Abstract
Human leukocyte antigen (HLA) class I loci are essential to an effective immune response against a wide variety of pathogenic microorganisms, and they represent the prototypes for genetic polymorphism that are sustained through balancing selection. The functional significance of HLA class I variation is better exemplified by studies involving HIV type 1 (HIV-1) than any other infectious organism. HLA class I molecules are essential to the acquired immune response, but they are also important in innate immunity as ligands for the killer cell immunoglobulin-like receptors (KIR), which modulate natural killer cell activity. Here we concentrate on the interaction between the HLA-B and KIR3DL1/KIR3DS1 genes, describe the effects of these loci on HIV disease, and discuss questions that remain unresolved.
Genetic variation affects HIV infection outcome
The influence of host genetic variation as a significant determinant for outcome to HIV infection is becoming increasingly well-accepted among the HIV scientific community. An extensive body of literature in this regard has focused on the gene encoding the major co-receptor for HIV, CCR5 [1–6], and the loci encoding its ligands [7,8]. The primary role (although not necessarily the only role) of these polymorphisms in altering HIV infection or disease progression has to do with viral entry into host cells. Most other genetic variants associated with HIV outcome identified to date occur within or near genes encoding molecules involved in the acquired or innate immune response. To some extent, this inclination could reflect a selection bias by researchers in this field because immune response genes are the logical first-line candidates in defense against any infectious disease and all but one study so far [9] have used candidate gene approaches to study effects of genetic variation on HIV disease. Still, it is not surprising that the genes most fundamental to the immune response and also most polymorphic in the human genome, human leukocyte antigen (HLA) class I, are front and center in terms of the effects of their polymorphism on HIV (Table 1). Their centrality in determining the inter-individual levels of protection against HIV has become further ingrained with the discovery of HLA class I as ligands for the killer cell immunoglobulin-like receptors (KIR), a polymorphic set of molecules that modulate natural killer (NK) cell activity [10].
Table 1.
HLA and KIR variants that influence HIV-1 infection and disease progression
| Gene | Genotype | Effect | Refs |
|---|---|---|---|
| HLA | HLA-A, HLA-B, HLA-C homozygosity | Accelerate disease progression | [71,72] |
| B*35Px | Accelerate disease progression | [73] | |
| B*57, B*27, Bw4 | Slower disease progression | [40,56,74–76] | |
| A1-B8-DR3 | Accelerate disease progression | [77] | |
| G*0105N | Protection against infection | [78,79] | |
| G*010108 | Increased risk of infection | [79] | |
| E*0103 | Protection against infection | [78] | |
| HLA-C-35 (35 kb upstream of HLA-C) | Lower viral load | [9] | |
| KIR | 2DS2 2DL2 | Faster rate of CD4 T cell decline | [58] |
| 3DS1 | Slower rate of CD4 T cell decline | [59] | |
| 3DS1 homozygosity | Protect against infection | [67] | |
| KIR+ | 3DS1+ Bw4–80I | Slower disease progression | [56] |
| HLA | Slower progression to opportunistic infections | [57] | |
| 3DL1+ B*57s | Slower disease progression | [80] | |
| 3DL1*h + Bw4–80I | Slower disease progression, lower viral load | [39] | |
| Absence of cognate HLA ligand for inhibitory KIR | Protect against infection | [81] |
NK cells are components of the innate immune system, representing the first line of defense against virally infected cells and tumor cells [11–14]. NK cells are actively inhibited by targets expressing self major histocompatibility complex (MHC) class I [15], but they reject cells that are missing class I [16,17]. Thus, downregulation of class I on tumor cells or virally infected cells, a mechanism that allows these cells to escape cytotoxic T lymphocyte recognition [18], theoretically confers vulnerability to NK cell-mediated extermination (i.e. ‘missing self’) [19], a concept that was recently validated in murine cytomegalovirus (MCMV) infection [20]. Furthermore, the activating receptor NKG2D, which recognizes autologous molecules upregulated by cellular stress (reviewed in [10]), has been implicated in NK cell killing of transformed cells [21], supporting the general concept of recognition of ‘induced self’ by NK cell receptors [22]. In addition, a subset of activating receptors allows NK cells to recognize ‘non-self,’ either MHC class I-like structures encoded by a virus [23,24] or MHC class I itself present on allogeneic cells [25–29]. Finally, activating NK receptors could also be able to detect a combination of MHC class I and virus, because the activating receptor Ly49P specifically recognizes MCMV-infected cells if they express the mouse MHC class I allele H-2Dk (i.e. ‘altered self’) [30]. Recognition of ‘missing self,’ ‘induced self,’ ‘non-self’ and ‘altered self’ might contribute to the HIV-specific NK cell response.
Among the human NK receptor families, only the KIR genes are highly polymorphic and therefore could explain to some extent the differential responses to viral infections and reproductive success across individuals. In rodents, the KLR gene family, termed Ly49, has been expanded and shares many functional and molecular genetic characteristics in common with KIR in humans [31]. Thus, the Ly49 family in mouse serves as an excellent model for shaping hypotheses regarding the biology of KIR in humans, and in particular, responsiveness to viral infection. In this review, we focus on recent genetic and functional data implicating variation at the HLA class I and KIR loci, in particular the HLA-B and KIR3DL1 and KIR3DS1 loci, as determinants in HIV disease outcomes.
Marriage of KIR and their HLA class I ligands
The KIR gene cluster maps to chromosome 19q13.4 and is not linked to the HLA class I loci on chromosome 6p21.3. Haplotypes of the KIR locus vary in the number and type of genes present, and all KIR genes show allelic polymorphism. Much of the variability in gene copy number has to do with the presence or absence of activating KIR. HLA-A, HLA-B, and HLA-C, which encode ligands for KIR, are exceptionally polymorphic as well, but each of these genes is present on every chromosome 6. The ligands for most KIR are unknown, but among those for which a ligand has been defined, the specificity is dependent on only a single or very few amino acids, such that a given KIR might recognize a large number of different HLA allotypes.
Among the various KIR genes, KIR3DL1 and KIR3DS1 (KIR3DL1/S1), which are alleles of the same locus, have been a major target of both evolutionary and disease association studies (particularly for HIV and AIDS) [32]. KIR3DL1/S1 is the only KIR gene encoding both inhibitory (KIR3DL1) and activating (KIR3DS1) receptors. Inhibitory KIR3DL1 allotypes bind HLA-B molecules with the Bw4 motif [33,34], an epitope present in the α-1 helix of the peptide binding groove of HLA-B that was first defined serologically [35]. About 40% of all HLA-B allotypes contain the Bw4 motif; all others contain the Bw6 motif, which is not a ligand for KIR3DL1. Position 80 of the Bw4 epitope is dimorphic and appears to affect its interaction with KIR3DL1 subtypes. In general, Bw4 allotypes with isoleucine at position 80 (Bw4–80I) appear to be better ligands for most of the KIR3DL1 allotypes tested [33,36,37]. However, direct [38] and indirect [39] data suggest that Bw4 allotypes with threonine at position 80 (Bw4–80T), particularly HLA-B*2705, are better ligands for other KIR3DL1 subtypes. This is worth careful consideration, because B*27 is a well-documented, protective allotype against AIDS progression, and most B*27 alleles contain the Bw4–80T motif [40–42]. Because expression of HIV nef (negative factor) induces downregulation of some HLA class I molecules, including HLA-B, this could be sensed via inhibitory KIRs, including KIR3DL1. In addition, the stronger the interaction between KIR3DL1 and HLA-B, the more dearly HLA-B will be missed by NK cells in this recognition of ‘missing self.’
More contentious is the putative receptor-ligand relationship between the activating KIR3DS1 and Bw4 allotypes, especially those with Bw4–80I. The initial hurdle was to show that KIR3DS1 is actually expressed on the cell surface, which was well established in 2007 by four groups [43–46]. Each of these groups also showed that KIR3DS1 confers activating signals to NK cells. The second hurdle has been to define the ligand for KIR3DS1, a more elusive task. Although KIR3DS1 shares >95% similarity with KIR3DL1 in its extracellular domain, assays measuring potential KIR3DS1 interactions with cell surface Bw4 or Bw4 tetramers folded with various viral peptides have not indicated binding of KIR3DS1 to Bw4 [43,44]. However, recent experiments (discussed below) indicate that KIR3DS1 can recognize ‘altered self’ in the context of HLA-B Bw4–80I [47], similar to the MCMV studies [30].
Layering of complexity: variegated expression of KIR
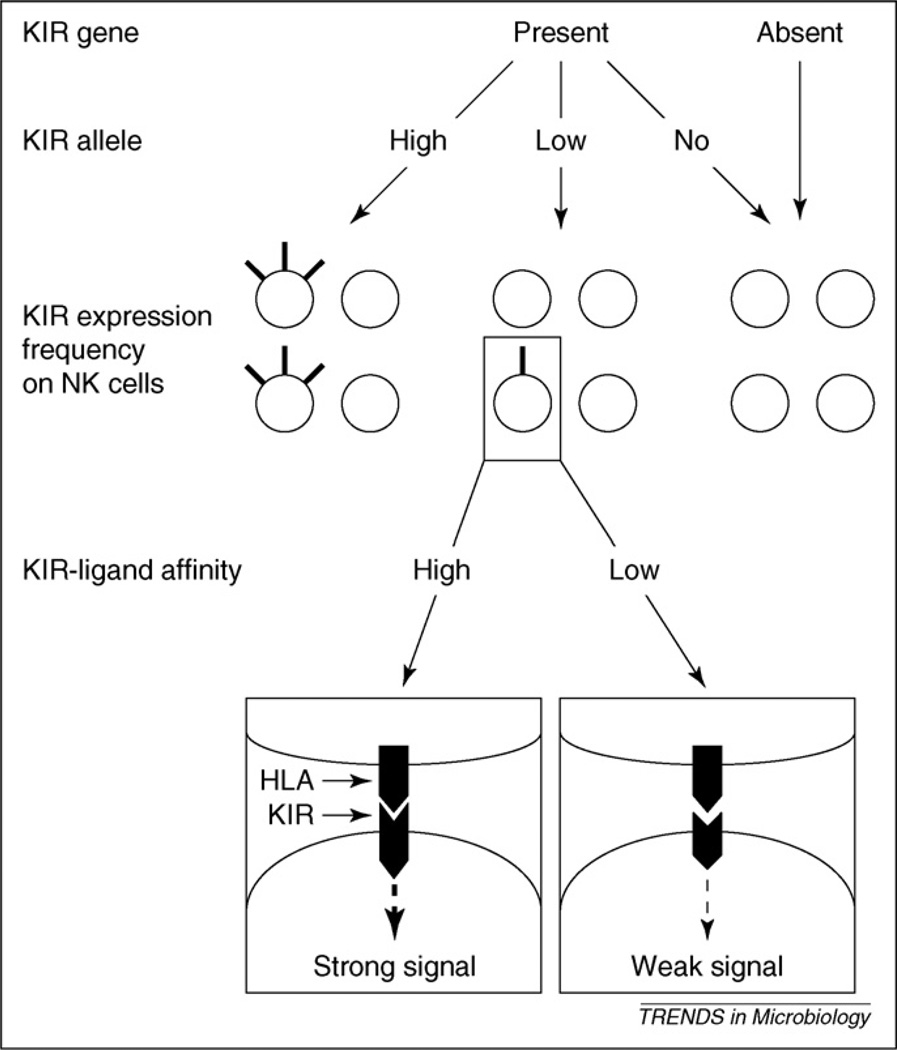
The ligand-receptor relationship between KIR and HLA is unusual compared to most other such pairings in the genome in that for any functionally relevant KIR-HLA pair, it is possible for one or the other or neither of these to be present in the genome of a given individual, both of which are functionally null. This situation results from the extreme polymorphism of both KIR and HLA loci, in addition to their physically unlinked status. On top of this, a given KIR is expressed on only some NK cell clones within an individual and different KIR can diverge in the frequency at which they are expressed within the NK cell population [48] as well as in the intensity with which they are expressed (Figure 1). Whether a KIR gene is expressed on a given NK cell clone is determined during NK cell maturation, and once acquired, the pattern of expression remains stable [48]. The frequency of KIR expression on NK cells is likely to be an important determinant in the strength of an NK cell response against viral infection, particularly because the number of NK cells expressing a given MHC-specific NK cell receptor has been shown to affect the degree of graft rejection [49]. Further study of the variability in frequency of expression across distinct KIR genes and alleles is timely and essential to understanding the relative importance of this variation in different diseases.
Figure 1.
Effects of KIR diversity on NK cell repertoire and function. Most KIR display presence or absence polymorphism (top). KIR alleles also differ in level and frequency of expression (high, low, or no), in which a nonexpressed allele (no) is likely to be equivalent to the absence of the corresponding KIR gene. Both types of polymorphism affect the contribution of the KIR in question to the overall NK repertoire, represented here by four NK cells for each situation. Some KIR allotypes are expressed at high levels on a large proportion of NK cells (left NK repertoire); others are expressed at lower levels by a small proportion of NK cells (center NK repertoire), and nonexpressed or absent alleles probably do not influence the NK repertoire (right NK repertoire). In addition, the affinity of a given KIR for its HLA class I ligands influences the strength of the signals received via the KIR. Note that this figure depicts the effect of a single KIR gene, and that the overall KIR repertoire on NK cells is an overlay of multiple KIR genes, each expressed largely independently of the others.
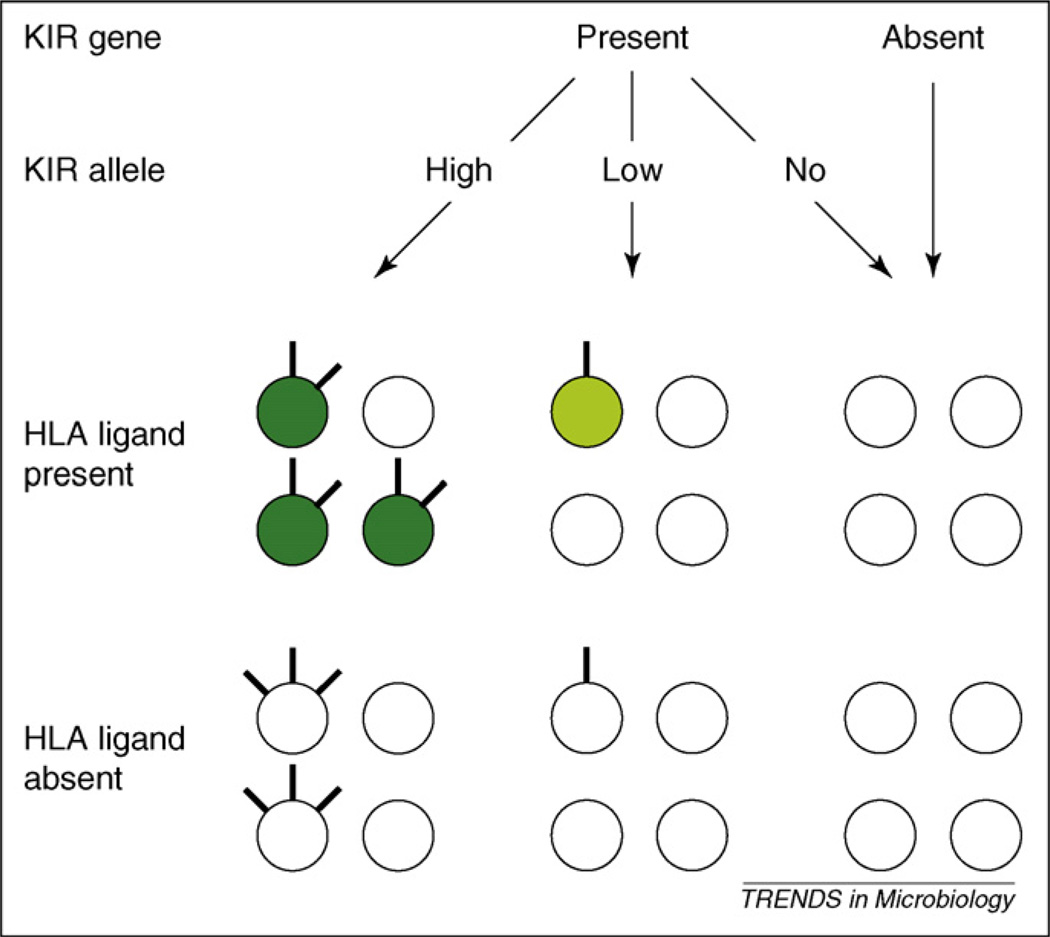
The rules governing acquisition of KIR by developing NK cells are largely unclear, but appear hard-wired in the genome [50], with a small but significant effect of the presence of HLA-encoded KIR ligands [51] (Figure 2). HLA class I does affect the functionality of NK cells: NK cells expressing inhibitory KIR specific for self HLA class I are more responsive upon stimulation through activating receptors than NK cells that do not express inhibitory KIR specific for self HLA class I [52–55] (Figure 2). In humans, this was shown experimentally by measuring interferon-γ (IFN-γ) production and CD107a mobilization (a surrogate marker for cytotoxic activity) by NK cells in response to targets expressing ligands for activating (but not inhibitory) NK receptors [52,54]. In these assays, KIR3DL1+ NK cells from Bw4/Bw4 individuals responded more vigorously than KIR3DL1− NK cells from Bw4/Bw4 individuals and KIR3DL1+ NK cells from Bw6/Bw6 individuals [55]. Thus it is plausible that the combination of KIR and HLA genes of an individual will determine the proportion of functionally mature NK cells and thereby the ability to recognize ‘missing self,’ ‘induced self,’ ‘non-self’ and ‘altered self.’
Figure 2.
Effects of inhibitory KIR-HLA interactions on NK cell responsiveness. Each KIR repertoire is represented by four NK cells, with individual NK cells expressing no, low or high levels of an inhibitory KIR on the cell surface. NK cells expressing a given inhibitory KIR in the presence of its HLA class I ligand (top left), are predicted to be more responsive to activating stimuli (indicated in dark green) than in the absence of its ligand (bottom left). In addition, the presence of HLA ligand increases the proportion of cells expressing the corresponding inhibitory KIR, but decreases the expression levels of this KIR on the NK cell surface. In the case of inhibitory KIR allotypes that are expressed at lower levels or frequencies (center), the effects of absence (bottom center panel) and presence of HLA ligand (top center panel) will be less pronounced. Note that this figure depicts the effect of a single KIR gene, and that the overall KIR repertoire on NK cells is an overlay of multiple KIR genes, each expressed largely independently of the others.
KIR3DS1: in the thick of HIV resistance
Activating KIR have been associated with several diseases [32] and the KIR3DS1 gene has engendered particular attention in HIV disease pathogenesis (Table 2). Although most genetic association studies agree that KIR3DS1, an allele that has a phenotypic frequency of ~39% in European Americans and 12% in African Americans, is probably protective against HIV, different conclusions regarding the requirement for the putative ligand Bw4–80I have emerged.
Table 2.
Multiple effects of KIR3DL1/S1 on HIV-1 infection and disease progression
| Type of study | Na | Results | Refs |
|---|---|---|---|
| KIR3DS1 + HLA-B Bw4–80I | |||
| Genetic | ~1000 | Slower disease progression | [56] |
| Genetic | 1184 | Slower progression to opportunistic infections but not malignancy | [57] |
| Genetic | 391 | Lower viral load | [57] |
| Cellular | 36 | Inhibition of HIV-1 replication | [47] |
| Genetic | 191 | Slower rate of CD4+ T cell decline but faster disease progression | [58] |
| KIR3DS1 | |||
| Genetic | 255 | Slower rate of CD4+ T cell decline | [59] |
| Functional | 60 | Increased IFN-γ production | [60] |
| Genetic | 384 | Homozygosity–protection against infection | [67] |
| KIR3DS1 expression | |||
| Functional | 70 | Higher levels of expression in HIV-1-exposed uninfected individuals | [70] |
Abbreviations: N, the number of individuals included in the study; IFN-γ, interferon-γ.
KIR3DS1 in combination with HLA-B Bw4–80I was associated with slow progression to AIDS in a group of ~1000 HIV seroconverters, and neither Bw4–80I without KIR3DS1 nor KIR3DS1 without Bw4–80I had any effect on disease progression [56]. There was a weak protective effect of this compound genotype on viral load, and a rather strong protective effect against opportunistic infections in 1184 individuals, which included the seroconverters used in the previous study and 109 additional seroprevalent individuals [57].
Smaller studies arrived at somewhat different conclusions. In a study of a group of 191 individuals [58], the opposite effect was observed: KIR3DS1 considered by itself and the compound genotype of KIR3DS1 + Bw4–80I was associated with rapid progression to AIDS, and this was statistically significant for KIR3DS1 + Bw4–80I. However, KIR3DS1 + Bw4–80I appeared to be protective in terms of CD4 decline relative to having KIR3DS1 alone, although this effect was not significant. Because only 34 individuals were positive for both KIR3DS1 and Bw4–80I, it is difficult to know whether the opposite effect of this compound genotype on CD4 decline and progression to AIDS could be at least partially due to power issues. Rather than a KIR3DS1 effect, this study [58] revealed a detrimental effect of KIR2DL2 and KIR2DS2 on CD4 T cell decline and progression to AIDS.
A study of 255 individuals indicated that KIR3DS1 independently associates with higher CD4 T cell counts, but does not have an effect on viral load levels [59]. On the other hand, Bw4–80I independently associated with lower viral loads, but had no effect on CD4 counts. The group with KIR3DS1+ Bw4–80I had lower viral loads and higher CD4 T cell counts, but not significantly in either case. The authors concluded that both KIR3DS1 and Bw4–80I are protective, but not in a synergistic manner.
As the genetic association studies did not reach entirely concordant results, it is of interest to consider functional studies on the interaction between KIR3DS1, HLA-B and HIV. Mouse data point to the possibility that activating NK receptors can detect a combination of MHC class I and a virus (i.e. ‘altered self’) [30]. Given the remarkable degree of convergent evolution between mouse Ly49 and human KIR in terms of function, it is not unreasonable to consider specific allotypes of HLA class I as ligands for activating KIR upon viral infection or some other cellular lesion.
The protective effect of KIR3DS1 + Bw4–80I was substantiated with functional data measuring inhibition of HIV replication in an autologous effector-target cell assay amongst a group of 36 individuals with various KIR-HLA genotypes [47], the first study to address the functional significance of a KIR-HLA genetic association. In this study, KIR3DS1 positive NK cells inhibited HIV replication in Bw4–80I positive T cells to a significantly greater extent relative to the three other putatively ‘null’ situations: KIR3DS1 positive NK cells in response to Bw4–80I negative T cells, KIR3DS1 negative NK cells in response to Bw4–80I positive T cells, or KIR3DS1 negative NK cells in response to Bw4–80I negative T cells. Based on the functional and genetic data, a model was proposed in which KIR3DS1 on NK cells interacts with Bw4–80I on HIV infected target cells, conferring a measurable level of control over the virus.
More recently, NK cells from HIV infected patients were tested for IFN-γ production and CD107a upregulation, both markers of activation, after stimulation with HLA class I negative targets [60]. The strongest effects were observed in assays measuring IFN-γ production, where NK cells from KIR3DS1 positive individuals showed greater production of IFN-γ compared to those without KIR3DS1. This was also the case for the KIR3DS1 + Bw4–80I positive group compared to those who did not have this compound genotype, and for those with KIR3DS1 + Bw4–80I compared to those who have KIR3DS1, but are missing Bw4–80I [60]. Statistics measuring strength of the effect of these genotypes were not provided in this study, but the strength of the response to class I deficient targets appeared greatest for the comparison of those with the compound genotype KIR3DS1+ Bw4–80I versus those without this genotype. Interestingly, KIR3DS1 + Bw4-80I positive individuals showed a rather strong IFN-γ response compared to those who have KIR3DS1, but are missing Bw4–80I. However, statistical power was limited in this comparison because the number of individuals in both groups were small (11 versus 9, respectively) and the difference was not significant. The authors concluded that KIR3DS1 associates with high NK cell functions in a manner that is independent of Bw4–80I, but an increase in sample numbers in the KIR3DS1+ Bw4–80I positive versus KIR3DS1 positive Bw4–80I negative comparison groups would help solidify or refute this conclusion. Together, these findings suggest that KIR3DS1 positive NK cells are intrinsically more active, an effect that might depend on HLA-B.
Biological science is rarely straightforward and even small differences in the way two different studies are performed (in terms of either experimental or analytical methodology) can affect the outcome, and therefore the conclusions of a study. However, one possibility is that KIR3DS1 binds certain Bw4 alleles, perhaps after modulation by HIV infection.
Inhibition translates into activation: KIR3DL1 subtypes in HIV disease
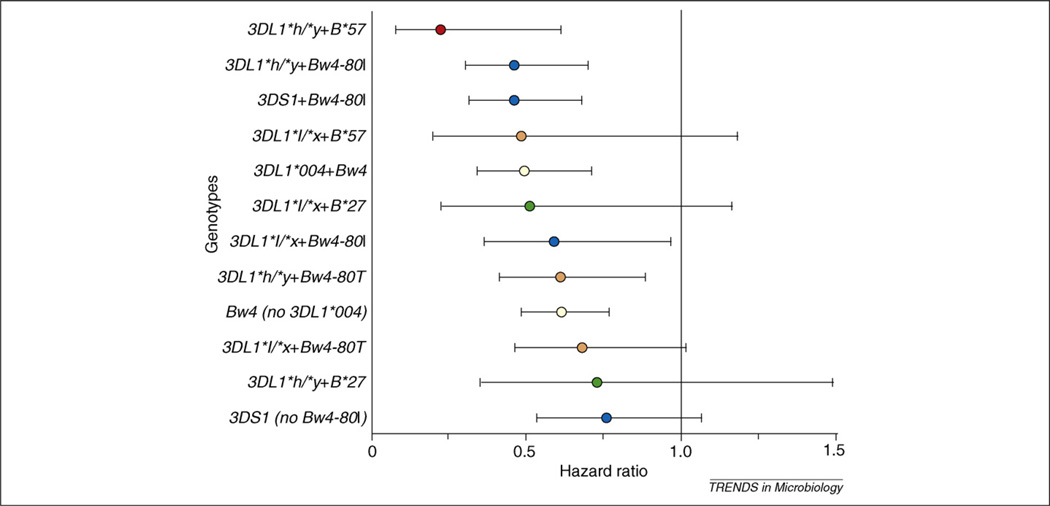
The inhibitory KIR3DL1 locus is among the most polymorphic of the KIR loci based on current data (http://www.ebi.ac.uk/ipd/kir/). KIR3DL1 allotypes differ in the level of their expression on cell surfaces [61,62] and these differences have functional repercussions [36,51]. It was proposed that the more inhibitory allotypes would associate with faster disease progression, because virtually all studies of KIR in HIV disease conclude that activating genotypes and phenotypes correlate with protection. To determine whether expression levels of KIR3DL1 might affect outcomes to HIV infection, alleles were grouped according to Gardiner et al. [61] into high (KIR3DL1*h), low (KIR3DL1*l), or no expression groupings [39]. HLA-B Bw4 molecules are known ligands for KIR3DL1 [34] and data indicate that the best ligands for the KIR3DL1 subtypes tested are the HLA-B Bw4–80I allotypes [33,36] (although not all KIR3DL1 or Bw4 subtypes have been tested), so those with Bw4–80I or Bw4–80T were grouped separately. Because KIR3DS1 + Bw4–80I was protective in these same cohorts, all individuals with this compound genotype were excluded from the analysis. To compare various KIR3DL1 + Bw4 groupings, individuals with Bw6/Bw6 (the alternative to Bw4 based on the dimorphic serological motif at position 77–83 of the HLA-B molecule) were used as the consistent reference group in all analyses because these individuals have no ligand for any KIR3DL1 subtype. Both disease progression analyses (n = 915) and viral load analyses (n = 901) were performed. Analysis of individual KIR3DL1 alleles revealed that only the unexpressed KIR3DL1*004 allotype in the presence of Bw4 had a protective effect. This raises the possibility that KIR3DL1*004 is indeed functional, perhaps by interaction with Bw4 intracellularly (as is the case for the interaction of KIR2DL4 and HLA-G [63]), or alternatively by altering the function of other receptors. Surprisingly, individuals possessing the highly expressed, highly inhibitory allotypes, KIR3DL1*h, along with Bw4–80I ligands had the lowest mean viral loads and progressed at a slower rate to AIDS relative to other KIR3DL1 + HLA-B genotypes [39]. The B*57 allele, which has the Bw4–80I motif, in combination with the KIR3DL1*h allotype showed stronger protection both in terms of AIDS progression and viral load than any other genetic variant identified to date (odds ratio = 0.1 for viral load and relative hazard ratio = 0.26 for progression to AIDS). B*27, the second most protective allele in the cohorts, contains the Bw4–80T motif, and interestingly, this allele showed greater protection in the presence of KIR3DL1*l, suggesting that B*27 (and perhaps other Bw4–80T allotypes) might have greater affinity for one or more of the KIR3DL1*l allotypes (Figure 3). Binding assays will be necessary to confirm or refute this possibility. Thus, level of expression and affinity of KIR3DL1 for the various Bw4 ligands could both contribute to functionality of these receptors on effector cells (Figure 2).
Figure 3.
Combinations of KIR3DL1/S1 and Bw4 protect against HIV-1 disease progression. Genotypes are ordered by degree of protection. Hazard ratios shown (colored dots) are relative to the Bw6/Bw6 control group. Lines through the dots represent 95% confidence intervals. Red dots, B*57 with 3DL1 genotypes; green dots, B*27 with 3DL1 genotypes; blue dots, Bw4–80I with 3DL1/S1 genotypes; orange dots, Bw4–80T with 3DL1 genotypes; yellow dots, Bw4 with or without 3DL1*004; black dot, 3DS1 without Bw4–80I.
So why would highly expressed, highly inhibitory KIR3DL1 subtypes confer, in general, relative protection against HIV? Well-substantiated models of NK cell repertoire development propose that NK cells must acquire sufficient inhibitory signals to quench autoreactivity through recognition of self class I ligands [48,64,65]. The larger the contribution of a given receptor-ligand pairing to NK cell inhibition under homeostatic conditions (as exemplified by the range expected to occur across high- and low-expressing KIR3DL1 subtypes [52,55]), the more potent a ‘missing self’ response will be when the interaction between that inhibitory receptor and its ligand is lost. The interaction could be lost upon viral infection if class I is downregulated, as is the case in HIV infection. In addition, as proposed above, the activation potential of the NK cell pool (i.e. its responsiveness to ‘missing self,’ ‘induced self,’ ‘non-self’ and ‘altered self’) is expected to correlate with the expression frequency and level of the KIR3DL1 allele and its affinity for the available HLA class I ligands.
The Cadillac of protection: preventing infection
The effect of genetic variation on HIV infection is poorly understood compared with its effect on outcomes to infection, such as disease progression or viral load levels. Apart from homozygosity for a deletion variant in the CCR5 co-receptor for HIV, CCR5Δ32, there are no polymorphisms that consistently show strong protection against HIV infection. Similar to the situation observed for AIDS progression, it is likely that there are many genetic variants that, when considered individually, have quite small effects on risk of HIV infection. Such variants collectively, however, could have substantial additive or epistatic effects.
The innate immune response probably weighs in when it comes to protection from HIV infection, at least in some cases. Indeed, NK cell activity in a group of 37 exposed uninfected (EU) intravenous drug users (IVDU) was significantly greater than that from 10 infected patients and 28 unexposed individuals [66]. The differences were striking for both NK cytolytic activity and cytokine production. Notably, NK cells from the HIV-infected patients showed similar measurements of NK cell activity before and after HIV infection, indicating that their relatively low levels of responses were intrinsic and not a result of HIV infection. Furthermore, low responsiveness was the norm, because the HIV-infected individuals and the unexposed individuals showed similar levels of response (with slightly better responses amongst the unexposed group). The data support the idea that strong NK cell responses could, under some circumstances, protect from HIV infection.
KIR3DS1 homozygosity was recently reported to be protective against HIV infection in a study involving 80 HIV-negative IVDU compared with individuals with primary HIV infection (odds ratio = 2.87) [67]. This protection was not dependent on Bw4–80I, suggesting once again that the ligand for KIR3DS1 is not Bw4–80I. As the authors discuss, it is also possible that KIR3DS1 is simply marking the true protective variant, which might confer recessive protection. In general, this study contrasts with the previous observation that two copies of KIR3DS1 in the absence of Bw4–80I associates rather strongly with rapid disease progression [56]. A more recent study from this group showed that the combination of KIR3DL1*h and HLA-B*57 was associated with a reduced risk of infection [68], which is in agreement with the effect of this compound genotype on disease progression [39]. Thus, genetic protection from infection and that from disease progression are not necessarily one and the same, and as for all genetic disease associations, functional data to address the basis of protection or susceptibility are warranted.
The presence or absence of specific KIR genes has been a primary focus in studies attempting to measure the potential influence of differential KIR activity in human disease [32] and functional studies are emerging to explain some of the genetic results [47,60,69]. A follow-up study of EU, HIV+, and unexposed Vietnamese individuals focusing on specific receptors has made clear that genetic presence or absence data provide only part of the KIR picture as a whole [70]. This study shows that the frequency of specific KIR transcript detection in peripheral blood mononuclear cells and the levels of those KIR transcripts in the NK cell population can differ significantly between HIV+ and EU individuals. These differences are not completely dependent on the presence of the given KIR gene in the genome of that individual. In general, NK cells from EUs as compared to those from HIV+ patients were characterized by patterns of KIR expression that are expected to result in NK cell activation. Of particular interest in the context of this review, there was a high KIR3DS1 to KIR3DL1 transcript ratio in the EUs. These differences between EUs and HIV+ groups in mRNA levels of certain KIR emphasize the need to consider not only the presence or absence of each KIR gene, but also expression levels, and allelic differences when studying effects of these loci on human disease.
Concluding remarks
Genetic association studies all indicate a role for KIR-HLA interactions in the control of HIV infection, but disagree on the exact nature of these associations. Generally, they demonstrate an association of KIR3DL1/S1 and HLA-B, either alone or in combination, with viral load, CD4 decline and/or progression to AIDS. More recent functional studies with NK cells expressing KIR3DS1 at least partially support a protective role for KIR3DS1 in HIV control, but fail to resolve the discrepancies among the genetic studies. The current challenge is to understand the molecular nature of the interactions among KIR, HLA and HIV. Such molecular studies could in turn help understand the divergent results obtained in the genetic association studies.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. JvB was supported by grant 0515 from the Landsteiner Foundation for Blood Transfusion Research.
References
- 1.Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 2.Liu R, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 3.Martin MP, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 4.McDermott DH, et al. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 5.Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 6.Zimmerman PA, et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, et al. Polymorphism in RANTES chemokine promoter affects HIV-1 disease progression. Proc. Natl. Acad. Sci. U. S. A. 1999;96:4581–4585. doi: 10.1073/pnas.96.8.4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanier LL. NK cell recognition. Annu. Rev. Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 11.Bancroft GJ. The role of natural killer cells in innate resistance to infection. Curr. Opin. Immunol. 1993;5:503–510. doi: 10.1016/0952-7915(93)90030-v. [DOI] [PubMed] [Google Scholar]
- 12.Biron CA, Brossay L. NK cells and NKT cells in innate defense against viral infections. Curr. Opin. Immunol. 2001;13:458–464. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 13.Robertson MJ, Ritz J. Biology and clinical relevance of human natural killer cells. Blood. 1990;76:2421–2438. [PubMed] [Google Scholar]
- 14.Trinchieri G. Biology of natural killer cells. Adv. Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Storkus WJ, et al. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc. Natl. Acad. Sci. U. S. A. 1989;86:2361–2364. doi: 10.1073/pnas.86.7.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoglund P, et al. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc. Natl. Acad. Sci. U. S. A. 1991;88:10332–10336. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karre K, et al. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- 18.Wiertz E, et al. Cytomegaloviruses use multiple mechanisms to elude the host immune response. Immunol. Lett. 1997;57:213–216. doi: 10.1016/s0165-2478(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 19.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 20.Voigt S, et al. Cytomegalovirus evasion of innate immunity by subversion of the NKR-P1B:Clr-b missing-self axis. Immunity. 2007;26:617–627. doi: 10.1016/j.immuni.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Guerra N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–580. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diefenbach A, Raulet DH. Strategies for target cell recognition by natural killer cells. Immunol. Rev. 2001;181:170–184. doi: 10.1034/j.1600-065x.2001.1810114.x. [DOI] [PubMed] [Google Scholar]
- 23.Arase H, et al. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 24.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biassoni R, et al. Role of amino acid position 70 in the binding affinity of p50.1 and p58.1 receptors for HLA-Cw4 molecules. Eur. J. Immunol. 1997;27:3095–3099. doi: 10.1002/eji.1830271203. [DOI] [PubMed] [Google Scholar]
- 26.Chewning JH, et al. KIR2DS1-positive NK cells mediate alloresponse against the C2 HLA-KIR ligand group in vitro. J. Immunol. 2007;179:854–868. doi: 10.4049/jimmunol.179.2.854. [DOI] [PubMed] [Google Scholar]
- 27.Foley B, et al. KIR2DS1-mediated activation overrides NKG2A-mediated inhibition in HLA-C C2-negative individuals. Int. Immunol. 2008;20:555–563. doi: 10.1093/intimm/dxn013. [DOI] [PubMed] [Google Scholar]
- 28.George TC, et al. Tolerance and alloreactivity of the Ly49D subset of murine NK cells. J. Immunol. 1999;163:1859–1867. [PubMed] [Google Scholar]
- 29.Stewart CA, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desrosiers MP, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat. Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson SK, et al. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol. Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 32.Kulkarni S, et al. The Yin and Yang of HLA and KIR in human disease. [10.1016/j.smim.2008.06.003];Semin. Immunol. 2008 doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cella M, et al. NK3-specific natural killer cells are selectively inhibited by Bw4- positive HLA alleles with isoleucine 80. J. Exp. Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gumperz JE, et al. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller CA, et al. Genetic and serological heterogeneity of the supertypic HLA-B locus specificities Bw4 and Bw6. Immunogenetics. 1989;30:200–207. doi: 10.1007/BF02421207. [DOI] [PubMed] [Google Scholar]
- 36.Carr WH, et al. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 37.Gumperz JE, et al. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell- inhibitory receptor. J. Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 38.Luque I, et al. Threonine 80 on HLA-B27 confers protection against lysis by a group of natural killer clones. Eur. J. Immunol. 1996;26:1974–1977. doi: 10.1002/eji.1830260845. [DOI] [PubMed] [Google Scholar]
- 39.Martin MP, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao X, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat. Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 41.Goulder PJ, et al. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 42.Phillips RE, et al. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature. 1991;354:453–459. doi: 10.1038/354453a0. [DOI] [PubMed] [Google Scholar]
- 43.Carr WH, et al. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J. Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Connor GM, et al. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 45.Pascal V, et al. Detection of KIR3DS1 on the cell surface of peripheral blood NK cells facilitates identification of a novel null allele and assessment of KIR3DS1 expression during HIV-1 infection. J. Immunol. 2007;179:1625–1633. doi: 10.4049/jimmunol.179.3.1625. [DOI] [PubMed] [Google Scholar]
- 46.Trundley A, et al. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 47.Alter G, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valiante NM, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 49.Johansson S, et al. Natural killer cell education in mice with single or multiple major histocompatibility complex class I molecules. J. Exp. Med. 2005;201:1145–1155. doi: 10.1084/jem.20050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shilling HG, et al. Genetic control of human NK cell repertoire. J. Immunol. 2002;169:239–247. doi: 10.4049/jimmunol.169.1.239. [DOI] [PubMed] [Google Scholar]
- 51.Yawata M, et al. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez NC, et al. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 55.Kim S, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. U. S. A. 2008;105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin MP, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 57.Qi Y, et al. KIR/HLA pleiotropism: protection against both HIV and opportunistic infections. PLoS Pathog. 2006;2:e79. doi: 10.1371/journal.ppat.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaudieri S, et al. Killer immunoglobulin-like receptors and HLA act both independently and synergistically to modify HIV disease progression. Genes Immun. 2005;6:683–690. doi: 10.1038/sj.gene.6364256. [DOI] [PubMed] [Google Scholar]
- 59.Barbour JD, et al. Synergy or independence? Deciphering the interaction of HLA Class I and NK cell KIR alleles in early HIV-1 disease progression. PLoS Pathog. 2007;3:e43. doi: 10.1371/journal.ppat.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Long BR, et al. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J. Virol. 2008;82:4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardiner CM, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 62.Thomas R, et al. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J. Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajagopalan S, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoglund P, et al. Host MHC class I gene control of NK-cell specificity in the mouse. Immunol. Rev. 1997;155:11–28. doi: 10.1111/j.1600-065x.1997.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 65.Raulet DH, et al. Specificity, tolerance and developmental regulation of natural killer cells defined by expression of class I-specific Ly49 receptors. Immunol. Rev. 1997;155:41–52. doi: 10.1111/j.1600-065x.1997.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 66.Scott-Algara D, et al. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J. Immunol. 2003;171:5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 67.Boulet S, et al. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS. 2008;22:595–599. doi: 10.1097/QAD.0b013e3282f56b23. [DOI] [PubMed] [Google Scholar]
- 68.Boulet S, et al. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS. 2008;22:1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 69.Ahlenstiel G, et al. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J. Clin. Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ravet S, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 71.Carrington M, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 72.Tang J, et al. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses. 1999;15:317–324. doi: 10.1089/088922299311277. [DOI] [PubMed] [Google Scholar]
- 73.Gao X, et al. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. EnglJ. Med. 2001;344:1668–1675. doi: 10.1056/NEJM200105313442203. [DOI] [PubMed] [Google Scholar]
- 74.Flores-Villanueva PO, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5140–5145. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaslow RA, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 76.Migueles SA, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Just JJ. Genetic predisposition to HIV-1 infection and acquired immune deficiency virus syndrome: a review of the literature examining associations with HLA. Hum. Immunol. 1995;44:156–169. doi: 10.1016/0198-8859(95)00034-8. [DOI] [PubMed] [Google Scholar]
- 78.Lajoie J, et al. Genetic variants in nonclassical major histocompatibility complex class I human leukocyte antigen (HLA)-E and HLA-G molecules are associated with susceptibility to heterosexual acquisition of HIV-1. J. Infect. Dis. 2006;193:298–301. doi: 10.1086/498877. [DOI] [PubMed] [Google Scholar]
- 79.Matte C, et al. Functionally active HLA-G polymorphisms are associated with the risk of heterosexual HIV-1 infection in African women. AIDS. 2004;18:427–431. doi: 10.1097/00002030-200402200-00008. [DOI] [PubMed] [Google Scholar]
- 80.Lopez-Vazquez A, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum. Immunol. 2005;66:285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Jennes W, et al. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 2006;177:6588–6592. doi: 10.4049/jimmunol.177.10.6588. [DOI] [PubMed] [Google Scholar]