Abstract
One of the persistent problems in biology is understanding how genetic variation contributes to phenotypic variation. Associations at many levels have been reported, and yet causal inference has remained elusive. We propose to rely on the knowledge of causal relationships established by molecular biology approaches. The existing molecular knowledge forms a firm backbone upon which hypotheses connecting genetic variation, transcriptional variation and phenotypic variation can be built. The sex determination pathway is a well-established molecular network, with the Yolk protein 1–3 (Yp) genes as the most downstream target. Our analyses reveal that genetic variation in expression for genes known to be upstream in the pathway explains variation in downstream targets. Relationships differ between the two sexes, and each Yp has a distinct transcriptional pattern. Yp expression is significantly negatively correlated with longevity, an important life history trait, for both males and females.
Keywords: systems biology, sex determination, ecdysone, aging, quantitative genetics, Yolk protein
Introduction
Genomic tools have enabled the simultaneous evaluation of many gene expression levels within a genome, making the study of transcriptional networks possible (Featherstone and Broadie, 2002; Guthke et al., 2005). There is evidence of genetic variation in expression networks (Tarone et al., 2005; Sieberts and Schadt, 2007; Nuzhdin et al., 2009), and genetic variation in transcription has been used as a systems biology tool for mapping large groups of genes underlying human diseases (Schadt et al., 2009a, 2009b) in a ‘top down' approach. These large networks help to pinpoint specific genes, but a problem remains as covariation in gene expression should not be confused with causality (Coffman et al., 2005; Lee et al., 2009). For example, gene expression levels will be correlated with each other when there is a causal relationship, such as when one gene is upstream of the other. Expression levels will also be correlated when two genes are regulated by a common factor (Jansen et al., 2009). When large numbers of genes are examined with small sample number of genotypes, the sheer complexity of organism-wide networks interfere with the identification of causal relationships (Coffman et al., 2005; Jansen et al., 2009). We propose a novel ‘bottom up' approach, relying on causal relationships among genes having been already established in prior molecular biological experiments. We apply this paradigm for natural small-effect mutations where the networks have been established using mutations of major effect. It remains an open question as to whether natural genetic variation recapitulates a molecular network structure and can be related to phenotypic variation (see Box 3 in Harshman and Zera (2007) and Stern (2000)). In addition, the nature of the response to expression variation (that is, linear versus nonlinear) may be important to network outcomes (Gjuvsland et al., 2007a, 2007b, 2007c). Here, we address these challenges using the Yolk protein (Yp) expression network as an exemplar.
The Yp expression network of Drosophila melanogaster (Figure 1) is well described (Ota et al., 1981; Belote et al., 1985; Kraus et al., 1988; Baker et al., 1989; Burtis et al., 1991; Abrahamsen et al., 1993; An and Wensink, 1995; Bownes et al., 1996; Cline and Meyer, 1996; Erdman et al., 1996; Li and Baker, 1998; Brodu et al., 2001; Hutson and Bownes, 2003; Kumar and Lopez, 2005; Billeter et al., 2006; Goldman and Arbeitman, 2007; Qi et al., 2007; Sanchez, 2008; Lebo et al., 2009). Consequently, causal relationships among genes in the network are known. Yp1–3 provide the major components of egg yolk (Bownes, 1992) and are located on the X chromosome. The Yp network has a number of distinct advantages (1) many mutants are not lethal and demonstrate intermediate phenotypes (Cline and Meyer, 1996), (2) dosage effects have been established with mutant experiments (that is, Li and Baker, 1998) and (3) a genetic component has been demonstrated for expression variation in all parts of the sex determination hierarchy, including mis-spliced isoforms of dsx (Tarone et al., 2005), that correlate with Yp1 expression in males. Accordingly, this set of well-established causal relationships among genes in the network regulating Yps, and quantitative responses to upstream variation in known causal factors, makes the Yp expression network an ideal system for evaluating the effects of natural variation from a systems biology perspective.
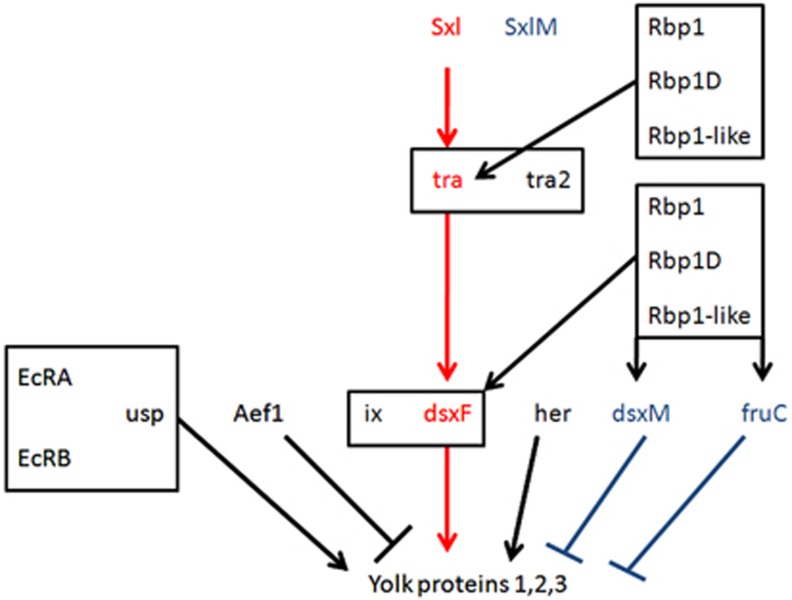
Figure 1.
A diagram of known regulatory interactions in the Yp expression network. Sxl, tra, tra2 and the Rbps are splicing factors, all other genes regulate expression. Arrows indicate activation of expression/splicing. Bars indicate inhibition of expression/splicing. Boxes indicate groups of physically interacting proteins. Red and blue represent alternative splices that are canonically considered to be expressed in a sex-specific manner (male, blue; female, red), though their expression is known to be leaky. Boxes represent genes that function together in complexes to influence downstream expression/splicing.
A detailed understanding of natural variation in the Yp network could also provide insight into several interesting biological problems. Several loci in the network, including all of the Yps, are located on the X chromosome, which is upregulated approximately twofold in Drosophila males compared with females (Hamada et al., 2005; Kind et al., 2008). Moreover, in the data analyzed here the X chromosome is enriched for genes that show additive genetic inheritance of gene expression in Drosophila males, but dominance variation in females (Wayne et al., 2007). This pattern may suggest nonlinear responses in the network (Gjuvsland et al., 2007a, 2007b, 2007c), variation in dosage compensation (Goldman and Arbeitman, 2007; Lebo et al., 2009) or a combination of both factors. In the Yp network, Sxl is the most upstream. Sxl is also the most upstream factor in the sex determination splicing cascade and it regulates dosage compensation (Cline and Meyer, 1996; Sanchez, 2008).
Manipulation of expression of genes in the Yp network has been demonstrated to affect indicators of fecundity such as fertility, hatch rates and egg numbers in females (Postlethwait and Shirk, 1981; Bownes et al., 1991; Terashima and Bownes, 2004). The Yps are the major component of eggs and their expression is a good indicator of females with high fertility and fecundity (Bownes et al., 1991; Terashima and Bownes, 2004). Reproduction is also known to result in a complex tradeoff with lifespan under some conditions (Rose, 1989; Djawdan et al., 1996; Partridge et al., 2005; Flatt et al., 2008; Toivonen and Partridge, 2009; Kenyon, 2010). There is also some evidence that would suggest Yps may have a role in longevity. Yps have been shown to bind ecdysone (Bownes et al., 1988), indicating that they may help relay hormonal signals that may in turn influence longevity (Simon et al., 2003; Tatar, 2004). Further, Yps are evolutionarily convergent with vitellogenins (Brandt et al., 2005; Tufail and Takeda, 2008, 2009) and recent evidence from studies of hymenoptera suggests that vitellogenins have a role in extending longevity (Brandt et al., 2005; Seehuus et al., 2006; Corona et al., 2007). In the hymenoptera, long-lived castes have been shown to express more vitellogenin than shorter-lived castes, which has been linked to a reduction in oxidative stress (Landis et al., 2004; Seehuus et al., 2006) as well as immune function (Amdam et al., 2004). For these reasons, studying natural expression variation in the Yp network may provide valuable information regarding the disparity in the inheritance of Drosophila gene expression levels between the sexes and could provide molecular mechanisms for life history tradeoffs (Chippindale et al., 2001).
The causal network, established by molecular biology techniques, combined with demonstrated links between large effect mutations and important life history traits provides the perfect backdrop upon which to test a ‘bottom up' approach to understanding how genetic variation contributes to phenotypic variation. If genetic variation in these networks exists, and is important, then variation in upstream genes will be associated with variation in downstream targets. If the network itself is important for life history traits then the most downstream targets (the Yps) will be associated with longevity. Further this backdrop allows us to ask structural questions about the system such as: (1) Do nonlinear responses reflect patterns of genetic variation in gene expression? (2) Are the relationships among genes in the Yp network the same in both sexes? (3) Does each Yp respond to variation in the same regulators? (4) Given the X-chromosomal positions of genes in the network (the Yps, usp, Rbp1-like and Sxl), is dosage compensation important to the male Yp network? Using a set of expression data from 72 genotypes, progeny from a diallel cross, Wayne et al. (2007), and longevity data from the same crosses, genetic variation in the Yp network and that impact on longevity is explored. The regulatory structure of the existing network is modeled using structural equation models (SEMs) derived from Wright's pathway analyses (Pearl, 2000). Structural equation modeling is distinct from regression modeling in that it is composed of a system of equations. In some equations a particular variable is independent, whereas others are dependent. In this ‘bottom up' approach, we establish a network of equations derived from molecular experiments that establish causal relationships among genes and then test whether the genetic variation is reflected in the relationships dictated by the molecular models.
Materials and Methods
Microarray gene expression data for the Yp network
Data from Wayne et al. (2007) were used for these analyses and results from this experiment for Yp network genes (Figure 1) can be observed in Table 1, Figure 2, Supplementary File 1 and Supplementary File 2. Reciprocal crosses of nine different inbred genotypes isolated from Winters, CA, USA were obtained and Agilent microarrays (Agilent, Santa Clara, CA, USA) were used to assay expression. At 3 days post-eclosion, RNA was isolated from whole carcasses of 10 males and 10 females derived from the full matrix of crosses among genotypes (minus the homozogotes). This scheme allows for the dissection of simple versus complex forms of inheritance. On a genome-wide scale, this experiment demonstrated a simple mode of gene expression inheritance for males (where offspring appear to express a gene at levels that are intermediate to parental expression levels), but a more complex pattern for females. Expression level data were derived from the natural log of microarray probe intensities after subtracting the mean background intensity and were only evaluated if the probe was determined to be expressed significantly beyond background levels (Wayne et al., 2007). Data entered into the SEMs were centered by dye and scaled such that the mean was zero. Maternal genotype effects were then regressed out of the data by extracting and evaluating the residuals from models of expression based on maternal genotype. Centering does not change the relationship between genes, but is important for multivariate depiction of the data (Neter et al., 1990).
Table 1. The proportion of crosses for which expression levels of a probe was not significantly above background levels.
| Gene | Probeuid | Males | Females |
|---|---|---|---|
| Aef1 | 1070 | 0.1 | 0 |
| dsxF | 6162 | 0.47 | 0.07 |
| dsxM | 12690 | 0 | 0.01 |
| EcRA | 5648 | 0.5 | 0.08 |
| EcRB | 14630 | 0.18 | 0.13 |
| fruC | 9294 | 0 | 0 |
| her | 4988 | 0 | 0 |
| ix | 5511 | 0.46 | 0.04 |
| Rbp1 | 7014 | 0 | 0 |
| Rbp1D | 12045 | 0 | 0 |
| Rbp1-like | 4731 | 0 | 0 |
| Sxl | 782 | 0 | 0 |
| SxlM | 824 | 0.11 | 0.19 |
| tra | 9174 | 0 | 0 |
| tra2 | 4734 | 0.03 | 0 |
| usp | 12140 | 0.03 | 0 |
| Yp1 | 13974 | 0 | 0 |
| Yp2 | 13101 | 0.88 | 0.01 |
| Yp3 | 2812 | 0 | 0 |
Probeuid indicates the unique identifier for a probe on the arrays associated with specific isoforms of genes in the genome. Males denote the proportion of males that exhibited expression that was statistically above background on the microarray for that probe. Females indicate the same information for the probe, but for females.
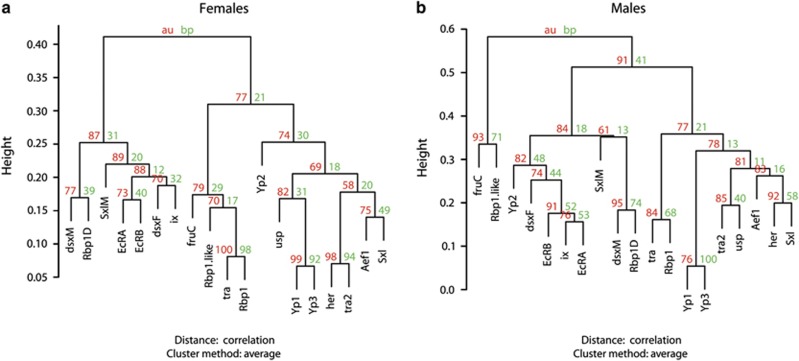
Figure 2.
Bootstrapped hierarchical clustering of Yp network transcripts in males and females. Red values reflect approximate unbiased probabilities determined through hierarchical bootstrapping. Green values reflect bootstrap values. (a=Females). Clustering of Rbp1/tra, Yp1/Yp3 and her/tra2 is significant. (b=Males). Clustering of Yp1/Yp3, fruC/Rbp1-like, Rbp1D/dsxM and EcRA/EcRB1/ix is significant.
Dosage compensation
Dosage compensation effects for genes in the Yp network were evaluated from two previously published data sets. The first study evaluated the genome-wide effects of msl-2 RNAi (Hamada et al., 2005). Results from the Yp network were extracted and assessed here. Also, the effect of functional Sxl in tra pseudomales on genes in the network was evaluated by comparing ratios of male/female expression to pseudomale/female expression with a t-test, as reported by the authors of that manuscript (Goldman and Arbeitman, 2007).
Genetic variation and heritability
Heritability estimates and the significance of variance components for each probe analyzed were available (Wayne et al., 2007), and can be found in Supplementary File 2. General combining ability (GCA) describes additive inheritance, specific combining ability (SCA) describes non-additive (dominance or epistasis) inheritance, and reciprocal values for these terms (RGCA, RSCA) denote inheritance of expression levels that are dependent on parental effects (Wayne et al., 2007). A nominal P-value of ⩽0.05 was considered significant. This approach may be liberal, in that some results that are significant may be false-positives. For this reason, we rely primarily on the consensus results across analytical approaches rather than individual tests.
The Yp network
The probes evaluated corresponded to loci known to affect Yp expression, sex determination genes and ecdysone receptor genes. The sex determination proteins encoded by dsxM and dsxF, ix, her and fruC have all been implicated in the regulation of Yps (Belote et al., 1985; Kraus et al., 1988; Abrahamsen et al., 1993; An and Wensink, 1995; Bownes et al., 1996; Hutson and Bownes, 2003; Goldman and Arbeitman, 2007; Lebo et al., 2009). The involvement of fru in Yp regulation has been implied recently, based on the observation that ectopic fruB expression, driven by a heat shock promoter, increases Yp expression and deletion of the male-specific fru P1 promoter increases expression of Yp (Goldman and Arbeitman, 2007; Lebo et al., 2009). These two pieces of evidence suggest different regulatory roles (positive and negative regulation). Accordingly, the effect of the fruC isoform on Yp expression was evaluated, as it is the least likely exon to be expressed by females through common promoters and it is expressed in the majority of cells that express the male-biased isoforms of fru (Goodwin et al., 2000).
In addition, probes from genes involved in ecdysone mediated regulation of Yps were evaluated and included EcR (the A and B1 isoforms), usp and Aef1 (An and Wensink, 1995; Bownes et al., 1996; Brodu et al., 2001; Hutson and Bownes, 2003). Likewise, the influence of concentration of the common exon of tra, the common exon of tra2, a common exon of Rbp1, a male-biased Rbp1 exon (referred to as Rbp1D) and Rbp1-like were evaluated as to their influences on dsx and fru splicing and mis-splicing in both sexes. The effects of the Rbp1 transcripts and a common (labeled here as Sxl) and male-biased (SxlM) exon of Sxl were considered as potential factors that could influence tra expression.
Structural equation models
The expression network was evaluated with SEMs (Pearl, 2000). Linear SEMs were developed using the known prior network structure (Figure 1). A confirmatory modeling procedure was used, where the regulatory relationships given by molecular approaches were used to derive a system of equations (Supplementary Files 3–9). These relationships where then tested to see whether genetic variation in the network was reflected in the given system of equations. Transcripts that were external variables were considered as covariates if they were isoforms of the same gene (that is, EcRA and EcRB), or if they were structurally and functionally similar (that is, Rbp1D and Rbp1-like, which are both male-biased splices of similar genes). One model was made for each Yp in each sex. Summaries of results can be found in Supplementary File 3 and Figure 3. Full reports from the analyses can be found in Supplementary Files 4–9. Connections between variables in the network were considered significant if they yielded a Q-value of <0.05 with an false discovery rate of 0.05 (Supplementary File 3). Relative expression values among genotypes were compared between sexes with histograms (Supplementary File 1) and by clustering of expression levels (Suzuki and Shimodaira, 2006). Bootstrap values (green) as well as approximate unbiased probabilities (red) are reported (Figure 2).
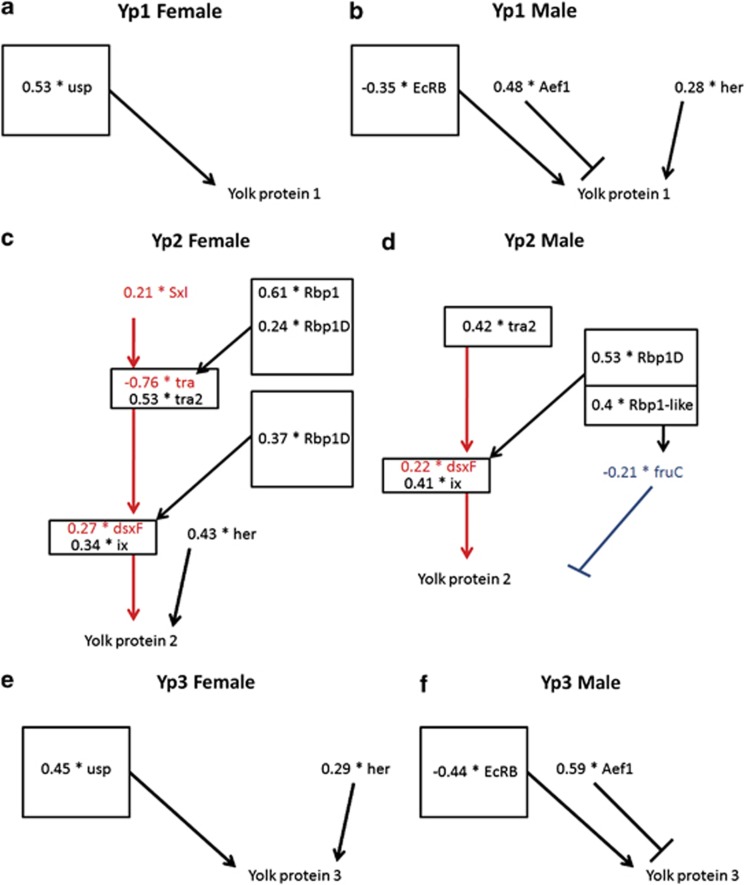
Figure 3.
SEM results for each Yp in both sexes. Arrows, bars and colors are the same as Figure 1 and represent predicted regulatory relationships. Only statistically significant correlations among transcript levels are noted for each Yp and sex. (a, c and e) Present results of Yp analyses in females, denoting Yp1, Yp2 and Yp3, respectively. (b, d and f) Present results of Yp analyses in males, denoting Yp1, Yp2 and Yp3, respectively. Numbers in front of genes names represent standardized parameter estimates for each response. Note that Yp2 was the only locus sensitive to expression variation of sex determination loci, whereas Yp1 and Yp3 were more responsive to variation in ecdysone signaling.
Regression models
Genes in the network were evaluated for nonlinearity of response using the mixed model: Yijkl=μ+di+sj+gk+gk2+sgjk+ɛijkl. For each of the relationships postulated in the network, the gene expression of the downstream gene was fit as the dependent variable (Y) and the upstream gene as the independent variable (g) with both a linear and quadratic term. The significance of quadratic term indicates a nonlinear response. The fixed effects of dye (d) and sex (s) and the interaction between the linear effect of the downstream gene and sex were also included in the model. Possible correlation among the observations due to the mating design was accounted for using a block diagonal structure for the error matrix with each dam having its own estimate of error. Full results are reported in Supplementary File 10. For these models a P-value of 0.05 was considered significant.
Longevity experiment
For each cross and sex the median age at which 15 individuals (per replicate) survived post-eclosion was used as a measure of longevity. Two independent replicates were performed for each cross and sex for a total of 288 measurements of cross averages. Samples where replicates were highly discordant and one of the replicates had values that were consistent with date entry error were removed. This left 248 samples for analysis, though it should be noted that the general pattern of our observations were unchanged by evaluating all 288. The model Yijk=μ+si+gj+sgij+ɛijk was fit where longevity, the dependent variable (Y), was the median time of death for sex i and genotype j. The mean of the array replicates for sex i and genotype j was used to estimate the effect of the Yp and possible correlation among the observations due to the mating design was accounted for using a block diagonal structure for the error matrix with each dam having its own estimate of error. Error was assumed to be normally distributed.
For the longevity experiments flies were reared on a Drosophila diet that consisted of the following components: water (92.45% v/v), unsulfured molasses (5.39% v/v), ethanol (1.54% v/v), propionic acid (0.62% v/v), cornmeal (9.24% v/w), torula yeast (7.70% w/v), agar (0.68% w/v) and Tegosept (0.31% w/v). Stocks were maintained at room temperature (22–24 °C). Adults were collected from each line as virgins and crossed in all pairwise combinations including reciprocal crosses. These crosses were made in cut bottles and eggs were harvested from the bottles and transferred to vials at ∼75 eggs per vial. Offsprings were collected from these vials and collected as virgins using light ether. These flies were transferred to cages at ∼30 newly eclosed virgin flies per cage for the lifespan assay. The cages consisted of quart deli containers with mesh on the lid for ventilation and two portals on opposite sides of the cylinder. One portal was a small circle cut into the side adapted as a fitting for fresh food vials. The other portal was a slit cut into an inner tube sewn onto the wall of the cylinder. An aspirator was inserted into the slit to collect dead flies from the cage. The lifespan assay was conducted at 25 °C, 12L: 12D. A fresh food vial replaced the previous vial in the cage every 4 days, and the dead flies collected and recorded until all flies in each cage were dead. Three cages were set up for each cross and all crosses were assayed at the same time. The entire lifespan assay was repeated once.
Results
Gene expression correlations
Expression was detected for all genes in this study (Table 1, (Wayne et al., 2007)). In males, the expression of dsxF, ix, EcRA and Yp2 was statistically indistinguishable from background in numerous crosses (Table 1). Expression levels of these genes were female biased (Supplementary File 1, Supplementary File 2).
Several groups of transcripts in the network were highly correlated. Unsupervised, hierarchical clustering of expression showed similar groupings in male and female profiles (Figure 2). Within sex-biased groups of genes (Wayne et al., 2007), there were several distinct modules of co-regulated genes (Supplementary File 2, Figure 2). Yp1 and Yp3 were co-expressed in a strongly supported cluster in both sexes (correlations were 0.95 and 0.93 in males and females, respectively; covariance estimates were 0.23 and 0.13, respectively). In males, the male-biased fruC transcript clustered with the male-biased splicing factor Rbp1-like (correlation: 0.67; covariance: 0.04) and dsxM clustered with the male-biased transcript of a splicing factor Rbp1 (correlation: 0.82; covariance: 0.05). Further, both isoforms of EcR significantly clustered with ix expression in males (average correlation: 0.83; average covariance: 0.06). In females, Rbp1 clustered with tra expression (correlation: 0.92; covariance: 0.11), whereas her clustered significantly with tra2 expression (correlation: 0.93; covariance: 0.1). Mixed-sex models were evaluated to identify statistical interactions between sex and expression. Table 2 indicates the many significant sex by gene interactions that were found in the network.
Table 2. Significant interactions of sex with gene expression levels in the Yp network.
| Regulator | NumDF | DenDF | F value | P-value | Model | Q-value |
|---|---|---|---|---|---|---|
| fruC*sex | 1 | 308 | 69.37496 | 2.71E−15 | Yp2 | 1.19E−13 |
| EcRB*sex | 1 | 304 | 44.70859 | 1.10E−10 | Yp2 | 2.41E−09 |
| Rbp1-like*sex | 1 | 306 | 43.36909 | 1.97E−10 | dsxF | 2.89E−09 |
| Rbp1D*sex | 1 | 308 | 31.11336 | 5.33E−08 | dsxF | 5.86E−07 |
| EcRA*sex | 1 | 308 | 26.40915 | 4.91E−07 | Yp2 | 4.33E−06 |
| her*sex | 1 | 306 | 17.38404 | 3.98E−05 | Yp2 | 0.000292 |
| Rbp1-like*sex | 1 | 306 | 14.9784 | 0.000133 | tra | 0.000836 |
| EcRA*sex | 1 | 308 | 13.71128 | 0.000253 | Yp3 | 0.001389 |
| ix*sex | 1 | 306 | 13.36864 | 0.000301 | Yp3 | 0.001422 |
| ix*sex | 1 | 306 | 13.22946 | 0.000323 | Yp2 | 0.001422 |
| Rbp1-like*sex | 1 | 306 | 12.80402 | 0.000402 | tra | 0.001608 |
| EcRA*sex | 1 | 308 | 12.26733 | 0.000529 | Yp1 | 0.00194 |
| ix*sex | 1 | 306 | 11.88957 | 0.000644 | Yp1 | 0.002178 |
| Rbp1-like*sex | 1 | 306 | 11.73646 | 0.000697 | fruC | 0.002189 |
| dsxF*sex | 1 | 308 | 10.70216 | 0.001192 | Yp2 | 0.003495 |
| Rbp1*sex | 1 | 308 | 10.57733 | 0.001272 | tra | 0.003498 |
| Rbp1D*sex | 1 | 308 | 10.24278 | 0.001515 | fruC | 0.003922 |
Abbreviations: FDR, false discovery rate; Yp, Yolk protein.
Regulator indicates the genes known to regulate the gene or splice in the model column. Q-values represent an FDR of 0.05.
Genetic variation and heritability
Heritability and genetic variance components were evaluated for genes in the network (Supplementary File 2) (Wayne et al., 2007). Heritability of expression was generally <0.1, reflecting the low amount of additive genetic variance in females. Less genetic variation was detected in males than females, with 8 of 19 genes in the analysis demonstrating significant genetic variation in males, whereas all 19 genes demonstrated significant genetic variation in females (Fisher's exact test, P=0.00012).
The entire network was assessed separately in males and females with SEMs based upon the known linkages among genes in this pathway (Supplementary File 3, Figure 3a–f; Pearl, 2000). SEMs demonstrated several interesting features. In males, variation in her, Aef1 and EcRB was correlated with variation in Yp1 expression. Similarly, Yp3 responded to variation in expression of Aef1 and her. However, variation in expression of Yp2 was significantly influenced by variation in ix, dsxF and fruC. Variation in dsxF expression was influenced by tra2 and Rbp1D levels, while fruC was significantly correlated with variation in Rbp1-like. In females, Yp1 and Yp3 were both correlated with expression levels of usp, but Yp3 also responded to variation in her. Yp2 variation in females was significantly influenced by dsxF, ix and her expression. Expression of dsxF was significantly correlated with tra, tra2 and Rbp1D expression and tra expression was, in turn, correlated with Sxl, Rbp1 and Rbp1D expression (Figure 3c and d).
Statistical analyses of the Yp network
The SEM analyses also revealed several interesting factors associated with the mis-splicing of transcripts. Previously, tra2 (a gene known to regulate dsx splicing) expression was observed to significantly correlate with dsxF levels in males (Tarone et al., 2005). The observation of a tra2 correlation with dsx mis-splicing was repeated in these analyses. Furthermore, Rbp1D was significantly correlated with dsxM levels in females and dsxF levels in males, whereas Rbp1-like levels in females significantly correlated with expression of the fruC transcript in females. All of these results indicate that fluctuations in the levels of specific splicing factors have downstream repercussions for the splicing of specific target transcripts.
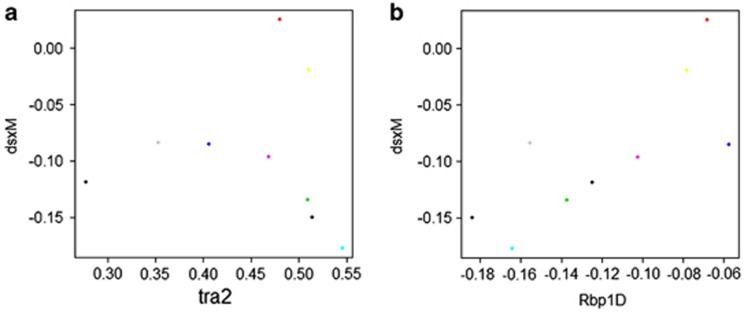
The results of the SEM analyses were then evaluated graphically and statistically to identify potential nonlinear responses in the network (Figure 4, Table 3). Several genes significantly correlated with expression levels of downstream targets and also exhibited linear responses within a sex with a small number of genotypes that clearly fell off of the line. However, when both sexes were observed together, these loci exhibited curved responses, with some overlap between the sexes observed among the outlier genotypes of both sexes (Figure 4a). The sexes clearly form two separate groups (Figure 4b and c) and the sexes were significantly different from each other (Table 2). Such patterns were typical of usp, Aef1, her, ix, dsxF and Rbp1-like. There was also a nonlinear response of dsxM in females, where the line means demonstrated a parabolic response to tra2, with the exception of two genotypes that expressed exceptionally high levels of Rbp1D, which was also significantly associated with mis-splicing (Figure 5). These responses potentially indicate a nonlinear switch-like behavior. To explicitly test the observation of nonlinear switches, a general model across sexes was used to test for nonlinear responses through the observation of significance in quadratic terms. These analyses identified 19 significant nonlinear responses between sexes (Table 3). However, within a sex all but one gene show linear responses.
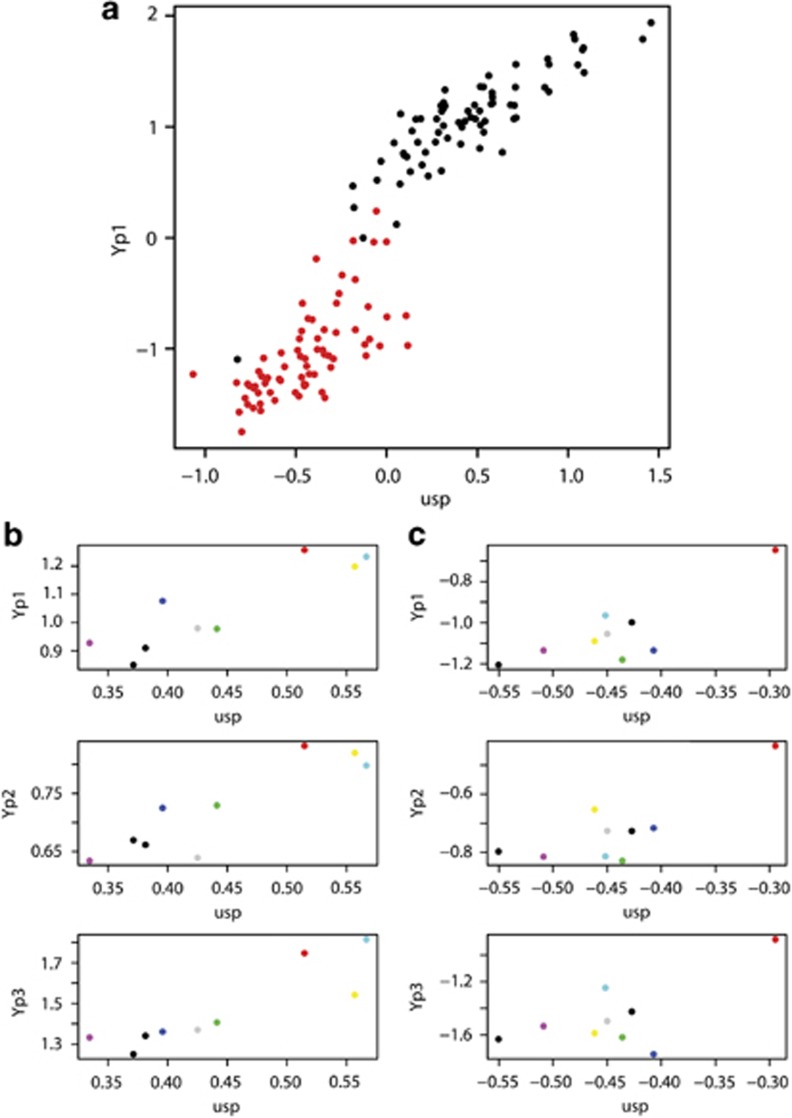
Figure 4.
Plots of Yp expression in terms of usp expression. These profiles were typical of usp, Aef1, her, ix, dsxF and Rbp1-like. (a) Yp1 expression as a function of usp expression from the diallel crosses displays linear responses within a sex, but a sigmoidal response between the sexes. Males are displayed in red and females in black. (b) Mean expression in females for each parental line, with each line assigned a different color that is the same in all. There were three lines that expressed high levels of usp, which also produced the highest Yp expression levels. These lines were much less likely to produce crosses that expressed Yps at intermediate levels. (c) Mean expression in males for each parental line, with each line assigned a different color. There was one line that expressed high levels of usp, which also produced the highest Yp expression levels. This line was much more likely to produce crosses that expressed Yps at intermediate levels.
Table 3. Significant nonlinear responses in the Yp network detected across sexes as determined by significant correlations of quadratic terms in a linear model with the transcript in the Model column.
| Regulator | NumDF | DenDF | F value | P-value | Model | Q-value |
|---|---|---|---|---|---|---|
| Rbp1*Rbp1 | 1 | 308 | 47.27511 | 3.44E−11 | dsxF | 1.51E−09 |
| tra*tra | 1 | 308 | 33.89814 | 1.46E−08 | dsxF | 3.21E−07 |
| Rbp1-like*Rbp1-like | 1 | 306 | 32.68229 | 2.58E−08 | dsxF | 3.78E−07 |
| Rbp1-like*Rbp1-like | 1 | 306 | 25.80027 | 6.60E−07 | tra | 6.68E−06 |
| usp*usp | 1 | 304 | 25.51169 | 7.60E−07 | Yp1 | 6.68E−06 |
| Aef1*Aef1 | 1 | 302 | 23.18078 | 2.34E−06 | Yp1 | 1.71E−05 |
| Aef1*Aef1 | 1 | 302 | 21.6355 | 4.94E−06 | Yp3 | 2.92E−05 |
| her*her | 1 | 306 | 21.31166 | 5.75E−06 | Yp2 | 2.92E−05 |
| her*her | 1 | 306 | 21.23315 | 5.98E−06 | Yp3 | 2.92E−05 |
| usp*usp | 1 | 304 | 18.15835 | 2.71E−05 | Yp3 | 0.000119 |
| her*her | 1 | 306 | 16.60402 | 5.87E−05 | Yp1 | 0.000235 |
| ix*ix | 1 | 306 | 15.91645 | 8.29E−05 | Yp2 | 0.000304 |
| Rbp1D*Rbp1D | 1 | 308 | 9.974343 | 0.001745 | tra | 0.005905 |
| tra2*tra2 | 1 | 308 | 8.971234 | 0.002965 | dsxF | 0.009319 |
| Rbp1*Rbp1 | 1 | 308 | 8.499873 | 0.003812 | tra | 0.011183 |
| Rbp1D*Rbp1D | 1 | 308 | 7.75046 | 0.005702 | tra | 0.01568 |
| Rbp1-like*Rbp1-like | 1 | 306 | 7.443663 | 0.006734 | tra | 0.017429 |
| dsxF*dsxF | 1 | 308 | 6.477048 | 0.011415 | Yp1 | 0.027904 |
| Rbp1-like*Rbp1-like | 1 | 306 | 5.863766 | 0.016037 | fruC | 0.037138 |
Abbreviations: FDR, false discovery rate; Yp, Yolk protein.
Q-values represent an FDR of 0.05.
Figure 5.
Mis-splicing of dsx in females is driven by tra2 and Rbp1D expression. (a) An evaluation of line means indicates that for seven of the nine genotypes, dsxM levels exhibit a parabolic response to tra2 expression. However, two genotypes express much higher levels of dsxM than the other genotypes. (b) These two strains express high levels of Rbp1D. These results indicate that an optimal level of tra2 (high or low levels) should produce the least mis-splicing in females, though some genotypes that express Rbp1D at high levels overcome this pattern, meaning interactions between these two regulators may be complex.
Dosage compensation
The effect of dosage compensation on the Yp network genes was evaluated with two data sets. The ‘Kuroda' data set (Hamada et al., 2005) is derived from a genome-wide evaluation of the effects of knocking out dosage compensation in a male cell line via RNAi of msl-2, which is necessary for dosage compensation. Any genes that were differentially expressed in this experiment are likely to be affected by dosage compensation, either directly or indirectly. These data indicated that tra, tra2, Rbp1, Aef1 and ix displayed stable and consistent differences in expression among RNAi and control treatments, which were statistically significant (t-test, P<0.05). In addition, the ‘Arbeitman' data set (Goldman and Arbeitman, 2007) measured expression differences between females and males and compared them to expression differences between females and tra pseudomales, which have two X chromosomes but are somatically male. These data showed significant differences among males and tra pseudomales for expression of Yp3 (t-test, P=0.01), suggesting that at least one Yp is influenced by the dosage compensation mechanism. Taken together, these data suggest that in both cell culture and in whole males, one or more components of the Yp network are significantly influenced by dosage compensation, which may be one reason for the discrepancy in gene expression inheritance observed between the sexes in this experiment.
Longevity
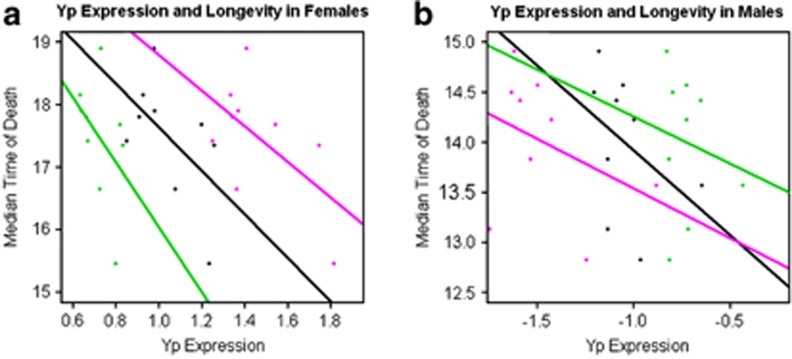
Recent data suggest that vitellogenin proteins, which are functionally similar to Yps, have effects on insect lifespans (Seehuus et al., 2006; Corona et al., 2007). All Yps were significantly correlated with median time of death in at least one sex, with females demonstrating stronger correlations than males (Figure 6a and b). When individual Yps were evaluated along with sex in a mixed linear model, sex significantly correlated with lifespan in all cases, with females living longer than males (P<0.0001). The median time of death was significantly correlated with Yp expression (Yp1 and Yp2; P=0.016 and 0.026, respectively). Median time of death was significantly associated with Yp3 expression (P=0.023), with a significant interaction between sex and Yp3 (P=0.026). Given the observation of an interaction between sex and Yp3 expression, correlations of that gene with lifespan were evaluated in each sex individually. This analysis demonstrated correlations with female (P=0.0004), but not male lifespan (P=0.71). In all analyses, high Yp expression levels were more likely to be found in individuals with shorter median times of death.
Figure 6.
Correlations of longevity with Yp expression. Regressions of line means for median time of death with Yp expression in females (a) and males (b). Yp1 expression levels are designated by the color black, Yp2 by green and Yp3 by magenta.
Discussion
Do nonlinear responses reflect patterns of genetic variation in gene expression?
Research by Gjuvsland et al. (2007a, 2007b, 2007c) would suggest that the large degree of non-additive genetic variation observed in females in this experiment (as compared with the predominantly additive inheritance of male gene expression) could be explained by nonlinear responses on additive inheritance. In such a case, females should exhibit more nonlinear responses in the network than males. The data presented here do not support the idea of within-sex nonlinear responses, with the exception of tra2 effects on dsxM expression in females. Accordingly, it is unlikely that nonlinearity explains differences in gene expression inheritance between the sexes.
However, there were numerous responses within the network that indicated a nonlinear, sigmoid, response if expression was observed between the sexes (Figure 4). This pattern indicates a switch-like response between sexes, with some genotypes more likely than others to produce intermediate expression levels that would fall on the nonlinear portion of the response. Note that these ‘switches' appear noisy, raising a question as to how unambiguous sex-specific regulation is maintained. These results clarify previous observations that variation in the sex determination pathway functions more like a dial than a switch (Tarone et al., 2005).
Does each Yp respond to genetic variation in the same regulators? Is dosage compensation important to the male Yp network?
The Yps are usually considered to respond to the same regulatory elements. However, each locus has its own position on the X chromosome and its own orientation with respect to the sequences that regulate their expression. Although each gene may be affected by the same set of transcription factors, the order of importance of any single regulator to a specific Yp may be unique to that Yp. There were clear differences among Yps responding to the same network variation. In both sexes, Yp1 and Yp3 were significantly correlated with each other more strongly than with Yp2 (Figure 2). It is particularly notable that this pattern was observed (in both sexes) given that Yp1 and Yp2 are considered to share the same regulatory sequence (though they read in opposite orientations from that region). In the SEMs, Yp1 and Yp3 also correlated with variation in ecdysone signaling transcripts (usp in females; EcrB and Aef1 in males), while Yp2 responded to variation in sex determination loci. These and other observed correlations within the network suggest that genetic variation in the Yp network affected specific targets differently.
Dosage compensation has been shown to decrease transcriptional noise in mammals (Yin et al., 2009), imposing a general pattern of simplicity of expression inheritance in males. In Drosophila, RNAi experiments with male tissues have been used to eliminate expression of two dosage compensation genes msl-2 and MOF, demonstrating a patchwork system where each locus on the X is upregulated to a slightly different degree, with some autosomal consequences (Hamada et al., 2005; Kind et al., 2008). We hypothesize that the variation in these dosage compensation responses among genotypes could have important consequences for the male Yp network. Indeed, experiments comparing genome-wide patterns of expression find many more genes differentially expressed between males and females than between females and tra pseudomales, which are not apparently engaged in dosage compensation (Goldman and Arbeitman, 2007).
Our assessment of published results from a genome-wide study of RNAi in male cell lines and a comparison of males to tra pseudomales is that dosage compensation may have a role in expression inheritance in the Yp network. Results from these studies indicate that Yp3, tra, tra2, ix, Aef1 and multiple isoforms of Rbp1are susceptible to dosage compensation effects in males. These transcripts fall within all of the major clusters of expression found in the network, and their response to knocking out the dosage compensation machinery indicates the potential for widespread effects of dosage compensation throughout the network, which may overwhelm the other inputs in the pathway, and account for the simple inheritance of male expression levels.
Does variation in Yp expression, an indicator of fecundity in females, correlate with variation in longevity?
Variation in the Yp network may shed light on the very complex tradeoff between fecundity and longevity (Chippindale et al., 2001; Partridge et al., 2005; Flatt et al., 2008; Toivonen and Partridge, 2009; Kenyon, 2010). We compared the state of the pathway with longevity phenotypes in the 72 genotypes and found extensive negative correlations of Yps on the median time of death for males and females. We focused on virgin flies that enabled the evaluation of longevity responses in the absence of interference from mating effects. In females, Yp mutants affect fertility, ovariole number and egg hatch rates (Postlethwait and Shirk, 1981; Bownes et al., 1991; Terashima and Bownes, 2004), therefore Yp expression is an indicator of fecundity. Tradeoffs between fecundity and lifespan are well described; therefore this correlation makes sense in females (Marden et al., 2003; Mukhopadhyay and Tissenbaum, 2007).
However, males have no ovaries and only express Yps at background rates, indicating that there may be non-germline mediated influences at have in this correlation. High Yp expression in males with short lifespans may mean that Yp expression is a marker of mis-expression of sex determination network genes, which could have negative repercussions for either sex. This potential effect is supported by the observation that overexpressing sex determination genes has negative effects on Drosophila lifespan (Shen et al., 2009). There is also a potential endocrine signaling function for Yps, related to the demonstrated ability of these proteins to bind ecdysone (Bownes et al., 1988) and the fact that they are known to be regulated by ecdysone signaling, which may also affect lifespan (Simon et al., 2003; Tatar, 2004). Another potential avenue by which male longevity may be directly affected by Yp expression is through immune function, which has also been linked to Yp levels (Amdam et al., 2004).
The potential causes of the observed negative correlation of Yps with longevity in male and female Drosophila are numerous and correspond to several distinct mechanisms that are considered critical to understanding mechanisms underlying tradeoffs between longevity and reproduction (Harshman and Zera, 2007). Functional information from hymenopteran vitellogenins would indicate that Yps may have a functional role in Drosophila longevity, possibly through influences on oxidative stress, juvenile hormone or immune function (Amdam et al., 2004; Landis et al., 2004; Seehuus et al., 2006). Note though that the effects here are opposite to what is known about effects of vitellogenins on lifespan of hymenoptera (Seehuus et al., 2006; Corona et al., 2007). Direct experimentation with Yp mutants and natural Yp variants, especially in males, will help to dissect the mechanism by which Yps negatively correlate with longevity in Drosophila. Further studies examining the relationship between genetic variation in Yp and longevity developed from independent populations of D. melanogaster and related species, as well as quantitative manipulations of Yp (for example, through RNAi), will be necessary before concluding that the relationship between Yp and longevity is causal.
Conclusions
The goal of this study was to test whether natural variation within a known gene expression network reflects known regulatory relationships and phenotypic associations. The Yp expression network was used as a model for this ‘bottom up' systems biology approach, as it is well-characterized molecularly. Variation in the network reflects many known interactions, though sex and individual Yps responded to genetic variation in different known factors. Yp expression also negatively correlated with longevity in both males and females, but to different degrees, indicating that systems-level analyses of this expression network may be a useful model system for unraveling molecular mechanisms of life history tradeoffs related to reproduction and lifespan.
Data Archiving
Data have been deposited at Dryad; doi:10.5061/dryad.49v70
Acknowledgments
We thank Peter Chang, Joe Dunham, Tom Turner, Maren Freisen, Michelle Arbeitman, Brad Foley, Ryan Bickel, JP Masly, Jennifer Brisson, Micky Eubanks and John Tower for helpful discussions and advice.
Authors contributions: The research was conceived by AMT and SVN. AMT and SVN conducted SEM analyses. LMM conducted the analyses of linear models. AMT analyzed results from RNAi and tra pseudomale experiments. LGH performed longevity experiments. AMT wrote the paper with major contributions from LMM, LGH and SVN. We acknowledge support from the NIH grants (R01GM077618 and S1R01GM077618-S1). AMT is supported by startup funds from the College of Agriculture and Life Sciences and from the Texas AgriLife Research at Texas A&M University.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abrahamsen N, Martinez A, Kjaer T, Sondergaard L, Bownes M. Cis-regulatory sequences leading to female-specific expression of yolk protein genes 1 and 2 in the fat body of Drosophila melanogaster. Mol Gen Genet. 1993;237:41–48. doi: 10.1007/BF00282782. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Simoes ZLP, Hagen A, Norberg K, Schroder K, Mikkelson O, et al. Hormonal control of the yolk precursor vitellogenin regulates immune function and longevity in honeybees. Exp Gerontol. 2004;39:767–773. doi: 10.1016/j.exger.2004.02.010. [DOI] [PubMed] [Google Scholar]
- An W, Wensink PC. Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J. 1995;14:1221–1230. doi: 10.1002/j.1460-2075.1995.tb07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Burtis K, Goralski T, Mattox W, Nagoshi R. Molecular genetic aspects of sex determination in Drosophila melanogaster. Genome. 1989;31:638–645. doi: 10.1139/g89-117. [DOI] [PubMed] [Google Scholar]
- Belote JM, Handler AM, Wolfner MF, Livak KJ, Baker BS. Sex-specific regulation of yolk protein gene expression in Drosophila. Cell. 1985;40:339–348. doi: 10.1016/0092-8674(85)90148-5. [DOI] [PubMed] [Google Scholar]
- Billeter JC, Rideout EJ, Dornan AJ, Goodwin SF. Control of male sexual behavior in Drosophila by the sex determination pathway. Curr Biol. 2006;16:R766–R776. doi: 10.1016/j.cub.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Bownes M. Why is there sequence similarity between insect yolk proteins and vertebrate lipases. J Lipid Res. 1992;33:777–790. [PubMed] [Google Scholar]
- Bownes M, Lineruth K, Mauchline D. Egg production and fertility in Drosophila depend upon the number of yolk protein gene copies. Mol Gen Genet. 1991;228:324–327. doi: 10.1007/BF00282485. [DOI] [PubMed] [Google Scholar]
- Bownes M, Ronaldson E, Mauchline D. 20-Hydroxyecdysone, but not juvenile hormone, regulation of yolk protein gene expression can be mapped to cis-acting DNA sequences. Dev Biol. 1996;173:475–489. doi: 10.1006/dbio.1996.0041. [DOI] [PubMed] [Google Scholar]
- Bownes M, Shirras A, Blair M, Collins J, Coulson A. Evidence that insect embryogenesis is regulated by ecdysteroids released from yolk proteins. Proc Natl Acad Sci USA. 1988;85:1554–1557. doi: 10.1073/pnas.85.5.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt BW, Zwaan BJ, Beekman M, Westendorp RGJ, Slagboom PE. Shuttling between species for pathways of lifespan regulation: A central role for the vitellogenin gene family. BioEssays. 2005;27:339–346. doi: 10.1002/bies.20161. [DOI] [PubMed] [Google Scholar]
- Brodu V, Mugat B, Fichelson P, Lepesant JA, Antoniewski C. A UAS site substitution approach to the in vivo dissection of promoters: interplay between the GATAb activator and the AEF-1 repressor at a Drosophila ecdysone response unit. Development. 2001;128:2593–2602. doi: 10.1242/dev.128.13.2593. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chippindale AK, Gibson JR, Rice WR. Negative correlation for adult fitness between sexes reveals ontogenic conflict in Drosophila. Proc Natl Acad Sci USA. 2001;98:1675–1675. doi: 10.1073/pnas.041378098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. Vive la difference: males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Coffman CJ, Wayne ML, Nuzhdin SV, Higgins LA, McIntyre LM. Identification of co-regulated transcripts affecting male body size in Drosophila. Genome Biol. 2005;6:R53. doi: 10.1186/gb-2005-6-6-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona M, Velarde RA, Remolina S, Moran-Lauter A, Wang Y, Hughes KA, et al. Vitellogenin, juvenile hormone, insulin signaling, and queen honey bee longevity. Proc Natl Acad Sci USA. 2007;104:7128–7133. doi: 10.1073/pnas.0701909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djawdan M, Sugiyama TT, Schlaeger LK, Bradley TJ, Rose MR. Metabolic aspects of the trade-off between fecundity and longevity in Drosophila melanogaster. Physiol Zool. 1996;69:1176–1195. [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Featherstone DE, Broadie K. Wrestling with pleiotropy: genomic and topological analysis of the yeast gene expression network. Bioessays. 2002;24:267–274. doi: 10.1002/bies.10054. [DOI] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D'Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, et al. Drosophila, germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci USA. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjuvsland AB, Hayes BJ, Meuwissen THE, Plahte E, Omholt SW. Nonlinear regulation enhances the phenotypic expression of trans-acting genetic polymorphisms. BMC Syst Biol. 2007a;1:32. doi: 10.1186/1752-0509-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjuvsland AB, Hayes BJ, Omholt SW, Carlborg O. Statistical epistasis is a generic feature of gene regulatory networks. Genetics. 2007b;175:411–420. doi: 10.1534/genetics.106.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjuvsland AB, Plahte E, Omholt SW. Threshold-dominated regulation hides genetic variation in gene expression networks. BMC Systems Biol. 2007c;1:52. doi: 10.1186/1752-0509-1-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin SF, Taylor BJ, Villella A, Foss M, Ryner LC, Baker BS, et al. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics. 2000;154:725–745. doi: 10.1093/genetics/154.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthke R, Moller U, Hoffmann M, Thies F, Topfer S. Dynamic network reconstruction from gene expression data applied to immune response during bacterial infection. Bioinformatics. 2005;21:1626–1634. doi: 10.1093/bioinformatics/bti226. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Park PJ, Gordadze PR, Kuroda MI. Global regulation of X chromosomal genes by the MSL complex in Drosophila melanogaster. Genes Dev. 2005;19:2289–2294. doi: 10.1101/gad.1343705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshman LG, Zera AJ. The cost of reproduction: the devil in the details. Trends Ecol Evol. 2007;22:80–86. doi: 10.1016/j.tree.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Hutson SF, Bownes M. The regulation of yp3 expression in the Drosophila melanogaster fat body. Dev Genes Evol. 2003;213:1–8. doi: 10.1007/s00427-002-0286-4. [DOI] [PubMed] [Google Scholar]
- Jansen RC, Tesson BM, Fu J, Yang Y, McIntyre LM. Defining gene and QTL networks. Curr Opin Plant Biol. 2009;12:241–246. doi: 10.1016/j.pbi.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Kenyon C. A pathway that links reproductive status to lifespan in Caenorhabditis elegans. Ann NY Acad Sci. 2010;1204:156–162. doi: 10.1111/j.1749-6632.2010.05640.x. [DOI] [PubMed] [Google Scholar]
- Kind J, Vaquerizas JM, Gebhardt P, Gentzel M, Luscombe NM, Bertone P, et al. Genome-wide analysis reveals MOF as a key regulator of dosage compensation and gene expression in Drosophila. Cell. 2008;133:813–828. doi: 10.1016/j.cell.2008.04.036. [DOI] [PubMed] [Google Scholar]
- Kraus KW, Lee YH, Lis JT, Wolfner MF. Sex-specific control of Drosophila melanogaster yolk protein 1 gene expression is limited to transcription. Mol Cell Biol. 1988;8:4756–4764. doi: 10.1128/mcb.8.11.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Lopez AJ. Negative feedback regulation among SR splicing factors encoded by Rbp1 and Rbp1-like in Drosophila. EMBO J. 2005;24:2646–2655. doi: 10.1038/sj.emboj.7600723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, Carrick J, et al. Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101:7663–7668. doi: 10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo MS, Sanders LE, Sun F, Arbeitman MN. Somatic, germline and sex hierarchy regulated gene expression during Drosophila metamorphosis. BMC Genomics. 2009;10:80. doi: 10.1186/1471-2164-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SI, Dudley AM, Drubin D, Silver PA, Krogan NJ, Pe'er D, et al. Learning a prior on regulatory potential from eQTL data. PLoS Genet. 2009;5:e1000358. doi: 10.1371/journal.pgen.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Baker BS. hermaphrodite and doublesex function both dependently and independently to control various aspects of sexual differentiation in Drosophila. Development. 1998;125:2641–2651. doi: 10.1242/dev.125.14.2641. [DOI] [PubMed] [Google Scholar]
- Marden JH, Rogina B, Montooth KL, Helfand SL. Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc Natl Acad Sci USA. 2003;100:3369–3373. doi: 10.1073/pnas.0634985100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Tissenbaum HA. Reproduction and longevity: secrets revealed by C. elegans. Trends Cell Biol. 2007;17:65–71. doi: 10.1016/j.tcb.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Neter J, Wasserman W, Kutner MH.1990Applied Linear Statistical Models: Regression, Analysis of Variance, and Experimental Designs3rd edn.Irwin: Homewood, IL [Google Scholar]
- Nuzhdin SV, Brisson JA, Pickering A, Wayne ML, Harshman LG, McIntyre LM. Natural genetic variation in transcriptome reflects network structure inferred with major effect mutations: insulin/TOR and associated phenotypes in Drosophila melanogaster. BMC Genomics. 2009;10:124. doi: 10.1186/1471-2164-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Fukunaga A, Kawabe M, Oishi K. Interactions between sex-transformation mutants of Drosophila melanogaster.1. Hemolymph vitellogenins and gonad morphology. Genetics. 1981;99:429–441. doi: 10.1093/genetics/99.3-4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Gems D, Withers DJ. Sex and death: what is the connection. Cell. 2005;120:461–472. doi: 10.1016/j.cell.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Pearl J. Causality: Models, Reasoning, and Inference. Cambridge University Press: Cambridge, UK; New York; 2000. [Google Scholar]
- Postlethwait JH, Shirk PD. Genetic and endocrine regulation of vitellogenesis in Drosophila. Am Zool. 1981;21:687–700. [Google Scholar]
- Qi JL, Su SH, Mattox W. The doublesex splicing enhancer components Tra2 and Rbp1 also repress splicing through an intronic silencer. Mol Cell Biol. 2007;27:699–708. doi: 10.1128/MCB.01572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR. Genetics of increased lifespan in Drosophila. Bioessays. 1989;11:132–135. doi: 10.1002/bies.950110505. [DOI] [PubMed] [Google Scholar]
- Sanchez L. Sex-determining mechanisms in insects. Int J Dev Biol. 2008;52:837–856. doi: 10.1387/ijdb.072396ls. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Friend SH, Shaywitz DA. OPINION A network view of disease and compound screening. Nat Rev Drug Discov. 2009a;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Friend SH, Shaywitz DA. A network view of disease and compound screening. Nat Rev Drug Discov. 2009b;8:286–295. doi: 10.1038/nrd2826. [DOI] [PubMed] [Google Scholar]
- Seehuus SC, Norberg K, Gimsa U, Krekling T, Amdam GV. Reproductive protein protects functionally sterile honey bee workers from oxidative stress. Proc Natl Acad Sci USA. 2006;103:962–967. doi: 10.1073/pnas.0502681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Ford D, Landis GN, Tower J. Identifying sexual differentiation genes that affect Drosophila life span. BMC Geriatr. 2009;9:56. doi: 10.1186/1471-2318-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AF, Shih C, Mack A, Benzer S. Steroid control of longevity in Drosophila melanogaster. Science. 2003;299:1407–1410. doi: 10.1126/science.1080539. [DOI] [PubMed] [Google Scholar]
- Stern DL. Perspective: evolutionary developmental biology and th problem of variation. Evolution. 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- Tarone AM, Nasser YM, Nuzhdin SV. Genetic variation for expression of the sex determination pathway genes in Drosophila melanogaster. Genet Res. 2005;86:31–40. doi: 10.1017/S0016672305007706. [DOI] [PubMed] [Google Scholar]
- Tatar M. The neuroendocrine regulation of Drosophila aging. Exp Gerontol. 2004;39:1745–1750. doi: 10.1016/j.exger.2004.06.024. [DOI] [PubMed] [Google Scholar]
- Terashima J, Bownes M. Translating available food into the number of eggs laid by Drosophila melanogaster. Genetics. 2004;167:1711–1719. doi: 10.1534/genetics.103.024323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivonen JM, Partridge L. Endocrine regulation of aging and reproduction in Drosophila. Mol Cell Endocrinol. 2009;299:39–50. doi: 10.1016/j.mce.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Tufail M, Takeda M. Molecular characteristics of insect vitellogenins. J Insect Physiol. 2008;54:1447–1458. doi: 10.1016/j.jinsphys.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Tufail M, Takeda M. Insect vitellogenin/lipophorin receptors: Molecular structures, role in oogenesis, and regulatory mechanisms. J Insect Physiol. 2009;55:87–103. doi: 10.1016/j.jinsphys.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Wayne ML, Telonis-Scott M, Bono LM, Harshman L, Kopp A, Nuzhdin SV, et al. Simpler mode of inheritance of transcriptional variation in male Drosophila melanogaster. Proc Natl Acad Sci USA. 2007;104:18577–18582. doi: 10.1073/pnas.0705441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S, Wang P, Deng W, Zheng H, Hu L, Hurst LD, et al. Dosage compensation on the active X chromosome minimizes transcriptional noise of X-linked genes in mammals. Genome Biol. 2009;10:R74. doi: 10.1186/gb-2009-10-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.