Abstract
Nonspecific association of serum molecules with short-interfering RNA (siRNA) nanoparticles can change their physiochemical characteristics, and results in reduced cellular uptake in the target tissue during the systemic siRNA delivery process. Serum albumin is the most abundant protein in the body and has been used to modify the surface of nanoparticles, to inhibit association of other serum molecules. Here, we hypothesized that surface modification of lipid-based nanoparticular siRNA delivery systems with albumin could prevent their interaction with serum proteins, and improve intracellular uptake. In this study, we investigated the influence of albumin on the stability and intracellular siRNA delivery of the targeted siRNA nanoparticles of a polymerizable and pH-sensitive multifunctional surfactant N-(1-aminoethyl) iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide] (EHCO) in serum. Serum resulted in a significant increase in the size of targeted EHCO/siRNA nanoparticles and inhibited cellular uptake of the nanoparticles. Coating of targeted EHCO/siRNA nanoparticles with bovine serum albumin at 9.4 μM prior to cell transfection improved cellular uptake and gene silencing efficacy of EHCO/siRNA targeted nanoparticles in serum-containing media, as compared with the uncoated nanoparticles. At a proper concentration, albumin has the potential to minimize interactions of serum proteins with siRNA nanoparticles for effective systemic in vivo siRNA delivery.
Keywords: multifunctional, lipid nanoparticles, RNA interference, pH-sensitive amphiphile, siRNA
Introduction
RNA interference is a promising therapeutic modality for silencing expression of disease-causing genes. It is a natural cellular process in which short-interfering RNA (siRNA) initiates complementary mRNA degradation, and thus inhibits synthesis of proteins.1,2 Several in vitro and in vivo studies have shown that siRNA therapeutics can silence gene expression in mammalian cells, nonhuman primates, and humans.3–6 Furthermore, siRNA can be tailored to knockdown expression of any gene, which highlights the potential of RNAi as a powerful tool for treating a variety of ailments.7 Unfortunately, siRNA’s negative charges prevent it from passing through the cell membrane to enter the cell cytoplasm. siRNA is also sensitive to serum and tissue nuclease degradation and is easily cleared from systemic circulation.1 These poor physiochemical and pharmaceutical properties compromise the therapeutic potential of siRNA. Therefore, effective delivery systems are needed to help siRNA to reach the cytoplasm of target cells.
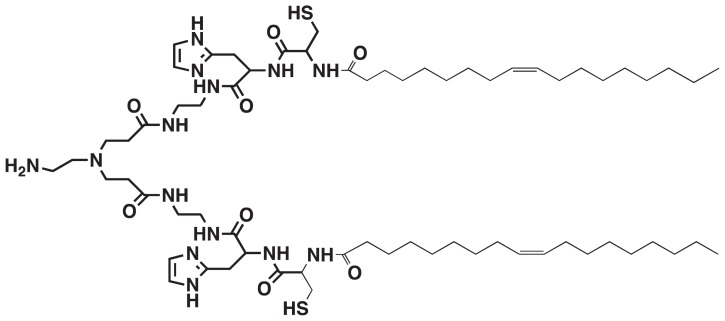
Recently, we designed and prepared a multifunctional polymerizable surfactant with pH-sensitive amphiphilicity, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide] (EHCO), for the systemic delivery of siRNA (Figure 1).8–10 The multifunctional properties present in EHCO make it an attractive and promising carrier for systemic siRNA delivery. EHCO comprises a protonatable amino head group, two cysteine residues, and two lipophilic tail groups. EHCO forms stable nanoparticles with siRNA via charge–charge interactions, lipophilic condensation of lipid tails, and formation of disulfide bonds. The pH-sensitive amphiphilicity of EHCO facilitates endosomal–lysosomal membrane disruption to allow dispersion of EHCO/siRNA nanoparticles into the cell cytoplasm. Reduction of disulfide bonds by glutathione in the cytoplasm facilitates dissociation of EHCO/siRNA nanoparticles to release siRNA. Our studies demonstrated that EHCO/siRNA nanoparticles were efficient in gene silencing, and most importantly were less cytotoxic than commercially available transfection agents.8–10 In addition, the thiol groups present on EHCO can be used to functionalize the surface of nanoparticles for efficient siRNA delivery. Polyethylene glycol (PEG) can be covalently attached to EHCO via a thioether bond to minimize nonspecific cellular uptake of EHCO/siRNA nanoparticles. Furthermore, the same coupling mechanism can be used to conjugate cell-specific ligands to EHCO, to improve the delivery of siRNA to a specific cell type.
Figure 1.
Chemical structure of N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide].
The gene-silencing efficacy of a systemically delivered siRNA depends on how effectively it crosses vascular endothelium at the target site and is internalized by the target cell. However, siRNA delivery efficiency can be influenced by the association of serum molecules with siRNA delivery systems, which alters their physiochemical properties. Serum molecules, such as immunoglobulins, complement proteins, and other serum proteins, adsorb on the surface of nanoparticles via nonspecific attractive forces.11 It is possible that adsorption of serum proteins alters the surface properties of siRNA nanoparticles, thus inhibiting their uptake into cells. Hydrophilic polymers and nonionic surfactants are commonly used to modify the surface of nanoparticles and prevent nonspecific adsorption of serum proteins.11,12 Several studies have shown that conjugation of PEG on the surface of nanoparticles has improved their blood circulation half-lives. 11,12 Recently, it has been reported that coupling albumin onto the surface of pegylated liposomes reduces association of serum proteins.13,14
In this study, we have investigated the effect of albumin on the cellular uptake of targeted EHCO/siRNA nanoparticles in the presence of serum. Albumin is the most abundant and multifunctional protein in the human body, and it is widely used as a component in the development of drug delivery systems because of its dysopsonic activity, biodegradability, and biocompatibility.15–19 Furthermore, albumin preferentially accumulates in tumor and inflamed tissues.15 Thus, we have hypothesized and investigated that the incorporation of bovine serum albumin into targeted EHCO/siRNA nanoparticles enhances their cellular uptake in the presence of serum, and hence improves the gene-silencing efficiency of the siRNA/EHCO nanoparticles.
Materials and methods
Materials
N-(1-Aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide] (EHCO) was synthesized as described previously.8 Cyclic c(RGDfK)-PEG (RGD-PEG-Mal), and cyclic c(RADfK)-PEG (RAD-PEG-Mal) conjugates with a maleimido end group were synthesized as described previously. 10 The size of PEG was 3.4 kDa. c(RGDfK), c(RADfK) and Mal-PEG-SCM were obtained from Peptides International Inc (Louisville, KY). RPMI-1640 medium and fetal bovine serum (FBS) were obtained from American Type Culture Collection (ATCC; Rockville, MD). Penicillin, streptomycin, Dulbecco’s phosphate-buffered saline (DPBS) with and without Ca2+ and Mg2+, and trypsin were obtained from Invitrogen (Life Technologies, Carlsbad, CA). MTT was purchased from Molecular Probes® (Life Technologies). Heparin, dimethyl sulfoxide, and bovine serum albumin (BSA), formaldehyde were obtained from Sigma Chemical (Sigma-Aldrich Co, St Louis, MO). Alexa Fluor 647 labeled anti-GFP-siRNA and anti1MIF siRNA was purchased from Qiagen (Venlo, The Netherlands). The sense sequence of siGFP is GCAAGCUGACCCUGAAGUUCAU and its antisense sequence is GAACUUCAGGGUCAGCUUGCCG. The sense sequence of siMIF is 5′-CCGCAACUACAGUAAGCUGdTdT-3′ and i t s ant i sense sequence i s 5′-CAGCUUACUGUAGUUGCGGdTdT-3′. siMIF was used for preparing different EHCO/siRNA nanoparticles for size determination and cytotoxicity studies.
Cell culture
Mouse mammary tumor 4T1 cells were obtained from ATCC. The 4T1 cells, 4T1 cells expressing firefly luciferase (4T1-Luc), and 4T1 cells expressing both firefly luciferase and green fluorescent protein (GFP) (4T1-GLuc) were maintained in cell culture growth media at 37°C in a humidified 5% CO2 atmosphere. The cell culture growth media was prepared by adding 10% FBS, 1% 5000 units/mL penicillin, and 1% 5000 μg/mL streptomycin to RPMI-1640 medium. The 4T1, 4T1-Luc cells were used for the determination of intracellular uptake of nanoparticles using flow cytometry, and for the evaluation of cytotoxicity of nanoparticles using MTT assay. 4T1-GLuc cells were used for determining intracellular uptake and gene silencing efficacy of nanoparticles using confocal microscopy.
Preparation and characterization of EHCO/siRNA nanoparticles
RGD-targeted EHCO/siRNA nanoparticles (N/P = 10) were prepared as follows. Initially, 2.7 μL EHCO (15 mM) was mixed with 2.7 μL RGD-PEG-Mal (1.5 μg/μL) and incubated at room temperature for 30 minutes on a shaker. The concentration of RGD-PEG-Mal relative to EHCO in solution was 2.5 mol-%. Subsequently, 21.3 μL siRNA (0.25 μg/μL) was added to the above solution and incubated at room temperature for 30 minutes on a shaker. The addition of 27 μL nanoparticles to 2 mL transfection media gave a final siRNA concentration of 200 nM. The volumes of siRNA, RGD-PEG-Mal, and EHCO were either scaled-up or scaled-down depending on the requirement of final concentration of siRNA in the nanoparticles and/or volume of transfection. Unmodified EHCO/siRNA nanoparticles, and EHCO/siRNA nanoparticles conjugated with mPEG, and mPEG-RAD were similarly prepared. In some cases, equal volumes of peptide-targeted EHCO/siRNA nanoparticles and either FBS or BSA were mixed and incubated for 30 minutes on a shaker at room temperature, before adding to the transfection media. siRNA tagged with fluorophores (Alexa Fluor® 647; Life Technologies) was used in the preparation of nanoparticles for confocal microscopy and flow cytometric analysis. The size of the nanoparticles was determined by dynamic light scattering with a Brookhaven Instruments Corp (Holtsville, NY) BI-200SM system equipped with a 5 mW helium neon laser with a wavelength output of 633 nm.
Cellular uptake of siRNA/EHCO nanoparticles
The cells 4T1 and 4T1-Luc were cultured in 6-well plates. Cells were then transfected with nanoparticles in the transfection media at 37°C in a humidified 5% CO2 atmosphere. RPMI media containing 0%, 10%, 25%, and 50% FBS were used as transfection media. Following 4 hours of transfection with nanoparticles, cells were washed twice with DPBS, trypsinized, collected, and fixed in 1% formalin. Samples were analyzed using a FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ). Software CellQuest (BD Biosciences) and WinList (Verity Software House Inc, Topsham, ME) were used to analyze the results. Untreated cells were used as a negative control for cells treated with different EHCO/siRNA nanoparticles. Alexa Fluor 647-tagged siGFP was used in the preparation of nanoparticles for quantifying cellular uptake of nanoparticles. The percentage of increase in cellular uptake of targeted nanoparticles was quantified by subtracting fluorescence in the untreated cell population from that in the treated cell population.
Cytotoxicity studies
The cytotoxicity of the targeted EHCO/siRNA nanoparticles coated with BSA was evaluated using an MTT cell proliferation assay kit (Molecular Probes) at 100 nM and 200 nM siRNA concentrations. Untreated cells, and cells treated with the targeted nanoparticles without the BSA coating were used as controls. MTT assay was performed according to the manufacturer’s protocol to determine the cytotoxicity of nanoparticles. 4T1 cells (104 per well) were cultured in 96 well plates. The following day, the cells were treated with the nanoparticles, and incubated for 24 and 48 hours at 37°C in a humidified 5% CO2 atmosphere, respectively. The medium was replaced by 110 μL fresh cell culture growth medium containing 10 μL 12 mM MTT and incubated at 37°C for 4 hours. Subsequently, 85 μL medium was removed from the wells and 50 μL dimethyl sulfoxide was then added to each well, mixed thoroughly, and incubated at 37°C for 10 minutes. The absorbance was read at 540 nm using a SpectraMax M5 microplate reader (Molecular Devices, Sunnyville, CA). The relative cell viability was calculated according to the equation ([Abs]Sample − [Abs]Blank)/([Abs]Control − [Abs]Blank) × 100%. Absorbance measured from the wells of untreated cells was used as a control, and absorbance measured from wells of no cells but containing MTT with fresh medium, was used as blank. Each condition was tested in triplicate.
Confocal microscopy
Confocal microscopic fluorescence imaging was used to qualitatively compare cellular uptake and gene-silencing efficiency of targeted EHCO/siRNA nanoparticles coated with and without BSA. 4T1-GLuc cells were cultured in 35 mm glass bottom culture dishes (Mat Tek Corporation, Ashland, MA). Cells were then transfected with the targeted EHCO/AF-647 siGFP nanoparticles coated with and without the BSA for 4 hours, and incubated at 37°C in a humidified 5% CO2 atmosphere. The transfection media was replaced 4 hours later by cell culture growth media, and incubated at 37°C in a humidified 5% CO2 atmosphere. After 48 hours, the samples were washed twice with DPBS, and replaced with fresh cell culture growth media. An Olympus confocal imaging system (IX81; Olympus Corp, Tokyo, Japan) was used to image the cells. Alexa Fluor-647-tagged siGFP was the siRNA used in the preparation of nanoparticles.
Statistics
One-way analysis of variance with Tukey–Kramer multiple comparisons test was used to evaluate the difference between treatment levels using NCSS and PASS software (NCSS LLC, Kaysville, UT). P values ≤ 0.05 were considered significantly different, unless otherwise specified. All error bars represent mean ± standard error.
Results and discussion
EHCO is a polymerizable multifunctional surfactant with pH-sensitive amphiphilicity (Figure 1) designed for the systemic delivery of nucleic acids.8–10 Our previous studies have demonstrated that EHCO forms stable nanoparticles with siRNA via electrostatic interaction and hydrophobic condensation. This resulted in efficient intracellular siRNA delivery and effective gene silencing with minimal cytotoxicity as compared with commercially available transfection agents. The other important feature is that sulfhydryl groups present in EHCO can be utilized for surface modification of the nanoparticles with cell-specific targeting moieties.
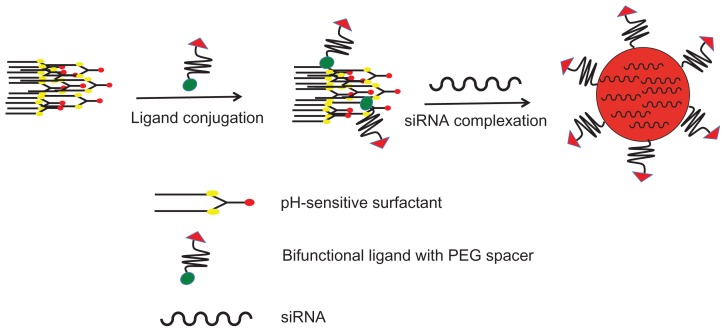
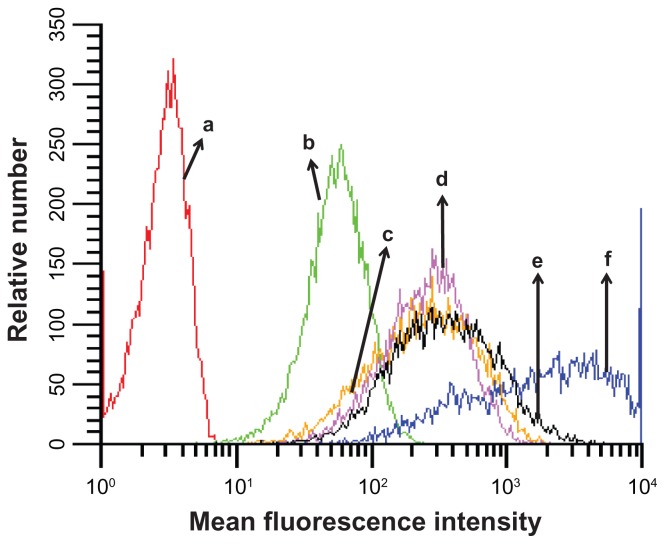
The preparation of the cyclic RGD targeted EHCO/siRNA nanoparticles with a bifunctional PEG spacer is illustrated in Figure 2. RGD peptide was introduced onto the surface of EHCO/siRNA nanoparticles by first conjugating EHCO with 2.5 mol-% RGD-PEG-Mal via a thioether bond, subsequently followed by complexation with siRNA. A 3400 Da PEG spacer was used in the preparation of nanoparticles to minimize nonspecific cellular uptake of targeted EHCO/siRNA nanoparticles. Unmodified EHCO/siRNA nanoparticles, and nontargeted EHCO/siRNA nanoparticles modified with arginine-alanine-aspartic acid (RAD)-PEG-Mal, and PEG-Mal, were prepared as control nanoparticles. The mean diameter of all the nanoparticles was in the range of 50–70 nm, as measured by dynamic light scattering (Table 1). The conjugation agents utilized for the modification of EHCO/siRNA nanoparticles, and sizes of the nanoparticles are reported in Table 1. The modification of the EHCO/siRNA nanoparticles with PEG-Mal, RAD-PEG-Mal and RGD-PEG-Mal only slightly increased the size of the particles. The uptake of these nanoparticles in 4T1 cells was first determined by flow cytometry, after transfection in serum free medium. Figure 3 shows the uptake of different EHCO and Alexa Fluor-647 tagged siRNA nanoparticles in 4T1 cells. The unmodified EHCO/siRNA nanoparticles exhibited high siRNA uptake in 4T1 cells. Modification of the nanoparticles with mPEG-Mal showed reduced nonspecific cellular uptake. However, modifying nanoparticles with RGD-PEG-Mal showed higher cellular uptake compared with nanoparticles modified with mPEG-Mal and RAD-PEG-Mal. This improved cellular uptake observed with the RGD-targeted nanoparticles was mainly due to the receptor-mediated endocytosis. The interactions between highly expressed integrin receptors (αvβ3 and α4β1) on 4T1 cells and their ligand (RGD) might facilitate cell surface retention and subsequent internalization via receptor-mediated endocytosis. These results were consistent with our previously published results in U87 glioblastoma cells.10
Figure 2.
Preparation of pegylated targeted EHCO/siRNA nanoparticles.
Abbreviations: EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; siRNA, short-interfering RNA; PEG, polyethylene glycol.
Table 1.
Conjugation agents and sizes of various EHCO/siRNA nanoparticles
| Type of EHCO/siRNA nanoparticle | Conjugation agent | Size range (nm) |
|---|---|---|
| Unmodified EHCO/siRNA nanoparticles | – | 53 ± 3.3 |
| Pegylated EHCO/siRNA nanoparticles | PEG-Mal | 66 ± 2.0 |
| RAD-targeted EHCO/siRNA nanoparticles | RAD-PEG-Mal | 58 ± 1.4 |
| RGD-targeted EHCO/siRNA nanoparticles | RGD-PEG-Mal | 56 ± 5.4 |
Note: The sizes of nanoparticles are presented as mean ± standard error (SE) of n = 3 different experiments.
Abbreviations: EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; siRNA, short-interfering RNA.
Figure 3.
Cellular uptake of different EHCO/siRNA nanoparticles. The cellular uptake of different nanoparticles was measured using flow cytometric analysis. The transfection was conducted in serum free media (RPMI-1640). AF-647 siGFP is the siRNA used in the preparation of nanoparticles. [AF-647 siGFP] = 100 nM; (a) untreated 4T1 cells; (b) siRNA; (c) PEG/EHCO/siRNA; (d) PEG-RAD/EHCO/siRNA; (e) PEG-RGD/EHCO/siRNA; (f) EHCO/siRNA.
Abbreviations: EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; siRNA, short-interfering RNA; PEG, polyethylene glycol.
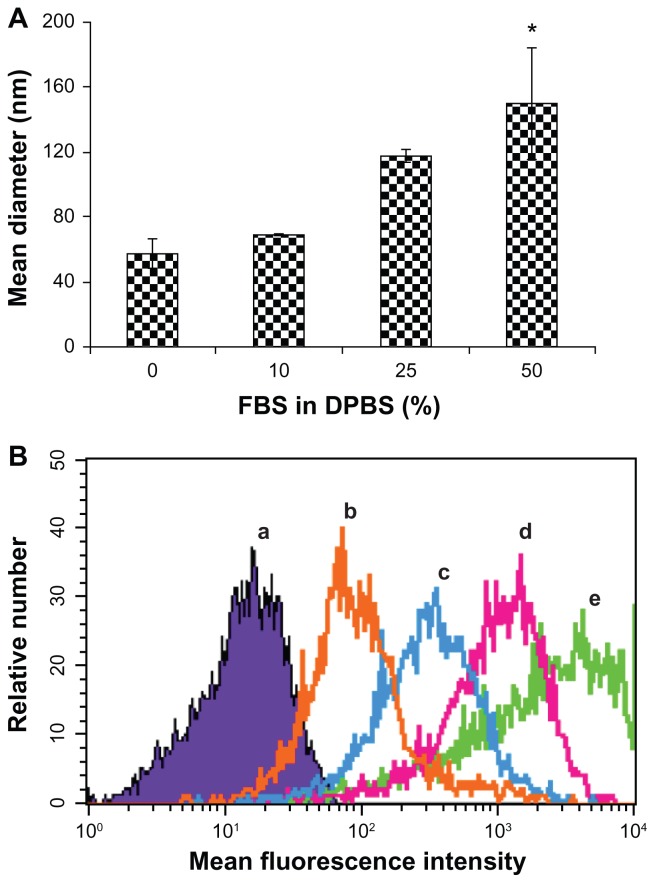
The therapeutic efficacy of an siRNA delivery system depends on the ability of the system to deliver siRNA into the target cells. Their ability to enter into cells can be altered when serum molecules interact with nanoparticles after intravenous administration. Such interactions can change the physiochemical properties of nanoparticles, such as size and stability. Therefore, we initially probed how the interactions of serum molecules with siRNA delivery systems influence their physiochemical properties and their uptake into cells, using in vitro assays. The effect of serum molecules on the stability and cellular uptake of the RGD-PEG-Mal modified EHCO/siRNA nanoparticles is shown in Figure 4. The size of the targeted EHCO/siRNA nanoparticles increased with the concentration of FBS in DPBS, as determined using dynamic light scattering (Figure 4A). The size of the nanoparticles increased from 57.4 ± 9.0 nm in DPBS to 149.7 ± 33.8 nm in 50% FBS in DPBS. This is possibly due to the adsorption of serum proteins on the targeted nanoparticles and the subsequent aggregation of the nanoparticles. The cellular uptake of the targeted nanoparticles decreased as the concentration of FBS increased in the transfection medium, as shown by flow cytometry (Figure 4B). The adsorption of serum proteins may alter the surface properties of the targeted nanoparticles and inhibit the receptor-mediated cellular uptake of the targeted nanoparticles. Although the biocompatible polymer PEG was incorporated as a spacer on the surface of nanoparticles to reduce the adsorption of serum proteins,11,12 it appears that PEG alone is not sufficient to prevent nonspecific adsorption of serum proteins on the targeted nanoparticles.
Figure 4.
Serum influences size and cellular uptake of targeted EHCO/siRNA nanoparticles. (A) Equal volumes of targeted EHCO/siRNA nanoparticles and FBS in DPBS at indicated concentrations were mixed and the size of the nanoparticles was measured using DLS ([siMIF] = 250 nM). The error bars represent mean ± standard error (SE); n = 3 experiments. *P ≤ 0.15, compared with 0 and 10% FBS in DPBS treatment groups. (B) 4T1-Luc cells were transfected with RGD-targeted EHCO/siRNA nanoparticles for 4 hours in transfection media containing 0, 10%, 25%, and 50% FBS in RPMI-1640 media and cellular uptake of the nanoparticles was analyzed using flow cytometric analysis ([AF-647 siGFP] = 200 nM; (a) untreated cells; (b) 50% FBS in RPMI-1640; (c) 25% FBS in RPMI-1640; (d) 10% FBS in RPMI-1640; (e) 0% FBS in RPMI-1640).
Abbreviations: EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; siRNA, short-interfering RNA; FBS, fetal bovine serum; DPBS, Dulbecco’s phosphate-buffered saline; DLS, dynamic light scattering.
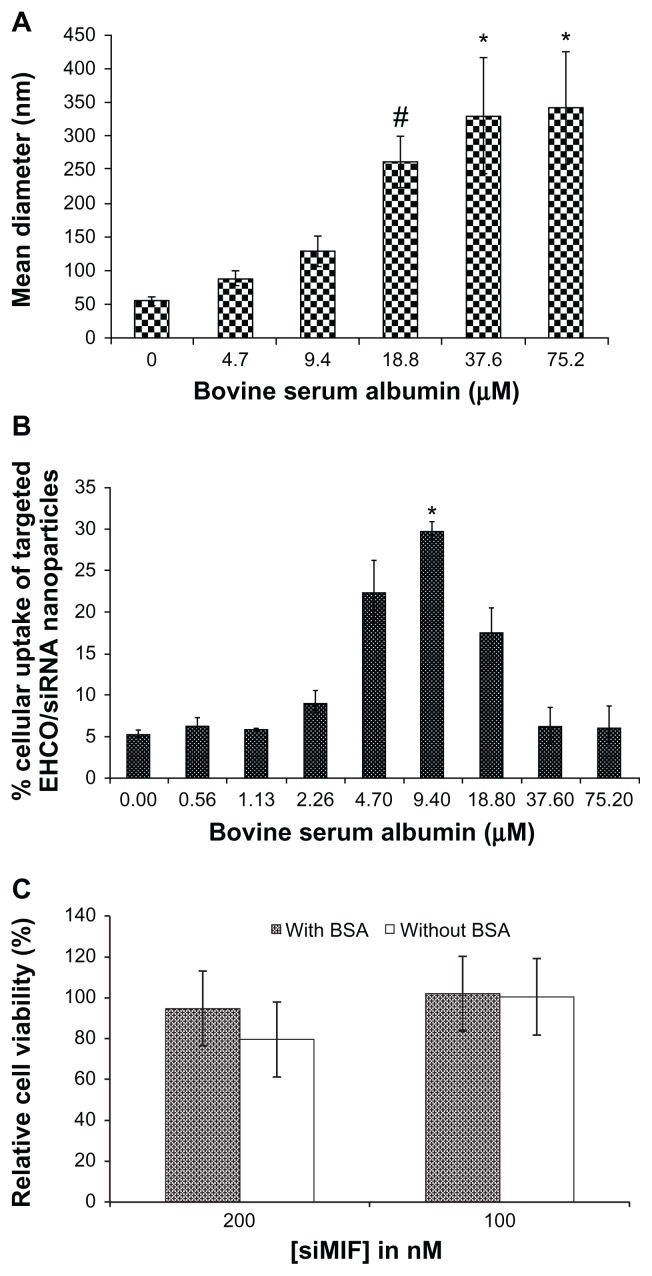
Minimizing such nonspecific interactions of serum molecules with targeted siRNA nanoparticles is critical for effective cellular internalization of siRNA delivery systems. In addition to its nonimmunogenic and nontoxic properties,15–19 albumin is inert to other serum molecules, and is used as a blocking agent in many in vitro molecular-and cellular-based assays. Also, it has been incorporated into various drug delivery systems.15–19 Therefore, we used BSA to modify the targeted EHCO/siRNA nanoparticles, and determined the effect of serum albumin on the stability and intracellular uptake of the targeted nanoparticles. The size of the nanoparticles increased with BSA concentration, as shown in Figure 5A. The size of the nanoparticles was 342.2 ± 84.0 nm at 75.2 μM BSA concentrations. Statistical analysis was performed with analysis of variance coupled with the Tukey–Kramer multiple comparison test, to determine how BSA coating influenced the size of nanoparticles. Significant aggregation of the nanoparticles was observed at or above 18.8 μM BSA concentrations (P ≤ 0.15). We also observed nanoparticle precipitation at BSA concentrations above 75.2 μM. The concentration of BSA also had a significant impact on the cellular uptake of the targeted EHCO/AF-647 siGFP nanoparticles treated with BSA in 4T1 cells in transfection medium containing 50% FBS (Figure 5B). The cellular uptake of the nanoparticles increased with BSA concentration up to 9.4 μM (P ≤ 0.05), and then decreased with further increase in BSA concentration. The decrease in the cellular uptake of the nanoparticles might be attributed to the formation of large particles (>250 nm) when the BSA concentration reached 18.8 μM or higher, which could not be endocytosed by the cells. BSA coating reduced the cytotoxicity of the targeted EHCO/siRNA nanoparticles at the high siRNA concentration, as shown by the MTT assay (Figure 5C).
Figure 5.
The effect of BSA on the size, cellular uptake and cytotoxicity of the targeted nanoparticles. (A) The size of targeted EHCO/siRNA nanoparticles measured by DLS indicates that BSA coating increases the size of nanoparticles. #P ≤ 0.15, compared with 0 μM BSA treatment group; *P ≤ 0.15, compared with 0, 4.7, and 9.4 μM BSA treatment groups. (B) Cellular uptake of targeted EHCO/siRNA nanoparticles coated with and without BSA in 4T1 cells was measured by flow cytometric analysis ([AF-647-siGFP] = 50 nM, 50% FBS in RPMI-1640). *P ≤ 0.05, compared with all other BSA treatment groups. (C) The viability of 4T1 cells was measured after they were transfected with the nanoparticles coated without and with BSA for 48 hours. BSA coating concentration was 9.4 μM.
Abbreviations: BSA, bovine serum albumin; EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; siRNA, short-interfering RNA; DLS, dynamic light scattering; FBS, fetal bovine serum.
It is known that the size and nature of the delivery system influences intracellular uptake of nanoparticles. However, the optimal size of nanoparticles for highest intracellular delivery needs to be determined for each delivery system. As shown in Figure 5, the nanoparticles coated with 9.4 μM BSA (about 130 nm) showed highest intracellular delivery of siRNA compared with nanoparticles coated with BSA concentrations above and below 9.4 μM BSA. At 18.8 μM or higher BSA concentrations, the cellular uptake of nanoparticles was reduced as nanoparticles aggregated and formed bigger particles, which could not be endocytosed by the cells. However, at BSA concentrations lower than 9.4 μM BSA, it is possible that BSA coating was not sufficient to prevent interference of serum molecules with nanoparticles. Such nonspecific interactions might have reduced intracellular uptake of nanoparticles. Therefore, for our targeted EHCO/siRNA delivery system, 9.4 μM BSA was the optimal coating concentration that prevented serum molecules interacting with nanoparticles, and helped improve intracellular delivery of siRNA in the presence of serum.
It is surprising that the sizes of siRNA nanoparticles did not show a similar trend in BSA-and FBS-containing buffers. A normal range of human serum albumin is 35–50 g/L or 520–750 μM. If serum albumin concentrations are similar among species, 10% FBS and 50% FBS might contain 52–75 μM and 260–375 μM albumin. Therefore, the size of nanoparticles coated with 75.2 μM BSA- and 10% FBS-(52–75 μM) containing buffers should be at the same order of magnitude. However, coating with 75.2 μM BSA resulted in 4.5 times bigger particles than coating with 10% FBS. Although the reason is unknown to us, we suspect that as serum contains other molecules such as immunoglobulins in addition to albumin, the actual molecules that first encounter nanoparticles could have major effect on the physiochemical properties of nanoparticles. Such complex molecular interactions might have first prevented serum album from interacting with nanoparticles in FBS-containing buffer. Furthermore, such serum molecules might have masked nanoparticles from interacting with target cells and thus inhibits internalization of the nanoparticles.
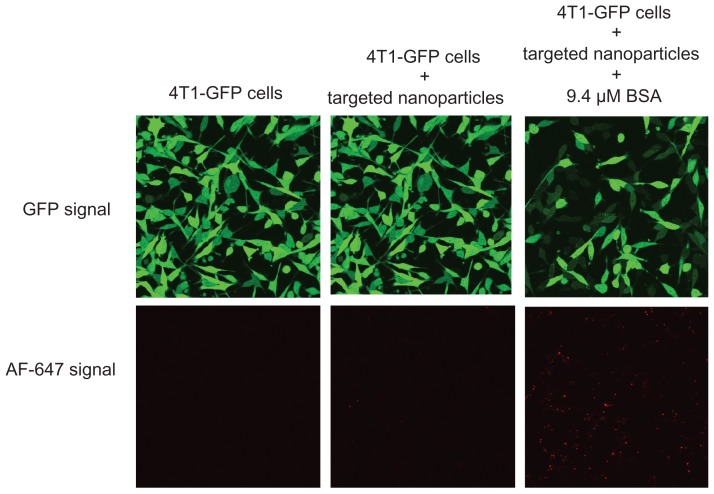
The cellular uptake and gene-silencing efficiency of the targeted EHCO/siRNA nanoparticles coated with BSA were further evaluated by confocal microscopy. 4T1-GLuc cells were transfected with the targeted EHCO/siRNA nanoparticles, containing AF-647 tagged siGFP, coated with and without 9.4 μM BSA for 4 hours in 50% FBS-containing medium. Untreated 4T1-GLuc cells were used as a negative control. After 4 hours, the transfection media were replaced with cell culture growth media, and then incubated at 37°C in a humidified 5% CO2 atmosphere for 44 hours. As shown in Figure 6, 4T1-GLuc cells treated with the targeted nanoparticles coated with BSA showed a simultaneous increase in the uptake of nanoparticles (red signal) and decreased GFP expression (green signal), while the cells treated with the targeted nanoparticles without the BSA coating showed low uptake and low gene-silencing efficiency. The cellular uptake and gene-silencing efficiency of the targeted EHCO/siRNA nanoparticles coated with albumin can be further improved by optimizing the molar ratios of EHCO and siRNA.
Figure 6.
BSA coating enhanced cellular uptake and gene-silencing efficiency of RGD-targeted EHCO/siRNA nanoparticles.
Notes: 4T1-GLuc cells were transfected with RGD-targeted nanoparticles ([AF-647-siGFP] = 50 nM) coated without and with BSA for 4 hours and then replaced with cell culture growth media. Images were taken using Olympus confocal microscopy at 40× magnification 48 hours later (Transfection media = 50% FBS in RPMI-1640).
Abbreviations: BSA, bovine serum albumin; EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; GFP, green fluorescent protein; siRNA, short-interfering RNA; FBS, fetal bovine serum.
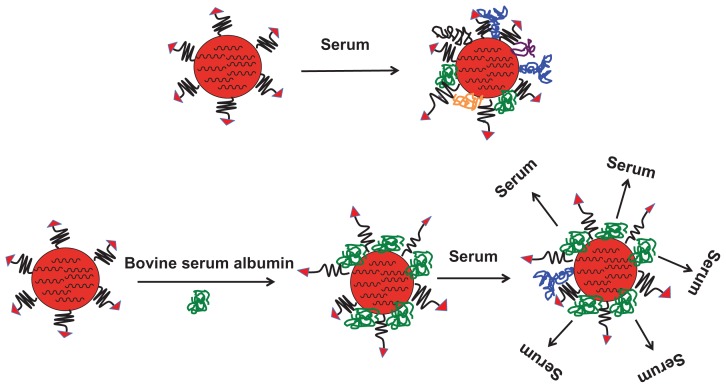
The possible mechanism for improved cellular uptake of the RGD-PEG-modified EHCO/siRNA nanoparticles coated with BSA is illustrated in Figure 7. When nanoparticles are exposed to serum, serum molecules adsorb onto the surface of nanoparticles, resulting in reduced cellular uptake. The extent of reduction in cellular uptake depends on what type of serum molecules adsorb, and how they orient on the surface of nanoparticles. The size and shape of serum molecules vary significantly from one another. For example, antibodies are globular proteins and their hydrodynamic diameter varies from 10 nm (IgG) to 35 nm (IgM), whereas fibronectins are rod-like molecules and each strand of fibronectin is 60 nm in length and 2.5 nm in diameter.20,21 Furthermore, serum molecules can polymerize in solution and form bigger molecules. It is also true that some serum molecules form complexes by interacting with other molecules. Therefore, it is highly complicated to predict what type of serum protein coats the nanoparticles. However, the type of serum molecule that adsorbs on the surface influences the size and stability of the nanoparticles, which can affect the cellular uptake of nanoparticles. Biocompatible polymer PEG is commonly used to modify the nanoparticle surface, to minimize serum protein coating onto the nanoparticles. As observed in this study, it seems that the PEG spacer (20 nm length) was not sufficient to block adsorption of serum molecules onto the EHCO/siRNA nanoparticles. Similar observations have previously been reported for pegylated liposomes and polystyrene nanoparticles. 13,14,19 We have shown in this study that the treatment of the targeted siRNA nanoparticles with 9.4 μM BSA (7.5 nm hydrodynamic diameter) has the potential to minimize serum protein coating, and to improve intracellular siRNA delivery and gene silencing, in serum containing medium. It has been reported that albumin might have endosomolytic activity,19 which could further facilitate nanoparticles’ escape from endosomal-lysosomal compartments. The unique dysopsonic and endosomolytic activities of albumin are attractive features that could contribute to improved in vivo gene delivery and gene-silencing efficiency of the targeted EHCO/siRNA nanoparticles. Thus, we reason that introduction of BSA on pegylated targeted EHCO/siRNA nanoparticles prevented adsorption of serum proteins. In addition, the incorporation of BSA between PEG-RGD chains will possibly create a brushlike conformation and favorable interactions of the targeting agent with integrins, and allow receptor-mediated endocytosis of targeted EHCO/siRNA nanoparticles.
Figure 7.
Illustration of a possible mechanism for improved cellular uptake of targeted EHCO/siRNA nanoparticles coated with BSA.
Abbreviations: EHCO, N-(1-aminoethyl)iminobis[N-(oleoylcysteinylhistinyl-1-aminoethyl)propionamide]; BSA, bovine serum albumin.
Albumin has been used for in vivo drug delivery. Previously, it has been shown that precoating of pegylated liposomes and polystyrene nanoparticles with albumin inhibited the association of opsonins on the surface of nanoparticles.13,14 When delivered intravenously, these albumin-coated particles exhibited decreased hepatic disposition and resulted in longer blood circulation times.13,14 In our study, we demonstrated that the precoating of lipid-based nanoparticles with 9.4 μM BSA prior to transfection in serum media overcame the inhibition of intracellular uptake of the lipid-based nanoparticles without BSA. Our study, together with previous studies, demonstrates that precoating of lipid-based nanoparticles with albumin at a proper concentration has the potential to decrease hepatic disposition, improve blood circulation times, and improve intracellular uptake of intravenously delivered nanoparticles. Currently, we are further investigating the effect of albumin modification on systemic in vivo siRNA delivery with the multifunctional delivery system.
Conclusion
In this study, we showed that serum proteins could coat targeted EHCO/siRNA nanoparticles and inhibit cellular uptake of the nanoparticles. However, pretreatment of targeted EHCO/siRNA nanoparticles with albumin at a proper concentration could minimize serum protein coating on the surface of the nanoparticles. It appears that coating of the EHCO/siRNA nanoparticles with 9.4 μM BSA reduced the cytotoxicity of the nanoparticles at a high siRNA concentration. The pretreatment of targeted EHCO/siRNA nanoparticles with 9.4 μM BSA prior to transfection improved cellular uptake and gene silencing efficacy of nanoparticles in serum-containing media. At a proper concentration, albumin has the potential to minimize opsonization of siRNA nanoparticles for effective systemic in vivo siRNA delivery.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kurreck J. RNA interference: from basic research to therapeutic applications. Angew Chem Int Ed Engl. 2009;48(8):1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann TS, Lee AC, Akinc A, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441(7089):111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 5.Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akinc A, Zumbuehl A, Goldberg M, et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat Biotechnol. 2008;26(5):561–569. doi: 10.1038/nbt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Love KT, Mahon KP, Levins CG, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107(5):1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang XL, Ramusovic S, Nguyen T, Lu ZR. Novel polymerizable surfactants with pH-sensitive amphiphilicity and cell membrane disruption for efficient siRNA delivery. Bioconjug Chem. 2007;18(6):2169–2177. doi: 10.1021/bc700285q. [DOI] [PubMed] [Google Scholar]
- 9.Wang XL, Xu R, Lu ZR. A peptide-targeted delivery system with pH-sensitive amphiphilic cell membrane disruption for efficient receptor-mediated siRNA delivery. J Control Release. 2009;134(3):207–213. doi: 10.1016/j.jconrel.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Wang XL, Xu R, Wu X, Gillespie D, Jensen R, Lu ZR. Targeted systemic delivery of a therapeutic siRNA with a multifunctional carrier controls tumor proliferation in mice. Mol Pharm. 2009;6(3):738–746. doi: 10.1021/mp800192d. [DOI] [PubMed] [Google Scholar]
- 11.Owens DE, 3rd, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307(1):93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 12.Li SD, Huang L. Pharmacokinetics and biodistribution of nanoparticles. Mol Pharm. 2008;5(4):496–504. doi: 10.1021/mp800049w. [DOI] [PubMed] [Google Scholar]
- 13.Furumoto K, Yokoe J, Ogawara K, et al. Effect of coupling of albumin onto surface of PEG liposome on its in vivo disposition. Int J Pharm. 2007;329(1–2):110–116. doi: 10.1016/j.ijpharm.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Yokoe J, Sakuragi S, Yamamoto K, et al. Albumin-conjugated PEG liposome enhances tumor distribution of liposomal doxorubicin in rats. Int J Pharm. 2008;353(1–2):28–34. doi: 10.1016/j.ijpharm.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Kratz F. Albumin as a drug carrier: design of prodrugs, drug conjugates and nanoparticles. J Control Release. 2008;132(3):171–183. doi: 10.1016/j.jconrel.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Xu R, Fisher M, Juliano RL. Targeted albumin-based nanoparticles for delivery of amphipathic drugs. Bioconjug Chem. 2011;22(5):870–878. doi: 10.1021/bc1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Low K, Wacker M, Wagner S, Langer K, von Briesen H. Targeted human serum albumin nanoparticles for specific uptake in EGFR-Expressing colon carcinoma cells. Nanomedicine. 2011;7(4):454–463. doi: 10.1016/j.nano.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Sebak S, Mirzaei M, Malhotra M, Kulamarva A, Prakash S. Human serum albumin nanoparticles as an efficient noscapine drug delivery system for potential use in breast cancer: preparation and in vitro analysis. Int J Nanomedicine. 2010;5:525–532. doi: 10.2147/ijn.s10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar CSSR. Biological and Pharmaceutical Nanomaterials. 1st ed. Weinheim, Germany: Wiley-VCH; 2006. [Google Scholar]
- 20.Foley JO, Nelson KE, Mashadi-Hossein A, Finlayson BA, Yager P. Concentration gradient immunoassay. 2. Computational modeling for analysis and optimization. Anal Chem. 2007;79(10):3549–3553. doi: 10.1021/ac062350v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacDonald DE, Markovic B, Allen M, Somasundaran P, Boskey AL. Surface analysis of human plasma fibronectin adsorbed to commercially pure titanium materials. J Biomed Mater Res. 1998;41(1):120–130. doi: 10.1002/(sici)1097-4636(199807)41:1<120::aid-jbm15>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]