Abstract
Members of hereditary nonpolyposis colon cancer (HNPCC) families harboring heterozygous germline mutations in the DNA mismatch repair genes hMSH2 or hMLH1 present with tumors generally two to three decades earlier than individuals with nonfamilial sporadic colon cancer. We searched for phenotypic features that might predispose heterozygous cells from HNPCC kindreds to malignant transformation. hMSH2+/− lymphoblastoid cell lines were found to be on average about 4-fold more tolerant than wild-type cells to killing by the methylating agent temozolomide, a phenotype that is invariably linked with impairment of the mismatch repair system. This finding was associated with an average 2-fold decrease of the steady-state level of hMSH2 protein in hMSH2+/− cell lines. In contrast, hMLH1+/− heterozygous cells were indistinguishable from normal controls in these assays. Thus, despite the fact that HNPCC families harboring mutations in hMSH2 or hMLH1 cannot be distinguished clinically, the early stages of the carcinogenic process in hMSH2 and hMLH1 mutation carriers may be different. Should hMSH2+/− colonocytes and lymphoblasts harbor a similar phenotype, the increased tolerance of the former to DNA-damaging agents present in the human colon may play a key role in the initiation of the carcinogenic process.
The sequence of genetic events required to transform normal human colonic epithelium into sporadic adenomas and carcinomas has been extensively documented (1, 2). The initial event frequently involves the functional inactivation of the tumor suppressor gene APC, which leads to the formation of an adenomatous polyp through deregulation of cell proliferation and differentiation. An adenoma then progresses to a carcinoma and eventually to metastatic cancer through the acquisition of further mutations in oncogenes and tumor suppressor genes, among them K-ras, DCC, SMAD4, and p53 (3).
In hereditary nonpolyposis colon cancer (HNPCC), a cancer predisposition syndrome linked to mutations in mismatch repair (MMR) genes (4, 5), the transformation process is morphologically similar but genetically distinct from sporadic disease (6). This is because inactivation of the MMR process results in a strong mutator phenotype, where DNA replication errors accumulate throughout the genome, particularly in microsatellites. The consequence of this phenomenon, known as microsatellite instability (MSI), is that genes involved in proliferative control that contain microsatellites, such as the TGFβ type II receptor (7) and BAX (8) genes, become mutated. MMR-deficient tumor cell lines differ from those of sporadic tumors in one additional aspect: they remain prevalently diploid and display only MSI, rather than undergoing gross genomic rearrangements apparent in sporadic colon cancers (9). Lastly, MMR-deficient cancer cells are resistant to killing by certain alkylating agents (10), probably because of their ability to tolerate the presence of modified bases in their DNA. MMR proteins were indeed implicated in the recognition and processing of “pseudomispairs” such as O6-methyl G/T (11). Recent evidence suggests that this tolerance may also be linked to a lack of DNA damage signaling to the apoptotic machinery (12–14).
HNPCC patients carrying mutations in the MMR genes hMSH2 or hMLH1 are typically diagnosed with colon cancer before the age of 50 (15). It is thought that the tumors in these individuals arise from cells where the second wild-type allele of the germline-mutated MMR gene has been inactivated by a somatic alteration. But how does this latter event come about? In cancer, wild-type alleles of genes are typically inactivated by one of two pathways: somatic mutation or loss of heterozygosity (LOH). The hMSH2 and hMLH1 genes are inactivated by both these mechanisms, albeit to different extents. It has been reported that the hMLH1 locus 3p21 is more prone to LOH (16), whereas inactivation of hMSH2 occurs predominantly through somatic mutation (17). We set out to search for the possible causes of this difference. To this end, we tested the sensitivity of hMSH2+/− or hMLH1+/− cells to the methylating agent temozolomide (TMZ), the steady-state levels of hMSH2 and hMLH1 proteins in these cells, and the efficiency of their extracts to recognize and repair base/base mismatches in vitro.
Materials and Methods
Cell Lines.
The lymphoblastoid cell lines were either established as described (18) or obtained from Bert Vogelstein (Johns Hopkins Oncology Center, Baltimore) or from the Finnish HNPCC databank. The cells were grown in RPMI medium 1640 supplemented with 20% FCS (GIBCO) and 2 mM glutamine (Sigma), in 5% CO2 at 37°C. Only cell lines with comparable growth rates and viability higher than 85% were included in this study.
TMZ Treatment.
TMZ (a gift of Schering-Plough) was dissolved in complete medium and protected from light. O6-benzylguanine (a gift of L. Lassiani, Institute of Pharmaceutical Chemistry, Trieste, Italy) was dissolved in ethanol (3 mg/ml). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) was dissolved in PBS (5 mg/ml). The effect of TMZ on the cell lines was evaluated by the tetrazolium salt method (19). The cells were suspended in complete medium (1.6 × 105 cells/ml) and dispensed in 50-μl aliquots into 96-well plates. O6-benzylguanine was then added (10 μl, final concentration 10 μM), and the cells were incubated for 2 h at 37°C. Fifty microliters of TMZ solution was added to final drug concentrations ranging from 1.95 μM to 1,000 μM, and a final O6-benzylguanine concentration of 5.5 μM. Cell growth was evaluated after 72 h. Control groups included cells incubated in complete medium alone or containing O6-benzylguanine. All samples were tested in quadruplicate. Twenty microliters of the MTT stock solution was then added, and the cells were incubated at 37°C for an additional 6 h. They were then lysed with a buffer (0.1 ml/well) containing 20% SDS and 50% N,N-dimethyl formamide, pH 4.7. After overnight incubation, the absorbance was read at 595 nm by using a 3,550-UV microplate reader (Bio-Rad). Cell sensitivity to drug treatment was expressed in terms of IC50. O6-methylguanine methyl transferase activity of O6-benzylguanine-treated cells was evaluated according to the method described by Morten and Margison (20). In all cell lines, enzyme activity was completely abrogated by 2-h exposure to O6-benzylguanine and remained undetectable until the end of the treatment.
Western Blot and Immunohistochemical Analysis.
The protein extracts (50 μg) were loaded on a 7.5% SDS/PAGE and electroblotted onto poly(vinylidene difluoride) membranes. The blots were first blocked in 0.1% Tween 20 in Tris-buffered saline (TBST) containing 5% nonfat dry milk for 1 h at 37°C and then incubated with the respective primary monoclonal antibodies (anti-hMSH2, no. NA26, Oncogene Science, 0.65 μg/ml; anti-hMLH1, no. 13271A, PharMingen, 0.09 μg/ml; anti-β-tubulin, no. N357, Amersham Pharmacia, 0.03 μg/ml) for 1 h at room temperature. After washing with TBST, the blots were incubated with horseradish peroxidase-conjugated sheep anti-mouse Ig (no. NXA 931, Amersham Pharmacia, 1:5,000) for 1 h at 20°C. The protein–antibody complexes were detected by enhanced chemiluminescence (Amersham Pharmacia). Prestained molecular weight marker (Bio-Rad) and protein samples from three normal and one negative controls were loaded alongside the heterozygous cell extracts on each gel. The intensity of the bands was quantified relative to β-tubulin by using the imagequant software of the Computing Densitometer (Molecular Dynamics). The levels of MMR proteins in the heterozygous cell extracts are reported as percentage of the mean value of the three controls. The differences between the protein levels in the control samples in the individual experiments were never greater than ±10%.
Pellets of cultured cells were washed in 0.9% NaCl and resuspended in warm (42°C) 1% Agar (Nobel) in 0.9% NaCl in a 2-ml Eppendorf tube. The cells were then centrifuged at 42°C (400 × g for 8–10 min), and the excess agar was removed. After cooling, both ends of the tube were cut off to leave a cylinder containing the pellet. This was placed in 10% neutral buffered formalin and left overnight at 4°C. The agar block was then removed from the tube, embedded in Paraplast, and sectioned into 6-μm slices. Immunohistochemical staining for hMSH2 and hMLH1 was carried out as described previously (21).
In Vitro MMR and Electrophoretic Mobility-Shift Assays.
Mismatch repair efficiency of the cell extracts was tested in vitro, by using an assay described previously (22, 23). The M13mp2 heteroduplexes tested contained either a single G/G or G/T mispair.
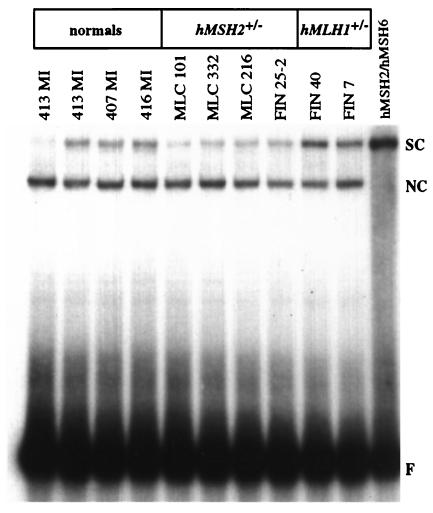
The electrophoretic mobility-shift assays were performed as described (24), except that each reaction contained 50 fmol of the 34-mer G/T oligo probe, 10 μg of the extract (Fig. 4), and 100 ng of poly d(I⋅C) nonspecific competitor (Amersham Pharmacia).
Figure 4.
Mismatch binding activities in extracts of selected lymphoblastoid cell lines analyzed in this study. The radiolabeled 34-mer double-stranded probes, either fully-complementary (left lane) or containing a single G/T mispair (remaining lanes) were incubated with equal amounts of the cell extracts listed (Top). The mixtures were separated by electrophoresis in a 6% nondenaturing polyacrylamide gel. F, free probe; SC, specific complex formed between the G/T probe and the hMSH2/hMSH6 present in the extracts; NC, nonspecific complex.
Statistical Analysis.
Data for each cell line were obtained from at least two independent experiments. Variability within and between cell lines was tested to assess reliability (intraclass correlation coefficient). Reliability of repeated measurements was good, because variability within a line was lower than variability between lines; intraclass correlation coefficients were: r = 0.454 for repair efficiency, r = 0.895 for protein expression, and r = 0.950 for IC50. Analysis of variance was used to test the differences between the groups and to calculate the best estimate of the pooled within-group standard deviation, which was used to calculate the 95% confidence intervals. Mann–Whitney and t tests were applied where necessary to assess the level of significance of the observed differences. Multiple linear regression was run to estimate the independent role of each of the explanatory variables. Receiver operator characteristic curve analysis was carried out to evaluate the accuracy of a test result. Statistical analyses were performed with stata (Stata, College Station, TX) and nanostat (Adelso, Rome) software.
Results
hMSH2+/− Lymphoblasts Display Tolerance to the Chemotherapeutic Methylating Agent Temozolomide.
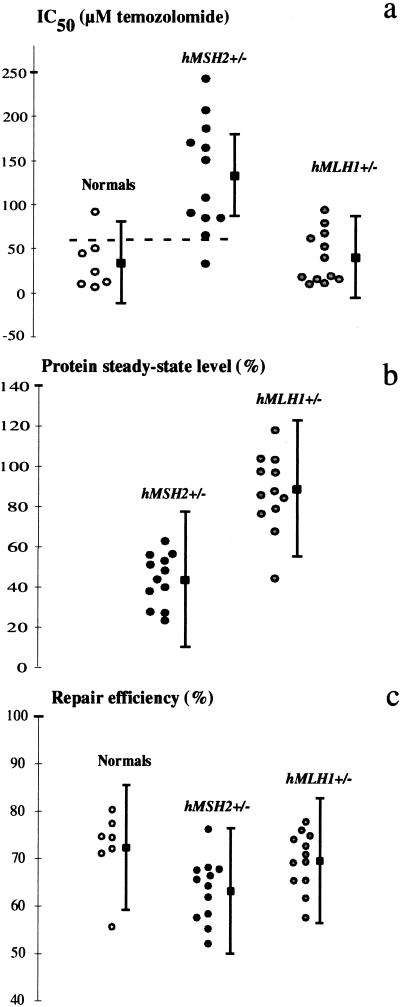
MMR-deficient cells are tolerant to the killing effects of methylating agents such as TMZ (25). We have taken advantage of this phenomenon to measure the sensitivity of the heterozygous lymphoblasts from HNPCC kindreds to the cytotoxic effects of TMZ. To this end, we generated a bank of immortalized lymphocytes from HNPCC index cases, i.e., hMSH2 or hMLH1 mutation carriers with early-onset colon cancer whose tumors displayed MSI. The patients originated from Finland, Switzerland, or the U.S., and all fulfilled the clinical criteria of HNPCC (26). The germline mutations identified in these families are listed in Table 1. Lymphoblastoid cell lines were established also from age-matched normal subjects without history of colon cancer. The cell lines were treated with the drug, and their IC50 values (drug concentrations at which cell growth is inhibited by 50%) were compared with those of the normal controls. To eliminate the differences in TMZ cytotoxicity because of variations in levels of methylguanine methyl transferase, the enzyme that converts O6-methylguanine back to guanine, we inhibited it with O6-benzylguanine. The mean IC50 values for each cell line are listed in Table 1 and their distribution is shown in Fig. 1a. The hMSH2+/− cells displayed IC50 values significantly higher than those observed in normal (P = 0.0014) and hMLH1+/− (P = 0.00016) cell lines. Because the IC50 values for TMZ treatment differed significantly between normal and hMSH2+/− groups, as confirmed by nonoverlapping confidence intervals, we tested the discriminating power of this parameter. Selecting IC50 = 60 (dashed line in Fig. 1a) as a cutoff value for discriminating between normal and hMSH2+/− cells, the sensitivity, specificity, and positive predictive value were 91.7, 85.7, and 91.7%, respectively. Among all the individuals tested, only one came out false negative (MLC 177; see below). We also identified one cell line that appeared false positive: the normal cell line 411 MI, which displayed an increased tolerance to TMZ and a reduced mismatch repair efficiency (see below). This test is not suitable for clinical diagnosis, as its reliability is currently too low, whereas the complexity of the immortalization procedure exceeds the competence of most diagnostic laboratories. Nevertheless, we were clearly able to identify an inverse correlation between the age at diagnosis of the hMSH2+/− patients and the sensitivity of their respective immortalized lymphocytes to TMZ (P < 0.05): for each 10 μM increase in IC50, the age at diagnosis decreased by approximately 1 year.
Table 1.
Summary of the genotypic and phenotypic features of the human lymphoblastoid cell lines
| Lymphoblasts | DNA change* | Exons | Coding change | % Repair Mean (SD) | Protein level† Mean (SD) | IC50 Mean (SD) | |
|---|---|---|---|---|---|---|---|
| hMSH2+/− | |||||||
| hMSH2 | |||||||
| FIN 25-2 | 2 bp ins | 12 | Frameshift | 76 (2) | 48 (6) | 85 (19) | |
| MLC 332 | 1981–2073 del | 12 | Frameshift | 68 (2) | 23 (1) | 186 (3) | |
| MLC 586 | Exon 7 del | 7 | Frameshift | 68 (7) | 39 (1) | 86 (18) | |
| MLC 177 | Exons 8–15 del | 8 to 15 | Frameshift | 67 (5) | 63 (1) | 33 (5) | |
| MLC 21 | Splice donor | 15 | Frameshift | 66 (4) | 37 (9) | 164 (32) | |
| MLC 216 | Splice donor | 5 | In-frame del | 65 (2) | 27 (11) | 66 (9) | |
| Sw 1587 | 1,148 C>T | 7 | Nonsense | 64 (5) | 56 (11) | 91 (9) | |
| FIN 106 | Splice acceptor | 9 | Frameshift | 62 (4) | 52 (1) | 108 (4) | |
| Sw 1383 | 3 bp del | 12 | Asn596 del | 58 (3) | 43 (6) | 151 (24) | |
| MLC 101 | Splice donor | 5 | In-frame del | 57 (10) | 27 (6) | 243 (36) | |
| MLC 442 | 173 bp ins | 7 | Frameshift | 55 (11) | 50 (2) | 207 (30) | |
| FIN 93 | 2 bp del | 10 | Frameshift | 52 (2) | 55 (11) | 170 (33) | |
| Mean | 63 | 43 | 132 | ||||
| hMLH1+/− | |||||||
| hMLH1 | |||||||
| FIN 40 | Splice acceptor | 14 | Frameshift | 78 (0) | 103 (0) | 16 (2) | |
| FIN 28 | 320 T>G | 4 | Missense (Ile107Arg) | 76 (1) | 68 (6) | 11 (4) | |
| FIN 8 | Splice donor | 12 | Frameshift | 75 (6) | 88 (11) | 67 (9) | |
| FIN 10 | 3.5 kb genomic del | 16 | In-frame del | 74 (3) | 97 (1) | 62 (6) | |
| FIN 7 | 1976 G>C | 17 | Missense (Arg659Pro) | 73 (3) | 103 (9) | 79 (4) | |
| 22 SD | 676 C>T | 8 | Nonsense | 71 (7) | 97 (14) | 16 (2) | |
| FIN 83 | 1975 C>T | 17 | Nonsense | 69 (3) | 117 (29) | 94 (1) | |
| Sw 1121 | Splice acceptor | 19 | Last exon del | 69 (1) | 84 (0) | 10 (3) | |
| 210 MI | Splice acceptor | 8 | Frameshift | 66 (6) | 86 (3) | 18 (3) | |
| FIN 26 | Splice acceptor | 6 | Frameshift | 65 (13) | 79 (11) | 40 (2) | |
| Sw 434 | 351 C>G | 4 | Missense (Thr117Arg) | 62 (14) | 76 (1) | 19 (5) | |
| Sw 1033 | 1 bp del | 16 | Frameshift | 57 (3) | 44 (3) | 52 (11) | |
| Mean | 70 | 87 | 40 | ||||
| Normals | |||||||
| FIN Co3 | 80 (7) | 12 (4) | |||||
| 413 MI | 77 (6) | 6 (2) | |||||
| 416 MI | 74 (0) | 9 (4) | |||||
| FIN Co2 | 74 (11) | 50 (8) | |||||
| 407 MI | 71 (8) | 45 (15) | |||||
| FIN Co1 | 71 (9) | 24 (5) | |||||
| 411 MI | 55 (5) | 92 (7) | |||||
| Mean | 72 | 34 |
Figure 1.
Heterozygous hMSH2 cells are phenotypically distinguishable from wild-type cells. Mean values of TMZ IC50 (a), hMSH2 and hMLH1 steady-state protein levels (b), and mismatch repair efficiency (c) for each cell line are shown, along with the group mean values (filled squares) ± SE. The hMSH2 and hMLH1 protein levels in b were expressed as percentage of the levels found in the normals (see Materials and Methods). The dashed line in a represents the arbitrary cutoff value set to distinguish between normal and tolerant lines (see Results).
hMSH2+/− Lymphoblasts Display Low Steady-State Levels of hMSH2.
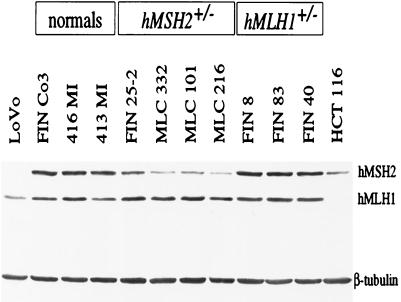
To test whether the increased tolerance of the hMSH2+/− lymphoblasts to TMZ was associated with reduced steady-state levels of hMSH2, we compared the amounts of this protein in the heterozygous and normal cell extracts by Western blotting. The amounts of hMSH2 and hMLH1 in the heterozygotes were compared with the mean levels observed in the normal controls. As shown in Table 1 and Fig. 1b, extracts of the hMSH2+/− lines showed a statistically significant (≈2-fold) decrease in the mean steady-state levels of hMSH2 as compared with normal controls (Fig. 2). The decrease in hMSH2 was accompanied by a similar decrease in the levels of hMSH6 in the hMSH2+/− cells (data not shown). This is because hMSH2 stabilizes hMSH6 against proteolysis in the heterodimeric mismatch binding factor hMuSα (27). One exception in this set were MLC 177 cells, which showed 63% of the normal values; interestingly, this line was also atypical in its sensitivity to TMZ, being more sensitive than other hMSH2+/− lines (see above).
Figure 2.
Western blot analysis of protein extracts from representative normal, hMSH2+/− and hMLH1+/− cell lines. The lower intensity of the hMSH2 bands in hMSH2+/− extracts is clearly apparent. Extracts of the colon cancer cell lines LoVo (hMSH2−/−) and HCT116 (hMLH1−/−) were used as negative controls. The proteins were separated in a 7.5% SDS polyacrylamide gel, and β-tubulin was used as an internal control (see Materials and Methods).
Multiple regression analysis showed a negative correlation between protein levels and IC50 values in the hMSH2+/− group. For each increase of 4% in protein level, the IC50 concentration decreased by about 10 μM when adjusted for repair efficiency (r2 = 0.5999; P < 0.05). In contrast to hMSH2+/− cells, the hMLH1+/− lymphoblasts displayed a mean steady-state hMLH1 levels of 86.7% as compared with the normal controls, a difference that was not statistically significant. This result did not change when we excluded the lines FIN 28, FIN 7, and Sw 434, which carry hMLH1 missense mutations. The mean steady-state level of hMLH1 in this group after the exclusion of the latter lines was 88.2%.
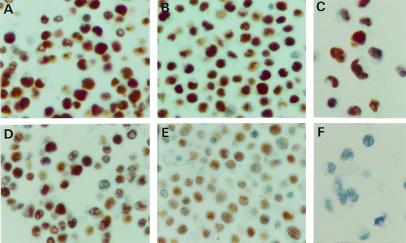
We wanted to exclude the possibility that the reduction in the hMSH2 levels observed in the Western blots was the result of our cell population being composed partly of cells carrying a full complement of the protein and a similar proportion of hMSH2 null cells. We therefore examined the hMSH2 and hMLH1 expression levels by immunohistochemistry. As shown in Fig. 3, the hMLH1 signal in a normal control line (A) was similar to that seen in a heterozygous hMSH2+/− line (B) and in the hMSH2 null line HEC59 (C). The hMSH2 signal in the normal control was similar in intensity to that seen with hMLH1 (D; see also A), and was absent from the negative control HEC 59 (F). However, in the hMSH2+/− cells, the hMSH2 signal was overall weaker in most cells (E). These results thus confirmed that the hMSH2+/− cells do indeed contain less hMSH2 than the hMSH2+/+ controls, as anticipated from the Western blots. This result is only qualitative and should be viewed as such. However, it further confirms that the increased resistance of the hMSH2+/− heterozygous cells to TMZ was indeed due to the halved gene dosage and thus the reduced capacity of addressing/signaling the methylating agent damage, rather than being due to the selection of highly-resistant hMSH2 null clones (i.e., cells expressing no hMSH2) from the total cell population.
Figure 3.
Immunohistochemical analysis of hMLH1 (Upper) and hMSH2 (Lower) levels. As shown (Upper), the normal control (A), the hMSH2+/− line MLC21 (B), and the hMSH2−/− line HEC59 (C) contain similar amounts of hMLH1. In contrast, the hMSH2+/− line MLC21 (E) contained substantially less hMSH2 than the normal control (D). No brown staining indicative of hMSH2 is apparent in the HEC59 line (hMSH2−/−) used as a negative control (F)
hMSH2+/− Cell Extracts Have a Reduced Mismatch Binding and Mismatch Correction Capacity.
Because hMSH2/hMSH6 heterodimer is the mismatch recognition factor in MMR (28, 29), we predicted that the base/base mismatch binding capacity of hMSH2+/− extracts would be impaired as a consequence of the reduced levels of hMSH2 in these cells. This was indeed the case, as shown by electrophoretic mobility-shift assay experiments (Fig. 4). On the basis of these findings, we decided to test the in vitro mismatch repair capacity of the extracts of our cell lines. Cells heterozygous in hMSH2 displayed a statistically significant decrease in repair efficiency as compared with normals (P = 0.019) or hMLH1+/− (P = 0.027) lines (Table 1 and Fig. 1c). A better distinction between normals and hMSH2+/− cells (P = 0.001) could be observed when the control line 411 MI was excluded from the analysis. Surprisingly, the repair efficiency of this “normal” line was ≈20% lower than that of the other controls. The reason underlying this phenomenon is unclear; sequencing of the hMSH2 gene, including the exon–intron boundaries, revealed four common polymorphisms (IVS1 + 9C→G, IVS6–10T→C, IVS9–9T→A and IVS12–6T→C), but no pathogenic germline mutations.
Multiple regression analysis showed that the repair efficiency and IC50 values were inversely correlated in the hMSH2+/− group: an increase of 3% in repair efficiency in vitro led to a decrease in IC50 of about 18 μM, when adjusted for protein level (r2 = 0.5999; P < 0.01). This inverse correlation was also found in the normal group by simple regression analysis: an increase of 3% in repair efficiency led to a decrease in IC50 of about 10 μM (r = 0.881; P < 0.01). However, no correlation was found between in vitro repair efficiency and steady-state levels of hMSH2, suggesting that the observed decreased hMSH2 levels were sufficient to ensure efficient mismatch repair in vitro, even under suboptimal conditions (shorter incubation times, lower temperatures, different salt and ATP concentrations) of the MMR assay.
The hMLH1+/− lymphoblast extracts displayed a mean repair efficiency that was not statistically different from that of the control cell lines but that was significantly higher than that of the hMSH2+/− cells (P = 0.027). We failed to differentiate between the hMLH1+/− and normal extracts, even when the conditions of our in vitro repair assay were suboptimal.
Discussion
We demonstrate that hMSH2+/− lymphoblasts are phenotypically distinguishable from wild-type cells in that they are significantly more tolerant to a methylating agent, a phenotype invariably linked to MMR deficiency (10). Analysis of immortalized hMSH2+/− lymphocytes (Fig. 3) and of their extracts (Fig. 2) revealed that the levels of hMSH2 and thus also of the hMSH2/hMSH6 mismatch binding factor (Fig. 4) were lower than in hMLH1+/− or in normal cells. However, the accompanying efficiency decrease of MMR in vitro (Fig. 1) is most likely too small to cause an increase in spontaneous mutation frequency. We base this prediction on the following findings. First, normal tissues of HNPCC patients do not display increased MSI. Similarly, four MSI markers in our hMSH2+/− lines were stable (data not shown). Second, expression of hMSH6 in hMSH6−/− HCT15 cells, which resulted in the restoration of hMutSα levels to only about 30% of the positive control, was largely sufficient to correct the MSI and MMR defect in these cells (30). Third, a diploid yeast strain with one disrupted MSH2 allele displayed only a very modest elevation in spontaneous mutation rates (31).
However, while reduced amounts of MMR proteins may be sufficient to cope with the correction of replication errors, this may not be the case when the mismatch repair system is saturated. Mouse embryonic stem cells carrying one disrupted Msh2 allele were reported to be more tolerant to low-dose rate ionizing radiation than wild-type controls (32). The HCT15 clone, in which expression of low levels of hMSH6 largely rescued its mutator phenotype, remained tolerant to the cytotoxic effects of methylating agents (30), presumably because the number of repair events triggered by the O6-methyl G/T mispairs was too low to trigger apoptosis. As shown in Fig. 1a, an analogous situation appears to exists in our hMSH2+/− lymphoblastoid cell lines: they were on average 4-fold less sensitive to TMZ than normal controls or hMLH1+/− cells. Considering that the mismatch repair deficient lymphoblastoid human cell line MT1 exhibited an 8-fold decrease in TMZ sensitivity (12), as compared with the mean of our control lines, the tolerance observed in the hMSH2+/− cells is highly significant. We can exclude the possibility that this effect was due to mutational inactivation of the wild-type allele during the treatment, which could have resulted in the selection of hMSH2−/− population; the cells were exposed to the drug for only 72 h, a time period too short to allow selection and phenotypic expression of hMSH2 null cells. Moreover, it was not due to selection of preexisting hMSH2 null cells in the treated population, because immunohistochemistry showed that the heterozygous cultures and the normal controls contained similar numbers of cells that failed to stain for hMSH2 (Fig. 3).
Although hMSH2+/− cells may not differ from control cells under normal conditions, their haploinsufficiency may demonstrate itself under conditions where the MMR system is saturated or overtaxed. Normal cells of the colonic epithelium are continuously exposed to DNA damaging agents, but because they begin to apoptose as they migrate through the colonic crypts, damaging their DNA might lead to earlier apoptosis and shedding, rather than to their transformation. In contrast, should the hMSH2+/− colonic cells behave similarly to their lymphocytic counterparts, they would be more resistant to killing by DNA-damaging agents (10). The unrepaired modifications may give rise to mutations during replication, some of which might affect the wild-type allele of the hMSH2 gene. This event would generate a MMR-deficient cell with a full-blown mutator phenotype and MSI, as well as an even higher tolerance to alkylating agents. This clone would be expected to proliferate further under the selective pressure of the colonic milieu and give rise to cells with mutations in genes that appear to play important roles in the genesis of HNPCC tumors such as TGFβ RII (7) or BAX (8). This hypothesis finds support in reports showing that MSI is present already in extremely small colonic polyps of HNPCC patients, which implies that both alleles of the respective MMR gene are inactivated very early in the tumorigenic transformation process (33, 34).
Our data showed that most of the hMLH1+/− cell lines behaved similarly to normal lymphoblasts. The observed phenotypic differences between hMSH2+/− and hMLH1+/− cells cannot be explained by the different nature of the germline mutations, which lead predominantly to truncated products. In the three lines that carry missense mutations in hMLH1, the mutated polypeptides were either shown (35) or predicted to be nonfunctional (36) (Table 1; see also Results). Thus, in the absence of a diminished MMR efficiency in hMLH1+/− cells, the wild-type allele of the hMLH1 gene may have to be inactivated differently from hMSH2. The chromosomal locus 3p21, which houses hMLH1, has been reported to be susceptible to loss of heterozygosity (37, 38) and has been predicted to house an as-yet-unidentified tumor suppressor gene (39–43). The ready transcriptional silencing of the hMLH1 locus through hypermethylation (44) represents another facile way of inactivating the wild-type hMLH1 allele. Moreover, such an epigenetic modification may bring about a change in chromatin conformation at the hMLH1 locus, which could, in turn, facilitate genomic rearrangement such as loss of heterozygosity.
de Wind et al. reported that embryonic stem (ES) cells from Msh2+/− mice were indistinguishable from wild-type cells when treated with N-methyl-N′-nitrosourea (45). The differences between the murine ES cells and the human lymphoblasts are real and may be due to the different apoptotic thresholds of the two cell types. Indeed, murine ES cells may not be representative of other cell types and tissues. In a later study, de Wind et al. (46) reported that mice heterozygous at the Msh2 locus displayed an increased incidence of tumors as compared with the normal controls. This demonstrated that even in the mouse model system, hMSH2+/− cells are not phenotypically normal, at least in some tissues.
It could be argued that the cell lines studied here are of lymphocytic origin and that our data may therefore be of little relevance to colon carcinogenesis. This possibility must be taken into consideration, and it is unfortunate that no human epithelial cells heterozygous at these MMR loci exist. However, immortalized lymphoblasts of HNPCC patients may yet turn out to be useful tools for the study of this syndrome. It is interesting to note that the age of disease onset in our hMSH2+/− cohort correlated with TMZ sensitivity, in that the most resistant lines originated from patients with the lowest age at diagnosis. The possibility that hMSH2 haploinsufficiency described above may play a role in the transformation process in HNPCC cannot therefore be disregarded at this time.
Acknowledgments
We thank Rick Boland and Minoru Koi for help with the lymphoblast immortalizations and Christine Hemmerle, Ritva Heider, and Helen Klingler for expert technical assistance. We are grateful to Bert Vogelstein and the Finnish HNPCC databank for the generous gift of immortalized lymphoblasts and to Alan Clark and Tom Kunkel (NIEHS, Research Triangle Park, NC) for the gift of the M13 phage. We also thank all our colleagues for many helpful discussions and Gray Crouse for critical reading of the manuscript. The generous financial support of the Swiss National Science Foundation to C.C., G.M., and J.J. is gratefully acknowledged.
Abbreviations
- HNPCC
hereditary nonpolyposis colon cancer
- MMR
mismatch repair
- MSI
microsatellite instability
- TMZ
temozolomide
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kinzler K W, Vogelstein B. Science. 1998;280:1036–1037. doi: 10.1126/science.280.5366.1036. [DOI] [PubMed] [Google Scholar]
- 2.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K W, Vogelstein B. Nature (London) 1996;379:19–20. doi: 10.1038/379019a0. [DOI] [PubMed] [Google Scholar]
- 4.Lynch H T, de la Chapelle A. J Med Genet. 1999;36:801–818. [PMC free article] [PubMed] [Google Scholar]
- 5.Jiricny J, Nyström-Lahti M. Curr Opin Genet Dev. 2000;10:157–161. doi: 10.1016/s0959-437x(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 6.Losi L, Ponz de Leon M, Jiricny J, Di Gregorio C, Benatti P, Percesepe A, Fante R, Roncucci L, Pedroni M, Benhattar J. Int J Cancer. 1997;74:94–96. doi: 10.1002/(sici)1097-0215(19970220)74:1<94::aid-ijc16>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, et al. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 8.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed J C, Perucho M. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 9.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 10.Karran P, Bignami M. BioEssays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 11.Duckett D R, Drummond J T, Murchie A I, Reardon J T, Sancar A, Lilley D M, Modrich P. Proc Natl Acad Sci USA. 1996;93:6443–6447. doi: 10.1073/pnas.93.13.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Atri S, Tentori L, Lacal P M, Graziani G, Pagani E, Benincasa E, Zambruno G, Bonmassar E, Jiricny J. Mol Pharmacol. 1998;54:334–341. doi: 10.1124/mol.54.2.334. [DOI] [PubMed] [Google Scholar]
- 13.Duckett D R, Bronstein S M, Taya Y, Modrich P. Proc Natl Acad Sci USA. 1999;96:12384–12388. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hickman M J, Samson L D. Proc Natl Acad Sci USA. 1999;96:10764–10769. doi: 10.1073/pnas.96.19.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wijnen J T, Vasen H F, Khan P M, Zwinderman A H, van der Klift H, Mulder A, Tops C, Møller P, Fodde R. N Engl J Med. 1998;339:511–518. doi: 10.1056/NEJM199808203390804. [DOI] [PubMed] [Google Scholar]
- 16.Hemminki A, Peltomäki P, Mecklin J P, Jarvinen H, Salovaara R, Nystršm-Lahti M, de la Chapelle A, Aaltonen L A. Nat Genet. 1994;8:405–410. doi: 10.1038/ng1294-405. [DOI] [PubMed] [Google Scholar]
- 17.Borresen A L, Lothe R A, Meling G I, Lystad S, Morrison P, Lipford J, Kane M F, Rognum T O, Kolodner R D. Hum Mol Genet. 1995;4:2065–2072. doi: 10.1093/hmg/4.11.2065. [DOI] [PubMed] [Google Scholar]
- 18.Luce M C, Marra G, Chauhan D P, Laghi L, Carethers J M, Cherian S P, Hawn M, Binnie C G, Kam-Morgan L N, Cayouette M C. Gastroenterology. 1995;109:1368–1374. doi: 10.1016/0016-5085(95)90600-2. [DOI] [PubMed] [Google Scholar]
- 19.Hansen M B, Nielsen S E, Berg K. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 20.Morten J E, Margison G P. Carcinogenesis. 1988;9:45–49. doi: 10.1093/carcin/9.1.45. [DOI] [PubMed] [Google Scholar]
- 21.Kuismanen S A, Holmberg M T, Salovaara R, Schweizer P, Aaltonen L A, de la Chapelle A, Nyström-Lahti M, Peltomäki P. Proc Natl Acad Sci USA. 1999;96:12661–12666. doi: 10.1073/pnas.96.22.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas D C, Roberts J D, Kunkel T A. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 23.Marra G, Chang C L, Laghi L A, Chauhan D P, Young D, Boland C R. Oncogene. 1996;13:2189–2196. [PubMed] [Google Scholar]
- 24.Jiricny J, Hughes M, Corman N, Rudkin B B. Proc Natl Acad Sci USA. 1988;85:8860–8864. doi: 10.1073/pnas.85.23.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karran P, Hampson R. Cancer Surv. 1996;28:69–85. [PubMed] [Google Scholar]
- 26.Vasen H F, Mecklin J P, Khan P M, Lynch H T. Dis Colon Rectum. 1991;34:424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 27.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Proc Natl Acad Sci USA. 1998;95:8568–8573. doi: 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 29.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 30.Lettieri T, Marra G, Aquilina G, Bignami M, Crompton N E, Palombo F, Jiricny J. Carcinogenesis. 1999;20:373–382. doi: 10.1093/carcin/20.3.373. [DOI] [PubMed] [Google Scholar]
- 31.Drotschmann K, Clark A B, Tran H T, Resnick M A, Gordenin D A, Kunkel T A. Proc Natl Acad Sci USA. 1999;96:2970–2975. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeWeese T L, Shipman J M, Larrier N A, Buckley N M, Kidd L R, Groopman J D, Cutler R G, te Riele H, Nelson W G. Proc Natl Acad Sci USA. 1998;95:11915–11920. doi: 10.1073/pnas.95.20.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacoby R F, Marshall D J, Kailas S, Schlack S, Harms B, Love R. Gastroenterology. 1995;109:73–82. doi: 10.1016/0016-5085(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 34.Shibata D, Peinado M A, Ionov Y, Malkhosyan S, Perucho M. Nat Genet. 1994;6:273–281. doi: 10.1038/ng0394-273. [DOI] [PubMed] [Google Scholar]
- 35.Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend S H, Kolodner R D, Ishioka C. Nat Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 36.Ban C, Yang W. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 37.Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D. Br J Cancer. 1999;79:1012–1017. doi: 10.1038/sj.bjc.6690162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wieland I, Ammermuller T, Bohm M, Totzeck B, Rajewsky M F. Oncol Res. 1996;8:1–5. [PubMed] [Google Scholar]
- 39.Chen L C, Matsumura K, Deng G, Kurisu W, Ljung B M, Lerman M I, Waldman F M, Smith H S. Cancer Res. 1994;54:3021–3024. [PubMed] [Google Scholar]
- 40.Chino K, Esumi M, Ishida H, Okada K. J Urol. 1999;162:614–618. [PubMed] [Google Scholar]
- 41.Daigo Y, Nishiwaki T, Kawasoe T, Tamari M, Tsuchiya E, Nakamura Y. Cancer Res. 1999;59:1966–1972. [PubMed] [Google Scholar]
- 42.Kok K, Naylor S L, Buys C H. Adv Cancer Res. 1997;71:27–92. doi: 10.1016/s0065-230x(08)60096-2. [DOI] [PubMed] [Google Scholar]
- 43.Roche J, Boldog F, Robinson M, Robinson L, Varella G M, Swanton M, Waggoner B, Fishel R, Franklin W, Gemmill R, Drabkin H. Oncogene. 1996;12:1289–1297. [PubMed] [Google Scholar]
- 44.Herman J, Umar A, Polyak K, Graff J, Ahuja N, Issa J, Markowitz S, Willson J, Hamilton S, Kinzler K, et al. Proc Natl Acad Sci USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 46.de Wind N, Dekker M, van Rossum A, van der Valk M, te Riele H. Cancer Res. 1998;58:248–255. [PubMed] [Google Scholar]