Abstract
Detailed genomic characterization of cancer specimens is required to identify all genes whose dysregulation contributes to tumorigenesis and/or tumor progression. These include amplification target genes, whose oncogenic functions derive from their overexpression in response to increased gene copy number, and which increasingly serve as therapeutic targets and predictive markers. We propose that identifying novel amplification target genes is becoming more challenging, and may require the comparative analysis of multiple studies mapping gene copy number changes and/or defining associations between gene copy number and expression. We therefore reviewed the array comparative genomic hybridization and single nucleotide polymorphism profiling literature to identify copy number increases that were restricted to chromosome 8q21 in human cancers, which were reported most frequently in breast cancer. We determined the minimal regions of overlap between gained regions and then examined which chromosome 8q21 genes were most frequently overexpressed, or otherwise supported, in individual studies. As these combined approaches supported the previously proposed amplification targets TCEB1, TPD52, and WWP1, the comparison of multiple genomic studies may therefore effectively predict candidate gene amplification targets, and prioritize these for further study.
Keywords: gene amplification, chromosome 8q21, array CGH, SNP profiling
Introduction
Gene amplification targets increasingly serve as cancer therapy targets, or otherwise highlight signaling pathways for therapeutic intervention.1 Amplification target genes are by definition increased in copy number in cancer tissue compared with normal somatic tissues, and are additionally expected to map within a minimal region of genomic gain, where they may be the sole gene within the region or amplicon.2 Amplification target genes are also expected to display significant positive associations between gene copy number and transcript and/or protein levels. Additional criteria further strengthen a gene’s candidature, such as associations between gene expression and clinical variables such as patient outcome, and phenotypes from candidate gene overexpression and/or knockdown studies in cultured cells or model organisms. 2 While fulfilling all these criteria may require years of investigation, confirmation that a gene maps within a minimal region of genomic gain, and is frequently overexpressed in response to increased gene copy number, is sufficient to justify further experiments to either support or refute that gene’s candidature.
Given the therapeutic targeting of defined amplification targets such as ERBB2, there is strong interest in identifying additional amplification target genes. It is therefore important to consider the likely attributes of amplification target genes that await discovery or recognition. Mutational studies first highlighted the “mountain and hill” concept, namely that few genes are frequently mutated in most specimens of a given cancer type and in multiple cancers (“mountains”), whereas many individual genes are mutated at lower frequencies (“hills”).3 An analogous situation is likely to pertain to gene amplification targets,4 where comparatively few genes are amplified frequently, and/or at high amplitude, in most cases of a given cancer type and/or across multiple cancers. In contrast, a greater number of genes may be less frequently gained and/or gained in only specific tumor types or within patient subpopulations. Such genes could also be gained at comparatively lower amplitudes, and/or show lesser associations with copy number. As for the case of gene mutations, it seems likely that most amplification “mountains” have been identified, and the “hills” remain to be discovered. It would be easy to dismiss “hills” as being of lesser importance, but their large numbers alone argue against this.3,4 Furthermore, “mountains” and “hills” may in fact map to common signaling pathways, where therapeutic targeting of more than one pathway member may be required for extinction of function. Identifying “hills” should therefore improve our understanding of defined signaling pathways, as well as provide new and unexpected insights into tumor initiation and progression.
It would be logical to predict that “hills” will be more difficult to identify than “mountains.” Gene copy number and expression studies frequently identify known amplification targets because these are readily identifiable within a single study and quickly recognized by researchers. It is also possible that some high-resolution and high-throughput genomic studies may be unwittingly biased towards identifying known, as opposed to novel, amplification targets. The availability of information regarding the copy number and/or expression of many individual genes encourages the use of high levels of statistical significance to prioritize candidates. However, as unknown amplification target genes may be less frequently gained, gained at lower amplitude, and/or show lesser associations between copy number and gene expression, such genes may fall below thresholds imposed for candidate gene identification.
We propose that the identification of novel amplification target genes may instead require the detailed comparison of the results of multiple studies. This could involve comparing the extents of genomic gain in order to identify minimal common regions or minimal regions of overlap, and then examining relationships between copy number and expression for genes within regions of interest. As a case study, we considered that it would be valuable to perform such an analysis for a chromosomal region where candidate amplification targets have been proposed, but are not universally recognized. We therefore reviewed the array comparative genomic hybridization (aCGH) and single nucleotide polymorphism (SNP) profiling literature reporting genomic gains in human cancers that were either largely or entirely restricted to human chromosome 8q21.
Chromosome 8q21 is a relatively gene-dense region encompassing 19.5 Mb (74,000,001-93,500,000 bp) and up to 91 genes. As will be outlined, chromosome 8q21 is frequently gained in different cancer types, and its gain has been ascribed prognostic significance. A number of amplification targets within this region have been proposed,5-8 but genomic studies can still refer to chromosome 8q21 as lacking a clear oncogene candidate.9 There has never been a systematic analysis of the genomic copy number literature to determine whether unbiased genomic screening studies support already proposed chromosome 8q21 amplification target genes, or whether other genes emerge as superior candidates. It is also not known whether the same or different chromosome 8q21 genes are likely to be targeted in different cancer types.
Early Reports of Chromosome 8q21 Gain in Cancer
The advent of CGH highlighted the frequent gain of the chromosome 8q arm in breast10-14 and prostate cancer,15,16 where this could be the most frequently gained genomic region.16-18 Gain of entire chromosomal arms may imply the existence of more than one target gene, and this was supported by chromosome 8q24, 8q22-q23, and 8q21 regions being found to be gained separately in breast cancers or cell lines.10 Similarly, minimal regions of overlap including chromosome 8q21 were reported in prostate cancer,15 osteosarcoma,19 ovarian clear cell carcinoma,20 and serous ovarian carcinoma.21 Furthermore, chromosome 8q21 was the most frequently gained genomic region in prostate cancer metastases,16 and chromosome 8q21-q23 was the most frequently gained region in familial breast cancers.22 These authors also associated chromosome 8q21-q23 gain with higher tumor grade, higher mitosis number, and increased Ki67 expression.22 Chromosome 8q21 gain was significantly associated with increased risk of death in a large breast cancer cohort,23 and was also significantly more frequent in estrogen receptor–positive breast tumors, which recurred after adjuvant tamoxifen and chemotherapy, and associated with reduced metastasis-free survival.24 There is thus substantial evidence that chromosome 8q21 may be gained independently of more distal chromosome 8q regions in multiple cancer types, and that chromosome 8q21 therefore harbors one or more genes with oncogenic function(s).
Does the Literature Support Existing or Novel Chromosome 8q21 Amplification Targets?
Based on cytogenetic evidence of chromosome 8q21 gain in breast and other cancers, 3 separate 8q21 amplification target genes have been proposed: elongin C or TCEB1 transcription elongation factor B (SIII), polypeptide 1 (TCEB1) at chromosome 8q21.11,6 tumor protein D52 (TPD52) at chromosome 8q21.13,5 and WW domain containing E3 ubiquitin protein ligase 1 (WWP1) at chromosome 8q21.3.7 In all cases, the reported effects of gene overexpression and/or knockdown obtained in targeted studies7,8,25,26 have supported the positive associations reported between gene copy number and expression.5-8,26
We wished to examine whether these or other genes were supported by the results of aCGH or SNP profiling analyses, particularly when these approaches were coupled with expression profiling analyses of the same samples. We therefore conducted literature searches to identify studies identifying chromosome 8q21 gain in cancer, using HighWire and Google search engines (search term “8q21 AND cancer”) performed until March 2012. This identified 22 studies that reported genomic gains in different cancers that were largely or entirely restricted to chromosome 8q21 (Table 1). The reported gained regions typically represented minimal regions of overlap derived from comparing multiple independent samples or cell lines. The reviewed studies predominantly examined breast tumors (n = 9), followed by prostate cancer (n = 3), collections of diverse cancer types and/or cell lines (n = 2), or hepatocellular carcinoma (n = 2), with single studies examining colorectal cancer, epithelial ovarian cancer, lung cancer, esophageal squamous cell carcinoma, small bowel carcinoid tumors, or osteosarcoma (Table 1). Eight of the reviewed studies presented the extents of genomic gains in the absence of gene expression data, whereas the remaining 14 studies also reported associations between gene expression and copy number. Gene expression and copy number associations reported by reviewed studies (Table 1) were compared with those of additional unbiased genomic studies, which reported associations between copy number and gene expression or other parameters for chromosome 8q21 genes, but did not define the extent of chromosome 8q21 gains (Tables 2 -4).
Table 1.
Genomic Studies Reporting Chromosomal Gains Largely or Entirely Restricted to Chromosome 8q21 in Cancer Specimens or Cell Lines
| Reference | Samples examined | Genomic platform(s) employed | Definition of genomic gains/ minimal regions of overlap | Copy number associations with gene expression presented? | Transcriptomic platform(s) employed |
|---|---|---|---|---|---|
| Man et al.29 | Osteosarcomas (n = 48, from 42 patients) | Spectral Genomics BAC | Statistics | No | N/A |
| van Duin et al.37 | Prostate cancers (n = 22), prostate cancer xenografts (n = 9), prostate cancer cell lines (n = 3) | Institutional BAC | Threshold | Yesa | N/A |
| Yao et al.30 | Ductal carcinoma in situ (n = 9), invasive ductal carcinomas (n = 18), lymph node metastases (n = 2) | Agilent cDNA | Threshold | Yesb | N/A |
| Jönsson et al.45 | Breast (cancer) cell lines (n = 11) | Institutional BAC | Threshold | Yes | Institutional oligonucleotide |
| Chin et al.33 | Primary operable invasive breast cancers (n = 171), breast cancer cell lines (n = 49) | Institutional oligonucleotide | Threshold | Yes | Institutional oligonucleotide |
| Rodriguez et al.32 | Breast cancer cell lines (n = 8) | Institutional BAC | Threshold | Yes | Invitrogen HEEBO oligonucleotide |
| Kim et al.34 | Prostate cancer specimens, localized (n = 18) or metastatic (n = 17) | Institutional cDNA | Threshold | Yes | Institutional cDNA |
| Weir et al.9 | Primary lung adenocarcinomas (n = 371) | Affymetrix SNP Sty I | Statistics (GISTIC) | No | N/A |
| Kulke et al.31 | Small bowel carcinoid primary tumors (n = 14) or metastases (n = 10), from 18 patients | Affymetrix 100K SNP | Threshold | No | N/A |
| Marchiò et al.42 | Micropapillary breast carcinomas (n = 12), invasive breast carcinomas of no special type (n = 24) | Institutional BAC | Threshold | No | N/A |
| Natrajan et al.43 | Grade III invasive breast carcinoma (n = 95) | Institutional BAC | Threshold | No | N/A |
| Woo et al.38 | Hepatocellular carcinoma (n = 15) | NimbleGen oligonucleotide | Statistics | Yes | NimbleGen oligonucleotide |
| Hu et al.56 | Esophageal squamous cell carcinoma (n = 30) | Affymetrix SNP Nsp I or Sty I | Threshold | Yes | Affymetrix GeneChip HG-U133A 2.0 |
| Kao et al.40 | Breast cancer cell lines (n = 52) | Institutional cDNA | Statistics | Yes | HEEBO oligonucleotide |
| Holcomb et al.39 | Castration-resistant prostate cancers (n = 54, from 14 patients), localized prostate cancers (n = 9) | Institutional BAC | Threshold | Yes | Institutional cDNA |
| Sayagués et al.44 | Primary colorectal carcinomas from patients who developed liver metastases (n = 23) | Affymetrix SNP Nsp I or Sty I | Threshold | No | N/A |
| Staaf et al.54 | HER2-positive breast cancers (n = 200) | BAC arrays, SCIBLU Genomics Centre | Statistics (GISTIC) | Yes | Published oligonucleotide and cDNA datasets |
| Abkevich et al.27 | Cancer cell lines (n = 178, 18 tissues of origin) | Affymetrix SNP Nsp I or Sty I, Agilent 244K CGH | Threshold | No | N/A |
| Ramakrishna et al.35 | Epithelial ovarian tumors: serous (n = 37), endometrioid (n = 14), mucinous (n = 7), clear cell (n = 9) | Affymetrix Genome-Wide Human SNP 6.0 | Threshold | Yes | Affymetrix Human Gene1.0 ST |
| Beroukhim et al.28 | Cancer tissue specimens (n = 2,520, representing 26 histological subtypes), cell lines (n = 541), melanoma short-term cultures (n = 70) | Affymetrix SNP mapping Sty I | Statistics (GISTIC) | No | N/A |
| Jia et al.41 | Hepatocellular carcinoma (n = 58 with paired normal samples) | Affymetrix Genome-Wide Human SNP 6.0 | Threshold | Yes | Affymetrix GeneChip HG-U133 Plus 2.0 |
| Guedj et al.46 | Breast cancers (n = 488) | CIT-CGH array (V6), BAC and PAC clones | Statistics | Yes | Affymetrix HG-U133 Plus 2.0, other published datasets |
Note: BAC = Bacterial artificial chromosome; SNP = single nucleotide polymorphism; CGH = comparative genomic hybridization; CIT = Cartes d’identité des tumeurs; PAC = P1-derived artificial chromosome; GISTIC = Genomic identification of significant targets in cancer; N/A = not applicable; HEEBO = Human exonic evidence based oligonucleotide.
RT-PCR for candidate genes.
Statistical analysis of breast cancer SAGE libraries.
Table 2.
Reported Associations between Gene Expression and Copy Number for Chromosome 8q21.11 Genes
| Reviewed studies reporting gene overexpression associated with defined increased copy number |
Other genomic studies supporting amplification target gene status |
||||
|---|---|---|---|---|---|
| Gene symbol | Chromosome 8 location (Mb) | Tumor | Reference | Tumor | Reference |
| RPESP/C8orf84 | 74.141-74.168 | Breast (t) | Chin et al.33 | Breast (c) | Hyman et al.53 |
| Prostate (t) | Kim et al.34 | ||||
| STAU2 | 74.624-74.821 | Breast (t) | Chin et al.33 | Breast (t) | Adélaïde et al.48 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Staaf et al.54 | ||
| UBE2W/FLJ11011 | 74.865-74.953 | Breast (t) | Chin et al.33 | Breast (c) | Hyman et al.53 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | André et al.50 | ||
| Breast (t) | Natrajan et al.51 | ||||
| Prostate (t) | Kim et al.34 | ||||
| TCEB1 | 75.021-75.046 | Breast (t) | Chin et al.33 | Breast (t) | Zhang et al.36 |
| Breast (c) | Rodriguez et al.32 | Breast DCIS (t) | Vincent-Salomon et al.67 | ||
| Prostate (t) | Kim et al.34 | Breast (t) | Natrajan et al.51 | ||
| Ovarian (t) | Ramakrishna et al.35 | Prostate (t) | Holcomb et al.39 | ||
| TMEM70 | 75.050-75.057 | Breast (t) | Chin et al.33 | Breast (t) | André et al.50 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Natrajan et al.51 | ||
| Breast (t) | Staaf et al.54 | ||||
| GDAP1 | 75.425-75.441 | Breast (t) | Chin et al.33 | Breast (t) | Adélaïde et al.48 |
| Prostate (t) | Kim et al.34 | Breast (t) | Natrajan et al.51 | ||
| PXMP3 | 78.055-78.075 | Breast (c) | Jönsson et al.45 | Breast (c) | Hyman et al.53 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (c) | Rodriguez et al.32 | ||
| Breast (t) | André et al.50 | ||||
| Breast (t) | Natrajan et al.51 | ||||
Note: Genes contained within minimal regions of overlap (Fig. 2) are shown in bold. t = tissue; c = cell lines; x = xenografts; DCIS = Ductal carcinoma in situ.
Table 3.
Reported Associations between Gene Expression and Copy Number for Chromosome 8q21.13 Genes
| Reviewed studies reporting gene overexpression associated with defined increased copy number |
Other genomic studies supporting amplification target gene status |
||||
|---|---|---|---|---|---|
| Gene symbol | Chromosome 8 location (Mb) | Tumor | Reference | Tumor | Reference |
| HEY1 | 80.838-80.842 | Breast (c) | Kao et al.40 | Breast (c) | Rodriguez et al.32 |
| Liver (t) | Jia et al.41 | ||||
| MRPS28 | 80.993-81.105 | Breast (t) | Chin et al.33 | Breast (c) | Rodriguez et al.32 |
| Breast (c) | Jönsson et al.45 | Breast (t) | André et al.50 | ||
| Liver (t) | Woo et al.38 | Breast (t) | Natrajan et al.51 | ||
| Breast (t) | Guedj et al.46 | Breast (t) | Staaf et al.54 | ||
| TPD52 | 81.109-81.155 | Prostate (t, c, x) | van Duin et al.37 | Breast (c, t) | Pollack et al.47 |
| Breast (c) | Jönsson et al.45 | Prostate (t) | Paris et al.49 | ||
| Breast (t) | Chin et al.33 | Breast (t) | Adélaïde et al.48 | ||
| Breast (c) | Rodriguez et al.32 | Breast (t) | André et al.50 | ||
| Breast (c) | Kao et al.40 | Breast (t) | Zhang et al.36 | ||
| Breast (t) | Guedj et al.46 | Hematopoietic (c) | Mattison et al.52 | ||
| Breast (t) | Natrajan et al.51 | ||||
| ZBTB10/RINZF | 81.561-81.597 | Prostate (t, c, x) | van Duin et al.37 | Breast (c) | Hyman et al.53 |
| Breast (c) | Kao et al.40 | Breast (c, t) | Pollack et al.47 | ||
| Ovarian (t) | Ramakrishna et al.35 | Breast (c) | Rodriguez et al.32 | ||
| Breast (t) | Zhang et al.36 | ||||
| Breast (t) | Staaf et al.54 | ||||
| FABP5 | 82.355-82.359 | Prostate (t) | Kim et al.34 | Prostate (t) | Paris et al.49 |
| Liver (t) | Jia et al.41 | ||||
| ZFAND1 | 82.776-82.796 | Breast (t) | Chin et al.33 | Breast (t) | André et al.50 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Natrajan et al.51 | ||
| Breast (t) | Guedj et al.46 | Breast (t) | Staaf et al.54 | ||
| CHMP4C | 82.807-82.834 | Breast (t) | Chin et al.33 | Breast (t) | Natrajan et al.51 |
| Ovarian (t) | Ramakrishna et al.35 | ||||
| Breast (t) | Guedj et al.46 | ||||
| SNX16 | 82.874-82.916 | Ovarian (t) | Ramakrishna et al.35 | Breast (t) | André et al.50 |
| Breast (c) | Kao et al.40 | ||||
| Breast (t) | Natrajan et al.51 | ||||
Note: Genes contained within minimal regions of overlap (Fig. 3) are shown in bold. t = tissue; c = cell lines; x = xenografts.
Table 4.
Reported Associations between Gene Expression and Copy Number for Chromosome 8q21.2 and 8q21.3 Genes
| Reviewed studies reporting gene overexpression associated with defined increased copy number |
Other genomic studies supporting amplification target gene status |
|||||
|---|---|---|---|---|---|---|
| Gene symbol | Chromosome 8q band | Chromosome 8 location (Mb) | Tumor | Reference | Tumor | Reference |
| E2F5 | 8q21.2 | 86.276-86.314 | Breast (t) | Chin et al.33 | Breast (t) | André et al.50 |
| Prostate (t) | Kim et al.34 | Breast (t) | Natrajan et al.51 | |||
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Staaf et al.54 | |||
| C8orf59 | 8q21.2 | 86.313-86.319 | Breast (t) | Chin et al.33 | Breast (t) | Natrajan et al.51 |
| Ovarian (t) | Ramakrishna et al.35 | |||||
| WWP1 | 8q21.3 | 87.424-87.549 | Breast (t) | Yao et al.30 | Breast (t) | Adélaïde et al.48 |
| Breast (t) | Chin et al.33 | Breast (t) | André et al.50 | |||
| Ovarian (t) | Ramakrishna et al.35 | Breast (c) | Kao et al.40 | |||
| Prostate (t) | Holcomb et al.39 | |||||
| Breast (t) | Staaf et al.54 | |||||
| FAM82B | 8q21.3 | 87.553-87.590 | Breast (t) | Chin et al.33 | Breast (t) | Adélaïde et al.48 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Staaf et al.54 | |||
| CPNE3 | 8q21.3 | 87.595-87.642 | Breast (t) | Chin et al.33 | Breast (t) | André et al.50 |
| Ovarian (t) | Ramakrishna et al.35 | Breast (t) | Natrajan et al.51 | |||
| Breast (t) | Staaf et al.54 | |||||
| NBN/NBS | 8q21.3 | 91.014-91.066 | Breast (c) | Hyman et al.53 | ||
| Breast (c) | Rodriguez et al.32 | |||||
| Breast (t) | André et al.50 | |||||
| Breast (t) | Natrajan et al.51 | |||||
| DECR1 | 8q21.3 | 91.082-91.133 | Breast (t) | Chin et al.33 | Breast (t) | André et al.50 |
| Breast (t) | Natrajan et al.51 | |||||
| Breast (c) | Rodriguez et al.32 | Breast (t) | Staaf et al.54 | |||
Note: Genes contained within minimal regions of overlap (Fig. 4) are shown in bold. t = tissue; c = cell lines; x = xenografts.
There are several issues to consider when comparing the results of genomic studies. The reviewed analyses used a variety of genomic platforms differing in resolution (Table 1), and data analysis (Table 1) and reporting varied both between laboratories and over time. Methods of data analysis could be broadly categorized as employing variable threshold criteria (in terms of amplitude, length, and/or frequency of genomic gain), or statistical methods, to identify genomic regions to be compared and/or reported (Table 1). In all cases, we compared genomic gains or regions of overlap as they were defined and reported by each individual study. However, while technical and analytical differences could affect the extent of gained genomic regions reported by individual studies, these should not impede the derivation of minimal regions of overlap across multiple studies. We also considered that it was valid to compare gains reported in different tumor types, as performed in some of the reviewed studies,27,28 because many gene amplification targets are implicated in more than one type of cancer.2 The possibility that reported gains may represent copy number variants needs to be considered, particularly as many of the reviewed studies were performed before the prevalence of copy number variants was appreciated. Genomic coordinates have also changed over time, so we identified the NCBI/hg build used to produce genomic coordinates for each reviewed study. The majority of these used the NCBI35/hg17 or NCBI36/hg18 builds, which showed identical coordinates for all compared regions of chromosome 8q21. In the case of 3 studies that used the earlier NCBI34/hg16 build,29-31 genomic coordinates were converted to NCBI36/hg18 coordinates using the convert function of the UCSC Genome Browser. All quoted genomic coordinates are according to the NCBI36/hg18 build. Finally, where necessary, we updated the gene nomenclature used to that accepted by HUGO.
Overview of Chromosome 8q21 Regions Gained in Cancer
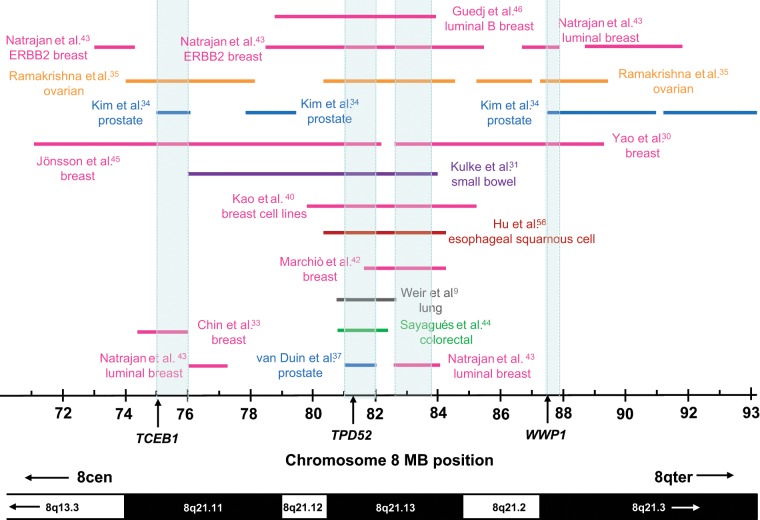
To first assess whether gains of different regions of chromosome 8q21 have been reported, we compared gains largely or entirely restricted to chromosome 8q21, which were at least 1 Mb in length (Fig. 1). Several regions of overlap emerged from the different studies and tumor types compared, at approximately 75 to 76 Mb, 81 to 82 Mb, 82.6 to 83.7 Mb, and 87.42 to 87.89 Mb, and these were predicted by the results of 4, 10, 9, and 4 studies, respectively (Fig. 1). Notably, 3 of these 4 regions of overlap include previously proposed amplification target genes, namely TCEB1 at chromosome 8q21.11 (75.021-75.046 Mb), TPD52 at chromosome 8q21.13 (81.110-81.246 Mb), and WWP1 at chromosome 8q21.3 (87.424-87.549 Mb).
Figure 1.
Summary of chromosome 8q21 copy number gains more than 1 Mb in length in cancer tissues or cell lines. Gained regions are indicated using horizontal lines according to the chromosome 8 coordinates below (in Mb), with the corresponding cytogenetic bands indicated on the lower ideogram. Arrows within cytogenetic bands indicate that these extend beyond the region shown. The study reporting each gained region is indicated to the left or right, or as space permitted, with colored lines and text highlighting studies examining particular cancer types. Regions of overlap between gained regions (shaded in light blue) were identified at 75 to 76 Mb (supported by 3 studies), 81 to 82 Mb (9 studies), 82.6 to 83.7 Mb (8 studies), and 87.42 to 87.89 Mb (4 studies). Three of these regions include the proposed amplification target genes TCEB1, TPD52, or WWP1, the 5′ positions of which are indicated using vertical arrows.
In order to refine these regions of overlap, we next compared gains of 1 Mb or greater with gains less than 1 Mb in length, typically reported in more recent studies. Gains further informing these regions of overlap will be described in the following sections.
Candidate Amplification Target Genes at Chromosome 8q21.11
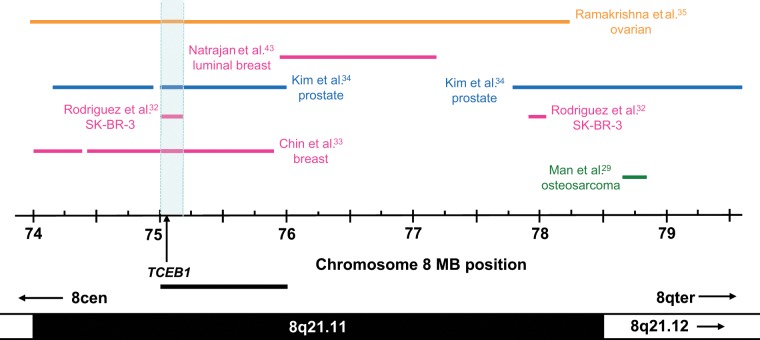
The addition of gains less than 1 Mb in length indicated that the region of overlap at 75 to 76 Mb (Fig. 1) could be refined to 75.01 to 75.22 Mb (Fig. 2) through a gain reported in SK-BR-3 cells.32 This region includes TCEB1 (75.021-75.046 Mb), a gene that was also supported by additional breast,33 prostate,34 and ovarian cancer studies,35 where TCEB1 was included within mapped amplicons and reported to be overexpressed (Table 2). While other genes within these amplicons were also overexpressed,33,34 no other chromosome 8q21.11 gene was supported by as many individual studies as TCEB1 (Table 2). Notably, Zhang et al.36 reported increased TCEB1 copy number and expression to be associated with poor breast cancer patient outcome. Thus, TCEB1 mapped within a minimal region of overlap supported by 4 independent studies and showed associations between copy number and expression in these and other studies (Table 2).
Figure 2.
Summary of chromosome 8q21.11 copy number gains in cancer tissues or cell lines. Gained regions are indicated using horizontal lines according to the chromosome 8 coordinates below (in Mb), with the corresponding cytogenetic bands indicated on the lower ideogram. Arrows within cytogenetic bands indicate that these extend beyond the region shown. The study reporting each gained region is indicated to the left or right, with colored lines and text highlighting studies examining particular cancer types. The minimal region of overlap (supported by 4 studies) is shaded in light blue and includes the proposed amplification target gene TCEB1, the 5′ position of which is shown using a vertical arrow. The position of the minimally gained region identified through consideration of gains of at least 1 Mb in length (Fig. 1) is shown as a horizontal bar below the chromosome 8 coordinate scale.
The minimal region of overlap including TCEB1 contrasted with a chromosome 8q21.12 region of overlap defined at 78.84 to 79.59 Mb (Fig. 1). The consideration of gains less than 1 Mb in length did not identify additional gains which contributed to or refined this region (Fig. 2), and the single gene that partially maps within this region (PKIA) was reported to show associations been copy number and gene expression in only one study.34
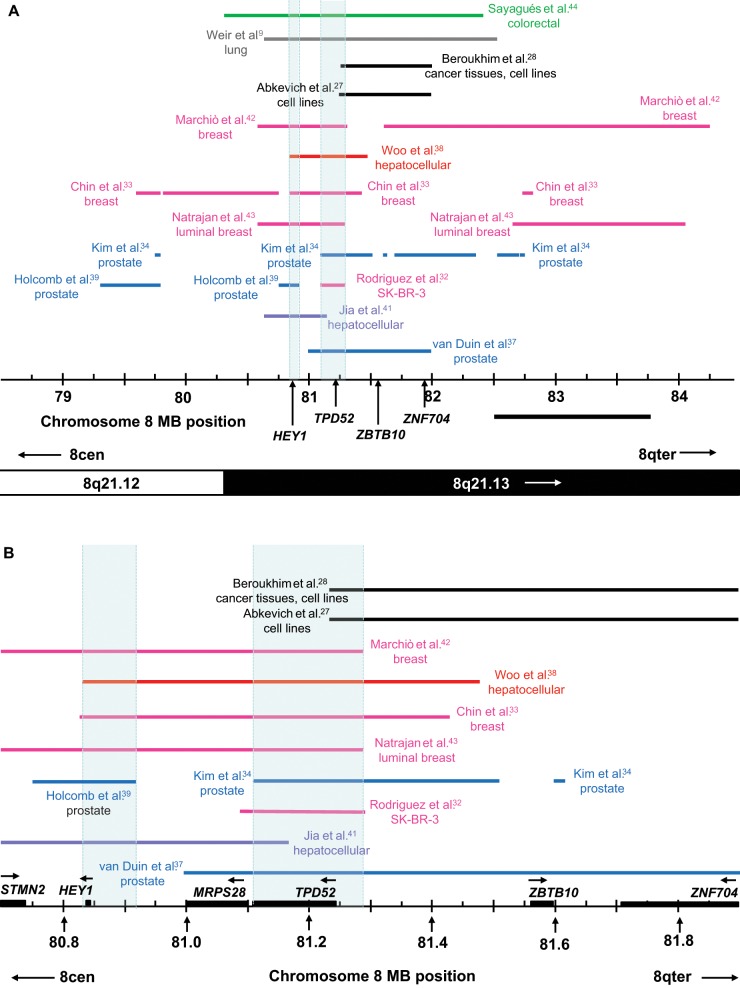
Candidate Amplification Target Genes at Chromosome 8q21.13
Some 13 studies reported gains largely restricted to chromosome 8q21.13 from 79 to 84 Mb (Fig. 3A). This region includes 2 of the regions of overlap shown in Figure 1. The more proximal of these was located at 81 to 82 Mb (Fig. 1) and was defined by gains in prostate cancer specimens, xenografts, and cell lines reported by van Duin et al.37 The addition of gains less than 1 Mb in length highlighted 2 smaller minimal regions of overlap (Fig. 3A). The most proximal of these was at 80.83 to 80.92 Mb, and was defined by the proximal extent of gain reported for hepatocellular carcinoma38 and breast cancer33 and the distal extent of gain reported by Holcomb et al.39 in prostate cancer (Fig. 3A). This region is included in overlapping gains reported by 8 studies (Fig. 3A). The single gene within this region is HEY1 (80.838-80.842 Mb) (Fig. 3B), which mapped to gained regions and was overexpressed in breast cancer cell lines40 and hepatocellular carcinoma41 (Table 3). Other studies reporting HEY1 gain did not examine gene expression,9,39,42-44 and only one other study reported an association between HEY1 expression and copy number32 (Table 3). From these combined results, HEY1 might be specifically targeted in hepatocellular carcinoma, but its role in other cancers is less clear.
Figure 3.
(A) Summary of chromosome 8q21.13 copy number gains in cancer tissues or cell lines. Gained regions are indicated using horizontal lines according to the chromosome 8 coordinates below (in Mb), with the corresponding cytogenetic bands indicated on the lower ideogram. Arrows within cytogenetic bands indicate that these extend beyond the region shown. The study reporting each gained region is indicated to the left or right, with colored lines and text highlighting studies examining particular cancer types. Minimal regions of overlap between gained regions (shaded in light blue) were identified at 80.83 to 80.92 Mb (supported by 8 studies) and 81.11 to 81.29 Mb (9 studies) and include HEY1 and TPD52, respectively. The ZBTB10 and ZNF704 genes included in the amplicons reported by Beroukhim et al.28 and Abkevich et al.27 are also shown, with the 5′ position of each gene being indicated using a vertical arrow. The position of a minimally gained region identified through the consideration of gains of at least 1 Mb in length (Fig. 1) was defined by the gain reported by van Duin et al.,37 whereas the second region is shown as a horizontal bar below the chromosome 8 coordinate scale. (B) Larger scale diagram of the minimal regions of overlap shown in A, highlighting the positions of all genes (as horizontal black boxes, with horizontal arrows to show the direction of transcription). The STMN2 and ZNF704 genes are incompletely contained within the region shown.
A second minimal region of overlap was identified at 81.11 to 81.29 Mb by comparing the results of 9 studies (Fig. 3A) and was defined by the proximal extent of gain reported by Kim et al.34 in prostate cancer and the distal extent of gain in SK-BR-3 cells32 and micropapillary42 or luminal breast cancer.43 Six of the 9 gained regions also included HEY1, but 3 others did not32,34,37 (Fig. 3A). The minimal region of overlap includes TPD52 (81.110-81.246 Mb) (Fig. 3B), first proposed as a chromosome 8q21 amplification target in breast cancer.5 While several studies reporting TPD52 gain did not examine gene expression,9,43,44 TPD52 was included in amplicons and overexpressed in breast32,33,40,45,46 and prostate cancer studies37 (Table 3). Other breast or prostate cancer studies also reported associations between increased TPD52 copy number and gene expression47-51 or poor patient prognosis.36 Increased TPD52 copy number in cancer cell lines was supported by the identification of viral integration sites upstream of Tpd52 in a mouse lymphoma model.52 However, while many studies supported TPD52 as an amplification target, others reporting TPD52 gain did not associate this with increased TPD52 expression.30,35,39,53
From the comparison of gains of 1 Mb or greater, a third region of overlap at chromosome 8q21.13 could be defined between 82.6 to 83.7 Mb (Fig. 1). However, the inclusion of gained regions of less than 1 Mb did not highlight a clear minimal region of overlap between these genomic coordinates (Fig. 3A). Similarly, no gene within this region was consistently amplified and overexpressed (Table 3).
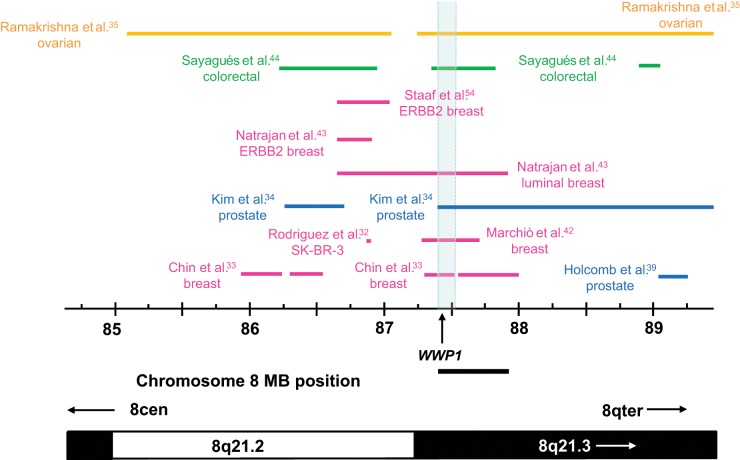
Candidate Amplification Target Genes at Chromosome 8q21.2-q21.3
The comparison of gains of at least 1 Mb in length highlighted a fourth region of overlap at 87.42 to 87.89 Mb, which was common to gains reported in breast,30,43 prostate,34 or ovarian cancers.35 This region was defined by the distal boundary of gain reported in luminal breast cancer,43 and the proximal boundary reported in prostate cancer34 (Fig. 1). When gains of less than 1 Mb were also considered, this region was supported by additional breast33,42 and colorectal cancer studies,44 and could be reduced to 87.42 to 87.53 Mb by the distal extent of gain reported by Chin et al.33 (Fig. 4).
Figure 4.
Summary of chromosome 8q21.2-q21.3 copy number gains in cancer tissues or cell lines. Gained regions are indicated using horizontal lines according to the chromosome 8 coordinates below (in Mb), with the corresponding cytogenetic bands indicated on the lower ideogram. Arrows within cytogenetic bands indicate that these extend beyond the region shown. The study reporting each gained region is indicated to the left or right, with colored lines and text highlighting studies examining particular cancer types. The minimal region of overlap (supported by 6 studies) is shaded in light blue and includes the proposed amplification target gene WWP1, the 5′ position of which is shown using a vertical arrow. The position of the minimally gained region identified through the consideration of gains of at least 1 Mb in length (Fig. 1) is shown as a horizontal bar below the chromosome 8 coordinate scale.
The 87.42- to 87.53-Mb region includes WWP1 (87.424-87.549 Mb), previously proposed as an amplification target in prostate and breast cancer.7,8 Overall, WWP1 mapped within defined amplicons and showed associated overexpression in breast30,33 and ovarian cancer studies35 (Table 4) and showed associations between expression and copy number in additional breast46,48,50,54 and prostate cancer studies39 (Table 4). WWP1 was also included within a 420-kb region of gain defined in a colorectal cancer cohort, where gene expression was not investigated44 (Fig. 4). It may be significant that increased WWP1 levels have been reported in colorectal cancer biopsies versus nontumor mucosa using antibody microarrays.55
Other regions of overlap at chromosome 8q21.2 and 8q21.3 were also considered, but these did not conclusively support additional target genes. A region of overlap at 86.62 to 86.89 Mb could be defined by a gained region reported in ERBB2-positive breast cancer 43 and was supported by 4 other studies (Fig. 4). However the REXO1L1 gene within this region was overexpressed in only one study that supported the region of overlap.35 Another region between 88.73 to 89.24 Mb was defined by the proximal extent of gain reported by Natrajan et al.43 and the distal extent of gain reported by Ramakrishna et al.35 and supported by 4 studies. However, the inclusion of gains less than 1 Mb in length did not refine this region further, and the single gene MMP16 that partially maps within this region was not supported by expression or other criteria.
Other Chromosome 8q21 Target Genes Predicted by Individual Studies
The study of Beroukhim et al.28 represented a landmark in terms of the numbers of cancer samples analyzed at a high level of genomic resolution, and from diverse tumor types. We therefore considered whether the findings of this single large study supported the combined predictions of all studies reviewed. Beroukhim et al.28 identified a peak region of amplification at chromosome 8q21.13 at 81.242 to 81.979 Mb (Fig. 3A). This was strikingly similar to that reported by Abkevich et al.27 at 81.240 to 81.974 Mb, which occurred in approximately 3% of the 178 cancer cell lines analyzed (Fig. 3A). These regions overlap with gains reported in breast cancer,43,46 breast cell lines,40 colorectal carcinoma,44 lung cancer,9 prostate carcinoma,37 epithelial ovarian cancer,35 esophageal squamous cell carcinoma,56 and small bowel carcinoid tumors31 (Figs. 1 and 3A). However, these regions did not contribute to the minimal regions of overlap predicted at chromosome 8q21.13 (Fig. 3A and 3B).
While Abkevich et al.27 proposed TPD52 as the relevant target gene, only the first alternatively spliced TPD52 exon was included within the defined regions of gain.27,28 Instead, these include the full gene sequences of ZBTB10 (81.561-81.597 Mb) and ZNF704 (81.713-81.949 Mb). We therefore considered whether ZBTB10 and/or ZNF704 genes could be supported as candidate gene amplification targets. ZNF704 overexpression in association with increased copy number was reported infrequently,35,51 and to date, this gene has not been functionally characterized. In contrast, ZBTB10 was overexpressed when increased in copy number in 3 studies,35,37,40 and other breast cancer studies have reported associations between ZBTB10 copy number and gene expression32,47,53,54 or patient prognosis36 (Table 3). Oncogenic functions are yet to be reported for ZBTB10, whose overexpression in MCF-7 cells arrested cell cycle progression and reduced estrogen receptor-a expression.57 ZBTB10 and ZNF704 may therefore represent passenger genes, and the inclusion of a TPD52 exon (and possibly upstream regulatory sequences) within the amplicons described by Abkevich et al.27 and Beroukhim et al.28 could indicate that these target TPD52. Passenger genes adjacent to target genes may alter the positions of minimal common regions such that these do not include the true target,58 so the comparison of results obtained by multiple studies may help to distinguish passengers from targets. Ultimately, direct experimentation will be required to determine whether ZBTB10 and/or ZNF704 present chromosome 8q21.13 passengers, or drivers of genomic gain.
Summary
We performed a literature analysis in order to predict candidate gene amplification targets at chromosome 8q21, using several approaches. Firstly, we performed 2-stage comparisons of chromosome 8q21 gains reported by different studies (Table 1) to highlight a number of regions of overlap between these (Figs. 1 -4). Where gene expression analyses were performed in parallel with amplicon mapping (Table 1), we then considered which genes included in defined 8q21 gains were overexpressed (Tables 2 -4). Finally, we considered which chromosome 8q21 genes showed associations between gene copy number and expression, or other parameters, in additional unbiased genomic studies (Tables 2 -4). These approaches highlighted 3 previously proposed amplification target genes TCEB1, TPD52, and WWP1 as mapping within minimal regions of overlap (Figs. 1 -3) and showing associations between copy number and expression in both the reviewed and additional studies (Tables 2 -4). Based upon human genomic copy number variants reported to date, it seems unlikely that increased copy number at these genes reflects germline copy number variations,59-62 although this possibility needs to be considered in future studies. Our findings therefore suggest that the detailed analysis of multiple studies may reliably predict candidate amplification targets, and that for common tumor types, the combined existing literature is likely to be helpfully predictive.
It is important to note that multiple forms of evidence were required to confidently predict candidate amplification targets. Some reviewed studies reported more than 10 discrete amplicons within chromosome 8q21, and many genes whose expression was upregulated.33,34 This led to some 22 chromosome 8q21 genes being supported by at least 3 of the studies considered (Tables 2 -4). Similarly, equal numbers of studies supported the chromosome 8q21.11 genes UBE2W, PXMP3, and TCEB1 if associations between gene copy number and expression or patient outcome were considered (Table 2). However when gene expression results obtained in association with amplicon mapping were considered, TCEB1 emerged as the stronger candidate (Table 2). This was consistent with this gene being located within the minimal region of overlap for gains at chromosome 8q21.11 (Fig. 2).
It was also notable that no chromosome 8q21 gene was unanimously identified as a putative amplification target by all relevant studies, and even ERBB2 was not strongly supported by associations between gene copy number and expression on occasions.40 Thus, the identities of amplification target genes may only emerge when the results of multiple studies are combined. Such an approach is clearly not infallible in that one region of overlap (Fig. 1) was not reproduced when gains of less than 1 Mb were considered (Fig. 3A and Table 3) and others (Figs. 2 and 4) did not target overexpressed genes. The latter could indicate the targeting of regulatory genomic regions as opposed to a coding gene, which could be further explored. However, our analyses suggest that comparative approaches may help prioritize genes for further study, and lead to more productive use of downstream resources. For example, careful predictions based upon genomic data may improve subsequent high-throughput gene inactivation assays, by allowing genes to be designated as either candidate drivers or passengers at the time of experimental design.
Future Perspectives
Biology has been recently transformed from a data-poor to a data-rich science.63 The possibilities allowed by this transition are enormous, but history suggests that our capacity for data generation is not immediately matched by our capacity for data analysis, particularly when these activities require different skill sets and tools. This gap translates to a failure to fully realize the opportunities allowed by technological advances, which becomes more significant when data generation comes at a high cost.
This gap may be widening through the use of next- generation sequencing techniques, where some 25,000 sequenced cancer genomes will be available for analysis in the coming 5 years.64 While next-generation sequencing has primarily been used to detect small sequence variations such as point mutations,64 this can also provide exact measures of copy number for individual genes. Unlike array-based approaches, genomic sequencing can also reveal the physical structure of DNA amplicons, and whether these include inversions, translocations, and/or insertions.65 For example, sequencing has recently revealed that SK-BR-3 cells contain a large 9.1-Mb tandem duplication at chromosome 8q21, which was also involved in highly amplified regions, and an interchromosomal translocation connecting chromosomes 8 and 17.65 However, the amplified chromosome 8q21 regions reported through sequencing65 were broader than those identified using array-based approaches.32 Thus, while array-based techniques may become less widely used for discovery, genomic profiling data should continue to invaluably complement sequencing data and contribute to its more rapid and accurate interpretation.
To facilitate the analysis of valuable genomic data by as many researchers as possible, data need to be very broadly accessible. Our analyses suggest that the aCGH and SNP profiling literature represents a rich and accessible source of information regarding the identities of critical genes. However, the potential of this literature may not have been fully exploited. While comparative literature analyses are available to all researchers, they are time intensive and can be made more so through variable and/or incomplete reporting of results, particularly for the genomic extent of gain or loss. Literature analyses are also facilitated by specialist knowledge of particular genomic regions, and not all researchers equipped with this knowledge will have the time or motivation to undertake these.
Electronic databases greatly help to distill information from the literature, and render this more accessible to a broader range of researchers. The particular databases that warehouse genomic data have been recently reviewed.66 However, databases compiling genomic aberrations reported by different studies may only provide information regarding whether a particular gene is gained or lost in cancer, without corresponding information regarding the genomic extent of gain or loss.66 While this approach may serve the needs of researchers interested in particular genes, this does not allow the merits of adjacent or neighboring genes to be compared, and may not allow driver and passenger genes to be distinguished. The Tumorscape database28 provides a comparative visual representation of the genomic extent of gains and losses reported for individual cases, and a statistical estimate of the significance of the gain or loss of individual genes. This database is also easily accessed by untrained users. However, differences between amplicons predicted by Beroukhim et al.28 and other studies reviewed here (Fig. 3A) highlight the value of comparing results obtained by different studies. There is currently no broadly accessible database that allows the comparison of amplicons and deletions reported by different investigators. Such a database would provide an invaluable resource for a broad range of cancer researchers, particularly if this also allowed cross-referencing to gene expression, mutation, and other data. Compared with the time required for literature-based analyses, such a database could greatly accelerate the identification of amplification target “hills” in genomic regions that lack broadly accepted candidates, and assist in the interpretation of next-generation sequencing data.
Acknowledgments
The authors thank Ms. Sarah Frost for critical reading of the article, anonymous reviewers for their constructive comments, and past and present group members and external collaborators for their support.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received the following financial support for the research, authorship, and/or publication of this article: Work in the Molecular Oncology Laboratory has been supported by the New South Wales Cancer Council (to J.A.B.), the National Health and Medical Research Council of Australia (to Y.C. and N.M.L.R.), and the CureCancer Foundation (to Y.C.) and by donations to the Children’s Cancer Research Unit and the Oncology Department of the Children’s Hospital at Westmead and the Kids Cancer Project.
References
- 1. Harris TJ, McCormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7:251-65 [DOI] [PubMed] [Google Scholar]
- 2. Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59-64 [DOI] [PubMed] [Google Scholar]
- 3. Wood LD, Parsons DW, Jones S, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-13 [DOI] [PubMed] [Google Scholar]
- 4. Leary RJ, Lin JC, Cummins J, et al. Integrated analysis of homozygous deletions, focal amplifications, and sequence alterations in breast and colorectal cancers. Proc Natl Acad Sci U S A. 2008;105:16224-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balleine RL, Fejzo MS, Sathasivam P, Basset P, Clarke CL, Byrne JA. The hD52 (TPD52) gene is a candidate target gene for events resulting in increased 8q21 copy number in human breast carcinoma. Genes Chromosomes Cancer. 2000;29:48-57 [DOI] [PubMed] [Google Scholar]
- 6. Porkka K, Saramäki O, Tanner M, Visakorpi T. Amplification and overexpression of Elongin C gene discovered in prostate cancer by cDNA microarrays. Lab Invest. 2002;82:629-37 [DOI] [PubMed] [Google Scholar]
- 7. Chen C, Sun X, Guo P, et al. Ubiquitin E3 ligase WWP1 as an oncogenic factor in human prostate cancer. Oncogene. 2007;26:2386-94 [DOI] [PubMed] [Google Scholar]
- 8. Chen C, Zhou Z, Ross JS, Zhou W, Dong JT. The amplified WWP1 gene is a potential molecular target in breast cancer. Int J Cancer. 2007;12:180-7 [DOI] [PubMed] [Google Scholar]
- 9. Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kallioniemi A, Kallioniemi O-P, Piper J, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci U S A. 1994;91:2156-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Isola JJ, Kallioniemi OP, Chu LW, et al. Genetic aberrations detected by comparative genomic hybridization predict outcome in node negative breast cancer. Am J Pathol. 1995;147:905-11 [PMC free article] [PubMed] [Google Scholar]
- 12. Ried T, Just KE, Holtgreve-Grez H, et al. Comparative genomic hybridization of formalin-fixed, paraffin-embedded breast tumors reveals different patterns of chromosomal gains and losses in fibroadenomas and diploid and aneuploid carcinomas. Cancer Res. 1995;55:5415-23 [PubMed] [Google Scholar]
- 13. Nishizaki T, DeVries S, Chew K, et al. Genetic alterations in primary breast cancers and their metastases: direct comparison using modified comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:267-72 [DOI] [PubMed] [Google Scholar]
- 14. Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi O-P. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177-84 [PubMed] [Google Scholar]
- 15. Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen RH. Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes Cancer. 1994;11:153-62 [DOI] [PubMed] [Google Scholar]
- 16. Cher ML, Bova GS, Moore DH, et al. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization. Cancer Res. 1996;56:3091-102 [PubMed] [Google Scholar]
- 17. Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Detection of DNA amplification in 17 primary breast carcinomas with homogenously staining regions by a modified comparative genomic hybridization technique. Genes Chromosomes Cancer. 1994;10:160-70 [DOI] [PubMed] [Google Scholar]
- 18. Courjal F, Theillet C. Comparative genomic hybridization of breast tumors with predetermined profiles of DNA amplification. Cancer Res. 1997;57:4368-77 [PubMed] [Google Scholar]
- 19. Tarkkanen M, Karhu R, Kallioniemi A, et al. Gains and losses of DNA sequences in osteosarcomas by comparative genomic hybridization. Cancer Res. 1995;55:1334-8 [PubMed] [Google Scholar]
- 20. Suehiro Y, Sakamoto M, Umayahara K, et al. Genetic aberrations detected by comparative genomic hybridization in ovarian clear cell adenocarcinomas. Oncology. 2000;59:50-66 [DOI] [PubMed] [Google Scholar]
- 21. Partheen K, Levan K, Osterberg L, Helou K, Horvath G. Analysis of cytogenetic alterations in stage III serous ovarian adenocarcinoma reveals a heterogeneous group regarding survival, surgical outcome, and substage. Genes Chromosomes Cancer. 2004;40:342-8 [DOI] [PubMed] [Google Scholar]
- 22. Melchor L, Alvarez S, Honrado E, et al. The accumulation of specific amplifications characterizes two different genomic pathways of evolution of familial breast tumors. Clin Cancer Res. 2005;11:8577-84 [DOI] [PubMed] [Google Scholar]
- 23. Rennstam K, Ahlstedt-Soini M, Baldetorp B, et al. Patterns of chromosomal imbalances defines subgroups of breast cancer with distinct clinical features and prognosis: a study of 305 tumors by comparative genomic hybridization. Cancer Res. 2003;63:8861-8 [PubMed] [Google Scholar]
- 24. Han W, Han MR, Kang JJ, et al. Genomic alterations identified by array comparative genomic hybridization as prognostic markers in tamoxifen-treated estrogen receptor-positive breast cancer. BMC Cancer. 2006;6:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jalava SE, Porkka KP, Rauhala HE, et al. TCEB1 promotes invasion of prostate cancer cells. Int J Cancer. 2009;124:95-102 [DOI] [PubMed] [Google Scholar]
- 26. Shehata M, Weidenhofer J, Thamotharampillai K, Hardy JR, Byrne JA. Tumor protein D52 overexpression and gene amplification in cancers from a mosaic of microarrays. Crit Rev Oncog. 2008;14:33-55 [DOI] [PubMed] [Google Scholar]
- 27. Abkevich V, Iliev D, Timms KM, et al. Computational method for estimating DNA copy numbers in normal samples, cancer cell lines, and solid tumors using array comparative genomic hybridization. J Biomed Biotechnol. 2010;2010:386870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463: 899-905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Man TK, Lu XY, Jaeweon K, et al. Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer. 2004;4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao J, Weremowicz S, Feng B, et al. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065-78 [DOI] [PubMed] [Google Scholar]
- 31. Kulke MH, Freed E, Chiang DY, et al. High-resolution analysis of genetic alterations in small bowel carcinoid tumors reveals areas of recurrent amplification and loss. Genes Chromosomes Cancer. 2008;47:591-603 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez V, Chen Y, Elkahloun A, Dutra A, Pak E, Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer. 2007;46:694-707 [DOI] [PubMed] [Google Scholar]
- 33. Chin SF, Teschendorff AE, Marioni JC, et al. High-resolution aCGH and expression profiling identifies a novel genomic subtype of ER negative breast cancer. Genome Biol. 2007;8:R215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim JH, Dhanasekaran SM, Mehra R, et al. Integrative analysis of genomic aberrations associated with prostate cancer progression. Cancer Res. 2007;67:8229-39 [DOI] [PubMed] [Google Scholar]
- 35. Ramakrishna M, Williams LH, Boyle SE, et al. Identification of candidate growth promoting genes in ovarian cancer through integrated copy number and expression analysis. PLoS One. 2010;5:e9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang Y, Martens JW, Yu JX, et al. Copy number alterations that predict metastatic capability of human breast cancer. Cancer Res. 2009;69:3795-801 [DOI] [PubMed] [Google Scholar]
- 37. van Duin M, van Marion R, Vissers K, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005;44:438-49 [DOI] [PubMed] [Google Scholar]
- 38. Woo HG, Park ES, Lee JS, et al. Identification of potential driver genes in human liver carcinoma by genomewide screening. Cancer Res. 2009;69:4059-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holcomb IN, Young JM, Coleman IM, et al. Comparative analyses of chromosome alterations in soft-tissue metastases within and across patients with castration-resistant prostate cancer. Cancer Res. 2009;69:7793-802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kao J, Salari K, Bocanegra M, et al. Molecular profiling of breast cancer cell lines defines relevant tumor models and provides a resource for cancer gene discovery. PLoS One. 2009;4:e6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jia D, Wei L, Guo W, et al. Genome-wide copy number analyses identified novel cancer genes in hepatocellular carcinoma. Hepatology. 2011;54:1227-36 [DOI] [PubMed] [Google Scholar]
- 42. Marchiò C, Iravani M, Natrajan R, et al. Genomic and immunophenotypical characterization of pure micropapillary carcinomas of the breast. J Pathol. 2008;215:398-410 [DOI] [PubMed] [Google Scholar]
- 43. Natrajan R, Lambros MB, Rodríguez-Pinilla SM, et al. Tiling path genomic profiling of grade 3 invasive ductal breast cancers. Clin Cancer Res. 2009;15:2711-22 [DOI] [PubMed] [Google Scholar]
- 44. Sayagués JM, Fontanillo C, Abad Mdel M, et al. Mapping of genetic abnormalities of primary tumours from metastatic CRC by high-resolution SNP arrays. PLoS One. 2010;5:e13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jönsson G, Staaf J, Olsson E, et al. High-resolution genomic profiles of breast cancer cell lines assessed by tiling BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2007;46: 543-58 [DOI] [PubMed] [Google Scholar]
- 46. Guedj M, Marisa L, de Reynies A, et al. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pollack JR, Sørlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adélaïde J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67:11565-75 [DOI] [PubMed] [Google Scholar]
- 49. Paris PL, Hofer MD, Albo G, et al. Genomic profiling of hormone-naive lymph node metastases in patients with prostate cancer. Neoplasia. 2006;8:1083-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Andre F, Job B, Dessen P, et al. Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin Cancer Res. 2009;15:441-51 [DOI] [PubMed] [Google Scholar]
- 51. Natrajan R, Weigelt B, Mackay A, et al. An integrative genomic and transcriptomic analysis reveals molecular pathways and networks regulated by copy number aberrations in basal-like, HER2 and luminal cancers. Breast Cancer Res Treat. 2010;121:575-89 [DOI] [PubMed] [Google Scholar]
- 52. Mattison J, Kool J, Uren AG, et al. Novel candidate cancer genes identified by a large-scale cross-species comparative oncogenomics approach. Cancer Res. 2010;70:883-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hyman E, Kauraniemi P, Hautaniemi S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240-5 [PubMed] [Google Scholar]
- 54. Staaf J, Jönsson G, Ringnér M, et al. High-resolution genomic and expression analyses of copy number alterations in HER2-amplified breast cancer. Breast Cancer Res. 2010;12:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Madoz-Gúrpide J, Cañamero M, Sanchez L, Solano J, Alfonso P, Casal JI. A proteomics analysis of cell signaling alterations in colorectal cancer. Mol Cell Proteomics. 2007;6:2150-64 [DOI] [PubMed] [Google Scholar]
- 56. Hu N, Wang C, Ng D, et al. Genomic characterization of esophageal squamous cell carcinoma from a high-risk population in China. Cancer Res. 2009;69:5908-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li X, Mertens-Talcott SU, Zhang S, Kim K, Ball J, Safe S. MicroRNA-27a indirectly regulates estrogen receptor-a expression and hormone responsiveness in MCF-7 breast cancer cells. Endocrinology. 2010;151:2462-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949-51 [DOI] [PubMed] [Google Scholar]
- 60. de Smith AJ, Tsalenko A, Sampas N, et al. Array CGH analysis of copy number variation identifies 1284 new genes variant in healthy white males: implications for association studies of complex diseases. Hum Mol Genet. 2007;16:2783-94 [DOI] [PubMed] [Google Scholar]
- 61. Ahn SM, Kim TH, Lee S, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pang AW, MacDonald JR, Pinto D, et al. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 2010;11:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schulze A, Downward J. Analysis of gene expression by microarrays: cell biologist’s gold mine or minefield? J Cell Sci. 2000;113:4151-6 [DOI] [PubMed] [Google Scholar]
- 64. Diamandis EP, Hudson T, Kallioniemi O, Liu ET, López-Otín C. Cancer genomes. Clin Chem. 2010;56:1660-4 [DOI] [PubMed] [Google Scholar]
- 65. Hillmer L, Yao F, Inaki K, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 2011;21:665-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Vincent-Salomon A, Lucchesi C, Gruel N, et al. Integrated genomic and transcriptomic analysis of ductal carcinoma in situ of the breast. Clin Cancer Res. 2008;14:1956-65 [DOI] [PubMed] [Google Scholar]