Abstract
DNA hypermethylation is a common epigenetic abnormality in colorectal cancers (CRCs) and a promising class of CRC screening biomarkers. We conducted a genome-wide search for novel neoplasia-specific hypermethylation events in the colon.
We applied methylation microarray analysis to identify loci hypermethylated in 17 primary CRCs relative to 8 non-neoplastic colonic tissues (NCs) from neoplasia-free subjects. These CRC-associated hypermethylation events were then individually evaluated for their ability to discriminate neoplastic from non-neoplastic cases, based on real-time quantitative methylation-specific PCR (qMSP) assays in 113 colonic tissues: 51 CRCs, 9 adenomas, 19 NCs from CRC patients (CRC-NCs), and 34 NCs from neoplasia-free subjects (control NCs).
A strict microarray data filtering identified 169 candidate CRC-associated hypermethylation events. Fourteen of these 169 loci were evaluated using qMSP assays. Ten of these 14 methylation events significantly distinguished CRCs from age-matched control NCs (p<0.05 by ROC curve analysis); methylation of VSX2 achieved the highest discriminative accuracy (83.3% sensitivity and 92.3% specificity, p<1E-6), followed by BEND4, NPTX1, ALX3, miR-34b, GLP1R, BTG4, HOMER2, ZNF583, and GJC1. Adenomas were significantly discriminated from control NCs by hypermethylation of VSX2, BEND4, NPTX1, miR-34b, GLP1R, and HOMER2 (p<0.05). CRC-NCs were significantly distinguished from control NCs by methylation of ALX3 (p<1E-4).
In conclusion, systematic, methylome analysis has identified ten novel methylation events in neoplastic and non-neoplastic colonic mucosae from CRC patients. These potential biomarkers significantly discriminate CRC patients from controls. Thus, they merit further evaluation in stool- and circulating DNA-based CRC detection studies.
Keywords: Colorectal cancer, colorectal adenoma, methylation, microarray, real-time methylation-specific PCR, biomarker
INTRODUCTION
In the United States, colorectal cancer (CRC) is the third most prevalent and the second most deadly cancer in both sexes(Jemal 2008). CRC is highly curable in its early, localized stages, with a 5-year survival rate exceeding 90%(Jemal 2008). Unfortunately, 61% of new cases are already advanced at the time of diagnosis(Jemal 2008). Delayed diagnosis occurs due to the asymptomatic nature of most early-stage CRCs; thus, the key to reducing deaths from CRC is periodic screening of the entire colon in the average-risk population (Kahi, et al. 2008). The current gold-standard method for the entire colon screening is colonoscopy (Kahi et al. 2008). However, invasive screening modalities, including colonoscopy, are not ideal for the application to asymptomatic population. Therefore, active investigations are now underway to discover noninvasive biomarkers, such as those found in stool, that could supplement or supplant colonoscopic screening.
Hypermethylation of CpG islands (CGIs) is a promising CRC biomarker with high potential for translation into non-invasive CRC detection modalities. CGI hypermethylation is a common epigenetic DNA abnormality that has been strongly linked to CRC (Fraga and Esteller 2007). CGI hypermethylation possesses several advantages as a biomarker: 1) hypermethylation at multiple CGIs often exists in adenomas, suggesting its potential utility in early detection (Kim, et al. 2006); 2) only one assay per locus is generally needed, in contrast to gene mutation that frequently require multiple assays due to the multiple mutational hotspots; and 3) quantitative methylation assays are applicable to low-integrity DNA commonly encountered in clinical specimens(Eads, et al. 2000; Uhlmann, et al. 2002). However, known cancer-specific methylation targets in the colon have in the past been identified based on their functional relevance to neoplastic progression, rather than their merit as biomarkers, partly due to the previous lack of genome-wide, high-resolution methodologies for the direct analysis of methylation.
Recent technological advances now offer the ability to perform high-throughput, direct assays of DNA methylation (Estecio, et al. 2007). In the current study, we employed a microarray-based direct scanning assay of DNA methylation to extensively search for CGI hypermethylation events, based purely on their performance as CRC biomarkers, for ultimate application to the average-risk population.
MATERIALS & METHODS
Patients and nucleic acid preparation
Sporadic CRC tissues were obtained during surgery. Adenomas were obtained during colonoscopy. All adenomas were ≥1 cm in diameter or exhibiting advanced histology (i.e., tubulovillous adenomas, villous adenomas, and adenomas with focal high-grade dysplasia). We excluded from the study recurrent CRC patients, polyposis- or inflammatory bowel disease (IBD)-associated CRC patients, and patients who ever underwent chemotherapy for CRC or other neoplasias prior to sampling.
Three types of non-neoplastic colonic mucosae (NCs) were used in this study: NCs from CRC patients (CRC-NCs), NCs from neoplasia-free subjects who were 40 years of age or older (control NCs), and NCs from neoplasia-free subjects who were younger than 40 years of age (young control NCs). Neoplasia-free subjects were those who underwent screening colonoscopy but presented no colonoscopic abnormalities and possessed no history of colonic neoplasia, IBD, or chemotherapy for any malignancies.
Tissue acquisition was conducted under a protocol approved by the institutional review board at the Johns Hopkins University (Baltimore, Maryland, U.S.A.). Written consent was obtained from all patients enrolled after full explanation of the purpose and nature of all procedures used. Genomic DNA was extracted from snap-frozen tissues using a DNeasy kit (Qiagen, Valencia, CA). Demographic data for cases studied in microarray/methylation-specific PCR (MSP) experiments and real-time quantitative MSP (qMSP) experiments are summarized in Table 1. All specimens interrogated in microarray experiments were also included in qMSP experiments. CIMP status of each tumor was determined based on qMSP measurement of the methylation status of five loci (RUNX3. SOCS1, NEUROG1, IGF2, and CACNA1G), as described previously (Weisenberger, et al. 2006). Neoplasias demonstrating methylation at ≥3 or <3 of the five loci were classified as CIMP-positive (+) or –negative (−), respectively.
Table 1.
Tissue demographic data.
| Assay | Microarray | qMSP | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Tissue type | Non-neoplastic | Neoplastic | Non-neoplastic | Neoplastic | |||||
|
| |||||||||
| Categories | control NC (CRC-free, ≥40y.o.) | CRC | young control NC (CRC- free, <40y.o.) | control NC (CRC-free, ≥40y.o.) | CRC-NC (CRC patient) | Adenoma | CRC (all) | Dukes AB CRC | Dukes CD CRC |
| Total number | 8 | 17 | 9 | 26 | 19 | 9 | 51 | 24 | 27 |
|
| |||||||||
| Age | |||||||||
|
| |||||||||
| mean | 67.5 | 63.2 | ***31.2 | 61.7 | 63.4 | 64.1 | 65.1 | 64.5 | 65.6 |
| SD | 8.5 | 10.8 | 7.1 | 12.6 | 8.7 | 8.9 | 11.1 | 13.3 | 9.0 |
| min.-max. | 55–77 | 36–80 | 23–39 | 43–80 | 36–75 | 51–77 | 36–87 | 36–87 | 51–81 |
|
| |||||||||
| Gender | |||||||||
|
| |||||||||
| F | 0 (0.0%) | 5 (29.4%) | **7 (77.8%) | 2 (7.7%) | 4 (21.1%) | 0 (0.0%) | 11 (21.6%) | 6 (25.0%) | 5 (18.5%) |
| M | 8 (100.0%) | 12 (70.6%) | 2 (22.2%) | 24 (92.3%) | 15 (78.9%) | 9 (100.0%) | 40 (78.4%) | 18 (75.0%) | 22 (81.5%) |
|
| |||||||||
| Site | |||||||||
|
| |||||||||
| L | 4 (50.0%) | 10 (58.8%) | 5 (55.6%) | 17 (65.4%) | 13 (68.4%) | 6 (66.7%) | 30 (58.8%) | 15 (62.5%) | 15 (55.6%) |
| R | 4 (50.0%) | 7 (41.2%) | 4 (44.4%) | 9 (34.6%) | 6 (31.6%) | 3 (33.3%) | 21 (41.2%) | 9 (37.5%) | 12 (44.4%) |
|
| |||||||||
| Histology | |||||||||
|
| |||||||||
| WD-WMD | NA | 0 (0.0%) | NA | NA | NA | NA | 11 (22.4%) | 6 (26.1%) | 5 (19.2%) |
| MD | NA | 9 (60.0%) | NA | NA | NA | NA | 26 (53.1%) | 11 (47.8%) | 15 (57.7%) |
| MPD-PD | NA | 6 (40.0%) | NA | NA | NA | NA | 11 (22.4%) | 5 (21.7%) | 6 (23.1%) |
| MUC | NA | 0 (0.0%) | NA | NA | NA | NA | 1 (2.0%) | 1 (4.3%) | 0 (0.0%) |
| TA | NA | NA | NA | NA | NA | 6 (66.7%) | NA | NA | NA |
| TA+HGD | NA | NA | NA | NA | NA | 1 (11.1%) | NA | NA | NA |
| TVA | NA | NA | NA | NA | NA | 2 (22.2%) | NA | NA | NA |
| UNK | 2 | 2 | 1 | 1 | |||||
|
| |||||||||
| Dukes stage | |||||||||
|
| |||||||||
| A | NA | 1 (5.9%) | NA | NA | NA | NA | 7 (13.7%) | 7 (29.2%) | NA |
| B | NA | 7 (41.2%) | NA | NA | NA | NA | 17 (33.3%) | 17 (70.8%) | NA |
| C | NA | 6 (35.3%) | NA | NA | NA | NA | 17 (33.3%) | NA | 17 (63.0%) |
| D | NA | 3 (17.6%) | NA | NA | NA | NA | 10 (19.6%) | NA | 10 (37.0%) |
|
| |||||||||
| CIMP | |||||||||
|
| |||||||||
| + | NA | 5 (29.4%) | NA | NA | NA | 0 (0.0%) | 5 (9.8%) | 2 (8.3%) | 3 (11.1%) |
| − | NA | 12 (70.6%) | NA | NA | NA | 9 (100.0%) | 46 (90.2%) | 22 (91.7%) | 24 (88.9%) |
|
| |||||||||
| MSI | |||||||||
|
| |||||||||
| H | NA | 5 (29.4%) | NA | NA | NA | 0 (0.0%) | 6 (11.8%) | 2 (8.3%) | 4 (14.8%) |
| L | NA | 4 (23.5%) | NA | NA | NA | 0 (0.0%) | 5 (9.8%) | 4 (16.7%) | 1 (3.7%) |
| S | NA | 8 (47.1%) | NA | NA | NA | 7 (100.0%) | 40 (78.4%) | 18 (75.0%) | 22 (81.5%) |
| UNK | 2 | ||||||||
UNK, unknown; NA, not applicable; SD, standard deviation; WD, well differentiated; WMD, well-to moderately differentiated; MD, moderately differentiated; MPD, moderately to poorly differentiated; PD, poorly differentiated; MUC, mucinous adenocarcinoma; TA, tubular adenoma; TVA, tubulovillous adenoma; HGD, high grade dysplasia. Statistically significant difference relative to control NC in respective assay: Single-, double-, and triple asterisks indicate significant difference from control NCs at p-level <.05 <.01, and <1E-6, respectively. No significant difference was observed between Dukes stage A/B CRCs versus Dukes stage C/D CRCs for age, gender, tumor site, histological differentiation, or CIMP status. Similarly, no significant difference was observed between CRCs versus adenomas for age, gender, tumor site, or CIMP status.
MCAM: methylated CpG island amplification (MCA) coupled with microarray analysis
MCAM was conducted according to a previously published protocol, using the isoschizomers SmaI and XmaI (Estecio et al. 2007). 244K Human CpG Island microarrays (Agilent Technologies, Santa Clara, CA) were employed as an array platform. Using this methodology, we were able to assess the methylation status of 34,396 SmaI-XmaI restriction fragments that covered to 50.4% of all CGIs in the genome. Ssst-treated fully methylated DNA was used as a control DNA. Normalized log2 array intensity ratio to control fully methylated DNA at each locus (referred to as “log2 array ratio” hereafter) was used to represent locus methylation level. We verified the robustness of this MCAM methodology as follows: two separate MCAM experimental batches of a specimen displayed markedly high reproducibility (R>0.99; Supplementary Figure 1A), and methylation measurements by MCAM and qMSP were significantly correlated (R>0.70; Supplementary Figure 1B). Further methodological details are described in Supplementary Methods.
Selection of candidate cancer-specific methylation targets based on the MCAM data
The criteria for autosomal cancer-specific methylation events in the colon were as follows: 1) mean log2 array ratio for CRCs greater than that for control NCs by more than 0.5 at t-test p<0.01; 2) no overlap in log2 array ratio between any CRCs versus any control NCs; 3) mean log2 array ratio for CRCs greater than the lower 95% confidence limits of mean normalized log2 array ratios for array normalization control probes (see supplementary Methods for description); 4) mean log2 array ratio for control NCs greater than the upper 95% confidence limits of mean log2 array ratios for normalization control probes.
Methylation-specific PCR (MSP)
MSP analyses were performed on pooled primary CRC-derived DNAs vs. pooled control NC-derived DNAs. Specimens analyzed by MSP were identical to those analyzed by MCAM. Thirty-seven cycles of PCR amplification were carried out, and PCR product quantity was measured by gel electrophoresis using a GelDoc XR system (BioRad, Hercules, CA). We verified both lack of amplification from unmethylated control DNA and efficient amplification from fully methylated control DNA. A given locus was classified as hypermethylated in CRC when the visualized PCR product from pooled CRCs was >5-fold more abundant than from pooled control NCs. Primer sequences are shown in Supplementary Table 1.
Real-time quantitative MSP (qMSP)
qMSP was performed using the same primer set as for MSP and a locus-specific TaqMan probe for each locus, as described previously(Mori, et al. 2006). The fraction of densely methylated DNA molecules at each locus (i.e., percent methylation, or, PMR) was calculated as described previously (Mori et al. 2006). TaqMan probe sequences are provided in Supplementary Table 1.
Statistical analyses
A p-value of less than 0.05 was used as the cut-off for statistical significance. Normalized MCAM data were assessed using Student’s t-tests, unless otherwise stated. qMSP data were analyzed using Mann-Whitney test, unless otherwise stated, due to their non-normal distribution. Receiver-operator characteristic (ROC) curve analysis was applied to evaluate the diagnostic performance of PMR data at each locus. ROC curves were generated using the PMR data for each locus as a continuous input variable. The non-parametric Delong Clarke-Pearson method was applied to compare areas under ROC curves (AUROCs; DeLong, et al. 1988). Forward stepwise discriminant analysis and five-fold cross validation were employed to generate diagnostic models based on methylation levels at multiple loci.
RESULTS
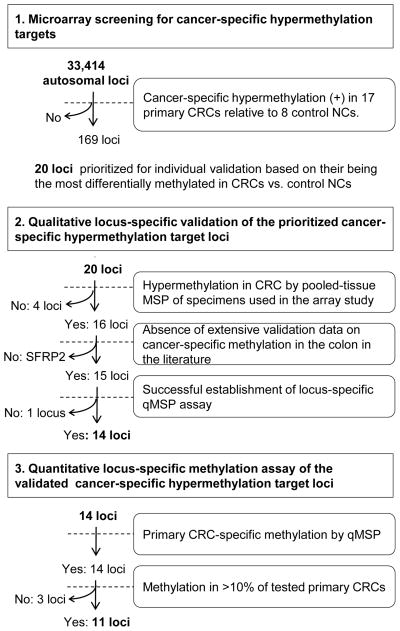
We conducted a genome-wide search for novel targets of CRC-specific hypermethylation by employing methylated DNA microarray-based scanning of primary CRCs followed by locus-specific qMSP-based validation (strategy outlined in Figure 1). A total of 33,414 autosomal CGI loci were interrogated. After performing qualitative validation in the tissue cohort that was used in the microarray analysis, quantitative validation was carried out in a larger tissue cohort utilizing locus-specific qMSP-based assays.
Figure 1.
Study outline.
Microarray scanning
Methylated DNA microarray analysis was performed using MCAM methodology (Estecio et al. 2007). Seventeen primary CRCs and 8 non-neoplastic colonic mucosae (NCs) from colonic neoplasia-free control subjects who were 40 years of age or older (control NCs) were analyzed (Table 1). We study aged control individuals in order to avoid mistakenly identifying age-associated hypermethylation targets as neoplasia-specific hypermethylation events. Matching non-neoplastic colonic tissues from CRC cases (hereinafter referred to as NC-CRC) were not used as controls, since these tissues may already carry hypermethylation events linked to an increased risk of carcinogenic progression due to a “field defect” (Belshaw, et al. 2010; Nosho, et al. 2009; Shen, et al. 2005; Svrcek, et al. 2010).
The majority of analyzed loci tended to be differentially methylated in CRCs relative to control NCs (p<.1: 18,892 of 33,414 analyzed autosomal loci). Cluster analyses of these 18,892 loci showed separation of CRCs from control NCs (Supplementary Figure 2). As expected based on previous publications (Estecio et al. 2007; Weisenberger et al. 2006), CIMP (+) and CIMP (−) CRCs clustered separately, with the exception of one CIMP (−) CRC that was methylated at two CIMP marker loci and clustered with CIMP (+) CRCs. We selected candidate autosomal loci for colonic neoplasia-specific methylation based on significant hypermethylation in CRCs relative to control NCs by a mean log2 array intensity ratio difference ≥0.5. In order to eliminate markers that would likely to exhibit low sensitivity and specificity in CRC diagnosis, we excluded loci whose methylation level overlapped between CRCs and control NCs (i.e., loci showing hypermethylation in CRC at which minimum log2 array ratio for CRCs is smaller than maximum log2 array ratio for control NCs, and vice versa). Based on these criteria, 169 loci were designated as candidate loci showing neoplasia-specific hypermethylation in colonic mucosae.
One of these 169 loci was SFRP2, a previously published target of cancer-specific methylation in the colon, whose methylation has been reported in 75–90% of stool DNAs from CRC patients by multiple groups (Huang, et al. 2007; Muller, et al. 2004; Nagasaka, et al. 2009; Wang and Tang 2008). The current MCAM study also confirmed significant hypermethylation of several other previously reported CRC methylation markers in CRCs relative to control NCs (such as RASSF2 and vimentin; Supplementary Table 2). However, unlike SFRP2, these loci demonstrated overlap in methylation levels between CRCs and control NCs in our study, and were therefore not included among the aforementioned 169 loci.
Individual qualitative validation of prioritized targets in a pilot pooled cohort
Twenty of these 169 candidate CRC-specific methylation target loci were prioritized for further individual validation based on having shown the largest differences between CRCs and control NCs and the smallest intra-group variance in array-based methylation levels (Supplementary Table 3). These 20 loci were then analyzed by qualitative MSP, using pooled DNA specimens for CRCs and control NCs that had been studied in microarray scanning experiments. Specimens were pooled in order to avoid exhaustion of limited clinical DNA resources. We reasoned that the previous and subsequent non-pooled analyses (i.e., microarray and qMSP assays) would eliminate false-positive findings caused by sample pooling (e.g., massive hypermethylation occurring in only a minority of CRCs). Hypermethylation in pooled CRCs vs. pooled control NCs was observed at 16 of the 20 analyzed loci: SFRP2, VSX2, BEND4, ALX3, NPTX1, GLP1R, HOMER2, GJC1, DOCK8, NME4, ZNF583, TMEM42, TTLL12, miR-34b, and MDFI (Supplementary Table 3). The miR34b locus flanks the region that is proximal to the BTG4 gene transcriptional start site and is hypermethylated in approximately 90% of primary CRCs (Toyota, et al. 2008).
Quantitative methylation assays of validated targets in a larger cohort
We then assessed methylation of the qualitatively validated CRC-specific methylation targets in a larger cohort using a quantitative methodology, qMSP. Two loci were eliminated prior to performing qMSP: MDFI, for failure to establish a successful qMSP assay; and SFRP2, for having already been established as a CRC detection marker (Huang et al. 2007; Muller et al. 2004; Nagasaka et al. 2009; Wang and Tang 2008). The 14 qMSP-tested loci comprised VSX2, BEND4, ALX3, NPTX1, GLP1R, HOMER2, GJC1, DOCK8, NME4, ZNF583, TMEM42, TTLL12, miR-34b, and BTG4 (i.e.., the previously analyzed miR34b-flanking region (Toyota et al. 2008)). The analyzed case-control cohort contained 113 specimens: 51 primary CRCs, 9 adenomas, 26 control NCs from non-neoplasia patients, 19 NCs from CRC patients (CRC-NCs), and 9 NCs from colon neoplasia-free cases who were younger than 40 years of age (young control NCs). The control NCs were analyzed as a base control group representing the target population for average-risk CRC screening. Case demographic data are shown in Table 1. There were no significant differences in case age, a well-established non-neoplastic methylation-promoting factor, between any groups except for the young control NCs.
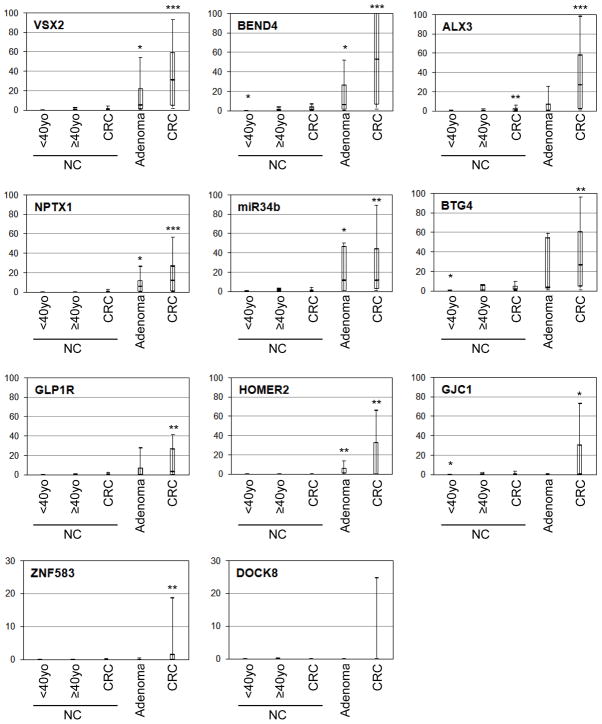
All 14 tested loci demonstrated varying degrees of hypermethylation in CRCs by qMSP assays. Significant hypermethylation in CRCs relative to control NCs was observed at all tested loci except DOCK8, NME4, TMEM42, and TTLL12 (Figure 2). These four loci demonstrated tumor-specific methylation in a minor subset of CRCs. NME4, TMEM42 and TTLL12 were methylated in less than 10% of the 51 CRCs, and methylation of these loci was observed only in CRCs that had been studied by MCAM. Thus, these three loci were eliminated from further analyses, leaving 11 loci for further study. No significant differences in methylation levels according to the gender, Dukes stage (AB vs. CD), or MSI status were observed at any of these 11 loci (data not shown). GJC1 was significantly more heavily methylated in proximal CRCs (median percent methylation, or PMR, 10.8%) than in distal CRCs (0.8%; p=0.02). CIMP (+) CRCs demonstrated significantly higher PMR levels than did CIMP (−) CRCs at ALX3, NPTX1, BTG4, GLP1R, HOMER2, DOCK8, and GJC1, although the majority of CIMP (−) CRCs were hypermethylated at all of these loci except DOCK8 (data not shown). DOCK8 was methylated in only 11 (25.6%) of 43 CIMP (−) CRCs, in contrast to CIMP (+) CRCs (4 of 5, or 80%; Fisher’s exact test, p=0.03).
Figure 2. Loci methylation levels for neoplastic and non-neoplastic colonic tissues.
These box plots represent the qMSP results of 51 CRCs and 9 adenomas, and 53 non-neoplastic colonic mucosal tissues (NCs, 8 young control NCs, 26 control NCs, and 19 CRC-NCs). Y-axis represents PMR value. Data on 11 loci that demonstrated methylation in at least one of the neoplastic tissues are shown. Median (bar), 25–75 percentile range (box), and 10–90 percentile range (whisker) of all informative specimens are displayed for each tissue category. Single-, double-, and tripleasterisks indicate significant difference from control NCs at p-level <.05 <.01, and <1E-6, respectively.
Significant hypermethylation in adenomas relative to control NCs was observed at BEND4, VSX2, NPTX1, miR34b, and HOMER2 (Figure 2). Only miR34b was methylated at equal levels in CRCs and adenomas (median PMR 10.9% vs. 11.4% for CRCs vs. adenomas, respectively; p=0.76). Remaining four loci were methylated at lesser degrees in adenomas than in CRCs, but these differences were insignificant. Tumor demographic data analyses were not performed for adenomas.
Notably, ALX3 was mildly but significantly hypermethylated in CRC-NCs relative to control NCs (median PMR 1.6% vs. 0.6% for NC-CRCs vs. control NCs, respectively; p=0.001; Figure 2 and Supplementary Figure 3). ALX3 methylation in CRC-NCs showed no significant association with age or corresponding CRC stage (data not shown). Methylation levels of NC from all CRC-free cases (viz., control NCs and young control NCs) at BEND4, GJC1, VSX2, and miR34b were significantly correlated with age (Spearman rank correlation R=0.55, 0.51, 0.39, and 0.38, respectively; p<0.05). However, differences between older and younger control NCs were small: median PMRs for old vs. young NCs were 0.3% vs. 0.0%, 0.1% vs. 0.0%, 0.3% vs. 0.0%, and 1.4% vs. 0.6%, for BEND4, GJC1, VSX2, and miR34b, respectively. These differences were smaller than those reported for classic age-dependent hypermethylation targets (e.g., N33 and ESR1; (Ahuja, et al. 1998; Issa, et al. 1994)). Association between gender and gene methylation was not assessed due to the small number of female control NC cases studied (n=2).
Evaluation of methylated loci as colonic neoplasia markers
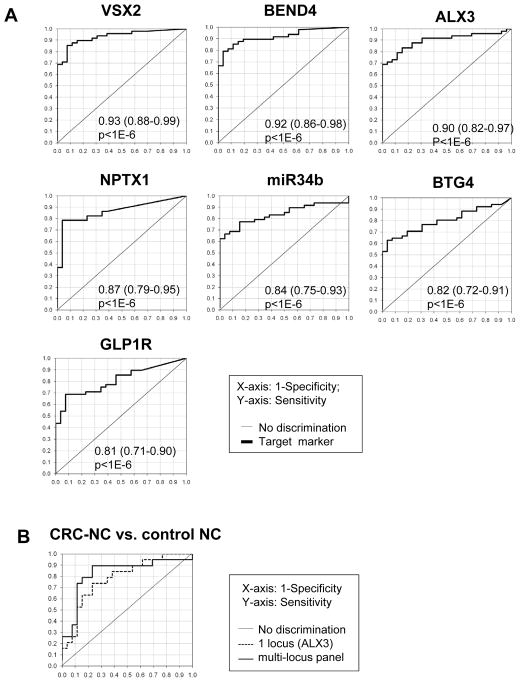
Next, we tested the 11 CRC-specific methylation targets for their abilities to distinguish colonic neoplasias from control NCs by employing ROC curve analysis. Methylation levels at all loci significantly distinguished CRCs from control NCs (p<.05; Table 2). VSX2 achieved the highest discriminative accuracy (the area under ROC curve, AUROC, 92.3, 83.3% sensitivity and 92.3% specificity; Figure 3A). BEND4, ALX3, NPTX1, miR34b, BTG4, and GLP1R also achieved particularly high diagnostic accuracy (AUROC>0.8, p<1E-6; Figure 3A). There was no statistically significant difference in AUROC between discrimination of Dukes AB vs. Dukes CD CRCs from control NCs for all but one locus: ALX3 discriminated Dukes AB CRCs significantly better than Dukes CD CRCs (p<0.03; Table 2). Five loci significantly distinguished adenomas from control NCs: VSX2, BEND4, NPTX1, miR34b, and HOMER2 (p<.05; Table 2), despite of our relatively small adenoma cohort size (n = 9). BTG4 also demonstrated weak discriminative capacity in this regard (p=0.09). Three loci were capable of significantly distinguishing CRC-NCs from control NCs: ALX3 (p=5.1E-5; Table 2). ZNF583 and BEND4 exerted similar significant discriminative abilities (p<.05), but the lower 95% confidence limit for their AUROCs did not exceed 0.5. Age did not significantly discriminate any diseased tissue classes from control NCs, as expected from our age-matched study enrollment strategy (data not shown). The use of a multi-locus methylation panel improved the discrimination of CRC-NCs from control NCs (AUROC 0.83; 95% CI 0.69–0.92) relative to the best-performing single locus (ALX3), although this improvement was insignificant (Figure 3B). The loci included in this multi-locus panel were ALX3, ZNF583, miR34b, and VSX2. The use of multi-locus methylation panels did not improve the discrimination of CRCs from NCs relative to the best-performing single locus (VSX2; data not shown).
Table 2.
The ROC curve analysis data for the discrimination from control NCs.
| Target tissue | CRC (n=51) | Dukes AB CRC (n=24) | Dukes CD CRC (n=27) | Adenoma (n=9) | CRC-NC (n=19) | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Loci | AUROC: mean (95% CI) | p-value | AUROC: mean (95% CI) | p-value | AUROC: mean (95% CI) | p-value | AUROC: mean (95% CI) | p-value | AUROC: mean (95% CI) | p-value |
| VSX2 | 0.93 (0.88–0.99) | <1.0E-6 | 0.94 (0.87–1.00) | <1.0E-6 | 0.92 (0.85–0.99) | <1.0E-6 | 0.74 (0.53–0.96) | 1.3E-02 | 0.56 (0.38–0.74) | 2.7E-01 |
| BEND4 | 0.92 (0.86–0.98) | <1.0E-6 | 0.92 (0.83–1.00) | <1.0E-6 | 0.92 (0.84–0.99) | <1.0E-6 | 0.74 (0.52–0.96) | 1.7E-02 | 0.65 (0.48–0.81) | 4.0E-02 |
| ALX3 | 0.90 (0.82–0.97) | <1.0E-6 | 0.97 (0.93–1.00) | <1.0E-6 | 0.83 (0.71–0.95) | <1.0E-6 | 0.57 (0.31–0.82) | 3.0E-01 | 0.78 (0.65–0.92) | 5.1E-05 |
| NPTX1 | 0.87 (0.79–0.95) | <1.0E-6 | 0.86 (0.75–0.97) | <1.0E-6 | 0.87 (0.77–0.97) | <1.0E-6 | 0.76 (0.56–0.96) | 5.2E-03 | 0.61 (0.45–0.77) | 9.1E-02 |
| miR34b | 0.84 (0.75–0.93) | <1.0E-6 | 0.87 (0.76–0.98) | <1.0E-6 | 0.81 (0.68–0.94) | 2.2E-06 | 0.76 (0.49–1.00) | 2.8E-02 | 0.47 (0.29–0.65) | 6.4E-01 |
| BTG4 | 0.82 (0.72–0.91) | <1.0E-6 | 0.85 (0.73–0.97) | <1.0E-6 | 0.79 (0.66–0.91) | 4.1E-06 | 0.67 (0.42–0.92) | 8.8E-02 | 0.54 (0.36–0.71) | 3.3E-01 |
| GLP1R | 0.81 (0.71–0.90) | <1.0E-6 | 0.84 (0.72–0.96) | <1.0E-6 | 0.78 (0.65–0.91) | 1.3E-05 | 0.61 (0.39–0.84) | 1.7E-01 | 0.55 (0.37–0.72) | 2.9E-01 |
| HOMER2 | 0.77 (0.68–0.86) | <1.0E-6 | 0.75 (0.62–0.87) | 6.2E-05 | 0.79 (0.67–0.90) | <1.0E-6 | 0.83 (0.66–1.00) | 7.2E-05 | 0.57 (0.43–0.70) | 1.7E-01 |
| ZNF583 | 0.69 (0.62–0.76) | <1.0E-6 | 0.70 (0.59–0.80) | 8.5E-05 | 0.68 (0.58–0.78) | 1.2E-04 | 0.56 (0.45–0.66) | 1.6E-01 | 0.58 (0.49–0.66) | 3.3E-02 |
| GJC1 | 0.66 (0.54–0.78) | 5.0E-03 | 0.72 (0.58–0.87) | 1.5E-03 | 0.60 (0.44–0.76) | 1.1E-01 | 0.38 (0.16–0.59) | 1.3E-01 | 0.55 (0.38–0.72) | 2.9E-01 |
| DOCK8 | 0.59 (0.49–0.68) | 4.0E-02 | 0.55 (0.43–0.67) | 1.9E-01 | 0.62 (0.50–0.74) | 2.7E-02 | 0.47 (0.35–0.59) | 3.1E-01 | 0.42 (0.35–0.49) | 9.8E-01 |
The ROC curve analysis results are shown for the detection of respective tissue class from control NCs. Mean AUROC and 95% CI are shown as well as the p-values corresponds to the comparison to non-discriminative curve.
Figure 3. The ROC curve-based assessment of methylation markers’ diagnostic accuracy.
A. The ROC curves representing the distinction of CRCs from control NC are shown for the seven loci demonstrating AUROC values >0.8. Mean and 95%CI of AUROC as well as p-value are shown in each panel. B. The ROC curves based on the multi-loci diagnostic panels are shown for the distinction of CRCs from control NCs. Gray solid line, and black solid line correspond to the profiles for the best single locus (ALX3) and the multi-locus panels (ALX3, ZNF583, miR34b, and VSX2), respectively.
DISCUSSION
This unbiased genome-wide methylomics scan identified 169 candidate hypermethylation targets in human primary CRCs. The validity of our method was supported by our finding significant hypermethylation ofpreviously reported genes undergoing hypermethylation in CRC, including SFRP2 (Huang et al. 2007; Muller et al. 2004; Nagasaka et al. 2009; Wang and Tang 2008). Individual qMSP assessment of systematically prioritized loci validated frequent hypermethylation in primary CRCs at 11 loci: VSX2, NPTX1, BEND4, ALX3, miR34b, BTG4, GLP1R, HOMER2, GJC1, DOCK8, and ZNF583. Infrequent but neoplasia-specific methylation was observed in 3 additional loci: NME4, TTLL12, and TMEM42. Hypermethylation at each of these 11 loci effectively discriminated CRCs from colonic mucosae of age-matched neoplasia-free cases (i.e., control NCs). Most of these loci exhibited high discriminative accuracy (i.e., AUROC>0.8 and p<1E-6), with VSX2 performing the best (AUROC=0.93). Multi-locus panels did not improve diagnostic accuracy relative to VSX2 alone, but combination with existing CRC detection markers could still be tested in future studies. Methylation levels of VSX2, NPTX1, BEND4, miR34b, and HOMER2 also significantly differentiated adenomas from control NCs (AUROC 0.74–0.83) and may constitute ideal markers for early-stage disease detection and/or risk stratification. The observed AUROC values for CRC and adenoma discrimination were very high even under current study conditions (i.e., use of age-matched control cases and lack of tumor cell enrichment by microdissection, which enhance methylation-based discriminative accuracy). Therefore, we believe that these loci merit a large scale independent validation study as well as study for their use as biomarkers for stool- and plasma-based CRC detection.
It is also notable that CRC cases, regardless of their CIMP status, were distinguished from age-matched neoplasia-free cases based on hypermethylation of non-neoplastic colonic mucosae at certain loci (such as ALX3). This finding is reminiscent of recent reports showing that CRC-associated hypermethylation target loci are mildly hypermethylated in non-neoplastic colonic mucosae from colonic neoplasia patients (Ahlquist, et al. 2008b; Belshaw, et al. 2008; Menigatti, et al. 2007; Worthley, et al. 2010). However, in these published reports, differential methylation of non-neoplastic mucosae was CIMP (+) neoplasia case-specific, or based on data from non-age-matched subjects. Our findings in non-neoplastic mucosae support the notion that CRC-associated hypermethylation initiates at an early, non-neoplastic stage, representing a widespread “field defect” (Belshaw et al. 2010; Nosho et al. 2009; Shen et al. 2005; Svrcek et al. 2010). These non-neoplastic mucosal methylation events should be clinically translatable into CRC risk prediction, by using non-neoplastic colonic or rectal mucosa as an analytic substrate. Moreover, CRC detection markers whose CRC-associated hypermethylation initiates at non-neoplastic stage may perform better in stool DNA-based tests than in primary tissue DNA-based tests, since stool DNA is derived from both non-neoplastic and neoplastic colonic mucosal cells. Further investigation of this concept is now indicated.
The current MCAM study also detected CRC-associated hypermethylation of multiple previously published CRC-specific methylation markers, including the most extensively studied methylation marker to date, vimentin (Ahlquist, et al. 2008a; Baek, et al. 2009; Chen, et al. 2005; Itzkowitz, et al. 2007; Li, et al. 2009). However, these markers, except for SFRP2, demonstrated methylation overlap between CRCs and NCs in our MCAM tissue cohort, and thus did not satisfy our selection criteria. Estecio et al. also performed MCAM on CRCs mainly focusing on CIMP class-based profiling, and reported hypermethylation of BARHL1 and RSHL1 (Estecio et al. 2007). Our MCAM study verified significant CRC-associated hypermethylation of BARHL1, but not of RSHL1. We designed our selection criteria to eliminate CRC-associated hypermethylation targets that were also moderately methylated in non-neoplastic colonic mucosae of neoplasia-free cases, since they would not be anticipated to perform well as stool biomarkers, due to normal DNA contamination in stool DNA. As proof-of-principle of the success of our strategy, the current candidates did not include previously reported targets exhibiting this type of methylation (e.g., SST and CAV1, which were previously identified in our own pharmacological unmasking study) (Mori et al. 2006).
The current study represents the first report of neoplasia-associated hypermethylation of VSX2, BEND4, GLP1R, HOMER2, GJC1, ZNF583, and NME4 in any tumor type. Among the other loci identified by our unbiased scanning strategy, cancer-specific hypermethylation at the miR-34b-BTG4 locus has been documented in multiple primary tumors, including CRC (Dong, et al. 2009; Kozaki, et al. 2008; Lujambio, et al. 2008; Toyota et al. 2008). Similarly, NPTX1 methylation has been reported in cancers of the pancreas and cervix (Hagihara, et al. 2004; Ongenaert, et al. 2008; Yang, et al. 2009). ALX3 methylation has been reported in neuroblastoma, and hypermethylation of another member of the same gene family, ALX4, showed promise as a CRC detection biomarker(Ebert, et al. 2006; Tanzer, et al. 2010; Wimmer, et al. 2002). Additionally, epigenetic downregulation of DOCK8 has been implicated in lung cancer (Takahashi, et al. 2006). These reports indicate that the cancer-associated hypermethylation of many loci is involved in malignancies arising from different cell lineages. Thus, the loci detected in the current study should also be explored for use as broad-spectrum malignancy biomarkers, especially in blood-based detection studies.
DNA hypermethylation overlapping gene promoter regions is often associated with abnormal transcriptional silencing (Fraga and Esteller 2007). The loci miR34b and BTG4 closely flank each other and overlap with a bidirectional promoter that can regulate the expression of both miR34b and BTG4 (reverse orientation; Toyota et al. 2008). Both genes exhibit promoter methylation-mediated gene silencing, along with tumor-suppressive properties, in vitro and in vivo (Lujambio et al. 2008; Toyota et al. 2008). Nevertheless, miR34b has been suggested as the principal transcript of this promoter in colonic epithelium (Toyota et al. 2008). Interestingly, miR34b was the only locus in the current study that was hypermethylated equally in both adenomas and CRCs. Taken together, these published and current findings suggest that de novo epigenetic silencing of miR34b is involved in the early stages of colorectal neoplastic progression.
Four additional CRC-specific hypermethylation targets were located within promoter regions: NPTX1, DOCK8, GLP1R, and ZNF583. ZNF583 and DOCK8 downregulation are associated with insensitivity to chemoradiotherapy in esophageal cancer (Maher, et al. 2009; Ogawa, et al. 2008). Thus, it is plausible that hypermethylation at one or more of these loci, in addition to miR34b, contributes to colonic neoplastic progression. Considering their links to chemosensitivity, ZNF583 and DOCK8 hypermethylation may also mark tumors with a poor prognosis or therapeutic response. A different cohort design will be necessary to further investigate these potentially important topics. NPTX1 and GLP1R are involved in endocrine pathways that have been linked to CRC. NPTX1 is downregulated by pharmacological inhibition of estrogen signaling, indicating that NPTX1 is a downstream effector of estrogen (Gomes, et al. 2011; Yasuhara, et al. 2008). Estrogen has been suggested to protect against CRC development (Hogan, et al. 2009; Lin, et al. 2011), and epigenetic inactivation of estrogen receptor alpha (ESR1) has been widely observed in colonic mucosae of aged individuals as well as in CRCs (Issa et al. 1994). NPTX1 downregulation is also associated with cell immortalization (Hiyama, et al. 2008), thus NPTX1 might contribute to the anticancer effects of estrogen. GLP1R signaling is linked diabetes mellitus by its ability to promote insulin production and dietary fat-induced insulin resistance (Ayala, et al. 2010). Notably, diabetes mellitus is associated with an elevated CRC risk (reviewed in (Larsson, et al. 2005)). GLP1R downregulation is induced by longstanding hyperglycemia and has been suggested to augment cellular responses to mitogenic signaling (Hadjiyanni, et al. 2010; Xu, et al. 2007). GLP1R is expressed in normal colon (Campos, et al. 1994), thus it is plausible that epigenetic downregulation of GLP1R is involved in the insulin-related carcinogenic mechanism. Further functional studies are indicated to evaluate the potential relevance of these two endocrine-related genes in colon carcinogenesis.
Three of the 14 qMSP-analyzed loci demonstrated only infrequent CRC-specific methylation. Furthermore, qualitative MSP failed to detect candidate CRC-associated hypermethylation at four of 20 loci that were identified and prioritized based on MCAM data. qMSP is a robust and sensitive methylation assay method that is applicable to a wide variety of target sequences and is directly translatable to clinical settings. However, qMSP (and, to a lesser degree, MSP) is less sensitive in detecting diffuse methylation events than is MCAM, because MCAM detection depends only on the methylation of CpGs within 6-base restriction enzyme recognition sites, while qMSP detection depends on the continuous methylation of multiple CpGs within a PCR amplicon. Additionally, some CpG dinucleotides assessed by MCAM could not be included in regions of interest for MSP assays, due to flanking sequence characteristics preventing adequate MSP amplification. This type of MSP assays might have failed to detect segmental methylation that was detectable by MCAM. Therefore, we speculate that presence of diffuse or segmental methylation contributed to this discrepancy between assays. Application of assay methods that can assess diffuse methylation (e.g., bisulfite pyrosequencing) to these loci may reveal additional CRC-associated hypermethylation targets.
The current study possesses some limitations due to cohort characteristics. This study did not assess methylation in association with chronic inflammation (e.g., IBD; Itzkowitz and Yio 2004). However, we considered IBD-associated hypermethylation to be unlikely to compromise the current study’s major focus, average-risk CRC screening: IBD patients undergo periodic endoscopic surveillance, making them unlikely to participate in average-risk screening (Itzkowitz, et al. 2005). The current study may also have failed to detect methylation markers unique to female CRC cases, since both our neoplastic and control cohorts were predominantly male due to patient demographics at the participating clinics. Additional potential confounding variables (e.g., folate intake, alcohol consumption, obesity, and race) were not addressed, because our control case number was not sufficiently large for this type of analysis. Thus, it will be desirable to further validate the current findings in an independent larger cohort. Additionally, including adenomas in the MCAM cohort could have further improved enrichment for novel markers that are highly methylated in adenomas.
In summary, the current study has successfully applied an unbiased, extensive genome-wide scanning strategy to discover neoplasia-specific methylation targets in the colon, identifying 169 candidate novel loci. Quantitative PCR-based analysis of prioritized loci in a larger patient cohort revealed that methylation events at 11 loci were accurate in distinguishing both neoplastic and non-neoplastic colonic mucosae of colonic neoplasia patients from control colonic mucosae of neoplasia-free patients. Two of these genes have been implicated in endocrine-related carcinogenesis. Methylation at these loci now merits further investigation in studies of independent cohort validation, stool- and plasma-based CRC detection, as well as in the evaluation of non-neoplastic mucosa for field defects, potentially indicating increased CRC susceptibility.
Supplementary Material
Acknowledgments
Funding: U01CA084986 (National Institute of Cancer; Y.M.), Senior Research Award (Crohn’s and Colitis Foundation of America; Y.M.), Wendy Will Case Fund (Y.M.), R01CA0133012 (National Cancer Institute; S.J.M.), and K08DK078046 (National Institute of Diabetes and Digestive and Kidney Diseases, J.H.K.).
We thank all nurses and technicians at the Johns Hopkins Greenspring Station Endoscopy Suite, the Johns Hopkins Outpatient Center Endoscopy Suite, and the Johns Hopkins Bayview Medical Center Operating Room for their assistance with sample acquisition. We also thank Stefan David, M.D., Bogdan C. Paun, M.D., Divya Singh, B.E., Ms. Christine Vazquez, Ms. Irina Khurana (Johns Hopkins University), and Mr. Julio David Vega (University of Maryland, Baltimore County) for their assistance with sample processing.
Abbreviations
- CRC
colorectal cancer
- NC
normal colonic mucosa
- CGI
CpG island
- CIMP
CpG island methylator phenotype
- MSI
microsatellite instability
- MSP
methylation-specific PCR
- qMSP
real-time quantitative MSP
- MCAM
methylated CpG island amplification coupled with microarray
- ROC
receiver-operator characteristic
- AUROC
area under ROC curve
- BEND4
BEN domain containing 4
- VSX2
visual system homeobox 2
- ALX3
ALX homeobox 3
- NPTX1
neuronal pentraxin I
- GLP1R
glucagon-like peptide 1 receptor
- HOMER2
homer homolog 2
- GJC1
gap junction protein, gamma 1
- ZNF583
zinc finger protein 583
- DOCK8
dedicator of cytokinesis 8
- TMEM42
transmembrane protein 42
- NME4
non-metastatic cells 4
- TTLL12
tubulin tyrosine ligase-like family, member 12
- CI
confidence interval
Footnotes
Declarations of interest: There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author Contributions: Study design (Y.M. and S.J.M.); Sample cohort development (Y.M., Y.C., J.Y., F.S.M., M.L., J.H.K., S.R.B., M.R.M., D.F.H., M.D.D., and S.J.M.); Experimental and analytical protocol development (Y.M., A.V.O., R.A., J.Y., and W.Y.); Bench-top experiments (Y.M., A.V.O., Y.C., D.L., and W.Y.); Data analysis (Y.M. and J.Y.); Manuscript preparation (Y.M., F.M.S., S.H., M.L., S.R.B., A.J.G., and S.J.M.).
Disclaimer.
This is not the definitive version of record of this article. This manuscript has been accepted for publication in Endocrine-Related Cancer, but the version presented here has not yet been copy edited, formatted or proofed. Consequently, the Society for Endocrinology accepts no responsibility for any errors or omissions it may contain. The definitive version is now freely available at http://erc.endocrinology-journals.org/content/18/4/465 or doi: 10.1530/ERC-11-0083 (2011), Society for Endocrinology.
References
- Ahlquist DA, Sargent DJ, Loprinzi CL, Levin TR, Rex DK, Ahnen DJ, Knigge K, Lance MP, Burgart LJ, Hamilton SR, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med. 2008a;149:441–450. W481. doi: 10.7326/0003-4819-149-7-200810070-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist T, Lind GE, Costa VL, Meling GI, Vatn M, Hoff GS, Rognum TO, Skotheim RI, Thiis-Evensen E, Lothe RA. Gene methylation profiles of normal mucosa, and benign and malignant colorectal tumors identify early onset markers. Mol Cancer. 2008b;7:94. doi: 10.1186/1476-4598-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–5494. [PubMed] [Google Scholar]
- Ayala JE, Bracy DP, James FD, Burmeister MA, Wasserman DH, Drucker DJ. Glucagon-like peptide-1 receptor knockout mice are protected from high-fat diet-induced insulin resistance. Endocrinology. 2010;151:4678–4687. doi: 10.1210/en.2010-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek YH, Chang E, Kim YJ, Kim BK, Sohn JH, Park DI. Stool methylation-specific polymerase chain reaction assay for the detection of colorectal neoplasia in Korean patients. Dis Colon Rectum. 2009;52:1452–1459. doi: 10.1007/DCR.0b013e3181a79533. discussion 1459–1463. [DOI] [PubMed] [Google Scholar]
- Belshaw NJ, Elliott GO, Foxall RJ, Dainty JR, Pal N, Coupe A, Garg D, Bradburn DM, Mathers JC, Johnson IT. Profiling CpG island field methylation in both morphologically normal and neoplastic human colonic mucosa. Br J Cancer. 2008;99:136–142. doi: 10.1038/sj.bjc.6604432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw NJ, Pal N, Tapp HS, Dainty JR, Lewis MP, Williams MR, Lund EK, Johnson IT. Patterns of DNA methylation in individual colonic crypts reveal aging and cancer-related field defects in the morphologically normal mucosa. Carcinogenesis. 2010;31:1158–1163. doi: 10.1093/carcin/bgq077. [DOI] [PubMed] [Google Scholar]
- Campos RV, Lee YC, Drucker DJ. Divergent tissue-specific and developmental expression of receptors for glucagon and glucagon-like peptide-1 in the mouse. Endocrinology. 1994;134:2156–2164. doi: 10.1210/endo.134.5.8156917. [DOI] [PubMed] [Google Scholar]
- Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, Platzer P, Lu S, Dawson D, Willis J, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–1132. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- Dong W, Tu S, Xie J, Sun P, Wu Y, Wang L. Frequent promoter hypermethylation and transcriptional downregulation of BTG4 gene in gastric cancer. Biochem Biophys Res Commun. 2009;387:132–138. doi: 10.1016/j.bbrc.2009.06.140. [DOI] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MP, Model F, Mooney S, Hale K, Lograsso J, Tonnes-Priddy L, Hoffmann J, Csepregi A, Rocken C, Molnar B, et al. Aristaless-like homeobox-4 gene methylation is a potential marker for colorectal adenocarcinomas. Gastroenterology. 2006;131:1418–1430. doi: 10.1053/j.gastro.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, Issa JP. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–1536. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Gomes GR, Yasuhara F, Siu ER, Fernandes SA, Avellar MC, Lazari MF, Porto CS. In vivo treatments with fulvestrant and anastrozole differentially affect gene expression in the rat efferent ductules. Biol Reprod. 2011;84:52–61. doi: 10.1095/biolreprod.110.085340. [DOI] [PubMed] [Google Scholar]
- Hadjiyanni I, Siminovitch KA, Danska JS, Drucker DJ. Glucagon-like peptide-1 receptor signalling selectively regulates murine lymphocyte proliferation and maintenance of peripheral regulatory T cells. Diabetologia. 2010;53:730–740. doi: 10.1007/s00125-009-1643-x. [DOI] [PubMed] [Google Scholar]
- Hagihara A, Miyamoto K, Furuta J, Hiraoka N, Wakazono K, Seki S, Fukushima S, Tsao MS, Sugimura T, Ushijima T. Identification of 27 5′ CpG islands aberrantly methylated and 13 genes silenced in human pancreatic cancers. Oncogene. 2004;23:8705–8710. doi: 10.1038/sj.onc.1207783. [DOI] [PubMed] [Google Scholar]
- Hiyama K, Tanimoto K, Nishimura Y, Tsugane M, Fukuba I, Sotomaru Y, Hiyama E, Nishiyama M. Exploration of the genes responsible for unlimited proliferation of immortalized lung fibroblasts. Exp Lung Res. 2008;34:373–390. doi: 10.1080/01902140802221912. [DOI] [PubMed] [Google Scholar]
- Hogan AM, Collins D, Baird AW, Winter DC. Estrogen and gastrointestinal malignancy. Mol Cell Endocrinol. 2009;307:19–24. doi: 10.1016/j.mce.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Huang Z, Li L, Wang J. Hypermethylation of SFRP2 as a potential marker for stool-based detection of colorectal cancer and precancerous lesions. Dig Dis Sci. 2007;52:2287–2291. doi: 10.1007/s10620-007-9755-y. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, 3rd, Sontag S, Johnson D, Skoletsky J, Durkee K, Markowitz S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–117. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Itzkowitz SH, Present DH Crohn’s & Colitis Foundation of America Colon Cancer in IBDSG. Consensus conference: Colorectal cancer screening and surveillance in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:314–321. doi: 10.1097/01.mib.0000160811.76729.d5. [DOI] [PubMed] [Google Scholar]
- Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol. 2004;287:G7–17. doi: 10.1152/ajpgi.00079.2004. [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Hao Y, Murray T, Thun MJ. Cancer statistics 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Kahi CJ, Rex DK, Imperiale TF. Screening, surveillance, and primary prevention for colorectal cancer: a review of the recent literature. Gastroenterology. 2008;135:380–399. doi: 10.1053/j.gastro.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Kim YH, Petko Z, Dzieciatkowski S, Lin L, Ghiassi M, Stain S, Chapman WC, Washington MK, Willis J, Markowitz SD, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–789. doi: 10.1002/gcc.20341. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Larsson SC, Orsini N, Wolk A. Diabetes mellitus and risk of colorectal cancer: a meta-analysis. J Natl Cancer Inst. 2005;97:1679–1687. doi: 10.1093/jnci/dji375. [DOI] [PubMed] [Google Scholar]
- Li M, Chen WD, Papadopoulos N, Goodman SN, Bjerregaard NC, Laurberg S, Levin B, Juhl H, Arber N, Moinova H, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol. 2009;27:858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KJ, Cheung WY, Lai JY, Giovannucci EL. The effect of estrogen versus combined estrogen-progestogen therapy on the risk of colorectal cancer. Int J Cancer. 2011 doi: 10.1002/ijc.26026. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher SG, Gillham CM, Duggan SP, Smyth PC, Miller N, Muldoon C, O’Byrne KJ, Sheils OM, Hollywood D, Reynolds JV. Gene expression analysis of diagnostic biopsies predicts pathological response to neoadjuvant chemoradiotherapy of esophageal cancer. Ann Surg. 2009;250:729–737. doi: 10.1097/SLA.0b013e3181bce7e1. [DOI] [PubMed] [Google Scholar]
- Menigatti M, Pedroni M, Verrone AM, Borghi F, Scarselli A, Benatti P, Losi L, Di Gregorio C, Schar P, Marra G, et al. O6-methylguanine-DNA methyltransferase promoter hypermethylation in colorectal carcinogenesis. Oncol Rep. 2007;17:1421–1427. [PubMed] [Google Scholar]
- Mori Y, Cai K, Cheng Y, Wang S, Paun B, Hamilton JP, Jin Z, Sato F, Berki AT, Kan T, et al. A genome-wide search identifies epigenetic silencing of somatostatin, tachykinin-1, and 5 other genes in colon cancer. Gastroenterology. 2006;131:797–808. doi: 10.1053/j.gastro.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Muller HM, Oberwalder M, Fiegl H, Morandell M, Goebel G, Zitt M, Muhlthaler M, Ofner D, Margreiter R, Widschwendter M. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363:1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- Nagasaka T, Tanaka N, Cullings HM, Sun DS, Sasamoto H, Uchida T, Koi M, Nishida N, Naomoto Y, Boland CR, et al. Analysis of fecal DNA methylation to detect gastrointestinal neoplasia. J Natl Cancer Inst. 2009;101:1244–1258. doi: 10.1093/jnci/djp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosho K, Kure S, Irahara N, Shima K, Baba Y, Spiegelman D, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620. e1601–1603. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa R, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Mori Y, Mori R, Tomoda K, Katada T, Harada K, et al. Identification of candidate genes involved in the radiosensitivity of esophageal cancer cells by microarray analysis. Dis Esophagus. 2008;21:288–297. doi: 10.1111/j.1442-2050.2007.00759.x. [DOI] [PubMed] [Google Scholar]
- Ongenaert M, Wisman GB, Volders HH, Koning AJ, Zee AG, van Criekinge W, Schuuring E. Discovery of DNA methylation markers in cervical cancer using relaxation ranking. BMC Med Genomics. 2008;1:57. doi: 10.1186/1755-8794-1-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Rosner GL, Xiao L, Hernandez NS, Vilaythong J, Houlihan PS, Krouse RS, Prasad AR, Einspahr JG, et al. MGMT promoter methylation and field defect in sporadic colorectal cancer. J Natl Cancer Inst. 2005;97:1330–1338. doi: 10.1093/jnci/dji275. [DOI] [PubMed] [Google Scholar]
- Svrcek M, Buhard O, Colas C, Coulet F, Dumont S, Massaoudi I, Lamri A, Hamelin R, Cosnes J, Oliveira C, et al. Methylation tolerance due to an O6-methylguanine DNA methyltransferase (MGMT) field defect in the colonic mucosa: an initiating step in the development of mismatch repair-deficient colorectal cancers. Gut. 2010;59:1516–1526. doi: 10.1136/gut.2009.194787. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kohno T, Ajima R, Sasaki H, Minna JD, Fujiwara T, Tanaka N, Yokota J. Homozygous deletion and reduced expression of the DOCK8 gene in human lung cancer. Int J Oncol. 2006;28:321–328. [PubMed] [Google Scholar]
- Tanzer M, Balluff B, Distler J, Hale K, Leodolter A, Rocken C, Molnar B, Schmid R, Lofton-Day C, Schuster T, et al. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PLoS One. 2010;5:e9061. doi: 10.1371/journal.pone.0009061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Wang DR, Tang D. Hypermethylated SFRP2 gene in fecal DNA is a high potential biomarker for colorectal cancer noninvasive screening. World J Gastroenterol. 2008;14:524–531. doi: 10.3748/wjg.14.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Wimmer K, Zhu XX, Rouillard JM, Ambros PF, Lamb BJ, Kuick R, Eckart M, Weinhausl A, Fonatsch C, Hanash SM. Combined restriction landmark genomic scanning and virtual genome scans identify a novel human homeobox gene, ALX3, that is hypermethylated in neuroblastoma. Genes Chromosomes Cancer. 2002;33:285–294. doi: 10.1002/gcc.10030. [DOI] [PubMed] [Google Scholar]
- Worthley DL, Whitehall VL, Buttenshaw RL, Irahara N, Greco SA, Ramsnes I, Mallitt KA, Le Leu RK, Winter J, Hu Y, et al. DNA methylation within the normal colorectal mucosa is associated with pathway-specific predisposition to cancer. Oncogene. 2010;29:1653–1662. doi: 10.1038/onc.2009.449. [DOI] [PubMed] [Google Scholar]
- Xu G, Kaneto H, Laybutt DR, Duvivier-Kali VF, Trivedi N, Suzuma K, King GL, Weir GC, Bonner-Weir S. Downregulation of GLP-1 and GIP receptor expression by hyperglycemia: possible contribution to impaired incretin effects in diabetes. Diabetes. 2007;56:1551–1558. doi: 10.2337/db06-1033. [DOI] [PubMed] [Google Scholar]
- Yang N, Eijsink JJ, Lendvai A, Volders HH, Klip H, Buikema HJ, van Hemel BM, Schuuring E, van der Zee AG, Wisman GB. Methylation markers for CCNA1 and C13ORF18 are strongly associated with high-grade cervical intraepithelial neoplasia and cervical cancer in cervical scrapings. Cancer Epidemiol Biomarkers Prev. 2009;18:3000–3007. doi: 10.1158/1055-9965.EPI-09-0405. [DOI] [PubMed] [Google Scholar]
- Yasuhara F, Gomes GR, Siu ER, Suenaga CI, Marostica E, Porto CS, Lazari MF. Effects of the antiestrogen fulvestrant (ICI 182,780) on gene expression of the rat efferent ductules. Biol Reprod. 2008;79:432–441. doi: 10.1095/biolreprod.107.067413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.