Abstract
Isopeptidases are essential regulators of protein ubiquitination and sumoylation. However, only two families of SUMO isopeptidases are at present known. Here, we report an activity-based search with the suicide inhibitor haemagglutinin (HA)-SUMO-vinylmethylester that led to the identification of a surprising new SUMO protease, ubiquitin-specific protease-like 1 (USPL1). Indeed, USPL1 neither binds nor cleaves ubiquitin, but is a potent SUMO isopeptidase both in vitro and in cells. C13orf22l—an essential but distant zebrafish homologue of USPL1—also acts on SUMO, indicating functional conservation. We have identified invariant USPL1 residues required for SUMO binding and cleavage. USPL1 is a low-abundance protein that colocalizes with coilin in Cajal bodies. Its depletion does not affect global sumoylation, but causes striking coilin mislocalization and impairs cell proliferation, functions that are not dependent on USPL1 catalytic activity. Thus, USPL1 represents a third type of SUMO protease, with essential functions in Cajal body biology.
Keywords: SUMO protease, ubiquitin-specific protease family, USPL1, zebrafish C13orf22l, Cajal body
Introduction
Reversible attachment of SUMO (small ubiquitin-related modifier) is an essential protein modification in all eukaryotic cells 1], [2]. Hundreds of proteins are regulated by transient sumoylation, which affects activity, localization and/or interactions of its targets. SUMO’s role as a molecular switch depends on rapid and tightly controlled cycles of sumoylation and desumoylation, and hence on both, conjugating enzymes and isopeptidases. E1, E2 and E3 ligases are involved in ATP-dependent conjugation of SUMO proteins to lysine side chains in targets. This results in mono-sumoylation and/or chain formation. Deconjugation is accomplished by SUMO-specific proteases. Well-characterized enzymes include two ubiquitin-like-specific proteases in yeast (Ulp1 and Ulp2) and six sentrin-specific proteases (SENP1, 2, 3, 5, 6 and 7) in mammals [3, 4]. Ulp and SENP proteases belong to the C48 family of cysteine proteases. They share a conserved catalytic domain that includes a His–Cys–Asp catalytic triad. Ulp/SENP proteases are not only required as isopeptidases, but also as carboxy-terminal hydrolases in SUMO maturation. Recently, a different protease family was implicated in desumoylation: two small proteins with a PPPDE domain (permutated papain fold peptidases of the double-stranded RNA viruses and eukaryotes) were found to desumoylate the transcription repressor BZEL [5]. These proteins, designated DeSI-1 and DeSI-2, seem to have restricted target specificity, as they fail to cleave sumoylated PML. Finally, the putative metalloprotease S.c. Wss-1 has been shown to exert desumoylation activity in vitro [6]. Whether it functions as a SUMO isopeptidase or a SUMO-directed ubiquitin isopeptidase in vivo is at present unclear.
The small number of known SUMO proteases stands in striking contrast to almost 100 different ubiquitin proteases belonging to five different families [7, 8]. The largest of these is the C19 cysteine protease family of ubiquitin-specific proteases (USPs) with more than 50 members [9, 10]. This encouraged us to embark on an activity-based search with suicide substrates (HA-SUMO-vinylmethylester) that should irreversibly crosslink with SUMO proteases. Similar substrates had been used, for example, for identification of active proteases of the ubiquitin family [11] or the characterization of known SUMO isopeptidases [12]. As shown below, this resulted in the unexpected identification of ubiquitin-specific protease-like 1 (USPL1) as a highly specific SUMO isopeptidase with an essential role both in human cells and in zebrafish.
Results and discussion
An activity-based search for novel SUMO isopeptidases
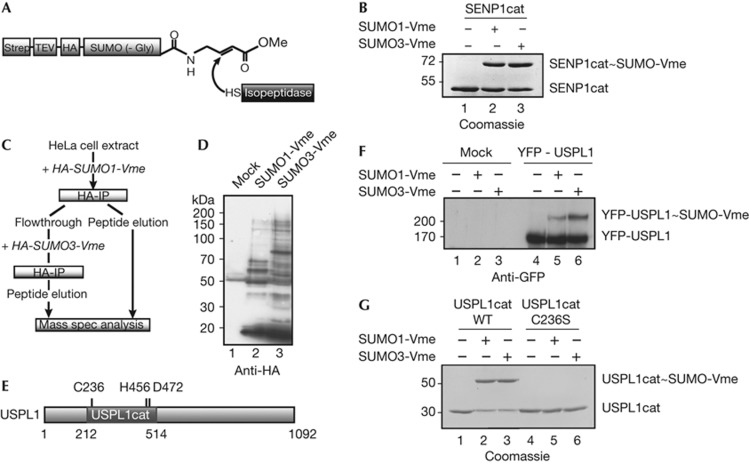
To search for new SUMO isopeptidases, we generated HA-tagged SUMO1 and SUMO3 derivatives ligated by intein chemistry to vinylmethylester (Fig 1A). SUMO1-Vme and SUMO3-Vme were both able to crosslink with the catalytic domain of SENP1 in vitro (Fig 1B), confirming functionality. Subsequently, we incubated HeLa lysates with increasing amounts of SUMO1-Vme or SUMO3-Vme, and enriched crosslinked proteins by anti-HA immunoprecipitation (IP). Number and size of the observed bands on immunoblot analysis suggested the existence of numerous SUMO isopeptidases (supplementary Fig S1 online). Finally, we carried out a large-scale consecutive IP experiment: 25 ml HeLa extract was incubated with HA-SUMO1-Vme, bound proteins were enriched by immobilized anti-HA antibodies and the supernatant was treated with HA-SUMO3-Vme. After a second anti-HA-IP, proteins were eluted from either beads and identified by mass spectrometry (Fig 1C,D). In addition to known isopeptidases, the most interesting candidate identified was USPL1 (swiss-prot/Q5W0Q7; Fig 1E). To confirm that USPL1 can be crosslinked to SUMO-Vme substrates, we transfected cells with YFP-USPL1 and added suicide substrates to cell extracts 24 h after transfection. Indeed,YFP-USPL1 reacts with SUMO1-Vme and with SUMO3-Vme (Fig 1F). We repeated the experiment with purified recombinant USPL1 catalytic domain (USPL1cat), and again observed covalent association with both suicide inhibitors (Fig 1G). Importantly, mutating USPL1’s catalytic cysteine (C236) to serine abolished the reaction (Fig 1G, lanes 5 and 6). Taken together, these findings provided the first evidence for a role of USPL1 as a SUMO protease.
Figure 1.
Identification of USPL1 as a SUMO isopeptidase candidate. (A) Model: suicide inhibitor Strep-TEV-HA-SUMO-vinylmethylester (SUMO-Vme) attacked by an isopeptidase. (B) SUMO-Vme is crosslinked to a known SUMO isopeptidase. The catalytic fragment GST-SENP1cat was incubated with SUMO1- or SUMO3-Vme. Reaction products were analysed by SDS–PAGE. (C) Sequential IP approach to identify new SUMO isopeptidases. (D) Analysis of peptide eluates (see C) by immunoblotting with anti-HA antibodies. (E) Schematic representation of USPL1 with catalytic domain and Cys–His–Asp triad. (F) Transfected YFP-USPL1 reacts with SUMO-Vme in HEK 293T extracts. Analysis was by immunoblotting with anti-GFP antibodies. (G) USPL1 requires its catalytic cysteine to react with SUMO-Vme. Recombinant USPL1cat (212–514) wt or C236S was incubated with SUMO1- or SUMO3-Vme. Reaction products were analysed by SDS–PAGE. cat, catalytic fragment; GFP, green fluorescence; Gly, glycine; GST, glutathione-S-transferase; HA, haemagglutinin epitope; IP, immunoprecipitation; Me, methyl; SENP1, SUMO/sentrin-specific protease 1; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; Strep, streptavidin; SUMO, small ubiquitin-related modifier; TEV, Tobacco Etch Virus protease cleavage site; USPL1, ubiquitin-specific protease like 1; Vme, vinylmethylester; WT, wild type; YFP, yellow fluorescence protein.
USPL1 binds SUMO but not ubiquitin
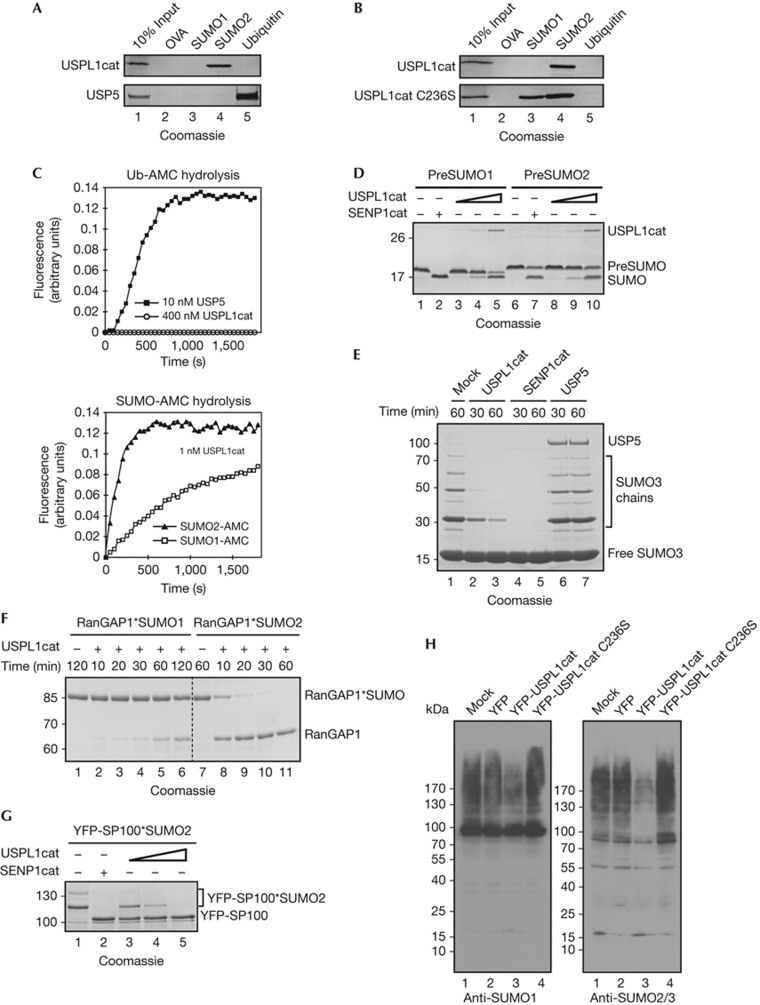
USPL1 is a poorly characterized 120-kDa member of the C19 cysteine protease USP family. Sequence similarity to conventional USPs is restricted to its catalytic domain, and highest for USP1 (19.5% residue identity). While USPL1’s catalytic domain contains the catalytic triad (C236, H456, D472, supplementary Fig S2 online), it has been assumed inactive (swiss-prot/Q5W0Q7; see also below). To test whether USPL1 can bind SUMO1, SUMO2 or ubiquitin non-covalently, we immobilized untagged mature Ubls on CNBr-activated sepharose and incubated them with USPL1cat. Full-length USP5 was included as a control for ubiquitin binding [13] and immobilized ovalbumin as a control for unspecific binding. Strikingly, USPL1cat interacted with SUMO2 but failed to bind ubiquitin (Fig 2A). Interaction with SUMO1 could be detected with USPL1cat C236S (Fig 2B), as this mutant bound SUMO proteins more stably than wild-type (wt). Together, these findings indicate that USPL1 interacts specifically with both SUMO proteins, but prefers SUMO2/3.
Figure 2.
USPL1 is a SUMO-specific protease. (A) USPL1 binds SUMO, not ubiquitin. Recombinant USPL1cat and USP5 were incubated with immobilized ovalbumin (OVA), SUMO1, SUMO2 and ubiquitin. Bound proteins were analysed by SDS–PAGE. (B) USPL1cat C236S binds to immobilized SUMO1 and SUMO2. Experiment as in A. (C) USPL1cat cleaves SUMO-, but not ubiquitin-AMC. Ubiquitin-, SUMO1- and SUMO2-AMC (0.25 μM) were incubated with USPL1cat or USP5, and released AMC was detected by fluorescence. (D) USPL1cat matures SUMO. PreSUMO1 or preSUMO2 (8 μM) were incubated with 20 nM SENP1cat or USPL1cat (40, 17 and 860 nM) and samples were analysed by SDS–PAGE. (E) USPL1cat cleaves SUMO3 chains. SUMO3 chains of 12.5 μM were incubated with 5 nM SENP1cat, 5 nM USPL1cat or 150 nM USP5 for the indicated times. Reaction products were analysed by SDS–PAGE. (F,G) USPL1 cleaves sumoylated targets. (F) Two micrometre RanGAP1*SUMO1 or RanGAP1*SUMO2 were incubated with 4 nM USPL1cat for indicated times. Reactions were analysed by SDS–PAGE. (G) YFP-SP100*SUMO2 was incubated with 5 nM SENP1cat or USPL1cat (4.3, 8.6 or 43 nM). Reaction products were analysed by SDS–PAGE. (H) USPL1cat deconjugates SUMO targets in cells. HEK 293T cells were transfected with YFP empty vector, YFP-USPL1cat wt or C236S, and analysed after 24 h by immunoblotting. AMC, 7-amino-4-methylcoumarin; OVA, ovalbumin; preSUMO, precusor of small ubiquitin-like modifier; RanGAP1, Ran GTPase-activating protein 1; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; SP100, nuclear autoantigen Sp100; SUMO, small ubiquitin-related modifier; Ub, ubiquitin; USP5, ubiquitin-specific protease 5.
USPL1 is a SUMO isopeptidase
To assess USPL1’s catalytic activities, we first tested for C-terminal hydrolase activity using the artificial substrates ubiquitin-, SUMO1- and SUMO2-AMC. As shown in Fig 2C, USPL1cat showed no activity for ubiquitin-AMC even at elevated concentrations (0.4 μM), but cleaved both SUMO-AMC substrates at 1 nM concentration. Rates were higher for SUMO2-AMC compared with SUMO1-AMC, consistent with USPL1’s preference in SUMO binding. USP5 served as a positive control for ubiquitin–AMC cleavage. Turning to physiological substrates, we tested whether USPL1cat exhibits SUMO maturation activity (Fig 2D). For this, 35, 170 and 860 nM USPL1cat were incubated with 8 μM preSUMO1 and preSUMO2. For comparison, we used 20 nM SENP1 catalytic fragment (SENP1cat), which is known to process both precursors [14]. USPL1 was indeed able to process preSUMO1 and preSUMO2, but was significantly less efficient than SENP1cat (Fig 2D). Notably, in contrast to SUMO–AMC, cleavage maturation of preSUMO1 and preSUMO2 was comparable for both SUMO isoforms. A likely interpretation is that the C-terminal extension of preSUMO1 is preferred over that of SUMO2. In conclusion, USPL1cat has some C-terminal hydrolase activity.
Next, we tested USPL1’s ability to function as a SUMO isopeptidase. First, we tested USPL1cat activity on SUMO3 chains (Fig 2E). SENP1cat and USP5 served as positive and negative controls. USPL1 indeed functioned as a SUMO isopeptidase, as revealed by the disappearance of SUMO3 chains. Finally, we tested whether USPL1cat is able to cleave substrates such as recombinant sumoylated RanGAP1 and YFP-Sp100 [15]. Indeed, both were rapidly demodified by USPL1 (Fig 2F,G). Comparison of cleavage rates for SUMO1- and SUMO2-modified RanGAP1 (2 μM substrate, 4 nM USPL1cat; Fig 2F) showed a clear preference of USPL1cat towards SUMO2. Quantitative analysis of USPL1cat on YFP-SUMO2*CFP-RanGAPtail (supplementary Fig S3A online) revealed that it is an efficient SUMO isopeptidase (kcat/KM=4 × 105 M−1 s−1), albeit two orders of magnitude less efficient than Senp2cat.
Finally, we tested whether USPL1cat targets sumoylated species in a cellular context. For this, we overexpressed YFP-USPL1cat wt and C236S mutant in HEK 293T cells and analysed cell extracts by immunoblotting (Fig 2H). Indeed, overexpression of USPL1cat caused a striking loss of SUMO2/3 conjugates. USPL1cat C236S on the other hand increased SUMO2/3 conjugates, indicating a dominant-negative effect. SUMO1 targets were affected less severely, consistent with in vitro findings. As expected, ubiquitylated species were not influenced by USPL1cat overexpression (supplementary Fig S3B online). Taken together, our findings demonstrate that USPL1 is a SUMO isopeptidase that prefers SUMO2/3 both in vitro and in vivo.
Zebrafish C13orf22l is also a SUMO isopeptidase
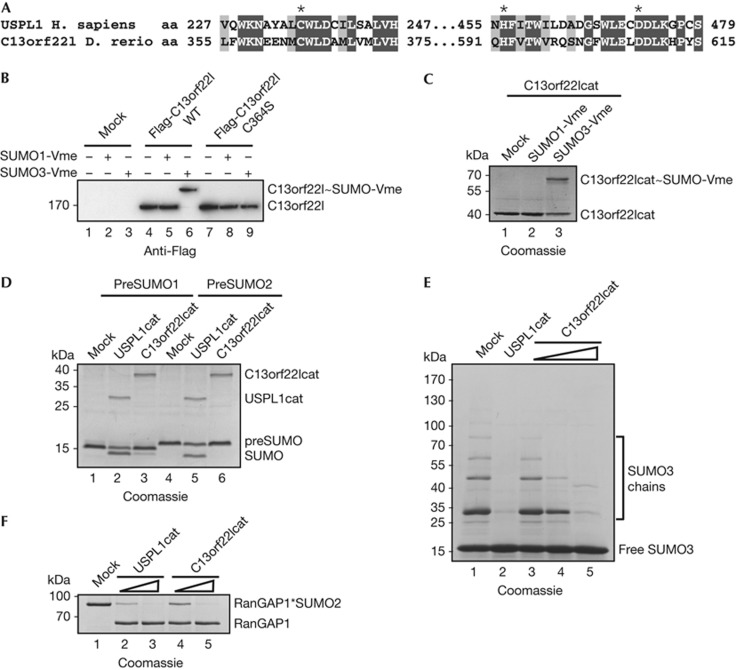
To expand our analysis of USPL1, we searched for homologues: based on available sequences, the USPL1 family is restricted to the metazoan kingdom, where it is found in species with and without bilateral symmetry (supplementary Fig S2 online). Intact USPL1 sequences are present in vertebrates, chordates and selected invertebrate phyla. The catalytic domain of USPL1 has been lost in several lineages, including insects, where proteins with similarity to the non-catalytic USPL1 amino terminus exist. Other phyla, such as nematodes, are devoid of USPL1-like sequences. We got especially interested in Danio rerio C13orf22l, as it had been identified in a mutagenesis screen as essential for early development [16]. Catalytic domains of C13orf22l and USPL1 share 34% sequence identity, including short regions of higher conservation (Fig 3A). Like USPL1, transfected wt Flag-C13orf22l and recombinant C13orf22lcat (aa 312–649) could be crosslinked to human SUMO3-Vme (Fig 3B,C). C13orf22lcat was highly active on SUMO2-AMC and did not cleave ubiquitin-AMC (supplementary Fig S4 online). It showed little activity in maturation assays involving human SUMO precursors (Fig 3D), but cleaved SUMO3 chains (Fig 3E) and SUMO2-modified RanGAP1 efficiently (Fig 3F). Taken together, zebrafish C13orf22l is a functional homologue of human USPL1.
Figure 3.
Zebrafish C13orf22l, a distant USPL1 homologue, is a SUMO-specific protease. (A) Alignment of human USPL1 and zebrafish C13orf22l highlighting conserved regions in the catalytic domain. * indicates residues of the catalytic triad. (B) C13orf22l reacts with SUMO3-Vme via its catalytic cysteine. Extract from untransfected, Flag-C13orf22l wt or C364S-transfected HEK 293T cells were labelled with SUMO1- and SUMO3-Vme, and analysed by immunoblotting. (C) Recombinant C13orf22lcat reacts with SUMO3-Vme. C13orf22lcat was incubated with SUMO1- or SUMO3-Vme. Reaction products were analysed by SDS–PAGE. (D) C13orf22lcat has little activity towards human SUMO propeptides. PreSUMO1 or preSUMO2 (8 μM) was incubated with 860 nM USPL1cat or 1 μM C13orf22lcat. Reaction products were visualized by SDS–PAGE. (E) C13orf22lcat cleaves human SUMO3 chains. SUMO3 chains (25 μM) were incubated with USPL1cat (8.6 nM) or C13orf22lcat (10, 50 or 250 nM). Reaction products were analysed by SDS–PAGE. (F) C13orf22lcat deconjugates human RanGAP1*SUMO2. RanGAP1*SUMO2 of 0.8 μM was incubated with USPL1cat (4.3, 21.5 nM) or C13orf22lcat (5, 25 nM). Products were analysed by SDS–PAGE. C13orf22l, zebrafish USPL1; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; SUMO, small ubiquitin-related modifier.
USPL1-specific residues are required for activity
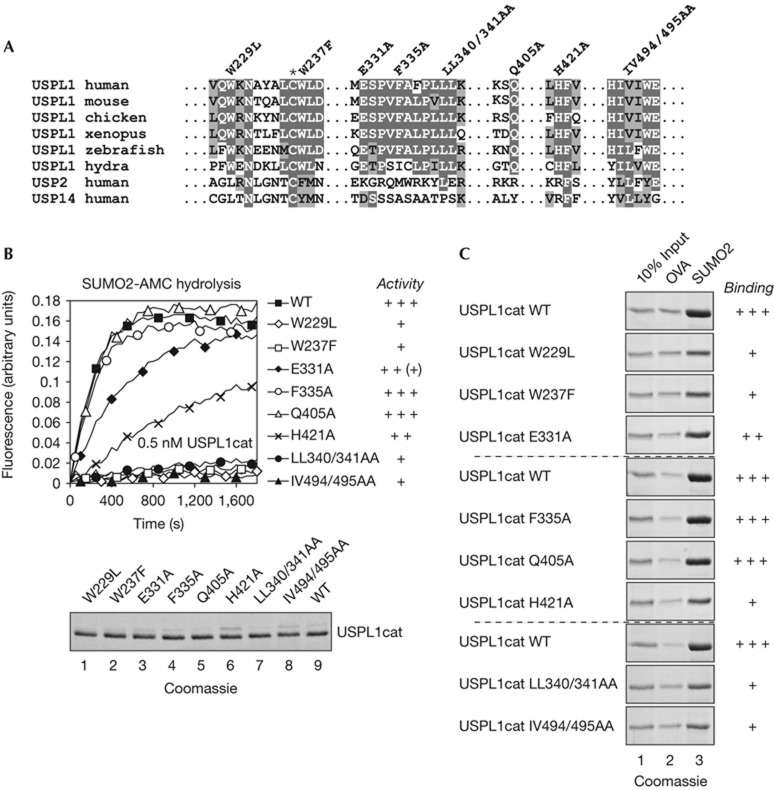
Our finding that USPL1 and C13orf22l are specific SUMO isopeptidases was unexpected, considering that the C48 family of Ulp/SENP proteases bears no substantial relationship to USPs [3, 17]. Moreover, SUMO and ubiquitin differ significantly in amino-acid composition and charge distributions [18]. However, USPL1 lacks several key residues implicated in ubiquitin binding (supplementary Fig S2 online), including a non-catalytic histidine [19] and residues of the QQD box [20, 21]. Aligning catalytic domains of USPL1 homologues from Hydra to Homo sapiens revealed clusters of conservation, some of which are specific for the USPL1 subfamily (Fig 4A; supplementary Fig S2 online). Within these clusters, W229, W237, Q405 and H421 are strictly conserved in USPL1 proteins, but different from residues in all other USPs; E331 is conserved in all USPL1s and absent in most other USPs; F335 and LL340/341 are localized in a hydrophobic patch specific for the USPL1 family; and IV494/495 are part of a hydrophobic patch similar in most USPs. To test contributions of these residues, we generated His-GST-TEV-USPL1cat variants and tested them for SUMO-AMC hydrolysis (Fig 4B) and SUMO binding (Fig 4C). Four mutants showed severe and two mutants moderate loss of activity. This was accompanied by severe or moderate reduction in binding, consistent with the idea that these conserved areas in USPL1 might be involved in SUMO recognition. Most striking was the effect of mutating W229 and W237 to residues found in USP2 and other ubiquitin-specific USPs (W229L and W237F): it dramatically impaired SUMO binding and cleavage. These findings are intriguing in light of structural studies of Ulp/SENP proteases [22, 23]: two tryptophanes close to the catalytic cysteine are needed to stably position the C-terminal diglycine motif of SUMO in the substrate tunnel and seem to allow trans–cis conversion of the scissile bond before cleavage. Whether USPL1 indeed shares these features with SENP proteases will have to await structural analysis.
Figure 4.
Identification of USPL1 residues required for SUMO binding and activity. (A) Alignment of USPL1 family members with human USP2 and USP14. * indicates catalytic cysteines. Mutations introduced in hUSPL1cat are indicated on top. (B) Top: His-GST-TEV-USPL1cat wt and indicated mutants (0.5 nM) were incubated with SUMO2-AMC (0.25 μM). Time-dependent release of AMC was detected by fluorescence. Bottom: SDS–PAGE of USPL1cat wt and mutants. (C) His-GST-TEV-USPL1cat wt and indicated mutants were applied to pulldown assays as described in Fig 2, using immobilized ovalbumin and SUMO2; lines separate sets of samples that were analysed on the same SDS gel. His, hexahistidine epitope tag; USP2, 5 and 14, ubiquitin-specific proteases 2, 5 and 14. AMC, 7-amino-4-methylcoumarin; OVA, ovalbumin; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; SUMO, small ubiquitin-related modifier; WT, wild type.
USPL1 is an essential Cajal body component
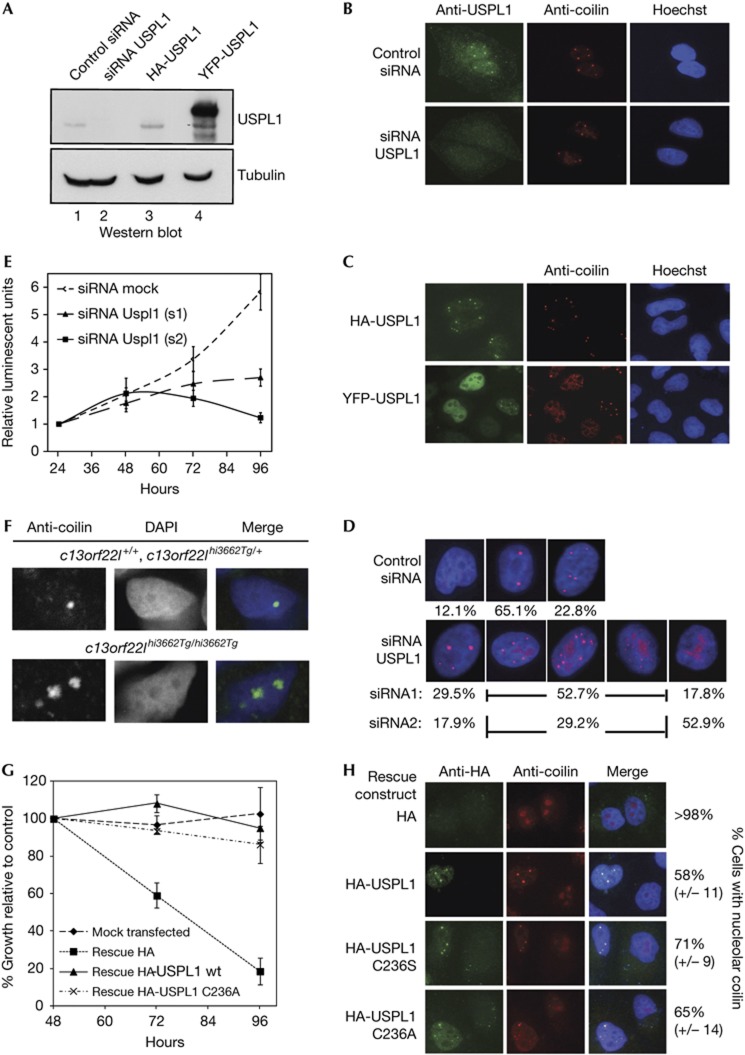
To gain insights into USPL1’s function, we generated affinity-purified antibodies and confirmed their specificity with short interfering RNA (siRNA) (Fig 5A). Immunoblotting, using recombinant USPL1 as a standard, indicates <5,000 USPL1 molecules per HeLa cell. To investigate USPL1’s endogenous localization, we turned to immunofluorescene. As shown in Fig 5B (upper panel), anti-USPL1 antibodies decorate nuclear bodies. Based on co-staining with anti-coilin, these are CBs. Specificity of staining was confirmed using USPL1 siRNA (Fig 5B, lower panel), and by transfection of tagged USPL1 (Fig 5C). While HA-USPL1 was found exclusively in CBs (Fig 5C, upper panel), YFP-USPL1 was present throughout the nucleoplasm when expressed at high levels (Fig 5C, lower panel). This points to limiting binding sites for USPL1 in CBs. CBs are highly dynamic structures that change in number, size and composition during cell cycle, development and stress [24, 25]. Functions associated with CBs reach from telomere maintenance to histone messenger RNA processing, but one of the most rate-limiting functions seems maturation and assembly of small nuclear ribonucleoprotein particles: depletion of coilin, which leads to early embryonic lethality in zebrafish, can be rescued by microinjection of mature human small nuclear ribonucleoprotein particles [26]. During our analysis, we noted that USPL1 knockdown alters the number and size of coilin-positive bodies (Fig 5D). While HeLa cells treated with control siRNA usually had 1–2 CBs, sometimes 3–7 CBs and rarely no CBs (Fig 5D, upper panel), USPL1 knockdown caused a marked change in CB size and number (Fig 5D, lower panel) that was accompanied by the appearance of coilin in nucleoli (Fig 5D, lower panel; supplementary Fig S5 online). At least 98% of all cells showed coilin partially or exclusively in the nucleolus (Fig 5D,H). This was neither accompanied by changes in coilin levels and appearance, nor by changes in global sumoylation (supplementary Fig S6 online). However, cells treated with siRNAs ceased to proliferate (Fig 5E), without obvious phenotypes. Considering that USPL1 disruption is embryonic lethal in zebrafish (strong necrotic phenotype in the central nervous system and eye at 2 days post fertilization (d.p.f.), death at 4–5 d.p.f. [16]), we tested whether the requirement of USPL1 for CBs was conserved in this organism: upon breeding of heterozygous mutants (c13orf22lhi3662Tg/hi3662Tg), we generated sections of the head region from wt and mutant embryos (supplementary Fig S7A online), fixed at 2 d.p.f., and stained with anti-coilin sera; indeed, many cells of c13orf22lhi3662Tg/hi3662Tg mutants revealed a striking accumulation of coilin in nucleoli (Fig 5F; supplementary Fig S7B online). In conclusion, C13orf22l is required for integrity of CBs in zebrafish embryos, indicating that this function of USPL1 is conserved.
Figure 5.
USPL1 is an essential Cajal body protein required for coilin localization and cell proliferation. (A) Affinity-purified anti-USPL1 recognize endogenous and transfected USPL1. Immunoblot analysis of HeLa cells transfected with nonspecific siRNA, siRNA against USPL1 (s2), pcDNA3.1-HA-USPL1 or pEYFP-USPL1. Tubulin was used as loading control. (B,C) Endogenous and transfected USPL1 colocalizes with coilin in Cajal bodies. (B) HeLa cells transfected with control siRNA or USPL1 siRNA s2 were stained with anti-USPL1 and anti-coilin. (C) HeLa cells transfected with pcDNA3.1-HA-USPL1 or pEYFP-USPL1 were stained with anti-HA and anti-coilin antibodies or observed directly (YFP). (D) Knockdown of USPL1 causes coilin mislocalization. HeLa cells were transfected with control siRNA or USPL1 siRNA, and stained with anti-coilin and Hoechst. Distribution of coilin was analysed in >200 cells per condition (three independent experiments). (E) Knockdown of USPL1 impairs HeLa cell proliferation. Analyses at indicated times were with a luminescent cell viability assay. Shown are means (±s.d.) of three independent experiments. (F) Transgenic c13orf22l zebrafish embryos show coilin mislocalization to the nucleolus. Labelling using anti-coilin sera (9EA2) on cryosections from the head of transgenic zebrafish mutants (c13orf22lhi3662Tg/hi3662Tg) and wt siblings (c13orf22l+/+, c13orf22lhi3662Tg/+) at 2 d.p.f. (G,H) Wt and inactive USPL1 variants rescue proliferation and coilin localization. Cells were transfected with USPL1 siRNA (s2), re-transfected with pcDNA3.1-HA-USPL1 variants and (G) analysed at indicated times with a colorimetric assay (normalized to control sample taken at the same times, error bars indicate s.d., five samples per time point), or (H) fixed and stained as indicated. Distribution of coilin was analysed in >200 cells per condition, in three independent experiments. DAPI, 4,6-diamidino-2-phenylindole; HA, haemagglutinin epitope; siRNA, short interfering RNA.
Finally, to test whether USPL1’s catalytic activity is required for cell proliferation and CB integrity, we retransfected siRNA-treated cells with wt and mutant HA-USPL1 variants, and analysed cell proliferation (relative to mock-treated cells; Fig 5G) and coilin distribution (Fig 5H). Strikingly, USPL1 inactive variants were as competent as wt USPL1 in rescue. These findings indicate that the new SUMO isopeptidase USPL1 has essential functions independent of catalysis. Whether these require USPL1’s ability to bind SUMO or depend on other unknown functions of USPL1 will be an intriguing question for future studies.
Methods
For plasmids, proteins, antibodies, oligos, synthesis of Strep-TEV-HA-SUMO1/3-Vme and detailed protocols see supplementary information online.
Nomenclature. SUMO1 (Smt3C; P63165), SUMO2 (Smt3A; also known as SUMO3, P55854) and SUMO3 (Smt3B; also known as SUMO2, P61956).
Identification of SUMO isopeptidases using HA-SUMO1/3-Vme. HeLa extracts (10 mg ml−1 protein) were prepared from frozen cells. For identification of SUMO-Vme-labelled proteins by mass spectrometry, 25 ml HeLa cell extract (250 mg proteins), 10 μg SUMO1-Vme and 10 μg SUMO3-Vme were used for consecutive IPs. SUMO-Vme-labelled proteins were enriched using anti-HA-agarose, and eluted with HA-peptide before immunoblotting or mass spectrometry.
Mass spectrometry. Mass spectrometric analyses were performed after in-gel digestion of the entire sample lane. Extracted peptides were sequenced by liquid chromatography-coupled tandem mass spectrometry on a Q-ToF Ultima (Waters) under standard conditions. Fragment spectra were searched against the NCBInr database using Mascot.
Binding and protease assays. For binding assays and generation of sumoylated model targets see supplementary information online. For fluorimetric hydrolase assays, recombinant enzymes were incubated with ubiquitin-, SUMO1- or SUMO2-AMC (BostonBiochem) in 384-well plates at 30 °C. Fluorescence was measured using a Fluoroskan Ascent FL (Thermo Scientific) with 380-nm excitation and 450-nm emission filters.
siRNA knockdown. siRNA was transfected using RNAiMAX (Invitrogen). For rescue, HeLa cells were transfected with siRNA-s2, and retransfected with siRNA-resistant pcDNA3.1 HA-USPL1, using Jet Prime reagent (for details see supplementary information online). Growth was measured either using the Cell Titre-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) or the CCK-8 cell counting kit (Dojindo).
Immunostaining of zebrafish sections. The transgenic fish line c13orf22lhi3662Tg/+ (AB/TU) was obtained from Zebrafish International Resource Centre (ZIRC). On breeding, mutant embryos (c13orf22lhi3662Tg/hi3662Tg) and wt siblings (c13orf22l+/+, c13orf22lhi3662Tg/+) were fixed at 2 d.p.f. Fourteen micrometre cryosections of the embryos were generated and the head region was stained with anti-zebrafish Coilin [26]. Imaging was carried out on a Leica TCS SPE confocal microscope.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Dr. Andreas Werner for reagents and advise, Anja Schreieck for excellent technical assistance, and Dr. Sina Barysch and Dr. Saskia Hutten (Dundee) for critical reading of the manuscript. We kindly acknowledge Dr. Karla Neugebauer (Dresden) for providing antibodies and the Zebrafish International Resource Centre (ZIRC; supported by grant P40 RR12546 from the NIH-NCRR) for providing zebrafish lines. Funding came from the EU NoE Rubicon (F.M. and S.S.), the A.v. Humboldt Foundation (fellowship to G.C.) and EMBO (E.M.).
Author contributions: S.S. and G.C. carried out most experiments and wrote the manuscript. L.K. identified USPL1 and carried out initial characterization. U.W. carried out experiments and contributed to writing, N.S.-V. carried out experiments. P.H. and J.W. guided zebrafish experiments, K.H. provided expert help with sequence analysis, H.U. provided mass spectrometric analysis. E.M. suggested the screening approach and generated suicide inhibitors in the group of H.O., who provided expert advise. F.M. guided the project and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11: 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Hay RT (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol 17: 370–376 [DOI] [PubMed] [Google Scholar]
- Yeh ET (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem 284: 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y (2012) DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep 13: 339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Chen CF, Brill SJ (2010) Wss1 is a SUMO-dependent isopeptidase that interacts genetically with the Slx5-Slx8 SUMO-targeted ubiquitin ligase. Mol Cell Biol 30: 3737–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10: 550–563 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem 78: 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Ye Y, Scheel H, Hofmann K, Komander D (2009) Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst 5: 1797–1808 [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM (2002) Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol 9: 1149–1159 [DOI] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD (2010) Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J 430: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Shanks JR, Komander D, Wilkinson KD (2008) Recognition of polyubiquitin isoforms by the multiple ubiquitin binding modules of isopeptidase T. J Biol Chem 283: 19581–19592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Au SW (2005) Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem J 386: 325–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Moutty MC, Moller U, Melchior F (2009) Performing in vitro sumoylation reactions using recombinant enzymes. Methods Mol Biol 497: 187–199 [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N (2004) Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci USA 101: 12792–12797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M, Salvesen GS (2008) DeSUMOylating enzymes--SENPs. IUBMB Life 60: 734–742 [DOI] [PubMed] [Google Scholar]
- Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J (1998) Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol 280: 275–286 [DOI] [PubMed] [Google Scholar]
- Quesada V, Diaz-Perales A, Gutierrez-Fernandez A, Garabaya C, Cal S, Lopez-Otin C (2004) Cloning and enzymatic analysis of 22 novel human ubiquitin-specific proteases. Biochem Biophys Res Commun 314: 54–62 [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Renatus M et al. (2006) Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 14: 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2006) Structural basis for SENP2 protease interactions with SUMO precursors and conjugated substrates. Nat Struct Mol Biol 13: 1060–1068 [DOI] [PubMed] [Google Scholar]
- Shen L, Tatham MH, Dong C, Zagorska A, Naismith JH, Hay RT (2006) SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat Struct Mol Biol 13: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI (2005) Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol 21: 105–131 [DOI] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG (2010) The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2: a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM (2010) Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 17: 403–409 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.