Abstract
R-loops are harmful structures with a negative impact on transcription and recombination during mitosis, but no information exists for meiosis. We used Saccharomyces cerevisiae and Caenorhabditis elegans THO mutants as a tool to determine the consequences of R-loops in meiosis. We found that both S. cerevisiae and C. elegans THO mutants show defective meiosis and an impairment of premeiotic replication as well as DNA-damage accumulation. Importantly, RNase H partially suppressed the replication impairment and the DNA-damage accumulation. We conclude that R-loops can form during meiosis causing replication impairment with deleterious results.
Keywords: R-loops, Saccharomyces cerevisiae , Caenorhabditis elegans , meiotic replication, genome instability
Introduction
Transcription has been shown to stimulate both mutation and recombination from bacteria to mammals [1], but the mechanisms underlying this phenomenon are still unknown. A particularly intriguing and physiologically relevant intermediate responsible for transcription-associated genetic instability is the R-loop, a co-transcriptionally formed structure between the nascent mRNA and the template DNA that displaces the non-template single-strand DNA [2]. Transcription-associated recombination in yeast mRNP biogenesis mutants, in vertebrate cells depleted of the ASF/SF2 splicing factor as well as in class-switch recombination at the Immunoglobulin (Ig) genes in vertebrates all have in common to be linked to R-loops [3, 4]. Recently, the importance of R-loops has been highlighted by several studies, including three different screening for genes involved in genome stability maintenance. Interestingly, they found a number of mutations that cause genome instability linked to R-loop formation [5, 6, 7, 8].
Eukaryotic factors with a role in preventing R-loop formation include the THO and THSC/TREX-2 protein complexes that have a role in mRNP biogenesis at the transcription–RNA export interface and on genome integrity [9]. Yeast cells lacking THO show a number of phenotypes, including a strong transcription-dependent hyper-recombination, which is partly mediated by R-loops [10, 11]. It has been shown that in hpr1Δ cells replication progression through transcribed DNA regions is impaired and the S-phase checkpoint is critical for their survival [12, 13].
Intriguingly, even though a number of studies have been devoted to the impact of transcription and R-loop formation in mitotic recombination, meiotic studies are unavailable. This is of particular interest given the impact that meiotic genome rearrangements might have in the progeny, in evolution or as a source of genetic diseases. C. elegans is an extremely amenable model system for meiosis studies that shares a high degree of conservation with humans. Each germline is spatially polarized in a distal-proximal manner with respect to mitotic proliferation and progression through meiotic prophase I. The mitotic dividing nuclei act as germline stem cells. Once nuclei enter the meiotic pathway and complete premeiotic S-phase, double-strand breaks (DSBs) are generated by SPO-11, chromosomes align and synapse, and recombination is largely completed by late pachytene. At diakinesis, chromosomes become highly condensed, forming six discrete bivalents held together by the chiasma, indicative of successful crossover recombination [14].
To evaluate whether R-loops could be formed in meiosis and the impact of these potential R-loops in the meiotic cell cycle, we used both yeast and C. elegans THO mutants as model systems. We describe that in both organisms R-loops lead to meiosis progression failure. We show that accumulation of R-loops in S. cerevisiae hpr1Δ leads to DNA breaks and DNA-damage checkpoint activation that causes meiosis impairment. Consistent with these observations, C. elegans thoc-2 mutants show impaired germline DNA replication due to the formation of R-loops. Our work provides evidence of the R-loop effects in meiosis and by extension in sexual reproduction.
Results
Meiotic defects in R-loop forming yeast
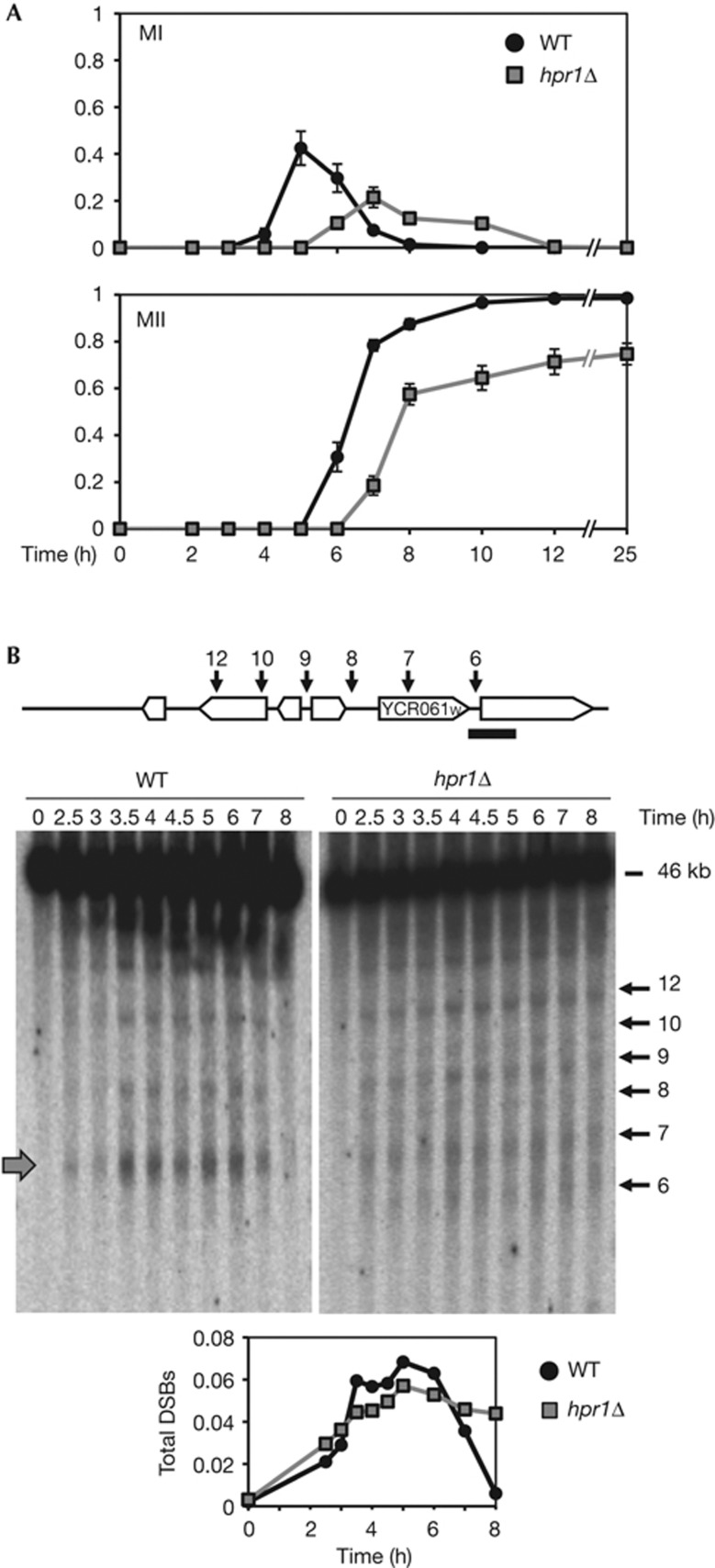
To assess the meiotic outcome of R-loop formation, we used S. cerevisiae hpr1Δ as a representative mutant that accumulates R-loops in mitosis. For this we constructed all yeast strains in the SK1 strain background to be able to synchronize meiosis [15]. hpr1Δ has reduced sporulation proficiency and decreased spore viability (56% viable spores in hpr1Δ versus 97% in wild-type (WT)), the fraction of tetrads with four viable spores being 16.5% in hpr1Δ versus 90% in WT cells (supplementary Table S1 online). The kinetics of nuclear divisions (MI and MII) was monitored in synchronous meiosis. In WT cells, MI started ∼4 h after transfer to sporulation media and was rapidly followed by MII (96.6% of cells have completed both divisions after 10 h; Fig 1A). In contrast, in hpr1Δ nuclear divisions were delayed and only 75% of cells reached the end of meiosis after 25 h in sporulation media (98.5% in WT cells), indicating a defect in meiosis progression in hpr1Δ.
Figure 1.
R-loop formation impairs meiosis progression. (A) Fraction of cells that completed meiosis I (MI) and II (MII) in HPR1 and hpr1Δ homozygous diploids. The means of cells±s.d. are shown. (B) Meiotic DSB formation in HPR1 and hpr1Δ diploids. Genomic DNA was taken after transfer to sporulation media and DSBs were scored by Southern probing with YCR065w. Numbered arrows indicate the position of described meiotic DSBs for this region (upper diagram). The grey arrow indicates a Spo11 DSB hotspot. Quantification of total DSBs activated during meiosis. Independent experiments were repeated three times with similar profiles. One is shown. DSB, double-strand break; WT, wild-type.
Next we determined whether meiotic DSB formation was altered in hpr1Δ cells. DSBs were analysed by Southern blot at the YCR054c–YCR065w chromosomal region (Fig 1B), which contains numerous DSB sites, including a strong hotspot in the YCR061w locus. No delay in the initiation of meiotic DSBs was observed, but DSBs persisted in hpr1Δ after 8 h, whereas in WT they are undetectable at that time (Fig 1B). Total DSBs showed similar levels in HPR1 and hpr1Δ cells, but the overall profile of meiotic DSBs in hpr1Δ is different from the WT, thus the cleavage of the YCR061w DSB hotspot (7-kb band) was lower in hpr1Δ (Fig 1B, grey arrow).
R-loops activate the DNA-damage checkpoint in meiosis
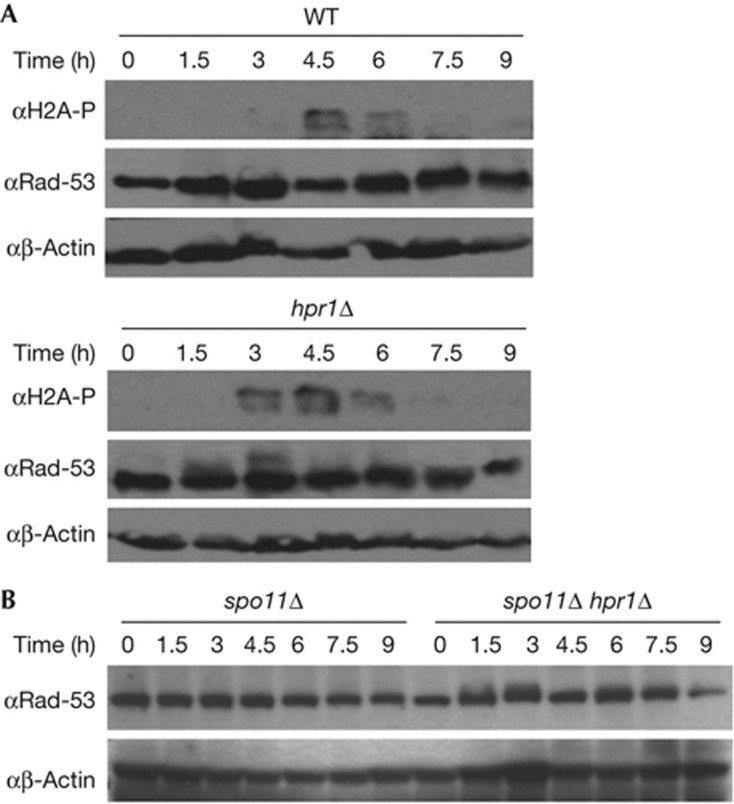
Next, we asked whether the meiosis defect was related to R-loop formation. First determining histone H2A phosphorylation as a measurement of accumulation of DNA breaks at different times after transfer to sporulation media by western blot (Fig 2A), we observed that hpr1Δ showed H2A phosphorylation signal at earlier time points (3 h) than the DSBs generated by Spo11 in WT cells (4.5 h).
Figure 2.
Meiotic DNA-damage and checkpoint activation in hpr1Δ cells. (A) Western analysis using anti-phosphorylation-H2A, anti-Rad53 and anti-β-actin antibodies for HPR1 and hpr1Δ homozygous diploids after transfer to sporulation media. (B) Western analysis using anti-Rad53 and anti-β-actin antibodies after transfer to sporulation media in spo11Δ and spo11Δ hpr1Δ homozygous diploids after transfer to sporulation media. WT, wild-type.
In contrast to mitosis, the meiosis-specific Mek1 kinase is activated in response to programmed meiotic DSBs instead of the canonical Rad53 DNA-damage checkpoint pathway unless there are persistent unrepaired or accidental DSBs [16]. Although in WT cells no Rad53 phosphorylation was observed, hpr1Δ cells showed Rad53 phosphorylation at 3 h. (Fig 2A). To examine if the early DSBs were Spo11-independent we generated the hpr1Δ spo11Δ double mutant and assess the phosphorylation state of Rad53. While no changes in the Rad53 phosphorylation pattern were observed in the spo11Δ single mutant, hpr1Δ spo11Δ double mutants showed Rad53 phosphorylation consistent with DSBs being Spo11-independent and able to trigger the DNA-damage response (Fig 2B). This increased level of DNA damage is most likely responsible for the incapacity of hpr1Δ cells to reach a 4C DNA content until 6 h in sporulation medium, whereas in WT cells replication is completed after 4.5 h (supplementary Fig S1A online).
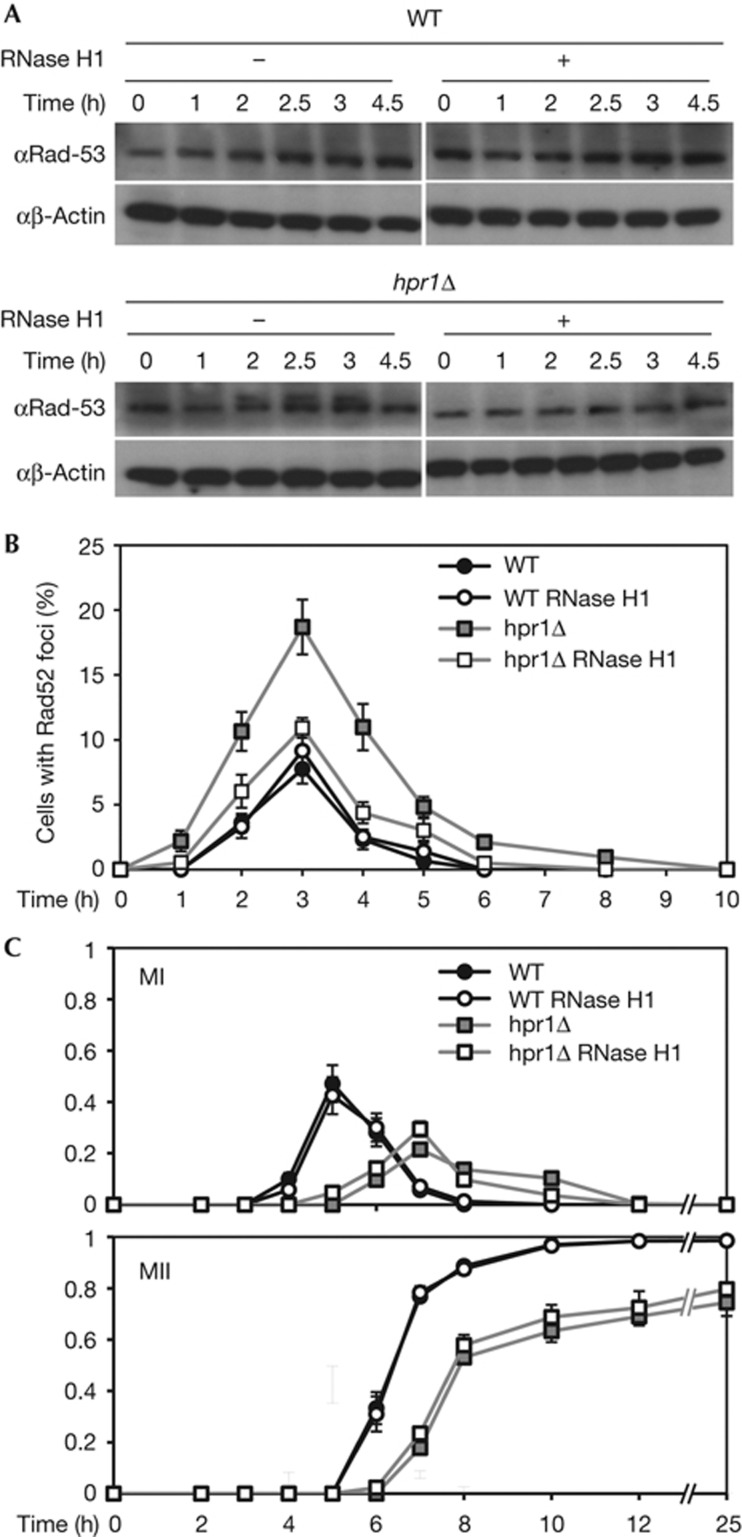
Finally, we took advantage of RNase H1 overexpression to specifically degrade DNA::RNA hybrids and partially suppress R-loop-dependent phenotypes, as previously shown in mitotic yeast cells [11] as a way to investigate whether R-loops were responsible for the observed DNA-damage checkpoint activation. We used the GAL1 promoter to overexpress RNase H1 (Fig 3). As shown in supplementary Table S2 online, galactose addition to the sporulation media did not interfere with meiosis progression, but to avoid carbon source issues we also confirmed the results using RNase H1 under the tet promoter (supplementary Fig S2 online). We overexpressed RNase H1 in the hpr1Δ diploid strains and analysed Rad53 phosphorylation in meiosis (Fig 3A; supplementary Figs S1B, S2A,B online). Whereas no changes in the Rad53 phosphorylation pattern were observed in the WT, RNase H1 overexpression abolished Rad53 phosphorylation in hpr1Δ cells. To correlate Rad53 phosphorylation with DSB formation, we monitored DSB levels by Rad52 foci accumulation during meiosis (Fig 3B). The levels of Rad52 foci were clearly higher in meiotic hpr1Δ cells with respect to the WT, and were reduced to WT levels on RNase H1 overexpression. Furthermore, fluorescence-activated cell sorting analyses showed that hpr1Δ cells pass through S phase slightly faster under RNase H1 overexpression (supplementary Figs S1B,S2B online). These results indicate that R-loops are responsible for premeiotic replication, increased DSB formation and meiosis progression impairment in S. cerevisiae THO mutants.
Figure 3.
Meiotic DNA-damage and checkpoint activation are R-loop dependent. (A) Western analysis using anti-Rad53 and anti-β-actin antibodies for HPR1 and hpr1Δ homozygous diploids after transfer to sporulation media with and without RNase H1 overexpression. (B) Quantification of Rad52 foci for HPR1 and hpr1Δ homozygous diploids after transfer to sporulation media with and without RNase H1 overexpression. The means of cells with foci±s.d. are shown. (C) Fraction of cells that completed meiosis I (MI) and II (MII) in HPR1 and hpr1Δ homozygous diploids with and without RNase H1 overexpression. The means of cells±s.d. are shown.
Finally, we analysed the kinetics of meiotic nuclear divisions (MI and MII) with and without RNase H1 overexpression. In WT cells, MI and MII show a similar profile under both conditions (98% of cells have completed the two divisions after 10 h; Fig 3C; supplementary Fig S2C online and supplementary Table S2 online). MI and MII nuclear divisions in hpr1Δ cells only showed a slight improvement on overexpression of RNase H1, with 79.7% of cells completing meiosis after 25 h in sporulation media versus 74.5% without RNase H1, whereas this value was 98.5% in WT cells. Therefore, RNase H1 overexpression rescues the defect in meiosis progression only slightly in hpr1Δ cells. Nevertheless, this rescue might be relevant because, as shown in supplementary Table S2 online, RNase H1 overexpression reduces spore viability even in the WT.
C. elegans THOC-2 is essential for fertility
To assess if R-loops could also hinder meiosis in metazoans we extended the analysis to C. elegans. Searching in the C. elegans genome for THO complex members we found several putative candidates, including thoc-2 (THO Complex subunit 2), the C. elegans orthologue of yeast THO2, predicted to encode a protein that displays a 55% and 40% amino-acid similarity with human THOC2 and yeast Tho2, respectively (supplementary Fig S3A online).
We investigated two thoc-2 deletion mutants, thoc-2(tm1310) and thoc-2(ok961), which result in premature stop codons and encode truncated THOC-2 products (supplementary Fig S3B,C online). We observed that both thoc-2 alleles conferred complete sterility (Ste phenotype), as the mutant strains laid no eggs (supplementary Table S3 online), indicating that thoc-2 is essential for C. elegans fertility. However, homozygous mutants produced by self-fertilization of heterozygous hermaphrodite parents are rescued beyond L4 stage by maternal contribution of thoc-2 mRNA, permitting the analysis of thoc-2 loss of function within adult tissues.
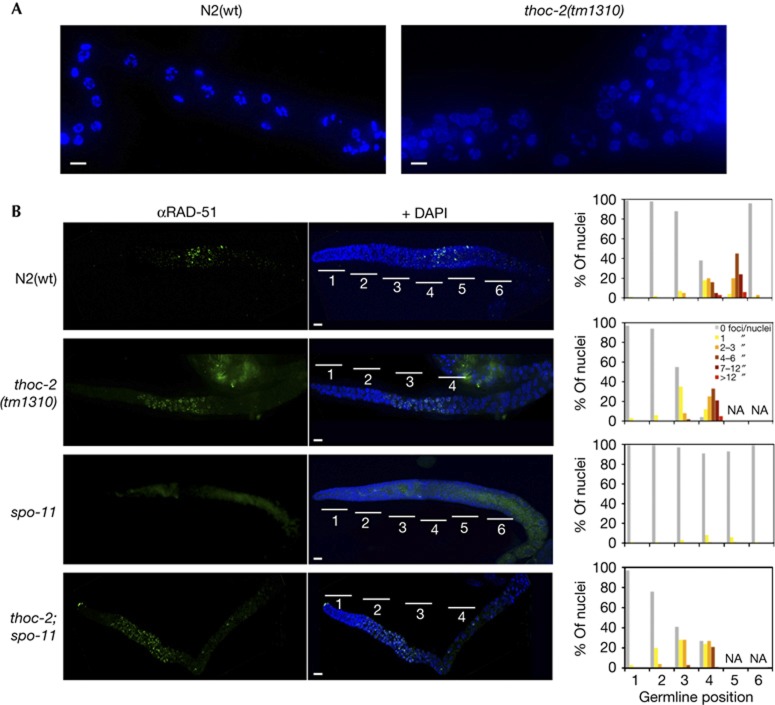
To determine whether the eggless phenotype was caused by a defect during meiosis, we analysed thoc-2 mutant germlines. Immunostaining with antibody against the SYP-1 core SC component [17] revealed that the SC assembled in meiotic prophase nuclei (supplementary Fig S4A online). 4,6-Diamidino-2-phenylindole (DAPI) staining in WT oocytes nuclei shows six discernible DAPI-stained bivalent chromosomes, indicative of successful crossover recombination (Fig 4A; supplementary Fig S4B online). In contrast, thoc-2 mutants have no distinguishable diakinesis region implying a meiosis progression defect.
Figure 4.
C. elegans thoc-2 mutants show meiosis failure and increased levels of SPO-11-independent RAD-51 foci. (A) Representative images of diplotene-diakinesis germline region of fixed germlines counterstained with DAPI. (B) Representative images of fixed germlines immunostained with anti-RAD-51 antibodies and counterstained with DAPI, and quantification of RAD-51 foci of germline nuclei. Zones 1–2 (mitosis), zone 3 (transition zone), zones 4–5–6 (pachytene). The number of nuclei was scored in 1-day post-L4 stage animals. At least 10 germlines per strain were scored for each experiment (n=30). Scale bar, 10 μm. DAPI, 4,6-Diamidino-2-phenylindole; NA, not applicable; WT, wild-type.
DNA damage in C. elegans thoc-2 meiosis
To examine if DSBs accumulated during meiosis in thoc-2 mutants, as occurred in S. cerevisiae, we analysed RAD-51 recruitment to DNA using α-RAD-51 antibodies [18] (Fig 4B; supplementary Figs S4C, S5 online). In N2(wt) worms, the levels of RAD-51 foci increased on entrance to the transition zone (zone 3), peaked at early mid-pachytene (zone 5) and disappeared by the end of pachytene (zone 6; Fig 4B; supplementary Fig S5 online). Upon entrance into meiosis (zone 3), thoc-2 mutants accumulated RAD-51 foci earlier than N2(wt) worms, and these foci were maintained until the end of the germline (Fig 4B; supplementary Fig S5 online), suggesting that the generation of meiotic DSBs is altered in thoc-2 mutants, as it occurred in yeast. As meiotic DSBs are generated by SPO-11 [19], we assayed whether the increased levels of DSBs in thoc-2 mutants were because of deregulation of the SPO-11 activity. We generated thoc-2;spo-11 double mutants and quantified RAD-51 foci. As can be seen in Fig 3 double mutants still showed high levels of RAD-51 foci indicating that the DBSs in thoc-2 mutants are independent of SPO-11 (Fig 4B).
As unrepaired meiotic DSBs cause an increase in germline cell corpses because of activation of a late pachytene DNA-damage checkpoint [20], we assayed whether the high levels of SPO-11-independent DSBs triggered the DNA-damage response. Apoptotic corpses in thoc-2 mutants increased 4.3- and 4.6-fold above the WT in thoc-2(tm1310) and thoc-2(ok961) mutants, respectively (supplementary Fig S4D online). Therefore, thoc-2 inactivation generates genome instability during meiosis in a SPO-11-independent manner, triggering apoptosis.
R-loops impair C. elegans meiotic replication
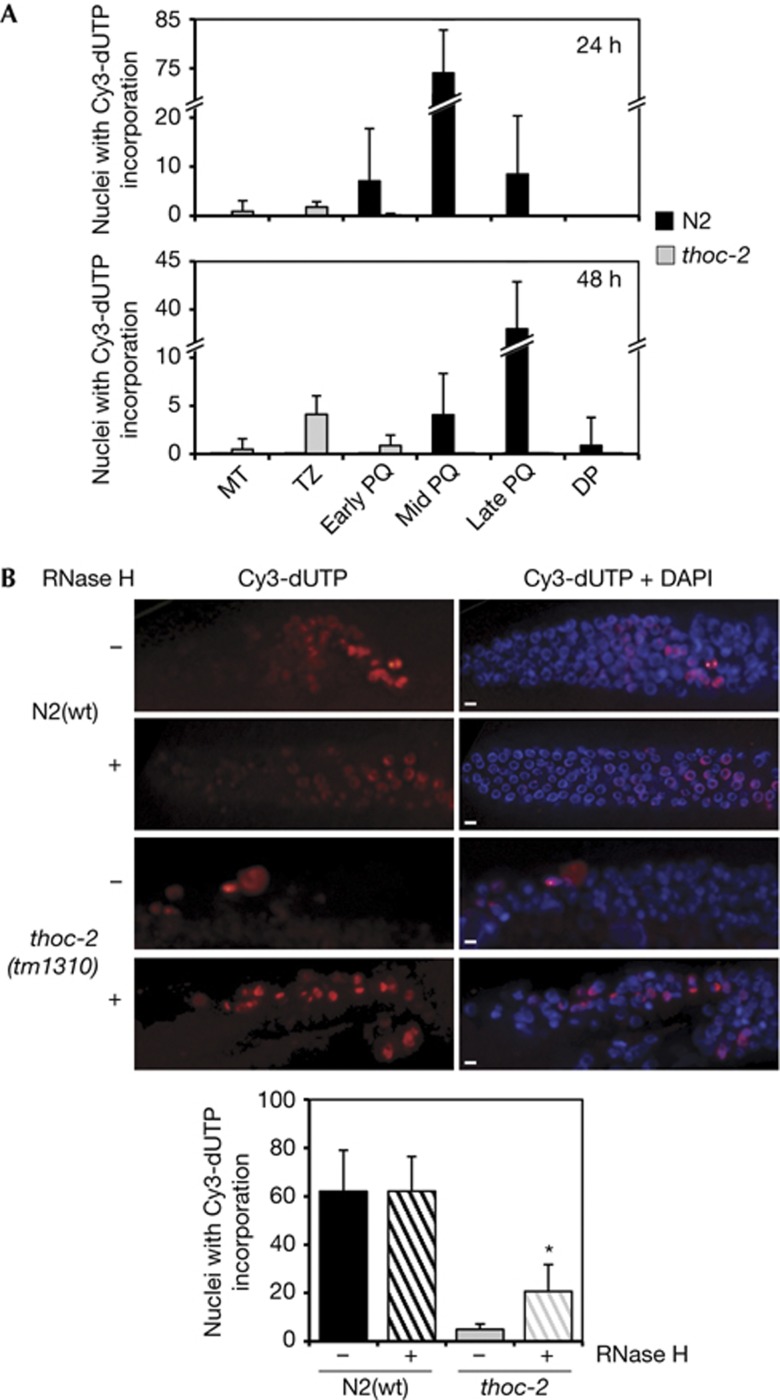
The observation of increased meiotic RAD-51–SPO-11-independent foci upon entrance to meiosis suggested that they arise from premeiotic replication defects. It is known that DNA replication failure leads to the occurrence of RAD-51 foci [20, 21], so we wondered whether the origin of genome instability in thoc-2 mutants occurs during germline DNA replication. To examine replication, we assessed incorporation of deoxyribonucleotides into the germline DNA as described [22]. Following microinjection of Cy3-dUTP into young adult germlines, worms were allowed to recover for 24 h and germlines were dissected, fixed and DAPI-stained. In N2(wt) worms, all labelled nuclei had progressed into pachytene showing Cy3-dUTP accumulation in nuclei located from early (eP) to late pachytene (lP; Fig 5A). However, in thoc-2 mutants most nuclei incorporating Cy3-dUTP were in the mitotic (MT) and transition zone (TZ). Only after 48 h of Cy3-dUTP microinjection a small fraction of such nuclei had progressed into early pachytene (Fig 5A). Therefore, premeiotic replication is impaired in thoc-2 mutants.
Figure 5.
Replication is impaired in C. elegans thoc-2 germlines and partially alleviated by RNase H microinjection. (A) Quantification of Cy3-dUTP incorporation 24 and 48 h after microinjection in the germline regions of mitosis (MT), transition zone (TZ), early-, mid- and late-pachytene (PQ) and diplotene (DT). Error bars indicate standard error of mean from at least 10 germlines for each experiment (n=30). (B) Representative images of fixed germlines of the indicated genotype of adult hermaphrodites 16 h after microinjection with Cy3-dUTP and with or without RNase H and quantification of Cy3-dUTP incorporation. Error bars indicate standard error of mean from at least 10 germlines for each experiment (n=15). Statistically significant differences with respect to N2(wt) (P<0.001) are indicated by an asterisk ‘*’ (Student’s t-test). Scale bar, 5 μm. DAPI, 4,6-Diamidino-2-phenylindole; WT, wild-type.
To assess whether replication defects in the thoc-2 mutant germline are linked to R-loops, as we observed in yeast, we co-microinjected Escherichia coli RNase H into the worms in our in vivo replication assay. Co-microinjection of RNase H did not affect the incorporation of Cy3-dUTP in N2(wt) worms, whereas in thoc-2 mutants caused a clear increase in Cy3-dUTP-labelled nuclei (Fig 5B). Therefore, R-loops form in thoc-2 mutants and are responsible for the premeiotic replication impairment observed in C. elegans.
Discussion
In this study, we asked whether R-loops can form in meiosis and if so, whether they have any effect in meiosis progression and genomic integrity. We analysed meiosis in a unicellular (S. cerevisiae) and a multicellular (C. elegans) model organism depleted of THO, an mRNP biogenesis factor that helps prevent the formation of co-transcriptional R-loops, as shown in mitotically dividing yeast cells [10, 11]. We show that S. cerevisiae THO mutants have a decrease in sporulation proficiency and spore viability. The results are similar in C. elegans where thoc-2 mutants show meiotic failure, indicating that the functional meiotic demand of THO is conserved. Moreover, C. elegans thoc-2 mutants show a clear increase in genome instability as determined by accumulation of RAD-51 foci after premeiotic replication. These DSBs are SPO-11 independent and trigger the pachytene checkpoint, as shown by the increase in apoptotic corpses. Likewise, in yeast hpr1Δ cells the Rad53 DNA–damage checkpoint is activated and DNA breaks are accumulated in hpr1Δ mutants earlier than in WT cells independent of Spo11, as deduced from H2A phosphorylation and increased Rad52 foci. This implies that in both yeast and C. elegans THO mutants, DNA breaks accumulate during meiotic replication.
Interestingly, S-phase checkpoint activation has been reported in vegetatively growing yeast THO mutants [12], demonstrating a conserved function of THO. The lethality observed in C. elegans and mice [23] are thus likely related to the incapacity of early embryonic cells to divide properly in a period in which the S phase and DNA cell division becomes critical. The relevance of DNA replication and repair functions to ensure genome integrity and proper S-phase progression is indicated by the embryonic lethality of knock-out mice for DSB repair genes such as MRE11, RAD51, BRCA1 and BRCA2 (for review see Hakem [24]).
In C. elegans thoc-2 mutants, premeiotic replication is impaired as detected by a reduction in the incorporation of Cy3-dUTP. This demonstrates that germline DNA replication is impaired in the absence of a functional THO complex, establishing a link between mRNA metabolism and premeiotic replication. Similar results were observed in yeast, as fluorescence-activated cell sorting analysis show a slowdown of meiotic replication. Impaired meiotic replication may be related to the ability of THO mutants to form co-transcriptional R-loops that become an obstacle for the replication fork either directly or indirectly (see review by Aguilera and Gomez-Gonzalez [3]). Notably, our results in yeast and C. elegans show that meiotic replication impairment, DNA-damage accumulation and checkpoint activation is suppressed by RNase H. Unfortunately, a complete recovery of meiosis proficiency was not possible on RNase H overexpression either in C. elegans or yeast cells. Therefore, we conclude that defective replication is caused by R-loops [3, 11, 12] and these are the cause of meiosis progression failure in both yeast and C. elegans THO mutants. In any case, we cannot exclude an R-loop-independent effect of THO depletion in late meiotic steps. Thus, we provide evidence that the absence of THO has a critical impact on meiotic genome integrity.
In summary, our work provides evidence that R-loops can be formed in meiosis and that they have important consequences in meiotic replication and genome stability, implying that RNA metabolism has an important influence in sexual reproduction and inheritance of pluricellular eukaryotes. THO and other related mRNP biogenesis factors with a role in preventing R-loop formation, such as THSC/TREX-2 [9, 11], likely become critical in meiosis. Consequently, this work opens new perspectives to our understanding of germline DNA replication and meiosis and the interface between transcription/mRNP biogenesis and genome instability.
Methods
All experimental procedures are detailed in supplementary information online. The nematode strains used were provided by the Caenorhabditis Genetics Center and are listed in supplementary information online. thoc-2(tm1310) was provided by S. Mitani of the National Bioresource Project for the nematode, Japan. SK1 isogenic derivatives yeast strains used are listed in supplementary information online. S. cerevisiae synchronous meiotic cultures were prepared as described previously [25]. For nuclear division profiles, aliquots were fixed with 70% (v/v) ethanol and stained with 0.1 μg/ml with DAPI. Mononucleate, binucleate and tetranucleate cells were scored by microscopy (at least 200 cells per time point). For meiotic DSB detection, genomic DNA was prepared in agarose plugs and digested with StuI (New England) and analysed by Southern blot. Western blots were performed as described [16]. For RNase H1 overexpression under GAL1 promoter, the appropriated strains were transformed with the pGAL::RNH1 [11] and the SPM was complemented or not with galactose (0.5%). To analyse Rad52 foci formation during meiosis, plasmid pWJ1344 carrying Rad52–YFP was used and described previously [12].
For antibody immunostaining, 1-day post-L4 adult gonads were treated as described [21]. Antibodies used are listed in supplementary information online. Direct incorporation of Cy3-dUTP into germline nuclei was performed as described [22]. After the appropriate exposure to the Cy3-dUTP, gonads were dissected, fixed and DAPI-stained. The total number of cells that incorporated Cy3-dUTP was determined for each dissected germline. Commercial E. coli RNase H (Invitrogen) was co-microinjected at 0.2 U/μl.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Shohei Mitani at the National Bioresource Project for kindly providing thoc-2(tm1310), the Caenorhabditis Genetic Center, A. Gartner, N. Kleckner, A. Villanueve provided C. elegans and yeast strains and reagents. We thank P. Askjaer and U. Vivegananthan for critical reading of the manuscript, and U. Galindo for technical assistance. This work was supported by grants from the Spanish Ministry of Science and Innovation (BFU2006-05260, BFU2010-16372 and Consolider Ingenio 2010 CSD2007-0015), the European Union (FEDER) and the Junta de Andalucía (BIO102 and CVI4567).
Author contributions: M.C.-P. and T.G.-M. performed the experiments, M.C.-P., T.G.-M. and A.A. designed the experiments and analysed and discussed the results, T.G.-M. and A.A. wrote the manuscript and all authors read it.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguilera A (2002) The connection between transcription and genomic instability. EMBO J 21: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilera A, García-Muse T (2012) R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 [DOI] [PubMed] [Google Scholar]
- Aguilera A, Gomez-Gonzalez B (2008) Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217 [DOI] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S (2011) Guanine repeat-containing sequences confer transcription-dependent instability in an orientation-specific manner in yeast. DNA Repair (Amst) 10: 953–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE et al. (2009) A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell 35: 228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischo HE, Gomez-Gonzalez B, Grzechnik P, Rondon AG, Wei W, Steinmetz L, Aguilera A, Proudfoot NJ (2011) Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell 41: 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Amon JD, Koshland D, Vuica-Ross M (2011) RNase H and multiple RNA biogenesis factors cooperate to prevent RNA:DNA hybrids from generating genome instability. Mol Cell 44: 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Chan YA, Minaker SW, Aristizabal MJ, Barrett I, Sipahimalani P, Kobor MS, Hieter P (2012) R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev 26: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Aguilera A (2007) Activation-induced cytidine deaminase action is strongly stimulated by mutations of the THO complex. Proc Natl Acad Sci USA 104: 8409–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Gomez-Gonzalez B, Felipe-Abrio I, Aguilera A (2009) The S-phase checkpoint is required to respond to R-loops accumulated in THO mutants. Mol Cell Biol 29: 5203–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A (2006) Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ (2007) Meiotic recombination in Caenorhabditis elegans. Chromosome Res 15: 607–621 [DOI] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Cao L, Kleckner N (1995) Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc Natl Acad Sci USA 92: 8512–8516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartagena-Lirola H, Guerini I, Manfrini N, Lucchini G, Longhese MP (2008) Role of the Saccharomyces cerevisiae Rad53 checkpoint kinase in signaling double-strand breaks during the meiotic cell cycle. Mol Cell Biol 28: 4480–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen AJ, Colaiacovo MP, McDonald K, Villeneuve AM (2002) Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev 16: 2428–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpi A, Pasierbek P, Gartner A, Loidl J (2003) Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16 [DOI] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM (1998) Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398 [DOI] [PubMed] [Google Scholar]
- Gartner A, Milstein S, Ahmed S, Hodgkin J, Hengartner MO (2000) A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol Cell 5: 435–443 [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ (2005) Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J 24: 4345–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A, Ellefson M, Villeneuve AM, Engebrecht J (2007) Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev Biol 308: 206–221 [DOI] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, Goodrich DW (2006) Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol 26: 4362–4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem R (2008) DNA-damage repair; the good, the bad, and the ugly. EMBO J 27: 589–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N (1990) Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell 61: 419–436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.