agents and HDAC Inhibitors in the Clinic
Given the interplay of the different epigenetic silencing mechanisms, dual targeting of aberrant DNA methylation and histone deacetylation has been pursued in AML. Several combination studies of a hypomethylating agent with a HDAC inhibitor have been reported. Gore et al studied AZA and PB in AML/MDS and observed a 14% CR rate in a group of 29 treated patients.94 Increased RNA expression associated with decreased promoter methylation of the P15 gene was observed in three of four responders, thereby providing the first definitive evidence in patients of a cause-effect relationship among administration of AZA, DNA hypomethylation, gene re-expression, and clinical response in patients with AML/MDS. Subsequently, Garcia-Manero et al reported a study of decitabine and valproic acid in AML/MDS, with a 19% CR rate seen in 54 patients.95 Similar results were found with combination of AZA with valproic acid.96 We recently reported a 16% CR rate in 25 patients enrolled in a study of decitabine with or without valproic acid in AML, administered at a different dose and schedule than those in the study previously reported.97 Interestingly, in our study, all of the CRs occurred in previously untreated patients older than 60 years and with complex karyotypes. Mechanistically, we demonstrated that re-expression of the estrogen receptor 1 (ESR1) gene, commonly silenced in AML/MDS, was significantly associated with clinical response (CR/CRi). Quantitative methylation studies showed that drug-induced hypomethylation within a unique region of the ESR1 promoter was the primary mechanism for re-expression of the gene. However, we did not find any benefit for increased (synergistic or additive) gene re-expression following the addition of tolerable doses of valproic acid to decitabine. Recent preliminary reports also have demonstrated feasibility of other novel combinations of epigenetic therapies including decitabine plus SAHA98 and AZA plus MGCD0103.99 A unifying and promising theme across several of the combination studies discussed above is markedly higher response rates in previously untreated, elderly patients who may not be candidates for intensive induction therapy. Altogether, these studies have shown the feasibility of combining different epigenetic targeting therapies and provide promise that further improvements on efficacy can be achieved. Whether combination of newer hypomethylating agents or HDAC inhibitors or treatment restricted to specific subsets of AML in which aberrant epigenetics plays a predominant leukemogenic role will lead to better clinical results remains an important question for further research.100
Acknowledgments
This work was supported by grants from the Leukemia Lymphoma Society (C.P.) and in part by NCI P30CA16058 (C.P.), CA93548 (C.P.) CA101956 (C.P.), CA102031 (G.M.). C.P. is a Leukemia Lymphoma Society Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–36. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 2.Caligiuri MA, Strout MP, Lawrence D, Arthur DC, Baer MR, Yu F, et al. Rearrangement of ALL1 (MLL) in acute myeloid leukemia with normal cytogenetics. Cancer Res. 1998;58:55–9. [PubMed] [Google Scholar]
- 3.Gilliland DG, Griffin JD. Role of FLT3 in leukemia. Curr Opin Hematol. 2002;9:274–81. doi: 10.1097/00062752-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Dohner K, Schlenk RF, Habdank M, Scholl C, Rucker FG, Corbacioglu A, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106:3740–6. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 5.Baldus CD, Liyanarachchi S, Mrozek K, Auer H, Tanner SM, Guimond M, et al. Acute myeloid leukemia with complex karyotypes and abnormal chromosome 21: Amplification discloses overexpression of APP, ETS2, and ERG genes. Proc Natl Acad Sci U S A. 2004;101:3915–20. doi: 10.1073/pnas.0400272101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108:3898–905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 7.Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102:1613–8. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 8.Chong S, Whitelaw E. Epigenetic germline inheritance. Curr Opin Genet Dev. 2004;14:692–6. doi: 10.1016/j.gde.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 10.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 11.Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, et al. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–7. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 13.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 14.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 15.Jost JP, Oakeley EJ, Zhu B, Benjamin D, Thiry S, Siegmann M, et al. 5-Methylcytosine DNA glycosylase participates in the genome-wide loss of DNA methylation occurring during mouse myoblast differentiation. Nucleic Acids Res. 2001;29:4452–61. doi: 10.1093/nar/29.21.4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann MJ, Schulz WA. Causes and consequences of DNA hypomethylation in human cancer. Biochem Cell Biol. 2005;83:296–321. doi: 10.1139/o05-036. [DOI] [PubMed] [Google Scholar]
- 17.Bestor TH, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–7. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 18.Recillas-Targa F, De La Rosa-Velazquez IA, Soto-Reyes E, Benitez-Bribiesca L. Epigenetic boundaries of tumour suppressor gene promoters: the CTCF connection and its role in carcinogenesis. J Cell Mol Med. 2006;10:554–68. doi: 10.1111/j.1582-4934.2006.tb00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–24. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 22.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayne EH, Allshire RC. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 2005;21:370–3. doi: 10.1016/j.tig.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–67. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 25.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- 26.Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 27.D'Alessio AC, Szyf M. Epigenetic tete-a-tete: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol. 2006;84:463–76. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- 28.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–12. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 30.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 31.Gibbons RJ, McDowell TL, Raman S, O'Rourke DM, Garrick D, Ayyub H, et al. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–71. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- 32.Dennis K, Fan T, Geiman T, Yan Q, Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 2001;15:2940–4. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu H, Geiman TM, Xi S, Jiang Q, Schmidtmann A, Chen T, et al. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–45. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–51. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol Cell Biol. 2007;27:7462–74. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Workman JL. Nucleosome displacement in transcription. Genes Dev. 2006;20:2009–17. doi: 10.1101/gad.1435706. [DOI] [PubMed] [Google Scholar]
- 38.Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci U S A. 2003;100:1775–80. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson K. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 41.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–71. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 42.Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, et al. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc Natl Acad Sci U S A. 2005;102:3336–41. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005;19:667–78. doi: 10.1016/j.molcel.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 44.Stratling WH, Yu F. Origin and roles of nuclear matrix proteins. Specific functions of the MAR-binding protein MeCP2/ARBP. Crit Rev Eukaryot Gene Expr. 1999;9:311–8. doi: 10.1615/critreveukargeneexpr.v9.i3-4.150. [DOI] [PubMed] [Google Scholar]
- 45.Ehrlich M. Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol. 2006;310:251–74. doi: 10.1007/3-540-31181-5_12. [DOI] [PubMed] [Google Scholar]
- 46.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–8. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 47.Smith LT, Otterson GA, Plass C. Unraveling the epigenetic code of cancer for therapy. Trends Genet. 2007;23:449–56. doi: 10.1016/j.tig.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 48.Baylin SB, Fearon ER, Vogelstein B, de Bustros A, Sharkis SJ, Burke PJ, et al. Hypermethylation of the 5' region of the calcitonin gene is a property of human lymphoid and acute myeloid malignancies. Blood. 1987;70:412–7. [PubMed] [Google Scholar]
- 49.Pfeifer GP, Steigerwald S, Boehm TL, Drahovsky D. DNA methylation levels in acute human leukemia. Cancer Lett. 1988;39:185–92. doi: 10.1016/0304-3835(88)90103-6. [DOI] [PubMed] [Google Scholar]
- 50.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Issa JP, Zehnbauer BA, Civin CI, Collector MI, Sharkis SJ, Davidson NE, et al. The estrogen receptor CpG island is methylated in most hematopoietic neoplasms. Cancer Res. 1996;56:973–77. [PubMed] [Google Scholar]
- 52.Herman JG, Civin CI, Issa JP, Collector MI, Sharkis SJ, Baylin SB. Distinct patterns of inactivation of p15INK4B and p16INK4A characterize the major types of hematological malignancies. Cancer Res. 1997;57:837–41. [PubMed] [Google Scholar]
- 53.Voso MT, Scardocci A, Guidi F, Zini G, Di Mario A, Pagano L, et al. Aberrant methylation of DAP-kinase in therapy-related acute myeloid leukemia and myelodysplastic syndromes. Blood. 2004;103:698–700. doi: 10.1182/blood-2003-07-2249. [DOI] [PubMed] [Google Scholar]
- 54.Seedhouse CH, Das-Gupta EP, Russell NH. Methylation of the hMLH1 promoter and its association with microsatellite instability in acute myeloid leukemia. Leukemia. 2003;17:83–8. doi: 10.1038/sj.leu.2402747. [DOI] [PubMed] [Google Scholar]
- 55.Melki JR, Vincent PC, Brown RD, Clark SJ. Hypermethylation of E-cadherin in leukemia. Blood. 2000;95:3208–13. [PubMed] [Google Scholar]
- 56.Issa JP, Zehnbauer BA, Kaufmann SH, Biel MA, Baylin SB. HIC1 hypermethylation is a late event in hematopoietic neoplasms. Cancer Res. 1997;57:1678–81. [PubMed] [Google Scholar]
- 57.Plass C, Yu F, Yu L, Strout MP, El-Rifai W, Elonen E, et al. Restriction landmark genome scanning for aberrant methylation in primary refractory and relapsed acute myeloid leukemia; involvement of the WIT-1 gene. Oncogene. 1999;18:3159–65. doi: 10.1038/sj.onc.1202651. [DOI] [PubMed] [Google Scholar]
- 58.Agrawal S, Hofmann WK, Tidow N, Ehrich M, van den Boom D, Koschmieder S, et al. The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood. 2007;109:3895–905. doi: 10.1182/blood-2006-08-040147. [DOI] [PubMed] [Google Scholar]
- 59.Esteller M. Profiling aberrant DNA methylation in hematologic neoplasms: a view from the tip of the iceberg. Clin Immunol. 2003;109:80–8. doi: 10.1016/s1521-6616(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 60.Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999;59:3730–40. [PubMed] [Google Scholar]
- 61.Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, Issa JP. Methylation profiling in acute myeloid leukemia. Blood. 2001;97:2823–9. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 62.Rush LJ, Dai Z, Smiraglia DJ, Gao X, Wright FA, Fruhwald M, et al. Novel methylation targets in de novo acute myeloid leukemia with prevalence of chromosome 11 loci. Blood. 2001;97:3226–33. doi: 10.1182/blood.v97.10.3226. [DOI] [PubMed] [Google Scholar]
- 63.International-Human-Genome-Sequencing-Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 64.Di Croce L, Raker VA, Corsaro M, Fazi F, Fanelli M, Faretta M, et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- 65.Liu S, Shen T, Huynh L, Klisovic MI, Rush LJ, Ford JL, et al. Interplay of RUNX1/MTG8 and DNA methyltransferase 1 in acute myeloid leukemia. Cancer Res. 2005;65:1277–84. doi: 10.1158/0008-5472.CAN-04-4532. [DOI] [PubMed] [Google Scholar]
- 66.Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Vire E, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–25. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 67.Villa R, Morey L, Raker VA, Buschbeck M, Gutierrez A, De Santis F, et al. The methyl-CpG binding protein MBD1 is required for PML-RARalpha function. Proc Natl Acad Sci U S A. 2006;103:1400–5. doi: 10.1073/pnas.0509343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tabe Y, Konopleva M, Kondo Y, Contractor R, Jin L, Ruvolo V, et al. PML-RARalpha and AML1-ETO translocations are rarely associated with methylation of the RARbeta2 promoter. Ann Hematol. 2006;85:689–704. doi: 10.1007/s00277-006-0148-7. [DOI] [PubMed] [Google Scholar]
- 69.Ohm JE, Baylin SB. Stem cell chromatin patterns: an instructive mechanism for DNA hypermethylation? Cell Cycle. 2007;6:1040–3. doi: 10.4161/cc.6.9.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–42. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 72.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–8. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 73.Opavsky R, Wang SH, Trikha P, Raval A, Huang Y, Wu YZ, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3:1757–69. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–33. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 75.Whitman SP, Ruppert AS, Marcucci G, Mrózek K, Paschka P, Langer C, et al. Long-term disease-free survivors with cytogenetically normal acute myeloid leukemia and MLL partial tandem duplication: a Cancer and Leukemia Group B study. Blood. 2007;109:5164–7. doi: 10.1182/blood-2007-01-069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dorrance AM, Liu S, Yuan W, Becknell B, Arnoczky KJ, Guimond M, et al. Mll partial tandem duplication induces aberrant Hox expression in vivo via specific epigenetic alterations. J Clin Invest. 2006;116:2707–16. doi: 10.1172/JCI25546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Whitman SP, Liu S, Vukosavljevic T, Rush LJ, Yu L, Liu C, et al. The MLL partial tandem duplication: evidence for recessive gain-of-function in acute myeloid leukemia identifies a novel patient subgroup for molecular-targeted therapy. Blood. 2005;106:345–52. doi: 10.1182/blood-2005-01-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the Cancer and Leukemia Group B. J Clin Oncol. 2002;20:2429–40. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- 79.Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–8. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 80.Fenaux P, Mufti G, Santini V, Finelli C, Giagounidis A, Schoch R, et al. Azacitidine (AZA) treatment prolongs overall survival (OS) in higher-risk MDS patients compared with conventional care regimens (CCR): results of the AZA-001 phase III study [abstract] Blood (ASH Annual Meeting Abstracts) 2007;110:817. [Google Scholar]
- 81.Fabre C, Gardin C, Mbida R-M, Quesnel Q, Dreyfus F, et al. Treatment of AML with azacytidine (AZA): current results of the French ATU program [abstract] Blood (ASH Annual Meeting Abstracts) 2007;110:1849. [Google Scholar]
- 82.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 83.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O'Brien S, Cortes J, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–7. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 84.Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–40. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 85.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 86.Cashen A, Schiller G, Larsen J, Cullen M, DiPersio J. Phase II study of low-dose decitabine for the front-line treatment of older patients with acute myeloid leukemia (AML) [abstract] Blood (ASH Annual Meeting Abstracts) 2006;108:1984. [Google Scholar]
- 87.Warrell RP, Jr, He LZ, Richon V, Calleja E, Pandolfi PP. Therapeutic targeting of transcription in acute promyelocytic leukemia by use of an inhibitor of histone deacetylase. J Natl Cancer Inst. 1998;90:1621–5. doi: 10.1093/jnci/90.21.1621. [DOI] [PubMed] [Google Scholar]
- 88.Warrell RP., Jr Arsenicals and inhibitors of histone deacetylase as anticancer therapy. Haematologica. 1999;84(Suppl EHA-4):75–7. [PubMed] [Google Scholar]
- 89.Gore SD, Weng LJ, Figg WD, Zhai S, Donehower RC, Dover G, et al. Impact of prolonged infusions of the putative differentiating agent sodium phenylbutyrate on myelodysplastic syndromes and acute myeloid leukemia. Clin Cancer Res. 2002;8:963–70. [PubMed] [Google Scholar]
- 90.Byrd JC, Marcucci G, Parthun MR, Xiao JJ, Klisovic RB, Moran M, et al. A phase 1 and pharmacodynamic study of depsipeptide (FK228) in chronic lymphocytic leukemia and acute myeloid leukemia. Blood. 2005;105:959–67. doi: 10.1182/blood-2004-05-1693. [DOI] [PubMed] [Google Scholar]
- 91.Garcia-Manero G, Yang H, Bueso-Ramos C, Ferrajoli A, Cortes J, Wierda WG, et al. Phase I study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid, SAHA) in patients with advanced leukemias and myelodysplastic syndromes. Blood. 2007 doi: 10.1182/blood-2007-06-098061. [DOI] [PubMed] [Google Scholar]
- 92.Kuendgen A, Strupp C, Aivado M, Bernhardt A, Hildebrandt B, Haas R, et al. Treatment of myelodysplastic syndromes with valproic acid alone or in combination with all-trans retinoic acid. Blood. 2004;104:1266–9. doi: 10.1182/blood-2003-12-4333. [DOI] [PubMed] [Google Scholar]
- 93.Gojo I, Jiemjit A, Trepel JB, Sparreboom A, Figg WD, Rollins S, et al. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109:2781–90. doi: 10.1182/blood-2006-05-021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 95.Garcia-Manero G, Kantarjian HM, Sanchez-Gonzalez B, Yang H, Rosner G, Verstovsek S, et al. Phase 1/2 study of the combination of 5-aza-2'-deoxycytidine with valproic acid in patients with leukemia. Blood. 2006;108:3271–9. doi: 10.1182/blood-2006-03-009142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Soriano AO, Yang H, Faderl S, Estrov Z, Giles F, Ravandi F, et al. Safety and clinical activity of the combination of 5-azacytidine, valproic acid, and all-trans retinoic acid in acute myeloid leukemia and myelodysplastic syndrome. Blood. 2007;110:2302–8. doi: 10.1182/blood-2007-03-078576. [DOI] [PubMed] [Google Scholar]
- 97.Blum W, Klisovic RB, Hackanson B, Liu Z, Liu S, Devine H, et al. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–91. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 98.Ravandi F, Faderl S, Deborah Thomas D, Burger J, Koller C, Garcia-Manero G, et al. Phase I study of suberoylanilide hydroxamic acid (SAHA) and decitabine in patients with relapsed, refractory or poor prognosis leukemia [abstract] Blood (ASH Annual Meeting Abstracts) 2007;110:897. [Google Scholar]
- 99.Garcia-Manero G, Yang A, Klimek V, Cortes J, Ravandi F, Newsome W, et al. Phase I/II study of MGCD0103, an oral isotype-selective histone deacetylase (HDAC) inhibitor, in combination with 5-azacitidine in higher-risk myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML) [abstract] Blood (ASH Annual Meeting Abstracts) 2007;110:444. [Google Scholar]
- 100.Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-{kappa}B-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood. 2007 Dec 14; doi: 10.1182/blood-2007-08-110171. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]