Abstract
Ewing sarcoma is a tumor of the bone and soft tissue caused by the expression of a translocation-derived oncogenic transcription factor, EWS/FLI. Overt metastases are associated with a poor prognosis in Ewing sarcoma, but patients without overt metastases frequently harbor micrometastatic disease at presentation. This suggests that the metastatic potential of Ewing sarcoma exists at an early stage during tumor development. We have therefore explored whether the inciting oncogenic event in Ewing sarcoma, EWS/FLI, directly modulates tumor cell features that support metastasis, such as cell adhesion, cell migration, and cytoarchitecture. We used an RNAi-based approach in patient-derived Ewing sarcoma cell lines. Although we hypothesized that EWS/FLI might induce classic metastatic features, such as increased cell adhesion, migration, and invasion (similar to the phenotypes observed when epithelial malignancies undergo an epithelial-to-mesenchymal transition during the process of metastasis), surprisingly, we found the opposite. Thus, EWS/FLI expression inhibited the adhesion of isolated cells in culture and prevented adhesion in an in vivo mouse lung assay. Cell migration was similarly inhibited by EWS/FLI expression. Furthermore, EWS/FLI expression caused a striking loss of organized actin stress fibers and focal adhesions and a concomitant loss of cell spreading, suggesting that EWS/FLI disrupts the mesenchymal phenotype of a putative tumor cell-of-origin. These data suggest a new paradigm for the dissemination and metastasis of mesenchymally derived tumors: these tumors may disseminate via a “passive/stochastic” model rather than via an “active” epithelial-to-mesenchymal type transition. In the case of Ewing sarcoma, it appears that the loss of cell adhesion needed to promote tumor cell dissemination might be induced by the EWS/FLI oncogene itself rather than via an accumulation of stepwise mutations.
Keywords: cell adhesion, cytoskeleton, Ewing sarcoma, EWS/FLI, metastasis
Cancer is the second leading cause of death worldwide. More than 90% of the deaths due to cancer are attributable to metastatic disease. Based on this clinical reality, it is widely argued that effective treatment of cancer will depend significantly on our ability to block the process of metastasis.1 Indeed, many therapeutic agents in development for cancer applications are designed to interfere with aspects of the metastatic cascade.
Because of the profound impact of metastasis on cancer outcomes, significant research focus has been applied to understanding the factors that influence metastatic spread of tumor cells. Work in this area has been concentrated on tumors of epithelial origin, in large part because carcinomas are the most common cancers. Years of investigation have led to a now widely accepted multistep model for tumor metastasis that involves an “active” process including an epithelial-to-mesenchymal transition with important morphologic changes that allow for release of cells from the primary tumor site, invasion of tumor cells into the surrounding tissue, movement of tumor cells into and out of blood or lymphatic vessels, and adaptation of tumor cells to microenvironmental conditions encountered at the ectopic site. Although different groups emphasize distinct aspects of the metastatic cascade, the process can generally be depicted as having 2 major phases: first, the translocation phase in which a tumor cell moves from the primary tumor site to a distant organ, and second, the colonization phase in which the tumor cells adapt and proliferate in the ectopic site to establish metastatic disease.2,3
Substantial mechanistic detail has been deciphered for how carcinomas achieve the capacity to navigate the translocation phase. For a cell within an epithelial-derived tumor, a critical step involves an epithelial-to-mesenchymal transition with associated loss of cell surface adhesion molecules, such as E-cadherin, that would normally tether the cell to its neighbors.4 The loss of E-cadherin is correlated with advanced metastatic disease in several types of epithelial-derived tumors.5,6 Moreover, the importance of E-cadherin loss for tumor metastasis has been directly demonstrated in several preclinical models. For example, disruption of E-cadherin expression in a mouse model of pancreatic cancer leads to reduced cell-cell adhesion as well as temporally accelerated tumor invasion and metastasis.7,8 In addition to loss of cell-cell adhesion, the carcinoma cells also acquire enhanced migratory and invasive potential that typically involves changes in expression of cytoskeletal proteins, integrins, and proteases that help dissolve the basement membrane and promote motility.9,10 These changes are thought to facilitate the movement of tumor cells into the vasculature, where they can be transported throughout the body. Ultimately, the tumor cells leave the vasculature and invade a new organ site, where significant adaptation and proliferation result in colonization and formation of metastases.
The dramatic changes in cell behavior that are necessary for metastasis must ultimately be driven by changes in gene expression programs within the tumor cells. It is envisioned that cancer cells navigate the substantial environmental challenges encountered along their metastatic journeys by acquisition of genetic and epigenetic changes beyond the initial oncogenic transformation events that influence cell properties such as adhesion, migration, and survival support progression to metastasis.11,12 Thus, metastasis is thought to represent an important later step during tumor progression. Consistent with the view that acquisition of metastatic potential occurs over an extended time period and involves multiple genetic steps, many cancers grow for years at the primary site prior to any evidence of dissemination and metastatic behavior.13-15
However, in apparent conflict with the multistep model of metastasis, some cancers are typically metastatic upon presentation, even under conditions where the primary tumor is small.16-18 These examples raise the possibility that a very early event, perhaps even the initiating oncogenic event, might simultaneously influence both cell proliferation and capacity for metastasis. As discussed above, the prevailing model for metastasis is derived from study of epithelial-derived tumor cells, which undergo an epithelial-to-mesenchymal transition during acquisition of metastatic potential. This raises interesting questions about whether nonepithelial tumors, such as sarcomas, may follow a different course on their path to metastasis.
Ewing sarcoma is an important example of a tumor that usually displays early evidence of metastatic dissemination. Ewing sarcoma is a small round blue cell tumor of the bone and soft tissue with peak incidence in teenage years. Ewing sarcoma represents the second most common bone tumor in children and adolescents. Although approximately two-thirds of patients with localized Ewing sarcoma can be cured with intensive chemotherapy, along with surgery and/or radiation therapy, patients with metastatic Ewing sarcoma have a very poor prognosis, with less than 20% 5-year disease-free survival.19 In the absence of chemotherapy, approximately 90% of patients die from disease following definitive surgery, suggesting that the vast majority of patients have micrometastatic disease at presentation.20,21 Furthermore, circulating tumor cells can often be identified in patients using RT-PCR or flow-cytometric techniques.22-29
The majority of Ewing tumors display an (11;22) (q24;q12) chromosomal translocation, encoding the chimeric transcription factor EWS/FLI,30-32 with other EWS fusion proteins associated with the remainder of Ewing sarcoma tumors.33 The EWS/FLI fusion protein links the amino terminus of EWS, a strong transcriptional activator, and the carboxy terminus of FLI1, a DNA-binding protein from the ETS family of transcription factors.31,34 EWS/FLI acts as a transcriptional regulator to modulate hundreds to thousands of genes expression, and ongoing EWS/FLI expression is required for maintenance of transformation.35-38
Although the role of EWS/FLI in transcriptional regulation is well-documented, little is understood about how EWS/FLI expression influences cell behaviors that contribute to the ultimate tumor phenotype in patients. Early studies relied on the stable expression of EWS/FLI in NIH3T3 murine fibroblasts.34,39-43 However, murine fibroblasts expressing EWS/FLI protein do not recapitulate the gene expression pattern of bona fide patient-derived Ewing sarcoma cell lines or tumors,44,45 highlighting the need for a cell-based model that more faithfully replicates what occurs in human Ewing sarcoma tumor cells. Efforts to develop a high fidelity experimental approach to study the consequences of EWS/FLI expression for cell behavior have been further complicated by the fact that the cell of origin that gives rise to Ewing sarcoma remains controversial.46-49
To circumvent these challenges and to achieve a cell culture model that reliably reflects the contextual properties of the patient tumor, we have used patient-derived Ewing sarcoma cells and stable RNA-interference to knock down EWS/FLI expression.35,38,50 This approach allowed us to study the effects of EWS/FLI on key properties related to the metastatic phenotype, such as cell adhesion, migration, invasion, and cytoarchitecture. Our initial hypothesis was that EWS/FLI might induce cellular features consistent with those observed in epithelial malignancies undergoing epithelial-to-mesenchymal transitions. Surprisingly, we observed the opposite phenotype, with EWS/FLI inhibiting cell adhesion, rather than promoting cell migration and invasion. Our results suggest that EWS/FLI expression might stimulate the dissemination of Ewing sarcoma cells by influencing cell adhesion and raises the possibility that sarcomas might metastasize via pathways that are distinct from epithelial malignancies.
Results
Efficient knockdown of EWS/FLI using RNA interference
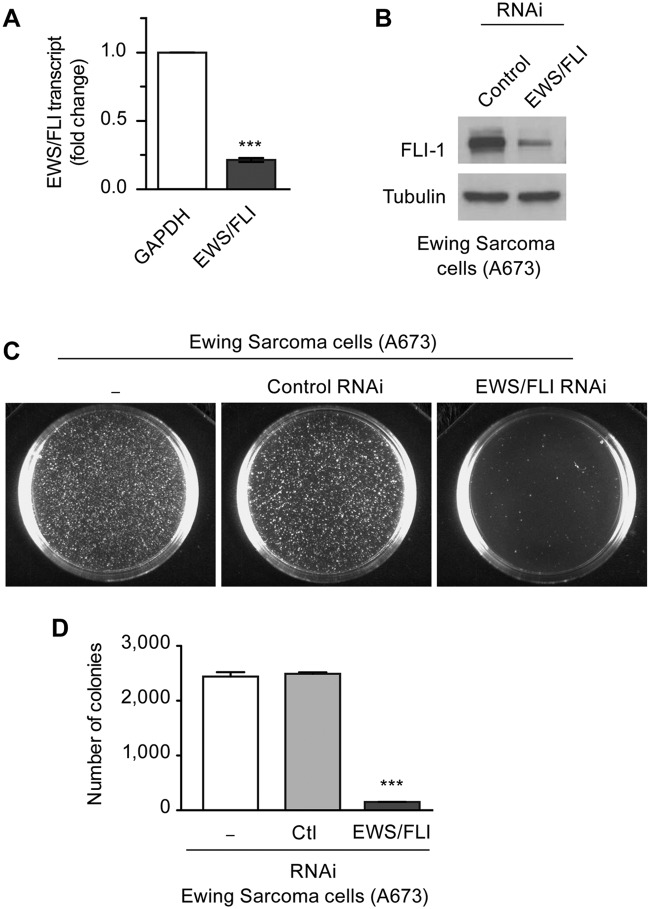
To characterize the impact of the EWS/FLI fusion oncoprotein on cell phenotype and behavior, we used a model system in which expression of endogenous EWS/FLI is knocked down by RNA interference (RNAi) in patient-derived Ewing sarcoma cells. Retroviral infection was used to introduce short-hairpin RNAs against luciferase as a control or against the 3′ untranslated region of EWS/FLI in A673 Ewing sarcoma cells.38 The A673 cell line was derived from a human tumor that was definitively diagnosed as Ewing sarcoma by molecular cytogenetic analysis51 and has been shown to establish tumors when introduced by subcutaneous injection into immunocompromised mice.38 RNAi mediated knockdown of EWS/FLI resulted in efficient reduction of EWS/FLI transcript (Fig. 1A). Because wild-type FLI is not expressed in Ewing sarcoma cells,38 the oncogenic fusion protein is the only protein targeted by the RNAi. Antibody directed against the carboxyl-terminus of FLI demonstrated a corresponding reduction in the level of EWS/FLI protein (Fig. 1B).
Figure 1.
Retroviral RNAi mediated knockdown of EWS/FLI in Ewing sarcoma cells. (A) Retroviral mediated knockdown of endogenous EWS/FLI fusion transcripts in Ewing sarcoma A673 cells was measured by semiquantitative RT-PCR, with GAPDH transcripts as control. Transcripts were normalized as fold change compared with control A673 cells. (B) Western immunoblot with FLI-1 antibody detected EWS/FLI fusion protein in A673 cells with control RNAi and significant reduction of EWS/FLI protein with EWS/FLI RNAi. Tubulin antibody was used to show equivalent protein loading. (C) In soft agar transformation assays of A673 cells, the number of colonies formed in 4 weeks was unaffected by control RNAi but significantly reduced by EWS/FLI RNAi (D). ***P < 0.001.
By monitoring growth in soft agar as a hallmark of oncogenic transformation, we observed nearly complete loss of colony forming efficiency when A673 cells were infected with EWS/FLI RNAi retroviruses versus those with control RNAi (Fig. 1C, D). We previously showed that EWS/FLI knockdown in A673 cells does not change cell proliferation or lead to growth arrest.38 Thus, the loss of capacity to grow in soft agar illustrates the requirement for ongoing expression of EWS/FLI to sustain anchorage-independent cell growth.
EWS/FLI expression inhibits cell adhesion and spreading
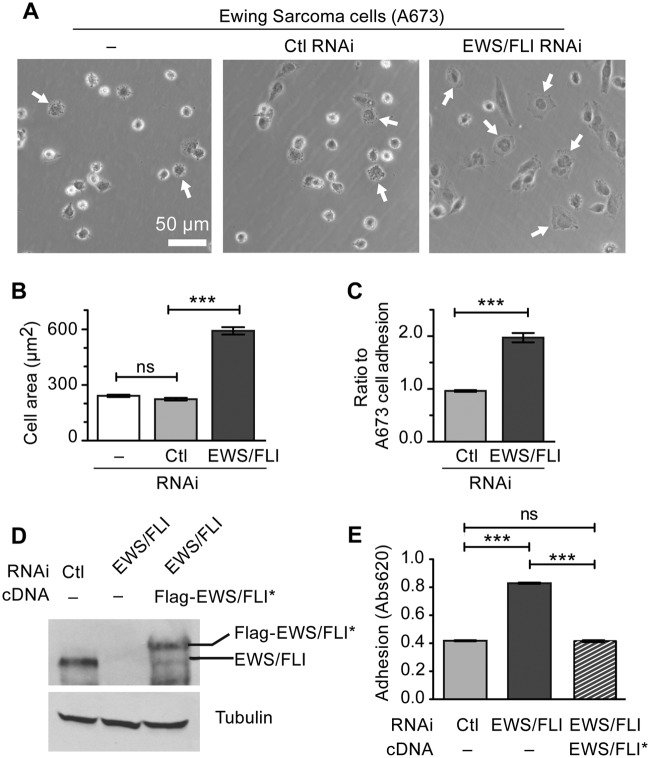
Since an early determinant of metastatic potential is the ability of tumor cells to exit the primary tumor mass, we first evaluated the impact of EWS/FLI expression on Ewing sarcoma cell adhesion. We analyzed the ability of cells to adhere and spread on fibronectin-coated tissue culture plastic over a 2-hour time period in the presence of serum. By visual inspection using phase contrast microscopy, we noted that Ewing sarcoma cells in which EWS/FLI expression was subject to RNA interference displayed more robust adhesion and spreading compared with Ewing sarcoma cells that retained EWS/FLI expression (Fig. 2A). Cell spreading requires the establishment of progressive, circumferential substratum adhesion zones as the cell periphery extends its reach. Direct measurement of cell area revealed that control Ewing sarcoma cells display a statistically significant reduction in their capacity to spread compared with Ewing sarcoma cells in which EWS/FLI expression is compromised (Fig. 2B), further suggesting that EWS/FLI induces a reduction in cell adhesion, which can be measured in vitro. Quantitative colorimetric adhesion assays confirmed these results, demonstrating that suppression of EWS/FLI expression results in a 200% increase in adherent cells compared with the parental A673 cells, whereas treatment with a control RNAi construct had no impact on cell adhesion (Fig. 2C).
Figure 2.
EWS/FLI expression abrogates cell adhesion and spreading. (A) Ewing sarcoma A673 cells alone or with control or EWS/FLI RNAi were plated on a tissue culture dish for 2 hours and then evaluated for cell spreading by phase contrast microscopy. Arrows indicate cells adhering to the substratum, spreading, and losing their phase brightness. (B) Measurement of A673 cell area following 2 hours adhesion showed that EWS/FLI RNAi enhanced cell spreading compared with cells with control RNAi. (C) In a colorimetric adhesion assay of cells plated for 2 hours, the compromised adhesiveness of cells with control RNAi compared with the EWS/FLI RNAi was quantitated and shown to be approximately 2-fold. (D) Western immunoblot for EWS/FLI protein in A673 cells with control RNAi or EWS/FLI RNAi were compared with EWS/FLI RNAi cells reconstituted with Flag-tagged EWS/FLI* cDNA that is resistant to the RNAi. The reconstituted EWS/FLI levels approached those seen in control A673 cells, and the tubulin signal shows equivalent protein loading. (E) Cell adhesion over 2 hours was measured by a colorimetric assay for A673 cells with control RNAi (light gray) and EWS/FLI RNAi (dark gray) and showed that EWS/FLI knockdown increased cell adhesion 2-fold over control cells. Reconstituted cells programmed to express RNAi-resistant EWS/FLI* (striped) exhibited adhesion that was not statistically different (ns) than control cells. ***P < 0.001.
To ascertain that these changes in cell adhesion and spreading are EWS/FLI-dependent and not due to an off-target effect of the RNAi, we performed a knockdown-rescue experiment. We reconstituted expression of EWS/FLI in Ewing sarcoma cells that had been subjected to oncogene knockdown; expression of EWS/FLI was achieved using a previously published Flag-EWS/FLI-cDNA construct that is insensitive to our RNAi construct (Fig. 2D).38 Re-expression of EWS/FLI induced a statistically significant reduction in cellular adhesion (Fig. 2E), thereby confirming that EWS/FLI expression influences cell adhesion. Cells that were programmed to re-express EWS/FLI after knockdown displayed adhesion properties comparable to the parental Ewing sarcoma cells (Fig. 2E).
EWS/FLI expression compromises in vivo cell adhesion
To assess whether the in vitro finding that EWS/FLI expression compromises cell adhesion is relevant in a physiological setting, we tested the ability of Ewing sarcoma cells harboring control or EWS/FLI-directed RNAi constructs to adhere in a mouse lung colonization assay. Equal numbers of differentially labeled fluorescent Ewing sarcoma cells that had been selected after infection with retroviruses harboring either control or EWS/FLI RNAi constructs were introduced into the circulation of NOD SCID mouse recipients via tail vein injection. In our initial experiments, control RNAi treated cells were labeled with the lipophilic carbocyanine dye, DiI, which displays an emission maximum of 565nm (red); EWS/FLI RNAi treated cells were labeled with DiO, which displays an emission maximum of 501nm (green). The appearance of fluorescently labeled cells in the lungs of the recipient mice was monitored by fluorescence microscopy of lung sections derived from animals sacrificed 24 hours after tumor cell inoculation.
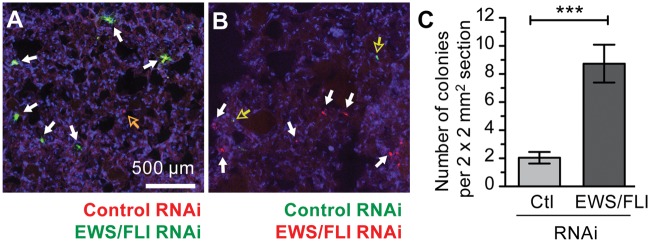
Consistent with the in vitro findings described above, A673 cells harboring the EWS/FLI knockdown construct (DiO labeled green fluorescent colonies in Fig. 3A) were found in greater abundance in the mouse lungs compared with cells harboring the control RNAi construct (DiI-labeled red fluorescent colonies in Fig. 3A). Six animals were examined for each RNAi construct. To control for potential differences in the lipophilic labels, such as fluorescence intensity or stability, we repeated this experiment, labeling the RNAi-treated Ewing sarcoma cells with the reciprocal dye, with similar results (Fig. 3B). Quantitative analysis was performed by counting the numbers of DiI- and DiO-labeled cell colonies in multiple lung sections as described in Materials and Methods. We found a statistically significant 4-fold reduction in the number of lung colonies derived from control Ewing sarcoma cells compared with those in which EWS/FLI expression was knocked down (Fig. 3C). These findings illustrate that cells expressing EWS/FLI are less adhesive in vivo, as measured by a lung colonization assay.
Figure 3.
EWS/FLI expression compromises cell adhesion in vivo. A673 cells with control RNAi were colored red (DiI) and cells with EWS/FLI RNAi were colored green (DiO), mixed in 1:1 ratio, and then injected into mouse lateral tail veins. Twenty-four hours later the lungs were harvested and cryosectioned, and the red and green cells were counted. (A) Maximum intensity projection of confocal images of lung sections showing differentially labeled A673 cells with control RNAi (red, hollow arrows) and EWS/FLI RNAi (green, solid white arrows). Nuclei are stained with DAPI (blue). (B) Lung sections from additional experiment of mice injected with the cell colors switched to avoid bias of dyes (green-control RNAi, red-EWS/FLI RNAi). (C) Cell colonies were counted for 4 mm2 sections in 15 representative sections and showed more cells with EWS/FLI RNAi (solid white arrows in A and B) accumulated in lung parenchyma compared with cells with control RNAi (hollow arrows in A and B). ***P < 0.001.
EWS/FLI expression adversely impacts both cellular migration and invasion
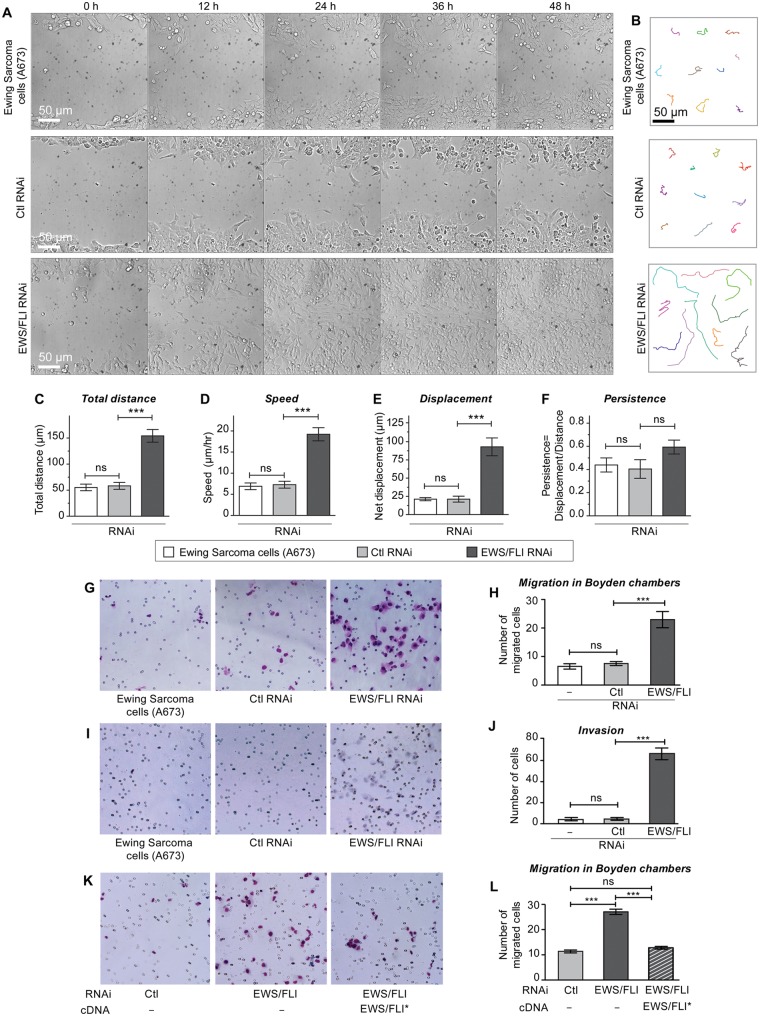
The finding that EWS/FLI expression caused decreased adhesion raised the possibility that cell migration might also be affected by EWS/FLI expression. Therefore, we next examined the impact of EWS/FLI expression on the inherent migratory and invasive ability of A673 Ewing sarcoma cells compared with A673 Ewing sarcoma cells in which EWS/FLI expression had been abrogated. Using a monolayer wound healing assay, we observed that the parental A673 Ewing sarcoma cells or A673 cells with control RNAi migrate more slowly than cells with EWS/FLI RNAi (Fig. 4A). Ewing sarcoma cells in which EWS/FLI expression was knocked down were able to reach the midline of the wound as early as 24 hours, whereas control RNAi expressing cells did not close the wound even after 48 hours.
Figure 4.
Expression of EWS/FLI results in reduced cellular migration and invasion. Ewing sarcoma A673 cells with control RNAi or EWS/FLI RNAi were evaluated for directed cell migration, random motility, chemotaxis, and invasion. (A) Cell migration into wounds scraped across cell monolayers was monitored by time lapse microscopy for 48 hours. A673 cells with EWS/FLI RNAi reached the midline of the wound within 24 hours, whereas the other A673 cells did not migrate effectively. (B) Random cell motility was tracked by time lapse microscopy (15-minute intervals for 8 hours), and representative cells tracks are shown. Analysis for total distance (C), speed (D), displacement (E), and persistence (F) was performed with MetaMorph software, and showed enhanced motility but no EWS/FLI-dependent difference in persistence in A673 cells with EWS/FLI RNAi. (G, H) Cells in serum-free media were plated onto Boyden transwell migration filters and then given 24 hours to migrate through pores (small circles on images) toward a high serum environment, fixed, and stained (blue). Compared with control cells, significantly more cells with EWS/FLI RNAi migrated. (I, J) Transwell invasion through Matrigel matrix (24 hours) indicated significant increase in cellular invasiveness upon knockdown of EWS/FLI. (K,L) A673 cells with control (light gray bar) or EWS/FLI RNAi (dark gray bar) were compared with EWS/FLI RNAi cells reconstituted with Flag-tagged EWS/FLI* cDNA (striped bar) that is resistant to the RNAi in transwell migration assays. Quantitation of the number of migrated cells indicated the EWS/FLI-dependence of this cell migration difference. ***P < 0.001, ns is not significantly different.
In an effort to dissect the mechanism underlying alteration in monolayer wound healing capacity associated with EWS/FLI expression, we performed time lapse microscopic analysis of individual cells to measure speed, distance, and displacement of cells over a 24 hour period in a random migration assay (Fig. 4B-F). By inspection of the motility tracks of individual cells, it was evident that expression of EWS/FLI inhibited the ability of the A673 cells to migrate on tissue culture plastic (Fig. 4B). Quantitative measurement of the individual cellular tracks revealed a statistically significant decrease in total distance traveled by control A673 cells compared with those in which EWS/FLI expression was knocked down (Fig. 4C). The mean cell velocity also reduced to 7 µm/h for control RNAi expressing cells compared with 19 µm/h for EWS/FLI RNAi expressing cells (Fig. 4D). The net displacement, defined as the length of a straight line drawn from the starting cell position to the ending position after the 8 hour migration interval had elapsed, was 92 µm for the A673 cells in which EWS/FLI was knocked down compared with 21 µm for the cells with control RNAi (Fig. 4E).
Cells can migrate randomly or in a directed fashion. To evaluate whether the expression of EWS/FLI also influences the directionality of Ewing sarcoma cell migration, we assessed the persistence of the migrating cells by calculating the ratio of net cell displacement to total path length travelled by the cell. Our analysis shows that A673 cells treated with either control RNAi or EWS/FLI RNAi exhibit comparable persistence (Fig. 4F). No statistically significant differences in distance, velocity, or net displacement were observed when comparing untreated A673 cells with those treated with control RNAi (Fig. 4C-F).
Consistent with the monolayer wound healing and single cell migration assays, we found that EWS/FLI expression in Ewing sarcoma cells also inhibited translocation through a porous membrane in a chemotactic Boyden Chamber assay in which cells are stimulated to migrate towards high serum gradient (Fig. 4G, H). Whereas EWS/FLI knockdown cells exhibited significantly increased capacity for invasion in a Matrigel invasion assay, expression of EWS/FLI, as occurred in the parental cell line, was associated with a striking lack of invasiveness by Ewing sarcoma cells (Fig. 4I, J). Confirmation that the effects of EWS/FLI knockdown on cell migration are due specifically to the loss of EWS/FLI was achieved by knockdown-rescue experiments in which we expressed an RNAi-insensitive EWS/FLI cDNA in the knockdown cells. We observed that expression of EWS/FLI causes reduced haptotactic cell migration in a Boyden chamber cell migration assay (Fig. 4K, L). Collectively, these results illustrate that EWS/FLI expression inhibits cell migration.
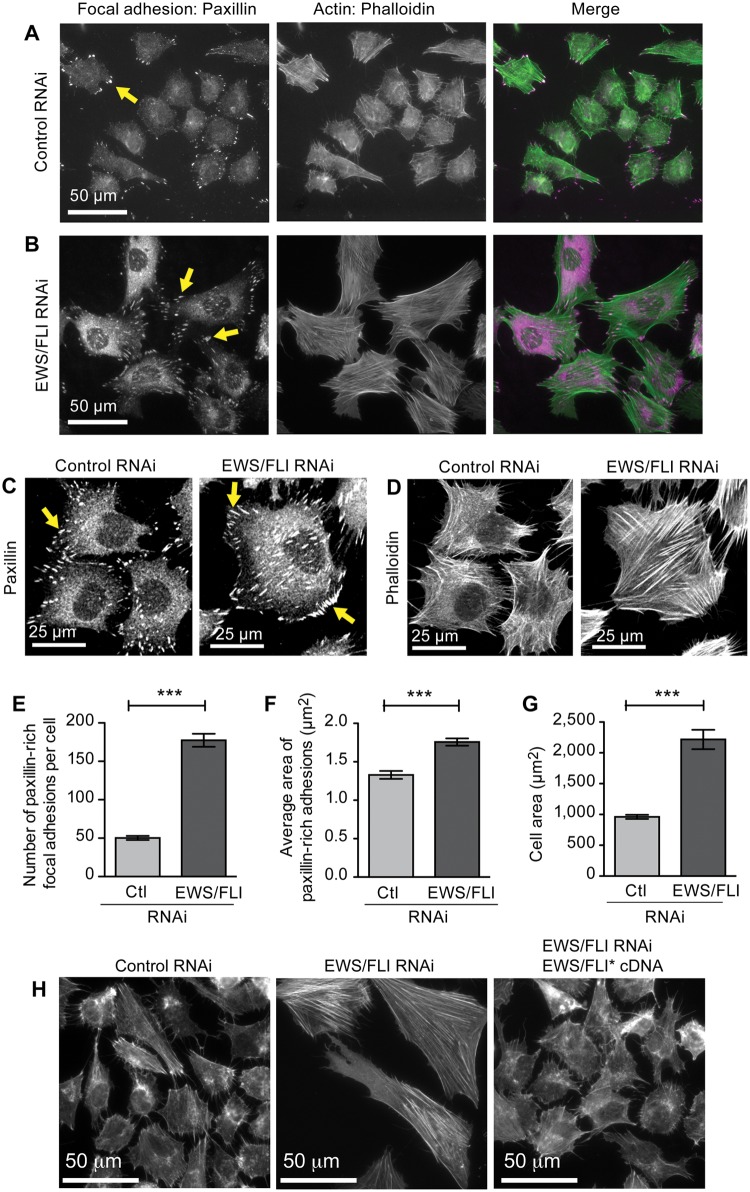
EWS/FLI expression induces deficiencies in the actin cytoskeleton and focal adhesions
The decreased adhesion and migration associated with EWS/FLI expression suggested that EWS/FLI expression might induce an underlying change in cytoarchitecture that is responsible for the observed alterations in cell behavior. To assess this possibility, we compared the cytoskeletal organization in A673 Ewing sarcoma cells and A673 cells in which EWS/FLI expression was knocked down by studying subcellular localization of cytoskeletal proteins. The cells were plated on fibronectin-coated coverslips in the presence of serum. Ewing sarcoma cells that express EWS/FLI displayed a small round cell morphology with thin and short actin stress fibers (Fig. 5A). In contrast, knocking down EWS/FLI resulted in cells with robust actin stress fibers that were distributed throughout the well spread Ewing sarcoma cells, seemingly consistent with a mesenchymal/fibroblastic morphology of a putative sarcoma cell-of-origin (Fig. 5B).
Figure 5.
EWS/FLI expression affects cellular morphology and cytoarchitecture. Widefield immunofluorescent images of A673 cells with (A) control RNAi or (B) EWS/FLI RNAi stained for focal adhesions (paxillin antibody) and for actin filaments (phalloidin) showed a striking EWS/FLI-dependent difference in cellular morphology, the actin cytoskeleton network, and focal adhesions. (C) Paxillin-rich focal adhesions and actin cytoskeletons (D) in cells with EWS/FLI or with EWS/FLI knockdown were analyzed. Both the number (E) and size (F) of focal adhesions were lower in A673 cells with control RNAi compared with cells with EWS/FLI RNAi, consistent with the compromised adhesion phenotype of Ewing sarcoma cells. (G) Cell area was measured in cells with control RNAi or EWS/FLI RNAi plated for 24 hours and then fixed and stained with phalloidin. Cell area of Ewing sarcoma A673 cells was significantly smaller than for cells with EWS/FLI knockdown. (H) Phalloidin-stained actin cytoskeleton in A673 cells with control RNAi, EWS/FLI RNAi, or EWS/FLI RNAi plus reconstituted Flag-EWS/FLI* showed that the loss of actin stress fibers is EWS/FLI-dependent. ***P < 0.001.
Actin stress fibers terminate at specialized regions of the cell membrane called focal adhesions, which support integrin-dependent adhesion to extracellular matrix proteins. Adherent mesenchymal cells typically display robust focal adhesions. The deficit in cell adhesion and actin stress fiber organization in Ewing sarcoma cells raised the possibility that expression of EWS/FLI might affect the establishment or maturation of focal adhesions. To test the impact of EWS/FLI expression on integrin-anchored focal adhesion sites, we compared focal adhesions in Ewing sarcoma cells treated with control or EWS/FLI directed RNAi by indirect immunofluorescence with primary antibody directed against the focal adhesion marker protein, paxillin (Fig. 5A,B,C,E,F).52 Quantitative analysis of focal adhesion number revealed that control A673 cells expressing EWS/FLI had fewer focal adhesions than A673 cells treated with EWS/FLI RNAi, displaying an average of 50 ± 15 focal adhesions per cell compared with 177 ± 50 focal adhesions for cells in which EWS/FLI expression was compromised (Fig. 5E). In addition to the reduced number of focal adhesions, cells with wild-type EWS/FLI expression had smaller focal adhesions, with average focal adhesion areas of 1.3 µm2 ± 0.3µm2 compared with an average focal adhesion area of 1.8 µm2 ± 0.3 µm2 for Ewing sarcoma cells treated with EWS/FLI RNAi (Fig. 5F).
Consistent with our observations that cells with EWS/FLI expression have compromised actin stress fibers and focal adhesions, these cells also exhibited an average cell area of 916 µm2 after 24 hours of plating, compared with an area of 2218 µm2 for A673 cells with reduced EWS/FLI expression (Fig. 5D, G). This finding complements the cell adhesion and cell spreading analysis presented in Fig. 2, which showed that Ewing sarcoma cells display deficits in adhesion and spread more slowly than Ewing sarcoma cells in which EWS/FLI expression is knocked down. Analysis of cell area 24 hours after plating illustrates that the Ewing sarcoma cells are not simply slow to spread; rather, they have lost their cytoskeletal organization and adhesive capacity, leading to a steady-state decline in cell area. As we demonstrated with the adhesion and motility measurements, EWS/FLI expression is specifically required for inducing the alterations in the actin cytoskeleton and cellular morphology observed in Ewing sarcoma cells. When an RNAi-insensitive variant of EWS/FLI cDNA is expressed in A673 sarcoma cells along with EWS/FLI RNAi, the cells lose their well-formed actin stress fibers and revert to small and round morphology (Fig. 5H).
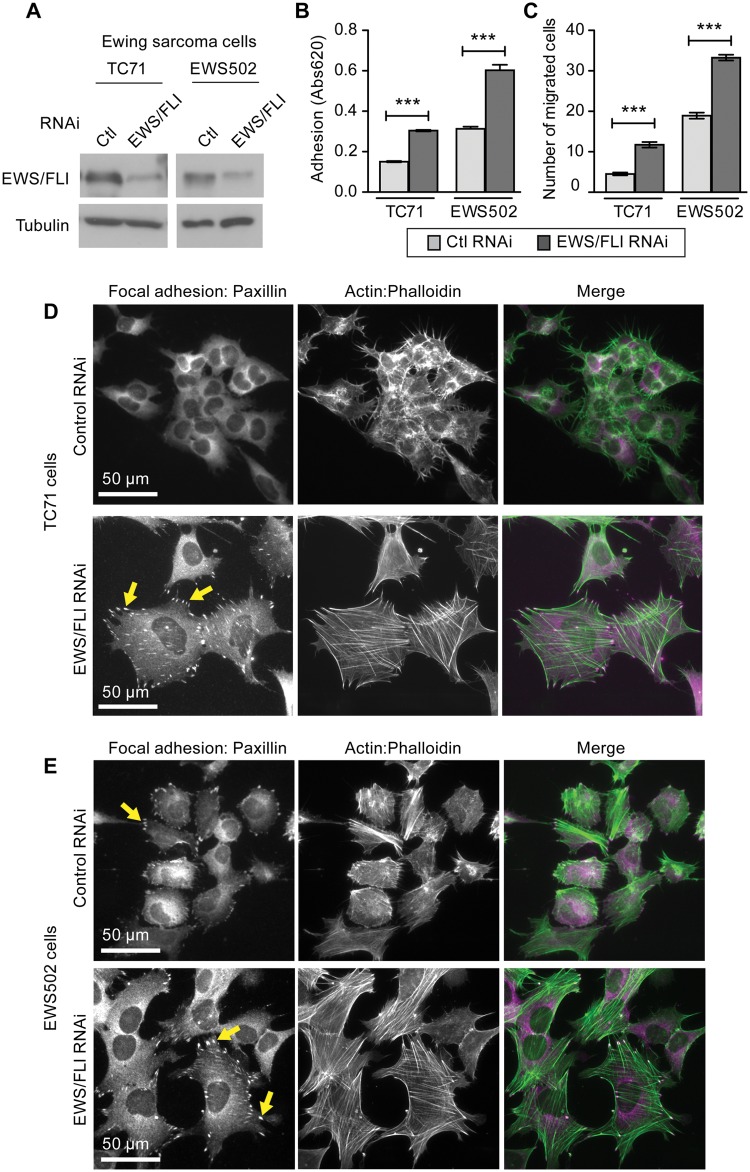
EWS/FLI-dependent reduction in cell adhesion, cell migration, and actin stress fibers is seen in several patient-derived Ewing sarcoma cell lines
To confirm that our findings using A673 cells were providing generalizable insights regarding the influence of EWS/FLI on cellular phenotype, we replicated our experiments using 2 additional primary tumor-derived Ewing sarcoma cell lines, TC71 and EWS502. Both of these cell lines displayed similar phenotypic changes upon RNAi based knockdown of EWS/FLI (Fig. 6A). Adhesion (Fig. 6B) and migration (Fig. 6C) of TC71 and EWS502 cells increased upon knockdown of EWS/FLI, illustrating that EWS/FLI expression abrogates both cell adhesion and motility. As we observed for A673 cells, EWS/FLI expression is associated with loss of actin filaments, reduced cell spreading, and compromised focal adhesions in both TC71 and EWS502 cells (Fig. 6D, E). It is striking that even with the baseline morphological heterogeneity among the Ewing sarcoma cell lines, all 3 display robust and parallel phenotypic responses to EWS/FLI knockdown, underscoring the fact that transformation by EWS/FLI induces dramatic changes in cell adhesion, migration, and cytoarchitecture.
Figure 6.
Ewing sarcoma cells TC71 and EWS502 exhibit EWS/FLI-dependent loss of adhesion, migration, and cytoarchitecture. (A) Cell lysates from Ewing sarcoma cell lines TC71 and EWS502 with retroviral mediated RNAi knockdown for control or EWS/FLI were evaluated by western immunoblot with FLI-1 antibody, and significant decreases in EWS/FLI expression were confirmed. (B) Ewing sarcoma cells with control RNAi (light gray bar) or EWS/FLI RNAi (dark gray bar) were seeded onto tissue culture plastic (uncoated (EWS502) or precoated with 10 µg/ml fibronectin (TC71)) and allowed to adhere for 2 hours, and then adherent cells were quantitated by a colorimetric assay. (C) Boyden chamber transwell inserts were precoated with fibronectin (5 µg/ml TC71; 1 µg/ml EWS502) and seeded with cells that were allowed 24 hours to migrate through the insert pores. EWS/FLI knockdown enhanced the number of migratory cells in both cell lines. (D) TC71 and (E) EWS502 cells with EWS/FLI or with EWS/FLI knockdown were stained for paxillin-rich focal adhesions and actin stress fibers (phalloidin). Cells with EWS/FLI RNAi displayed more pronounced focal adhesions and actin stress fibers. ***P<0.001
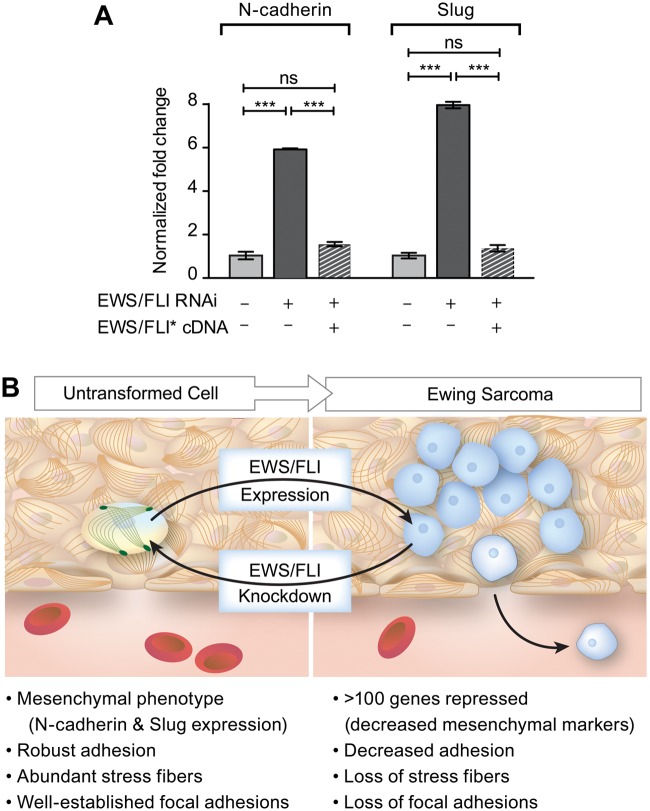
The loss of adhesion, migration, and actin cytoskeletal integrity induced by EWS/FLI expression represents a constellation of effects that is generally consistent with a loss of mesenchymal cell features. To explore whether the expression of well-established mesenchymal markers is affected by EWS/FLI, we analyzed previously published gene expression profiles derived from Ewing sarcoma cells and cells in which EWS/FLI expression was compromised.36,38 We noted significant upregulation of mRNAs encoding mesenchymal markers upon knockdown of EWS/FLI in A673 cells. We performed an independent biological validation of 2 of these genes, N-cadherin and Slug, using qRT-PCR (Fig. 7A). Our results confirmed that the presence of EWS/FLI is associated with low expression of both N-cadherin and Slug in Ewing sarcoma cells and that knockdown of EWS/FLI expression using RNA interference results in 6- to 8-fold upregulation of transcripts encoding these mesenchymal markers. Upon expression of RNAi-resistant EWS/FLI cDNA, the levels of N-cadherin and Slug transcripts were reduced.
Figure 7.
Model for EWS/FLI-dependent coupling of transformation to acquisition of potential for tumor cell dissemination. (A) RT-PCR analysis of mesenchymal markers, N-cadherin and Slug, in Ewing sarcoma cells programmed to express control RNAi, EWS/FLI RNAi, or EWS/FLI RNAi and an RNAi-insensitive EWS/FLI* construct. ***P<0.0001, ns is not statistically different. (B) In this model, EWS/FLI-induced oncogenic transformation simultaneously causes downregulation of cellular adhesion. Consistent with this model, EWS/FLI knockdown caused cells to return to a more normalized mesenchymal phenotype, including expression of mesenchymal markers, robust adhesion, pronounced stress fibers, and well-established focal adhesions. We postulate that these events prime the tumor cells for dissemination and may account for the early acquisition of metastatic potential in Ewing sarcoma.
Discussion
Metastatic disease is an important tumor phenotype that portends poor prognosis for patients. For patients whose primary site of tumor development is adequately controlled with current local therapies (mainly surgery and/or radiation therapy), relapse at distant sites indicates that undetected metastatic disease (termed micrometastasis) was present before the primary tumor was controlled. In the case of epithelial malignancies (carcinomas), the development of metastatic disease is thought to represent a stepwise progression of tumor development, following genetic and/or epigenetic alterations. These alterations are thought to culminate in the development of an epithelial-to-mesenchymal transition (EMT), with the result that tumor cells acquire the ability to escape the primary tumor, adhere, migrate, and invade through the extracellular matrix and ultimately to extravasate into the blood or lymphatic system.1,12
Sarcomas are mesenchymally derived tumors of less certain origin than many carcinomas. The process of metastatic spread is less well understood for sarcomas compared with carcinomas, and whether an “EMT-equivalent” process occurs that allows for increased cellular adhesion, migration, and invasion is unknown. Furthermore, the stepwise progression model of metastatic spread occurring as a late phenotype in tumor development does not adequately explain tumors that exhibit very early metastatic (or micrometastatic) spread. We therefore evaluated Ewing sarcoma as an example with a clear propensity for early metastatic and micrometastatic spread. This also allowed us to evaluate the role of the critical oncoprotein, EWS/FLI, in modulating metastatic-relevant phenotypes in this disease.
Here we report that EWS/FLI expression has a profound effect on cell adhesion, reducing capacity for adhesion and associated cellular spreading, both in vitro and in vivo. Analysis of focal adhesion architecture and actin cytoskeletal organization revealed that EWS/FLI expression results in loss of actin stress fibers and reduced size and number of focal adhesions, 2 structural changes that likely account for the functional deficit in cell adhesion. Reciprocal changes in cellular morphology have also been observed following expression of EWS/FLI in human mesenchymal stem cells, which causes the conversion of a well-spread, fibroblast-like shape to a rounded cell morphology.53 Somewhat surprisingly, given the highly metastatic nature of Ewing sarcoma and results from previous studies,39,54 we found that EWS/FLI expression also resulted in reduced cell migration, as assayed in single cell migration studies, wound healing assays, and transwell migration studies. Similarly, we found that EWS/FLI reduces the ability of Ewing sarcoma cells to invade Matrigel matrices. These findings suggest the possibility that active promotion of cell migration is not critical to drive the metastatic potential of Ewing sarcoma.
These data stand in direct contrast to our initial hypothesis, that EWS/FLI might stimulate metastasis by inducing increased cellular adhesion, migration, and invasion, and therefore prompt a reevaluation of how Ewing sarcoma might navigate the processes required for metastatic development. Our data (and prior data from other investigators) are consistent with the notion that Ewing sarcoma arises from a mesenchymal cell type (perhaps a mesenchymal stem or progenitor cell).48,53 In this view, a mesenchymal cell of origin would display strong adhesion to extracellular matrix proteins. The cell would thus be tethered to the extracellular matrix via these strong adherent properties that would constrain inappropriate release of locally resident cells. As illustrated in the model in Fig. 7B, EWS/FLI expression results in downregulation of several hundred genes (including mesenchymal markers), decreased adhesion, and significant cytoskeletal changes with loss of actin stress fibers and focal adhesions. Reciprocally, RNAi-based knockdown of EWS/FLI causes a tumor-to-mesenchymal-transition, where Ewing sarcoma cells are “normalized” to a more mesenchymal phenotype. This is demonstrated in our study by increased expression of N-cadherin and Slug, the restoration of robust actin stress fibers and focal adhesions, and increased cell adhesion. The loss of cell adhesion induced by EWS/FLI expression could promote dissemination of the tumor cells. The histological features of the capillary beds in the bone marrow, which display a fenestrated endothelium and discontinuous basement membrane, would be expected to promote permissive transfer of nonadherent tumor cells into the circulation. If tumor cell dissemination is a rate-limiting step in progression to metastasis, EWS/FLI-dependent abrogation of adhesion may be a powerful way to promote the early metastasis observed in this disease. Such a model might be considered “stochastic” or “passive” in contrast to the “active” process observed in epithelial tumors undergoing an EMT.
An interesting precedent for such a stochastic/passive scenario, in which loss of adhesion might be permissive for exit of cells from bone marrow, is found in the case of erythroid differentiation. In this case, mammalian reticulocytes are tethered in the bone marrow early in the erythroid differentiation process by binding to fibronectin. At later stages along the path toward maturation to erythrocytes, the cells lose adhesion to fibronectin, and it has been proposed that this allows passive release of the mature cells from the interstitial matrix of the bone marrow into the circulation.55-57 From this perspective it is interesting to note that approximately 85% of Ewing sarcoma cases arise in and around the bone (usually with extension into the marrow cavity).58 Thus, Ewing sarcoma cells typically arise in locations that have ready access to the intravascular space. In this environment, tumor cells may not require enhanced migratory or invasive capacity to access the vasculature on the path to metastasis but rather would simply require downregulation of cell adhesion, as occurs upon expression of the EWS/FLI oncoprotein.
Although the loss of cellular adhesion that is promoted by EWS/FLI expression may help to explain how Ewing sarcoma cells are so effectively disseminated from the site of the primary tumor, it is less clear how this reduced adhesion might influence the ability of the circulating tumor cells to take up residence and survive in a secondary site. Our experiments show that Ewing sarcoma cells in which EWS/FLI is knocked down are more likely to seed the lung when introduced into the circulation compared with the parental cell line. Nevertheless, the parental Ewing sarcoma cells are also effective in colonizing the lung. It is possible that Ewing sarcoma cells, even though less adhesive than their nontransformed counterparts, have adequate capacity to adhere and grow in the lung. Alternatively, it has been shown that tissue microenvironment can influence tumor biology, including oncogene expression.59,60 In the case of Ewing sarcoma, growth factor availability can influence the expression of EWS/FLI itself,61 raising the possibility that microenvironmental cues might modulate the expression of EWS/FLI and thus provide a mechanism for Ewing sarcoma cells to adapt and modulate their cellular phenotypes in a location-restricted fashion.
An important conclusion from our studies is that the primary oncogene (EWS/FLI) that induces Ewing sarcoma directly modulates a cell’s adhesive and cytoarchitectural phenotypes. This again stands in contrast to current models suggesting that tumor metastasis is a later effect resulting from an accumulation of secondary mutations or epigenetic changes during tumor progression. Importantly, our EWS/FLI reconstitution studies further signified that these phenotypic changes are EWS/FLI-dependent. Our data suggest that EWS/FLI serves an even larger role than previously envisioned for Ewing sarcoma tumorigenesis. It has previously been demonstrated that ongoing EWS/FLI expression is important for oncogenic “transformation” (in this case defined as the ability to grow as colonies under anchorage-independent conditions and as the ability to form tumors when injected into immunodeficient mice).37,38 Downstream targets, such as IGFBP3, NKX2.2, and NR0B1, are absolutely required for this phenotype.35-38 Subsequent studies have demonstrated that EWS/FLI also modulates drug resistance via the regulation of GSTM4 expression.62 The current study adds to this growing list of EWS/FLI-regulated phenotypes by demonstrating that the fusion protein modulates cell adhesion, migration, and invasiveness and thus might affect the metastatic phenotype.
In conclusion, we suggest that the transformed growth properties of Ewing sarcoma are induced in tandem with changes in cell behavior, such as reduced adhesion, that would promote tumor cell dissemination. The molecular mechanism by which EWS/FLI expression triggers the changes in adhesion, motility, and cytoskeleton reported here is not currently understood. EWS/FLI is a chimeric transcription factor that induces many alterations in the expression of a large number of genes.37,38,63 Other than a few EWS/FLI targets that have been extensively studied, such as NR0B1, NKX2.2, and Caveolin1,35,36,64 little is understood about the physiological influence of EWS/FLI on complex cell behaviors, but it is likely that EWS/FLI exerts its influence via both direct and indirect transcriptional targets. Inspection of the genes whose expression is modulated by EWS/FLI revealed numerous genes that encode cytoskeletal and cell adhesion factors (our unpublished results). Future work will be required to identify the critical factors that are modulated downstream of EWS/FLI to affect features such as cytoskeletal integrity and cell adhesion of the tumor cells. These studies will be a key to enable better understanding of the genesis of Ewing sarcoma cell behavior.
Materials and Methods
Reagents
Antibodies were used for western immunoblots and indirect immunofluorescence microscopy as per manufacturer’s instructions: FLI-1 rabbit antibody C-19 (sc-356X Santa Cruz Biotechnology, Santa Cruz, CA), Paxillin mouse antibody (P13520 Transduction Laboratories, San Jose, CA), β-tubulin mouse monoclonal antibody clone 2-28-33 (T 5293 Sigma-Aldrich, St. Louis, MO), HRP-conjugated antibodies for immunoblots (GE Healthcare, Pittsburgh, PA), and the Alexa-Fluor antibodies and phalloidin for microscopy (Molecular Probes/Invitrogen, Eugene, OR). Prolong Gold anti-fade reagent with DAPI (Invitrogen (Gibco, Life Technologies, Grand Island, NY), catalog #P-36931) was used for mounting lung sections. Fibronectin and Mowiol (Sigma-Aldrich, St. Louis, MO) and Vybrant Cell-Labeling solutions DiI and DiO (Molecular Probes/Invitrogen, Eugene, OR) were used as per manufacturer’s instructions.
Cell culture
Ewing sarcoma cell lines A673, TC71, and EWS502 were grown in DMEM media with 10% FBS, RPMI media with 10% FBS, and RPMI media with 15% FBS, respectively, as previously published.38
RNA interference
Knockdown experiments used the luciferase-RNAi (as control RNAi) and EF-2-RNAi (as EWS/FLI RNAi) constructs previously described.38 Constructs were made in pSRP retroviral vector with an H1 promoter to allow expression of sh-RNAs and a puromycin resistance marker. The retrovirally infected cells were selected in 2 µg/ml puromycin containing DMEM media for 2 days prior to use and maintained in selection for up to 5 weeks.
RT-PCR analysis
Total RNA was extracted from cell pellets using QIAgen RNeasy mini kit (QIAgen, Valencia, CA, catalog #74174). Using previously established primers,38 EWS/FLI and glyceraldehyde-3-phosphate dehydrogenase were amplified and detected using iScript One-Step RT-PCR kit with SYBR green (BIO-RAD, Hercules, CA, catalog #170-8893) for quantitative analysis. Specific primers were designed to analyze N-cadherin (forward primer 5′-GAGCAGTGAGCCTGCAGATTTTAAG G-3′ and reverse primer 5′-CCTTTGTAGGTGGCCACTGTGC-3′) and Slug (forward primer 5′-GCCAAACTACAGCGAACTGGACAC-3′ and reverse primer 5′-GCTTTCTGAGCCAC TGTGGTCC-3′) as mesenchymal markers.
Western blots
Whole cell lysates in RIPA buffer with (Complete Mini-EDTA free) protease inhibitor cocktail tablets (Roche Diagnostics GmbH, Indianapolis, IN catalog #11836153001) were electrophoresed on 10% SDS-PAGE gels transferred onto nitrocellulose membranes. Proteins were detected with primary antibodies (see Reagents) and horseradish peroxidase-conjugated antibodies and enhanced chemiluminescence (GE Healthcare, Buckinghamshire, UK).
Soft agar assays
Soft agar assays were performed as described,65 in short, with 1.6% SeaPlaque GTG agarose (Lonza, Rockland, ME, catalog #50111) and Iscove’s modification Eagle’s media, penicillin/streptomycin, and glutamine making a 0.35% agar underlayer with or without 2 µg/ml puromycin. Over this layer, a layer of cells was seeded at a density of 1 × 105 cells per 6 cm plate in 0.35% agar supplied with media of similar composition as the underlayer mentioned above. Cells were grown at 37°C and 5%CO2 for 3 to 4 weeks and imaged, and colonies were counted. Assays were performed 3 times with A673, TC-71, and EWS 502 cells.
Adhesion and spreading assays
Ewing Sarcoma cells (A673 and EWS-502) were seeded at 100,000, 300,000, or 500,000 density per well in a non-ECM coated 24 well plate. TC-71 cells were seeded onto a 24-well plate precoated with 5 µg/ml fibronectin. Cells were allowed to adhere to the dish for 2 hours at 37°C in their respective media, washed 3 times with PBS to remove nonadherent cells, fixed (3.7% formaldehyde for 15 minutes), washed (PBS for 5 minutes), and stained with Toluidene Blue for 1 hour. Excess stain was removed, and then cells were washed, air dried overnight, and dissolved (500 µl of 2% SDS solution). O.D. was measured in duplicates at 620 nm.66
For the spreading assay, after 2 hours of adhesion, cells were fixed and imaged (LucPlan 40X LWD objective (NA = 0.6) on an Olympus IX70 microscope with an Olympus CCD camera and Andor acquisition system). Cell area was measured for 10 representative fields of cells using Metamorph Imaging v7.5 software.
In vivo lung adhesion assay
This assay was adapted from a lung metastasis assay.67 The A673 cells with control RNAi and with EWS/FLI RNAi were differentially labeled with lipophilic dyes Vybrant DiI (red) and DiO (green) and diluted to 1 × 106 stained cells per 100 µl. The 2 solutions were mixed in equal amounts and verified as 1:1 ratio by FACS. The mixed cell suspension (200 µl) was then injected into the lateral tail veins of 3 NOD-SCID mice (stock #1303, Jackson Laboratories, Sacramento, CA). After 24 hours, these mice were sacrificed, and the lungs were prepared by perfusion with 4% paraformaldyde and frozen in OCT for cryopreservation. Lungs were cryosectioned (16-20 µm), mounted on slides using Prolong Gold anti-fade reagent with DAPI, and examined by fluorescent microscopy (Nikon A1R laser scan confocal acquisition on a Ti inverted microscope and Plan Fluor 40× Oil DIC H N2 objective). A large 3 field × 3 field (2 mm × 2 mm) mosaic image was created with each field having nine 2-µm optical sections (NIS Elements v3 software). The red and green colonies were then counted along with DAPI staining, and normalized values are reported for 15 representative mosaic images. The experiments were performed 3 times, including once with the dyes switched for the respective cell types to avoid any dye-based bias. Experiments were performed following approval from the University of Utah Institutional Animal Care and Use Committee.
Migration assays: monolayer wound healing assay
A monolayer of confluent Ewing sarcoma cells (A673) was generated by seeding 1 × 106 cells per well in a 6-well dish. Cells were imaged in DMEM/F/12-HEPES media lacking Phenol Red (Invitrogen) supplemented with 10% FBS and maintained at 37°C (LIS) for microscopy. A micropipette tip was scraped across the monolayer to create a “wound.” Migration into the wound was then monitored by time-lapse microscopy at 15-minute intervals for 48 hours (inverted Nikon TE300 microscope, Plan Fluor ELWD 20x objective) and by Ludl XY stage (Ludl Electronic Products). Images were captured with Andor DV885 EMCCD camera and Andor IQ imaging software (Andor Technologies).66
Random cell motility assay
A673 cells were seeded at a density of 25,000 cells/well in a non-ECM coated 6-well dish. Cells were imaged at 15-minute intervals for 24 hours as described for the wound healing assay above. For generating cell tracks and analysis, the Manual tracking plugin in ImageJ software was used.
Haptotactic cell migration assay
Ewing sarcoma cells (A673 and EWS502) were seeded (30,000 cells) in serum-free media onto 24-well cell culture insert membranes (polyethylene terephthalate) with 8-µm pores (Becton Dickinson, San Jose, CA). These inserts were precoated overnight at 4°C with 1 µg/ml fibronectin solution. For TC-71 cells, the inserts were coated with 5 µg/ml fibronectin. Inserts with cells were placed in a 24-well plate of media supplemented with 10% FBS, and cells were allowed to migrate for 24 hours.66 Nonmigratory cells were scraped off the top chamber, and migratory cells on the bottom surface were fixed in 100% methanol for 15 minutes and then stained overnight at 4°C with 1:20 modified Giemsa Stain. Chambers were washed in water, and images were captured for 5 representative fields per well (Olympus IX70 microscope, Luc Plan 20x phase objective, NA = 0.45, Olympus CCD camera and Andor IQ acquisition software). Migratory cells were scored for 3 individual wells per cell line. Three biological replicates were done.
Invasion assay
BioCoat Matrigel transwell invasion chambers with 8-µm pores (BD Biosciences, Bedford, MA catalog #354480) in 24-well plates were seeded with 150,000 cells for a 24-hour invasion assay as described above for transwell migration assays.
Immunofluorescence studies
Ethanol sterilized coverslips in 12-well plates were coated with 10 µg/ml fibronectin overnight at 4°C, and then Ewing sarcoma cells were seeded (75,000 cells/well) and allowed to adhere for 24 hours. Cells were fixed (15 minutes in 3.7% formaldehyde), washed for 5 minutes in PBS, and permeabilized in 0.2% Triton X-100/PBS. Cells were incubated with paxillin antibody (1:100) for 1 hour at 37°C followed by a 15-minute PBS wash, stained with AlexaFluor secondary antibody (1:200) and AlexaFluor-phalloidin (1:100) for 1 hour at 37°C, washed, and mounted in Mowiol medium.66 Cell images were captured using a Zeiss Axioskop2 mot plus microscope with a 40× plan NA 0.75 NeoFluor objective, Zeiss Axiocam MR camera, and Zeiss Axiovision v4.8.1 software (Carl Zeiss MicroImaging, Inc.).
Analysis of adhesions and cell spreading
Immunofluorescence microscopy of fixed cells used Nikon A1R Ti inverted microscope, 60× Oil Plan Apo DIC N2 (NA 1.4) objective, and Nikon Elements v3 software. Cell area was measured using the trace tool to identify the boundary of cells, a threshold was set using the actin/phalloidin staining, and region of interest areas were recorded (Metamorph v7.5 software). To count and measure the size of the paxillin-rich focal adhesions, single-cell images were first processed in Image J to despeckle and remove the noise, and background was subtracted using rolling ball (radius 10). Using Metamorph imaging v7.5 software, image threshold was set in these single cell images, and integrated morphometric analysis was performed to count the number of focal adhesions and measure the area of the thresholded region to represent size of focal adhesions.
Acknowledgments
We thank Dr. Chris Rodesch at the University of Utah Cell Imaging and Microscopy Core Facility for help with fluorescent imaging, image processing, and analysis. We thank Savita Sankar at Huntsman Cancer Institute for providing cells for the reconstitution studies. We acknowledge Diana Lim for help with manuscript preparation.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the NIH (R01 GM50877 to M.C.B. and R01 CA140394 to SLL) and the Huntsman Cancer Foundation. The Cancer Center Support Grant (2 P30 CA042014) awarded to the Huntsman Cancer Institute provided developmental funds and Shared Resources critical to this project.
References
- 1. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-64 [DOI] [PubMed] [Google Scholar]
- 3. Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274-84 [DOI] [PubMed] [Google Scholar]
- 4. Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14: 818-29 [DOI] [PubMed] [Google Scholar]
- 5. Guilford P, Hopkins J, Harraway J, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402-5 [DOI] [PubMed] [Google Scholar]
- 6. Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5:101-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perl AK, Wilgenbus P, Dahl U, et al. A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature. 1998;392:190-3 [DOI] [PubMed] [Google Scholar]
- 8. Frixen UH, Behrens J, Sachs M, et al. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816-26 [DOI] [PubMed] [Google Scholar]
- 10. Rathinam R, Alahari SK. Important role of integrins in the cancer biology. Cancer Metastasis Rev. 2010;29:223-37 [DOI] [PubMed] [Google Scholar]
- 11. Kumar S, Weaver VM. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Talmadge JE, Fidler IJ. AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Res. 2010;70:5649-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Eyles J, Puaux AL, Wang X, et al. Tumor cells disseminate early, but immunosurveillance limits metastatic outgrowth, in a mouse model of melanoma. J Clin Invest. 2010;120:2030-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759-67 [DOI] [PubMed] [Google Scholar]
- 15. Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893-5 [DOI] [PubMed] [Google Scholar]
- 16. Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6:339-51 [DOI] [PubMed] [Google Scholar]
- 17. Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. APMIS. 2008;116:754-70 [DOI] [PubMed] [Google Scholar]
- 18. Ramaswamy S, Ross KN, Lander ES, et al. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49-54 [DOI] [PubMed] [Google Scholar]
- 19. Spraker HL, Price SL, Chaturvedi A, et al. The clone wars—revenge of the metastatic rogue state: the sarcoma paradigm. Front Oncol. 2012;2:2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dahlin DC, Coventry MB, Scanlon PW. Ewing’s sarcoma: a critical analysis of 165 cases. J Bone Joint Surg Am. 1961;43-A:185-92 [PubMed] [Google Scholar]
- 21. Wang CC, Schulz MD. Ewing’s sarcoma; a study of fifty cases treated at the Massachusetts General Hospital, 1930-1952 inclusive. N Engl J Med. 1953;248:571-6 [DOI] [PubMed] [Google Scholar]
- 22. Dubois SG, Epling CL, Teague J, et al. Flow cytometric detection of Ewing sarcoma cells in peripheral blood and bone marrow. Pediatr Blood Cancer. 2010;54:13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peter M, Magdelenat H, Michon J, et al. Sensitive detection of occult Ewing’s cells by the reverse transcriptase-polymerase chain reaction. Br J Cancer. 1995;72:96-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pfleiderer C, Zoubek A, Gruber B, et al. Detection of tumour cells in peripheral blood and bone marrow from Ewing tumour patients by RT-PCR. Int J Cancer. 1995;64:135-9 [DOI] [PubMed] [Google Scholar]
- 25. Zoubek A, Kovar H, Kronberger M, et al. Mobilization of tumour cells during biopsy in an infant with Ewing sarcoma. Eur J Pediatr. 1996;155:373-6 [DOI] [PubMed] [Google Scholar]
- 26. West DC, Grier HE, Swallow MM, et al. Detection of circulating tumor cells in patients with Ewing’s sarcoma and peripheral primitive neuroectodermal tumor. J Clin Oncol. 1997;15:583-8 [DOI] [PubMed] [Google Scholar]
- 27. de Alava E, Lozano MD, Patino A, et al. Ewing family tumors: potential prognostic value of reverse-transcriptase polymerase chain reaction detection of minimal residual disease in peripheral blood samples. Diagn Mol Pathol. 1998;7:152-7 [DOI] [PubMed] [Google Scholar]
- 28. Fagnou C, Michon J, Peter M, et al. Presence of tumor cells in bone marrow but not in blood is associated with adverse prognosis in patients with Ewing’s tumor. Societe Francaise d’Oncologie Pediatrique. J Clin Oncol. 1998;16:1707-11 [DOI] [PubMed] [Google Scholar]
- 29. Fidelia-Lambert MN, Zhuang Z, Tsokos M. Sensitive detection of rare Ewing’s sarcoma cells in peripheral blood by reverse transcriptase polymerase chain reaction. Hum Pathol. 1999;30:78-80 [DOI] [PubMed] [Google Scholar]
- 30. Aurias A, Rimbaut C, Buffe D, et al. Translocation involving chromosome 22 in Ewing’s sarcoma: a cytogenetic study of four fresh tumors. Cancer Genet Cytogenet. 1984;12:21-5 [DOI] [PubMed] [Google Scholar]
- 31. Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162-5 [DOI] [PubMed] [Google Scholar]
- 32. Turc-Carel C, Philip I, Berger MP, et al. Chromosome study of Ewing’s sarcoma (ES) cell lines: consistency of a reciprocal translocation t(11;22)(q24;q12). Cancer Genet Cytogenet. 1984;12:1-19 [DOI] [PubMed] [Google Scholar]
- 33. Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204:351-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. May WA, Lessnick SL, Braun BS, et al. The Ewing’s sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol. 1993;13:7393-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kinsey M, Smith R, Lessnick SL. NR0B1 is required for the oncogenic phenotype mediated by EWS/FLI in Ewing’s sarcoma. Mol Cancer Res. 2006;4:851-9 [DOI] [PubMed] [Google Scholar]
- 36. Owen LA, Kowalewski AA, Lessnick SL. EWS/FLI mediates transcriptional repression via NKX2.2 during oncogenic transformation in Ewing’s sarcoma. PLoS One. 2008;3:e1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Prieur A, Tirode F, Cohen P, et al. EWS/FLI-1 silencing and gene profiling of Ewing cells reveal downstream oncogenic pathways and a crucial role for repression of insulin-like growth factor binding protein 3. Mol Cell Biol. 2004;24:7275-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith R, Owen LA, Trem DJ, et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing’s sarcoma. Cancer Cell. 2006;9:405-16 [DOI] [PubMed] [Google Scholar]
- 39. Amsellem V, Kryszke MH, Hervy M, et al. The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp Cell Res. 2005;304:443-56 [DOI] [PubMed] [Google Scholar]
- 40. Jaishankar S, Zhang J, Roussel MF, et al. Transforming activity of EWS/FLI is not strictly dependent upon DNA-binding activity. Oncogene. 1999;18:5592-7 [DOI] [PubMed] [Google Scholar]
- 41. Song Y, Maul RS, Gerbin CS, et al. Inhibition of anchorage-independent growth of transformed NIH3T3 cells by epithelial protein lost in neoplasm (EPLIN) requires localization of EPLIN to actin cytoskeleton. Mol Biol Cell. 2002;13:1408-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lessnick SL, Braun BS, Denny CT, et al. Multiple domains mediate transformation by the Ewing’s sarcoma EWS/FLI-1 fusion gene. Oncogene. 1995;10:423-31 [PubMed] [Google Scholar]
- 43. May WA, Gishizky ML, Lessnick SL, et al. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90:5752-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Braunreiter CL, Hancock JD, Coffin CM, et al. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing’s sarcoma. Cell Cycle. 2006;5:2753-9 [DOI] [PubMed] [Google Scholar]
- 45. Hancock JD, Lessnick SL. A transcriptional profiling meta-analysis reveals a core EWS-FLI gene expression signature. Cell Cycle. 2008;7:250-6 [DOI] [PubMed] [Google Scholar]
- 46. Cavazzana AO, Miser JS, Jefferson J, et al. Experimental evidence for a neural origin of Ewing’s sarcoma of bone. Am J Pathol. 1987;127:507-18 [PMC free article] [PubMed] [Google Scholar]
- 47. Ewing J. Diffuse endothelioma of bone. Proc NY Pathol Soc. 1921;21:17-24 [Google Scholar]
- 48. Tirode F, Laud-Duval K, Prieur A, et al. Mesenchymal stem cell features of Ewing tumors. Cancer Cell. 2007;11:421-9 [DOI] [PubMed] [Google Scholar]
- 49. Kovar H. Context matters: the hen or egg problem in Ewing’s sarcoma. Semin Cancer Biol. 2005;15:189-96 [DOI] [PubMed] [Google Scholar]
- 50. Owen LA, Lessnick SL. Identification of target genes in their native cellular context: an analysis of EWS/FLI in Ewing’s sarcoma. Cell Cycle. 2006;5:2049-53 [DOI] [PubMed] [Google Scholar]
- 51. Martinez-Ramirez A, Rodriguez-Perales S, Melendez B, et al. Characterization of the A673 cell line (Ewing tumor) by molecular cytogenetic techniques. Cancer Genet Cytogenet. 2003;141:138-42 [DOI] [PubMed] [Google Scholar]
- 52. Turner CE. Paxillin and focal adhesion signalling. Nat Cell Biol. 2000;2:E231-6 [DOI] [PubMed] [Google Scholar]
- 53. Riggi N, Suva ML, Suva D, et al. EWS-FLI-1 expression triggers a Ewing’s sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176-85 [DOI] [PubMed] [Google Scholar]
- 54. Herrero-Martin D, Osuna D, Ordonez JL, et al. Stable interference of EWS-FLI1 in an Ewing sarcoma cell line impairs IGF-1/IGF-1R signalling and reveals TOPK as a new target. Br J Cancer. 2009;101: 80-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Eshghi S, Vogelezang MG, Hynes RO, et al. Alpha4beta1 integrin and erythropoietin mediate temporally distinct steps in erythropoiesis: integrins in red cell development. J Cell Biol. 2007;177: 871-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Patel VP, Lodish HF. Loss of adhesion of murine erythroleukemia cells to fibronectin during erythroid differentiation. Science. 1984;224:996-8 [DOI] [PubMed] [Google Scholar]
- 57. Patel VP, Ciechanover A, Platt O, et al. Mammalian reticulocytes lose adhesion to fibronectin during maturation to erythrocytes. Proc Natl Acad Sci U S A. 1985;82:440-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kimber C, Michalski A, Spitz L, et al. Primitive neuroectodermal tumours: anatomic location, extent of surgery, and outcome. J Pediatr Surg. 1998;33:39-41 [DOI] [PubMed] [Google Scholar]
- 59. Shibue T, Weinberg RA. Metastatic colonization: settlement, adaptation and propagation of tumor cells in a foreign tissue environment. Semin Cancer Biol. 2011;21:99-106 [DOI] [PubMed] [Google Scholar]
- 60. Sterling JA, Edwards JR, Martin TJ, et al. Advances in the biology of bone metastasis: how the skeleton affects tumor behavior. Bone. 2011;48:6-15 [DOI] [PubMed] [Google Scholar]
- 61. Girnita L, Girnita A, Wang M, et al. A link between basic fibroblast growth factor (bFGF) and EWS/FLI-1 in Ewing’s sarcoma cells. Oncogene. 2000;19:4298-301 [DOI] [PubMed] [Google Scholar]
- 62. Luo W, Gangwal K, Sankar S, et al. GSTM4 is a microsatellite- containing EWS/FLI target involved in Ewing’s sarcoma oncogenesis and therapeutic resistance. Oncogene. 2009;28:4126-32 [DOI] [PubMed] [Google Scholar]
- 63. Kauer M, Ban J, Kofler R, et al. A molecular function map of Ewing’s sarcoma. PLoS One. 2009;4:e5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tirado OM, Mateo-Lozano S, Villar J, et al. Caveolin-1 (CAV1) is a target of EWS/FLI-1 and a key determinant of the oncogenic phenotype and tumorigenicity of Ewing’s sarcoma cells. Cancer Res. 2006;66:9937-47 [DOI] [PubMed] [Google Scholar]
- 65. Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1:393-401 [DOI] [PubMed] [Google Scholar]
- 66. Hoffman LM, Jensen CC, Kloeker S, et al. Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J Cell Biol. 2006;172:771-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Padua D, Zhang XH, Wang Q, et al. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66-77 [DOI] [PMC free article] [PubMed] [Google Scholar]