Abstract
Purpose
The purpose of this study was to demonstrate a novel protein-based magnetic resonance imaging (MRI) contrast agent that has the capability of targeting prostate cancer and which provides high-sensitivity MR imaging in tumor cells and mouse models.
Procedure
A fragment of gastrin-releasing peptide (GRP) was fused into a protein-based MRI contrast agent (ProCA1) at different regions. MR imaging was obtained in both tumor cells (PC3 and H441) and a tumor mouse model administrated with ProCA1.GRP.
Results
PC3 and DU145 cells treated with ProCA1.GRPs exhibited enhanced signal in MRI. Intratumoral injection of ProCA1.GRP in a PC3 tumor model displayed enhanced MRI signal. The contrast agent was retained in the PC3 tumor up to 48 h post-injection.
Conclusions
Protein-based MRI contrast agent with tumor targeting modality can specifically target GRPR-positive prostate cancer. Intratumoral injection of the ProCA1 agent in the prostate cancer mouse model verified the targeting capability of ProCA1.GRP and showed a prolonged retention time in tumors.
Keywords: MRI, Contrast agents, Prostate cancer, Molecular imaging, Relaxivity
Introduction
Magnetic resonance imaging (MRI) is a powerful, noninvasive, clinical diagnostic tool that provides high-resolution images without the limitation of tissue depth. More than one third of clinical MRI applications rely on the administration of contrast agents [1, 2]. However, the progress of imaging of disease markers using MRI contrast agents is largely limited by the following: the relatively low relaxivity (<5–10 mM−1 s−1) of conventional, small chelator-based contrast agents; low levels of biomarker expression on the cell surfaces; and pharmacokinetics of contrast agents [3, 4]. Developing MRI contrast agents that can specifically target various biomarkers, which would allow for real-time imaging of biological events at the molecular level, has drawn enormous research attention [5, 6]. Approaches include the use of peptides or monoclonal antibodies as ligands for targeting coupled with supermagnetic nano-particles, as well as liposomes loaded with a high number of paramagnetic metal ions for contrast enhancement [7, 8]. Short peptide fragments derived from affinity proteins have been widely used for targeting over-expressed receptors in disease cells for diagnosis and treatments; this method has an advantage of increased tissue penetration due to its relatively small size compared to the whole antibodies [4, 9–11]. However, the conjugates of ligands and nanoparticles or liposomes typically are large complexes; their in vivo applications are limited due to numerous delivery barriers and affinity of short peptides that are often less favorable than whole antibodies. Therefore, in order to overcome the limitations of low sensitivity and specificity, the development of MRI contrast agents that possess high relaxivities and targeting capabilities for molecular imaging of disease biomarkers is imperative.
We have recently developed a protein-based MRI contrast agent (ProCA1, previously named as CA1.CD2) by de novo design of Gd3+ binding site(s) into a stable host protein, domain 1 of rat CD2, with a strong metal binding affinity and selectivity, as well as greater enhancement in relaxivity [12]. Here, we report further advancement of this protein-based MRI contrast agent, which now contains an insertion of a gastrin-release peptide (GRP), and its capability of producing an MR image of prostate cancer. GRP acts through its cell surface receptors (GRPR) which occur in very high levels in prostate cancer, small cell lung cancer, and pancreatic cancer cells [13–16]. This new MRI contrast agent is designed to target GRPR using the peptide of ten amino acids from the C-terminal of GRP. The C-terminal of GRP has a loop structure which has essential binding properties and biological functions [17, 18]. Our results suggest that the GRP sequence grafted in the scaffold protein (ProCA1.GRP(52)) has a better targeting capability than that fused to the C-terminal (ProCA1.GRPC). Most importantly, our results show that the newly designed protein-based MRI contrast agent can specifically target prostate cancer cells and prostate cancer tumor models which express the GRP receptor.
Materials and Methods
Designing and Modeling of Targeted Protein-Based MRI Contrast Agent
The targeting sequence, GRP, was added to the different sites of ProCA1 to construct new targeted protein-based MRI contrast agents. The modeling of our protein-based MRI contrast agents was constructed by Modeler 9v3 software [19]. Three-dimensional structure of the designed metal binding protein (1T6W) was applied as the template for modeling protein structures [12]. Gastrin-releasing peptide was grafted in the C-terminal or between Ser52 and Gly53 of the protein contrast agent to construct ProCA1. GRPC and ProCA1.GRP(52), respectively. Finally, the model structures were drawn with PyMOL (Delano Scientific LLC, Palo alto, CA, USA).
Construction of Protein Contrast Agents and Protein Purification
A flexible linker (GGSGG) and ten amino acids of the C-terminal of GRP were inserted into the plasmid pGEX2t-ProCA1 at different regions by PCR [20]. The plasmids pGEX2t-ProCA1.GRPN, pGEX2t-ProCA1.GRPC, and pGEX2t-ProCA1.GRP(52) were transfected into BL21-DE3 cells. All proteins were expressed and purified using a modified protocol for CD2 variants [21].
Tryptophan Fluorescence Spectroscopy and Metal Binding Study
The conformational analysis of the engineered proteins was investigated by fluorescence using established methods [22]. The fluorescence spectrum of the tryptophan in the proteins was measured in the emission region of 300–400 nm (excited at 282 nm). The metal binding properties were investigated with an established protocol for Tb3+ fluorescence [21].
Enzyme-Linked Immunosorbent Assay
Three different cell types (PC3, DU145, and H441) were used in this experiment. Cells (2×104) were seeded in a 96-well plate and cultivated for 16 h. The medium was replaced by 250 µl fresh medium with 5% FBS the next day. ProCA1, ProCA1.GRPC, and ProCA1.GRP(52) were incubated with cells for 1 h at different concentrations. A CD2 antibody was used as the primary antibody. An HRP conjugated goat anti-mouse IgG was used as the secondary antibody. The absorbance intensity was detected by the FLUOstar OPTIMA plate reader at an absorbance wavelength of 490 nm.
Quantitative Analysis of Gd Isotope
ProCA1.GRPC and ProCA1.GRP(52) were pre-incubated with 153Gd at room temperature. Collection of DU145, PC3, and H441 cells (1×107) was followed by incubation with 10 µM of 153Gd- ProCA1.GRPC and 153Gd-ProCA1.GRP(52) for 1 h. Cell pellets were washed with TBST three times, for 5 min each time. Finally, cell pellets were collected and 153Gd radioactivity was measured using a gamma counter.
Fluorescence Imaging of Cells
DU145, PC3, and H441 cells (8×104) were seeded in four-well chambers (BD) at 37°C overnight. On the second day, the medium was changed. Contrast agents (ProCA1, ProCA1.GRPC, and ProCA1.GRP(52)) were incubated with the cells at different time points at 37°C. Subsequently, the cells were washed with 1× PBS three times and treated with 3% formaldehyde for 15 min. After the cells were rinsed in 1× PBS, 0.2% Triton X-100 was added to permeabilize the cells for 10 min. The cells were washed and four drops of Image-IT Fx signal enhancer were applied to each sample to block unspecific binding. After three washes, the CD2 antibody was used as a primary antibody. Goat anti-mouse IgG conjugated with Alexa Fluor 488 was used as the detection antibody. The fluorescence image was acquired using 488 nm and UV lasers.
MR Imaging of Cells
DU145, PC3, and H441 cells (1×107) were collected and washed with serum-free medium. Different contrast agents (gadodiamide (Omniscan), Gd-ProCA1, Gd-ProCA1.GRPC, and Gd-ProCA1. GRP(52)) were incubated with cells at 37°C for 1 h. Gd-DTPA was used as a reference. After incubation, the cells were washed three times with ice-cold 1× TBS, then harvested in Eppendorf tubes which are placed in a rack. T1-weighted MR images were acquired using an inversion recovery sequence (TR=4,000 ms, TE =12 ms, IR=500 ms) on a 3.0-T MRI (Siemens trio) scanner.
Measurements of r1 and r2 Relaxivities
The T1 and T2 relaxation times of our protein contrast agents were measured at 3 T. T1 was determined using inversion recovery sequence, and T2 was determined by using a multi-echo Carr–Purcell–Meiboom–Gill sequence. Different concentrations of contrast agents were placed in Eppendorf tubes. The tubes were then placed on a rack. Finally, the rack was placed in the MRI scanner for measurement of relaxation times. r1 and r2 were calculated by , where T1s and T2s are relaxation times with contrast agent and T1c and T2c are relaxation times without contrast agent. C is the concentration of contrast agent in millimolar.
Animal MR Imaging of Tumor-Bearing Mice
PC3 and H441 cells (1×106) were injected into both flanks of nude mice (n=10). After 4–5 weeks, tumors grew around 0.5 cm. The contrast agents, Gd-ProCA1 and Gd-ProCA1.GRP(52) (140 µM, 20 µl), were directly injected into the tumors of the mice. Mice were scanned on a 4.7-T MRI scanner (Varian Unity) using a dedicated rodent coil. During MR scan, the mouse was under anesthesia with 1.5% isoflurane and kept warm with a heated pad. MR images were acquired by T1- and T2-weighted fast spin echo sequences (TR=2 s, TE=0.022 s, and ESP = 0.01 s) with field of view of 3×3 cm, matrix of 256×256, and slice thickness of 1 mm. The tumors were dissected after MRI experiments. Tissue from the tumor and muscle were homogenized and extracts were examined by using the polyclonal antibody against the CD2.
Measurement of Tumor MRI Signal Intensity
The images were analyzed using a program of Image J 1.42 (downloaded from the web site http://rsb.info.nih.gov/ij/). Five regions of interest (ROIs) in both positive and negative tumors of each mouse were selected. Signal intensities from each ROI were measured and then averaged to obtain the mean intensity of the tumor. The relative intensity was calculated by dividing the mean intensity of positive tumor with that of negative tumor. After subtracting the initial intensity, the relative intensity was normalized to obtain the normalized relative change [12, 23–25].
Toxicity Studies of Protein Contrast Agent In Vitro and In Vivo
The toxicity study of our contrast agent ProCA1.GRP(52) was performed at both cell and animal levels. Cytotoxicity of our contrast agents in the cells was analyzed by MTT assay (Chemicon Inc.). PC3 and H441 cells (104) were grown under normal growth conditions (cell medium containing 10% fetal bovine serum) in 96- well plates. Cells were incubated with different concentrations of contrast agent or saline phosphate buffer for 48 h. After incubation, a standard MTT assay was applied to assess the cell viability. The absorbance at 570 nm was measured using VICTOR III microplate reader. Two groups of mice with PC3 and H441 tumor growing were used for toxicity studies in vivo. One group of mice received saline buffer as control. After 48 h of 100 mg/kg of contrast agent injection, mice were euthanized and blood was collected to analyze the clinical biochemistry, including alanine transaminase by a commercially available source (RADIL research Animal Diagnostic Laboratory).
Immunoblot Analysis of Cell and Tissue Extracts
After MR imaging, cell extracts were made freshly using RIPA cell lysis buffer (Pierce) and tumor extracts were made using Tissue PE LB buffer (GBioscience). The protein concentration of extracts was determined using Bradford assay (Bio-rad). Lysate protein was loaded into prepared sodium dodecyl sulfate gel and then transferred to nitrocellulose membrane. After blotting, the membrane was incubated with primary antibody (Anti-CD2 antibody) at 4°C overnight and then incubated with horseradish peroxidase conjugated secondary antibody at room temperature for 1 h. The blotting signal was detected using SuperSignal West Dura Extended Duration Substrate (Pierce). GADPH was used as loading reference.
Statistical Analysis
Statistical analysis was performed on both determination of r1 and r2 relaxivity of our protein contrast agents and MR imaging in tumor-bearing mice using Student’s t test, respectively. Three samples of each protein contrast agents and Gd-DTPA were used for the measurement of r1 and r2 relaxivity. The mean and standard deviation of r1 and r2 values of each contrast agent were obtained from the measured values. Gd-DTPA was applied as a reference for the statistical analysis to get p value of each protein contrast agent. Ten mice were used in MR imaging of tumor-bearing mice. The imaging results were analyzed with ImageJ to obtain the normalized relative intensity change for each tumor-bearing mouse. Then, control and treatment groups were compared by performing an analysis of variance. Finally, results were presented as mean ± SD.
Results
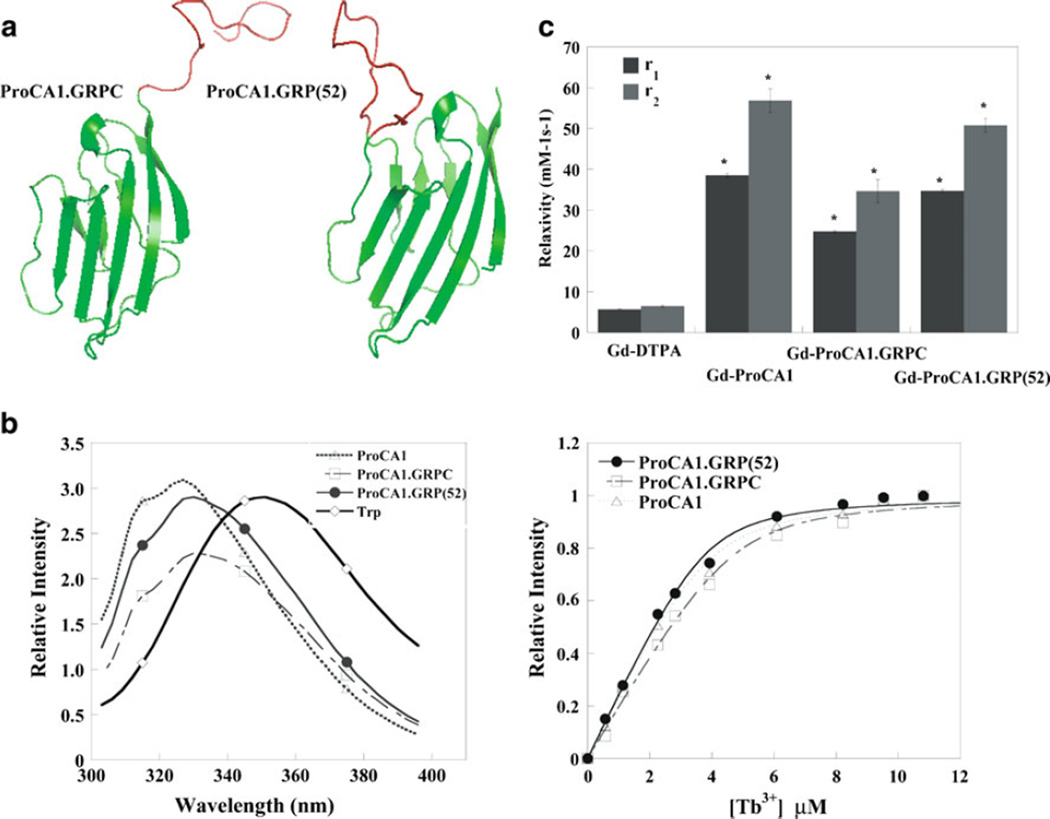
We have previously reported a protein-based MRI contrast agent (ProCA1) with high relaxivity. ProCA1 was created by designing a metal binding site in domain 1 of CD2 [12]. Our previous studies demonstrated that the introduction of the EF-hand metal binding loop into CD2 does not alter its overall native structure [26]. We used a similar approach to add the targeting sequence, GRP, at the different regions of ProCA1 to construct new targeted protein-based MRI contrast agents. We have created three targeting contrast agents at different regions of ProCA1. The truncated GRP was grafted into an exposed loop of the ProCA1 using a flexible linker sequence (ProCA1.GRP(52); Fig. 1a). As a comparison, the GRP peptide was also fused to ProCA1 at either the N or C-terminal (ProCA1.GRPN and ProCA1. GRPC). Fusion of the GRP peptide at the N-terminal significantly decreases the bacterial expression level and stability of the protein (data not shown). Therefore, no further study of ProCA1.GRPN was pursued. Our results show that C-terminal fusion or grafting of GRP peptide into ProCA1 did not change the overall protein folding and metal binding properties, as demonstrated by the Trp fluorescence spectroscopy and metal binding studies of Tb-FRET (Fig. 1b). Further characterization of the relaxivities of the ProCA1 variants indicated that the addition of ten amino acids of GRP sequence into ProCA1 did not change the overall protein metal binding properties. They showed a more than eightfold enhanced T1 relaxivity at the field strengths of 3.0 T, which is consistent with our result reported previously (Fig. 1c) [12].
Fig. 1.
a Modeled structures of engineered contrast agents. b Fluorescence spectroscopy and metal binding properties of contrast agents. Insertion of additional peptide from GRP will not affect protein folding and the metal binding properties of contrast agents. c Relaxivities (r1: gray bar and r2: black bar) of contrast agents at 3.0 T (*p<0.05).
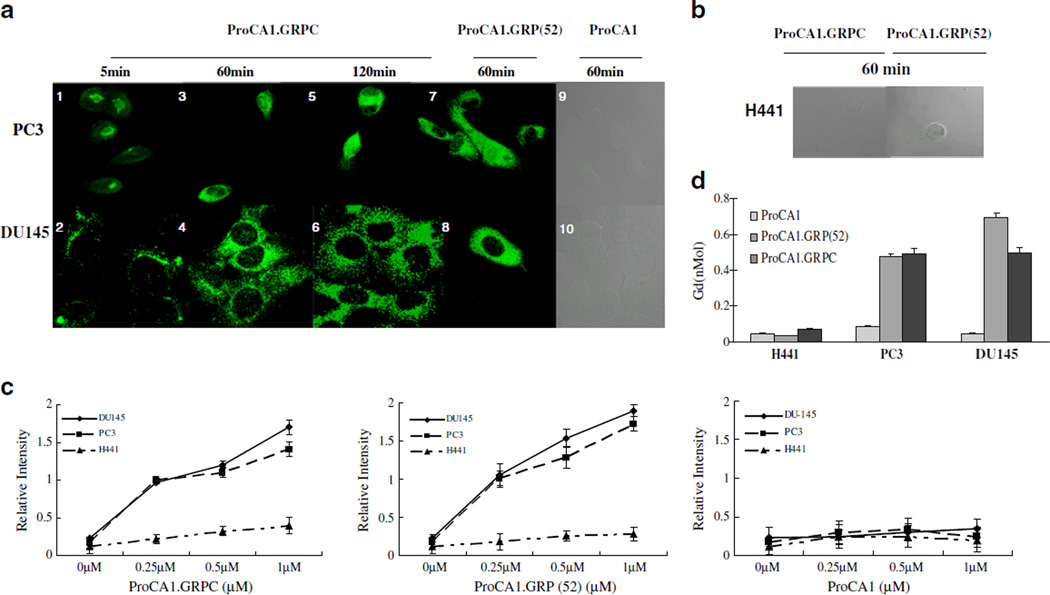
We next examined whether ProCA1.GRPC or ProCA1. GRP(52) can target cancer cells that express GRPR. We first carried out cell binding analyses by immunostaining with three cancer cell lines PC3, DU145, and H441 that express different levels of GRPR: l×105, ~4×105, and ~ 200 receptors/cell, respectively [27]. The immunostaining of ProCA1 using a polyclonal antibody against CD2 showed an increase in the staining intensities of ProCA1.GRPC and ProCA1.GRP(52) bound to both DU145 and PC3 cells when incubated for 2 h. Binding of the proteins to the cells triggered receptor-mediated endocytosis within 5 min, and a majority of the proteins had entered the cells after a 120-min incubation in both DU145 and PC3 cells (Fig. 2a). No binding was detected in either DU145 or PC3 cells treated by ProCA1. Strikingly, the protein was stable after internalization at 120 min, indicating that the designed proteins withstood protein degradation by endocytosis. No binding was detected in H441 treated by ProCA1.GRPC or ProCA1. GRP(52) (Fig. 2b). Binding of ProCA1.GRPC and ProCA1. GRP(52) to the selected three cell lines was further examined by enzyme-linked immunosorbent assay (ELISA) measurement (Fig. 2c). Results support the data obtained from the immunoassays, indicating that ProCA1.GRPC and ProCA1.GRP(52) had bound to PC3 and DU145 cells strongly, while no significant detectable binding was observed with H441 cells. ProCA1.GRP(52) showed higher binding capability with GRPR-positive cancer cells compared to ProCA1.GRPC. Cellular-bound Gd3+ was measured to further verify the binding of contrast agents to GRPR expression cells. The amount of Gd3+ was quantified by γ-counting the trace isotope 153Gd3+ in the Gd–protein complexes (Fig. 2d). The quantification of cellular-bound Gd3+ provided additional evidence that Gd- ProCA1.GRPC and Gd-ProCA1.GRP(52) can target DU145 and PC3 cells. Cells treated by ProCA1.GRP (52) showed stronger ELISA and Gd3+ radioactivity compared to that treated by ProCA1.GRPC, which supports our hypothesis that peptide sequence grafted in the scaffold protein had a better targeting capability than when fused to the C-terminal.
Fig. 2.
a, b Time-dependent uptake of ProCA1, ProCA1.GRPC, and ProCA1. GRP(52) in different cancer cells. DU145 and PC3 with different expression levels of GRPR showed uptake of contrast agents into the cells. Accumulation of contrast agents in the cells exhibited enhanced fluorescence. c ELISA analyzed the binding of contrast agents and cancer cells expressing different levels of GRPR. d Quantitative study of Gd3+ isotope by incubating 153Gd-labeled proteins and cancer.
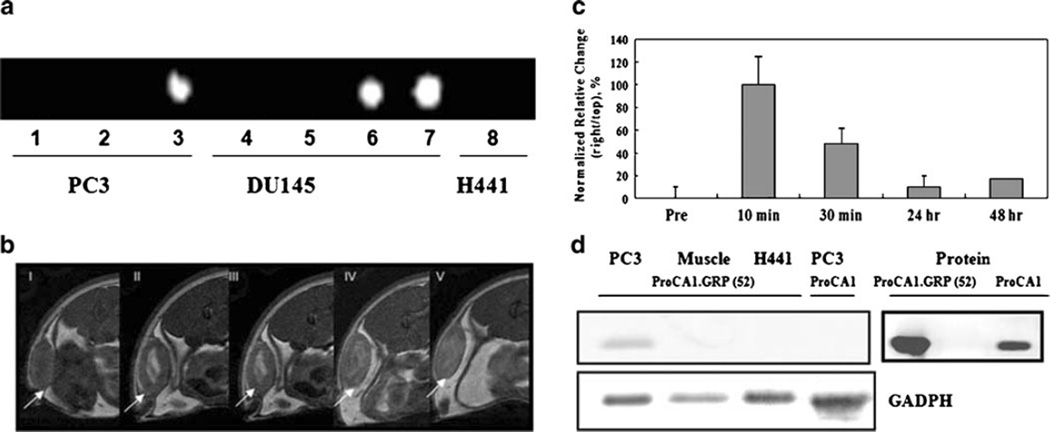
To test whether our designed proteins are applicable for targeted imaging, we first carried out MR imaging of cancer cells that were incubated with the protein contrast agents. PC3 and DU145 cells with high numbers of GRPR showed brighter imaging in the presence of Gd- ProCA1.GRPC and Gd-ProCA1.GRP(52), as demonstrated by T1-weighted spin echo imaging (Fig. 3a). In contrast, no similar contrast enhancement was observed in H441 cells. As we expected, cells treated with Gd-DTPA (as reference) and Gd-ProCA1 without the targeting moiety produced no contrast enhancement. Further, immunoblotting analysis of the protein extracts made from cell pellets treated by our agents revealed that ProCA1.GRPC and ProCA1.GRP(52) proteins were intact after MR imaging (Electronic supplementary material). These results support our hypothesis that insertion of the GRP targeting sequence to our protein contrast agent has the potential to provide molecular imaging of cancer cells via receptor-mediated recognition.
Fig. 3.
a MR imaging of cancer cells at 3.0 T (inversion recovery sequence, TR=4,000 ms, TE=12 ms, IR=500 ms). PC3 cells and DU145 cells (107) with GRP receptor expression were incubated with different contrast agents, respectively. (Gd-DTPA(1 and 4), Gd-ProCA1(2 and 5), Gd-ProCA1.GRPC(3 and 6), and Gd-ProCA1.GRP(52) (7 and 8)). H441 cells without GRP receptor expression were incubated with Gd-ProCA1.GRP(52) (8). Gd-DTPA without targeting was used as reference. b T1-weighted fast spin echo images obtained from a 4.7-T MRI scanner show contrast enhancement in tumors after intratumoral injection of contrast agents in mouse tumor models (fast spin echo sequences, TR=2 s, TE=0.022 s, and ESP = 0.01 s, Slice thickness= 1 mm). I Pre-injection; II 10 min after injection; II 30 min after injection; III 24 h after injection; IV 48 h after injection. c MRI signal intensities from selected ROIs were analyzed by ImageJ (n=5, p<0.05). d Immunoblotting analysis of tissue extracts. CD2 antibody was used as primary antibody to detect the protein contrast agent. ProCA1.GRP(52) was detected in PC3 tumor, but not H441 tumor. ProCA1 without targeting can not be detected in PC3 tumor.
To investigate the toxicity of the contrast agent in both cells and animals, cell viability was analyzed after incubating PC3 and H441 cells with the agent for 48 h. In addition, clinical biochemistry analyses of animal blood samples after administration of the contrast agent were performed. After 48 h of incubation with our contrast agent, no cytotoxicity was observed for both H441 and PC3 cell lines revealed by MTT assay (ESM Fig. SI-3 and Table SI-1). Similarly, 2 days after administration of our contrast agent with a dosage threefold higher than that used in animal imaging studies, no significant acute toxicity of our contrast agents was observed.
To examine whether the targeted MRI contrast enhancement can be detected in the tissue in vivo, we used tumor xenograft models. To this end, PC3 and H441 tumors (~0.5 cm) were grown with immunodeficient mice. The contrast agent Gd-ProCA1 and Gd-ProCA1.GRP(52) (140µm, 20µl) were directly injected into the tumor. MR images were recorded at different time points (pre-scan and 10 min, 30 min, 24 h, and 48 h after the injection) to follow the change of MRI contrast in various regions. The observed contrast enhancements in the H441 tumor started to decrease after 30 min post-injection and reached almost background level within 24 h (Electronic supplementary material). The PC3 tumor maintained a strong contrast enhancement until 48 h post-injection (Fig. 3b, c). The targeting capability of our contrast agents was further demonstrated by immunoblotting analyses of tumor tissue extracts (Fig. 3d). Our protein was detected in the PC3 tumor, but not in the H441 tumor. This result indicates that this prostate cancer targeting MRI contrast agent generated a strong GRPR targeted contrast enhancement both in cancer cells and in the prostate cancer xenograft model. Retention of the contrast over time is likely due to specific targeting and interaction between the targeting moiety and the GRPR over-expressed cancer cells.
Discussion
Our results clearly show that the grafting of the GRP targeting moiety led to specific targeting of the receptor (GRPR) and accumulation of Gd-binding proteins, which resulted in a significant GRPR-dependent contrast enhancement in MR imaging. The protein contrast agent had three important properties that may be advantageous in MR imaging of prostate cancer as well as other biomarkers in vivo: (1) ProCA1.GRP(52) demonstrated better metal binding affinity and targeting capability than ProCA1.GRPC, suggesting that a grafted targeting sequence with a better defined loop structure facilitates the protein stability and is required for the metal binding and receptor binding. This finding demonstrates an effective development in imaging and therapeutic agents that overcomes the lack of defined native conformation of peptide fragments. (2) Binding of the designed protein to the cell surface GRPR led to the accumulation of the contrast agent in cells. Interestingly, calculating the amounts of bound Gd3+ from γ-counting revealed that the Gd3+ ions were bound to cells at ~0.5 fmol Gd/cell. Assuming that 107 cells make a volume of 50– 100 µl, this binding capacity led to the accumulation of Gd3+ at approximately 20–50 µM in the cell pellets. This local concentration is sufficient to produce a strong MRI contrast with high relaxivity [28]. Our protein contrast agent remained intact for up to 2 h after internalization in the cell. One plausible explanation is that the host protein, domain 1 of CD2, has been shown to withstand acidic conditions, remaining fully folded at a pH as low as 2 [29]. Thus, it is conceivable that the protein contrast agent was not denatured by the low pH conditions in endosomes after endocytosis. (3) It was clear that the agent that was injected into the GRPR negative tumor H441 was washed away within 24 h, while the agent injected into the GRPR-positive tumor PC3 was retained at the tumor site for up to 48 h. Immunoblotting using antibody against parental protein CD2 shown in Fig. 3d further demonstrated that our contrast agent can be retained in PC3 tumor, but not in H441 tumor. These results indicate that our protein contrast agent has excellent penetration properties in GRP receptor positive tumors.
In principle, the MRI contrast enhancement and the off rate of the contrast agent from over-expressed cancer biomarkers at the tumor environment should proportionally reflect the binding capability of the target contrast agents to the cancer cells. Intratumoral injection allows us to directly monitor the molecular recognition capability in situ, avoiding the complication from dynamic process associated with systematic delivery. Our animal MR imaging by intratumoral injection shown in Fig. 3 clearly indicates that our developed protein MRI contrast agent has the capability to target the cell surface receptor and led to GRPR-specific MRI contrast enhancements. The experiments would provide the excellent guidance for MR imaging GRPR by systemic administration.
Currently, assessment of GRP receptor levels without invasive biopsy is difficult. Even with biopsy samples, determination of the GRP receptor expression levels and patterns in prostate cancers is still a challenge. Development of an MR imaging methodology that will allow for noninvasive semi-quantitative measurement of GRP receptor levels in prostate cancer will have a great implication in prostate cancer diagnosis and prognosis. The GRP receptor has been validated as an important cancer therapy target. Many therapeutic strategies have been developed to target GRP receptor [11, 30–32]. Our results may have strong clinical implications for cancer diagnosis and therapeutics since a high density of GRPR was found in the prostate, small cell lung, and pancreatic cancer cells. Introduction of an effective imaging method to monitor the changes in GRP receptor levels will enable us to improve and monitor cancer treatments.
Conclusion
In this study, we demonstrated that our developed contrast agents grafted with the GRP fragment were able to target prostate cancer cells that over-express GRP receptors. MRI of both cells and prostate cancer xenograft model with our contrast agents showed the GRP receptor targeting capability of our contrast agents. Only the GRPR-positive cells treated with GRPR targeting contrast agents showed a greater than tenfold enhancement in MRI signal, which demonstrated that our contrast agents have high specificity for GRPRpositive cells. The high sensitivity of our contrast agent for MR imaging is demonstrated by the capability in detection of GRPR expressed cancer cells at a number as low as 107, which is applicable in detecting the tumor at sizes <0.3 cm [33]. Finally, our results may have strong clinical implications for cancer diagnosis and therapeutics since a high density of GRPR was found in the prostate, small cell lung, and pancreatic cancer cells.
Supplementary Material
Acknowledgments
We thank Dan Adams, Birgit Neuhaus, Dr. Christie Cater, Jie Jiang, Dr. Leland Chung for their assistance. This work is supported in part by research grants from NIH CA118113 to Zhi-Ren Liu and NIH GM62999 and EB007268 to Jenny J Yang.
Abbreviations
- MRI
Magnetic resonance imaging
- GRP
Gastrin-releasing peptide
- GRPR
Gastrin-releasing peptide receptor
- ELISA
Enzyme-linked immunosorbent assay
- HRP
Horse-radish peroxide
- TBST
Tris-buffered saline and Tween-20
- RIO
Region of interest
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11307-010-0342-9) contains supplementary material, which is available to authorized users.
References
- 1.Sherry AD, Woods M. Chemical exchange saturation transfer contrast agents for magnetic resonance imaging. Annu Rev Biomed Eng. 2008;10:391–411. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caravan P, Ellison JJ, McMurry TJ, Lauffer RB. Gadolinium(III) chelates as MRI contrast agents: structure, dynamics, and applications. Chem Rev. 1999;99:2293–2352. doi: 10.1021/cr980440x. [DOI] [PubMed] [Google Scholar]
- 3.Tweedle MF. Peptide-targeted diagnostics and radiotherapeutics. Acc Chem Res. 2009;42:958–968. doi: 10.1021/ar800215p. [DOI] [PubMed] [Google Scholar]
- 4.Yoo B, Pagel MD. An overview of responsive MRI contrast agents for molecular imaging. Front Biosci. 2008;13:1733–1752. doi: 10.2741/2796. [DOI] [PubMed] [Google Scholar]
- 5.Mulder WJ, Strijkers GJ, Griffioen AW, van Bloois L, Molema G, et al. A liposomal system for contrast-enhanced magnetic resonance imaging of molecular targets. Bioconjug Chem. 2004;15:799–806. doi: 10.1021/bc049949r. [DOI] [PubMed] [Google Scholar]
- 6.Cyrus T, Lanza GM, Wickline SA. Molecular imaging by cardiovascular MR. J Cardiovasc Magn Reson. 2007;9:827–843. doi: 10.1080/10976640701693766. [DOI] [PubMed] [Google Scholar]
- 7.Branca RT, Chen YM, Mouraviev V, Galiana G, Jenista ER, et al. iDQC anisotropy map imaging for tumor tissue characterization in vivo. Magn Reson Med. 2009;61:937–943. doi: 10.1002/mrm.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins DJ, Padhani AR. Dynamic magnetic resonance imaging of tumor perfusion. Approaches and biomedical challenges. IEEE Eng Med Biol Mag. 2004;23:65–83. doi: 10.1109/memb.2004.1360410. [DOI] [PubMed] [Google Scholar]
- 9.Stephen RM, Gillies RJ. Promise and progress for functional and molecular imaging of response to targeted therapies. Pharm Res. 2007;24:1172–1185. doi: 10.1007/s11095-007-9250-3. [DOI] [PubMed] [Google Scholar]
- 10.Artemov D, Mori N, Ravi R, Bhujwalla ZM. Magnetic resonance molecular imaging of the HER-2/neu receptor. Cancer Res. 2003;63:2723–2727. [PubMed] [Google Scholar]
- 11.Zhang X, Cai W, Cao F, Schreibmann E, Wu Y, et al. 18F-labeled bombesin analogs for targeting GRP receptor-expressing prostate cancer. J Nucl Med. 2006;47:492–501. [PubMed] [Google Scholar]
- 12.Yang JJ, Yang J, Wei L, Zurkiya O, Yang W, et al. Rational design of protein-based MRI contrast agents. J Am Chem Soc. 2008;130:9260–9267. doi: 10.1021/ja800736h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez N, Moody TW, Igarashi H, Ito T, Jensen RT. Bombesin-related peptides and their receptors: recent advances in their role in physiology and disease states. Curr Opin Endocrinol Diabetes Obes. 2008;15:58–64. doi: 10.1097/MED.0b013e3282f3709b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun B, Halmos G, Schally AV, Wang X, Martinez M. Presence of receptors for bombesin/gastrin-releasing peptide and mRNA for three receptor subtypes in human prostate cancers. Prostate. 2000;42:295–303. doi: 10.1002/(sici)1097-0045(20000301)42:4<295::aid-pros7>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 15.Uchida K, Kojima A, Morokawa N, Tanabe O, Anzai C, et al. Expression of progastrin-releasing peptide and gastrin-releasing peptide receptor mRNA transcripts in tumor cells of patients with small cell lung cancer. J Cancer Res Clin Oncol. 2002;128:633–640. doi: 10.1007/s00432-002-0392-8. [DOI] [PubMed] [Google Scholar]
- 16.Grady EF, Slice LW, Brant WO, Walsh JH, Payan DG, et al. Direct observation of endocytosis of gastrin releasing peptide and its receptor. J Biol Chem. 1995;270:4603–4611. doi: 10.1074/jbc.270.9.4603. [DOI] [PubMed] [Google Scholar]
- 17.Shin C, Mok KH, Han JH, Ahn JH, Lim Y. Conformational analysis in solution of gastrin releasing peptide. Biochem Biophys Res Commun. 2006;350:120–124. doi: 10.1016/j.bbrc.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Heimbrook DC, Boyer ME, Garsky VM, Balishin NL, Kiefer DM, et al. Minimal ligand analysis of gastrin releasing peptide. Receptor binding and mitogenesis. J Biol Chem. 1988;263:7016–7019. [PubMed] [Google Scholar]
- 19.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Yang W, Wilkins AL, Ye Y, Liu ZR, Li SY, et al. Design of a calcium-binding protein with desired structure in a cell adhesion molecule. J Am Chem Soc. 2005;127:2085–2093. doi: 10.1021/ja0431307. [DOI] [PubMed] [Google Scholar]
- 21.Yang W, Wilkins AL, Li S, Ye Y, Yang JJ. The effects of Ca2+ binding on the dynamic properties of a designed Ca2+-binding protein. Biochemistry. 2005;44:8267–8273. doi: 10.1021/bi050463n. [DOI] [PubMed] [Google Scholar]
- 22.Ye Y, Lee HW, Yang W, Shealy SJ, Wilkins AL, et al. Metal binding affinity and structural properties of an isolated EF-loop in a scaffold protein. Protein Eng. 2001;14:1001–1013. doi: 10.1093/protein/14.12.1001. [DOI] [PubMed] [Google Scholar]
- 23.Roberts HC, Roberts TP, Brasch RC, Dillon WP. Quantitative measurement of microvascular permeability in human brain tumors achieved using dynamic contrast-enhanced MR imaging: correlation with histologic grade. AJNR Am J Neuroradiol. 2000;21:891–899. [PMC free article] [PubMed] [Google Scholar]
- 24.Hawnaur JM, Zhu XP, Hutchinson CE. Quantitative dynamic contrast enhanced MRI of recurrent pelvic masses in patients treated for cancer. Br J Radiol. 1998;71:1136–1142. doi: 10.1259/bjr.71.851.10434907. [DOI] [PubMed] [Google Scholar]
- 25.Ye F, Wu X, Jeong EK, Jia Z, Yang T, et al. A peptide targeted contrast agent specific to fibrin–fibronectin complexes for cancer molecular imaging with MRI. Bioconjug Chem. 2008;19:2300–2303. doi: 10.1021/bc800211r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye Y, Shealy S, Lee HW, Torshin I, Harrison R, et al. A grafting approach to obtain site-specific metal-binding properties of EF-hand proteins. Protein Eng. 2003;16:429–434. doi: 10.1093/protein/gzg051. [DOI] [PubMed] [Google Scholar]
- 27.Jensen JA, Carroll RE, Benya RV. The case for gastrin-releasing peptide acting as a morphogen when it and its receptor are aberrantly expressed in cancer. Peptides. 2001;22:689–699. doi: 10.1016/s0196-9781(01)00380-1. [DOI] [PubMed] [Google Scholar]
- 28.Nunn AD, Linder KE, Tweedle MF. Can receptors be imaged with MRI agents? Q J Nucl Med. 1997;41:155–162. [PubMed] [Google Scholar]
- 29.Maniccia AW, Yang W, Li SY, Johnson JA, Yang JJ. Using protein design to dissect the effect of charged residues on metal binding and protein stability. Biochemistry. 2006;45:5848–5856. doi: 10.1021/bi052508q. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Li ZB, Cao Q, Liu S, Wang F, et al. Small-animal PET of tumors with (64)Cu-labeled RGD-bombesin heterodimer. J Nucl Med. 2009;50:1168–1177. doi: 10.2967/jnumed.108.061739. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Niu G, Wang F, Chen X. (68)Ga-labeled NOTA-RGDBBN peptide for dual integrin and GRPR-targeted tumor imaging. Eur J Nucl Med Mol Imaging. 2009;36:1483–1494. doi: 10.1007/s00259-009-1123-z. [DOI] [PubMed] [Google Scholar]
- 32.Ho CL, Chen LC, Lee WC, Chiu SP, Hsu WC, et al. Receptor-binding, biodistribution, dosimetry, and micro-SPECT/CT imaging of 111In-[DTPA(1), Lys(3), TyR4)]-bombesin analog in human prostate tumor-bearing mice. Cancer Biother Radiopharm. 2009;24:435–443. doi: 10.1089/cbr.2008.0616. [DOI] [PubMed] [Google Scholar]
- 33.Brennan MJ. Some reflections on research. J Assoc Phys Ment Rehabil. 1963;17:171. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.