SUMMARY
Argonaute proteins lie at the heart of the RNA-induced silencing complex (RISC), wherein they use small RNA guides to recognize targets. Initial insight into the architecture of Argonautes came from studies of prokaryotic proteins, revealing a crescent-shaped base made up of the amino-terminal, PAZ, middle, and PIWI domains. The recently reported crystal structure of human Argonaute-2 (hAgo2), the “slicer” in RNA interference, in complex with a mixed population of RNAs derived from insect cells provides insight into the architecture of a eukaryotic Argonaute protein with defined biochemical and biological functions. Here, we report the structure of human Ago2 bound to a physiologically relevant microRNA, microRNA-20a, at 2.2 Å resolution. The miRNA is anchored at both ends by the Mid and PAZ domains and makes several kinks and turns along the binding groove. Interestingly, miRNA binding confers remarkable stability on hAgo2, locking this otherwise flexible enzyme into a stable conformation.
INTRODUCTION
A universal feature of RNAi-related pathways is their reliance on Argonaute family proteins, which bind small RNAs and use these as guides to select their silencing targets (Joshua-Tor and Hannon, 2011). Early work suggested association between Argonaute and small RNAs within RISC (Hammond et al., 2001). This was confirmed by studies of the isolated PAZ (P-element induced wimpy testes [PIWI], Argonaute and Zwille) domain, whose structure revealed an oligonucleotide-binding fold that engaged the 3′ ends of small RNA guides (Lingel et al., 2003, 2004; Ma et al., 2004; Song et al., 2003; Yan et al., 2003). Their 5′ ends occupy a pocket within the Argonaute Mid domain, which may confer 5′ end nucleotide preferences upon some Ago family members (Boland et al., 2010; Frank et al., 2010; Ma et al., 2005; Mi et al., 2008; Parker et al., 2005).
A critical insight came with the realization that Argonaute proteins were directly responsible for small RNA-directed silencing, embodying the endonucleolytic “slicer” activity of RISC (Song et al., 2004). The PIWI domain resembled an RNase H enzyme, a relationship that could not be inferred from primary sequence but which was apparent from similarity of their tertiary structures. This type of enzyme was consistent with the known biochemistry of the slicing reaction, which produced 5′ phosphate and 3′ hydroxyl termini (Martinez and Tuschl, 2004; Schwarz et al., 2004). It also pointed to key aspartate residues within a presumed Argonaute active site (Liu et al., 2004; Song et al., 2004). Reconstitution experiments with purified components verified that the minimal complex needed for target cleavage is hAgo2, a guide strand, and Mg2+ (MacRae et al., 2008; Rivas et al., 2005). The Argonaute active site is proximal to a large positively charged groove, which could accommodate the small RNA guide and its base-paired target, placing the scissile bond directly adjacent to the active site (Song et al., 2004). The two other domains in the structure, the N-terminal (N) domain and the Mid domain are apparent in all Argonaute structures solved to date.
A series of structures of the bacterial Thermus thermophilus Argonaute (TtAgo) bound to various combinations of DNA guide strands and RNA and DNA target strands (Wang et al., 2008b; Wang et al., 2009; Wang et al., 2008c) provided a great deal of insight into the conformational changes in both the protein and the RNA that accompany the action of the slicer enzyme. Two structures with substrates of different lengths (Wang et al., 2009) illustrate the range of motions that the PAZ domain can undergo in the catalytic cycle. Moreover, they reveal the disassociation of the 3′ end of the guide from the PAZ domain upon target binding as first proposed by Zamore and colleagues based upon their biochemical characterization of RISC (Schwarz et al., 2004).
Our understanding of the architecture of full-length Argonaute proteins has until recently been based on structures of prokaryotic Argonautes of unknown biochemical and biological function. Insight into the architecture of eukaryotic Argonautes initially came from individual protein domains, including early studies of human and Drosophila PAZ domains (Lingel et al., 2003, 2004; Ma et al., 2004; Song et al., 2003; Yan et al., 2003) and structures of Mid domains from the Neurospora crassa Argonaute QDE-2 (Boland et al., 2010) and from hAgo2 (Frank et al., 2010). These revealed a structural basis for 5′-nucleotide recognition of the guide strand and for the sequence bias toward uridine monophosphate that is observed in RNAs bound to eukaryotic Argonautes. Subsequently, the structure of the Mid-PIWI lobe of QDE-2 was determined (Boland et al., 2011), hinting to a conserved mode of nucleic acid binding between prokaryotic and eukaryotic Argonautes. Recently, Schirle and Macrae (2012) reported the crystal structure of full-length human Argonaute-2 bound to a mixed population of RNA guides. This study revealed the architecture of hAgo2, showing that the overall domain organization and mode of small RNA binding is similar to that of prokaryotic Argonautes. These findings also begin to establish a structural basis for aspects of Argonaute biology unique to eukaryotes, such as binding of GW family proteins.
Having successfully developed an approach to purify hAgo2 devoid of a nucleic acid partner, we were interested in characterizing the structure of Argonaute-2 bound to a physiologically relevant miRNA. Here, we report the structure of the full-length hAgo2 in complex with miR-20a, to 2.2 Å resolution. We find that the domain organization is strikingly similar to that seen in prokaryotic proteins, as also noted by Shirle and Macrae (2012). However, hAgo2 presents a more open conformation, largely due to the PAZ domain sitting further from the remainder of the protein. The miRNA is bound at both ends—the 5′ end to the Mid domain with some contributions from the PIWI domain, and the 3′ end to the PAZ domain. The seed sequence is in a narrow portion of the RNA binding groove, and several kinks caused by Argonaute residues protruding into the groove control the trajectory of the RNA along the protein. As the miRNA threads along the protein, it interacts with every domain of hAgo2 including its linkers. We find that the RNA confers remarkable stability on the enzyme most likely as a consequence of these interactions being spread along the entire protein. Together with the recent report by Schirle and Macrae (2012), our findings provide key insights into a mammalian Argonaute protein of essential biological functions.
RESULTS
Recombinant hAgo2 Copurifies with Small RNAs
For crystallographic studies, we isolated hAgo2 from baculovirus-infected SF9 cells. Through the course of our purification, hAgo2 showed an unusually low A280/A260 ratio, indicating the potential presence of copurifying nucleic acids. We extracted RNA from hAgo2 fractions at different stages of our purification process and analyzed these by denaturing PAGE. We noted a strong band of ~20 nucleotides (Figure S1A available online), suggesting that hAgo2 might have become loaded with endogenous small RNA species in insect cells.
To investigate the nature of the bound RNAs, we used standard small RNA cloning and followed this with deep sequencing on the Illumina platform. hAgo2 bound small RNAs show a non-Gaussian distribution with a strong enrichment for RNAs of 20 nucleotides (Figure S1B). SF9 cells are derived from the fall armyworm, Spodoptera frugiperda. Because we lack an assembled, annotated genome for this organism, we sequenced small RNA populations from SF9 for comparison to those bound to recombinant hAgo2.
SF9 is thought to be derived from Spodoptera female germ cells. In accord with this notion, we see two general size classes of small RNAs in libraries designed to capture species of 19–29 nucleotides (Figure S1C). One peak, comprising ~19–23 nucleotides is likely to represent endogenous siRNAs and microRNAs. This is supported by comparisons to Drosophila, which reveal multiple shared miRNAs that appear mainly in SF9 cells as 23 nucleotide species. A second peak encompassing ~25–28 nucleotides likely represents armyworm piRNAs, though we failed to find substantial enrichment among these for small RNAs that could be unambiguously mapped to known Drosophila transposons.
To determine whether recombinant hAgo2 was being loaded with bona fide SF9 small RNAs, we compared hAgo2-bound species to the various size categories of SF9 small RNAs. Because we lack a genome against which to filter for only mapping reads, we considered those RNAs represented by more than 10 reads to reduce spurious background in the sequencing. In hAgo2 these comprised ~49,000 different sequences. Of the SF9 small RNAs in the 20–21 nucleotide size range, sequences matching small RNAs in hAgo2 accounted for ~80% of all reads. In the 22–23 nt size range, 86% of all sequence reads could be found among hAgo2 bound species. In contrast, for presumptive piRNAs, only 17% of reads could be accounted for by hAgo2 bound species, and this might be an overestimate because in other insects, piRNAs and endogenous siRNAs often overlap because of their enrichment for transposon content.
We conclude that hAgo2 samples endogenous SF9 small RNA populations. Given the tight size distribution of hAgo2-bound RNAs, it must either select strongly for variants of SF9 small RNAs that are 20 nt in length or initially acquire longer small RNAs that are subsequently trimmed to a size optimal for hAgo2 occupancy. Support for the former hypothesis can be derived from experiments in which a 21 nt human miRNA was coexpressed in SF9 cells with hAgo2. In this case, the size distribution profile shifted toward the 21 nt range when hAgo2 became loaded with this abundant, overexpressed RNA (data not shown).
hAgo2 is normally found in association with a small RNA population that is enriched for U at the 5′ end. In part, this reflects a bias in human miRNAs, but it is also driven by a higher affinity of the 5′-end binding pocket in the Mid domain for U and A residues (Boland et al., 2010). When we analyzed the small RNAs selected by hAgo2 from SF9 cells, we did note a 5′ nucleotide bias with U and A being strongly preferred over C and G (Figure S1D). This bias was slightly greater than that seen in 20 nt SF9 small RNAs (Figure S1E) and in total SF9 small RNA populations (data not shown).
Because our goal was to analyze the structure of hAgo2 in complex with a single, human microRNA, we attempted to exchange bound endogenous species for synthetic RNAs. Though this was unsuccessful, our experiments did indicate the presence of a fraction of hAgo2 that was not loaded with an endogenous RNA and that could accept an exogenously added species.
hAgo2 Is Stabilized by Small RNA Binding
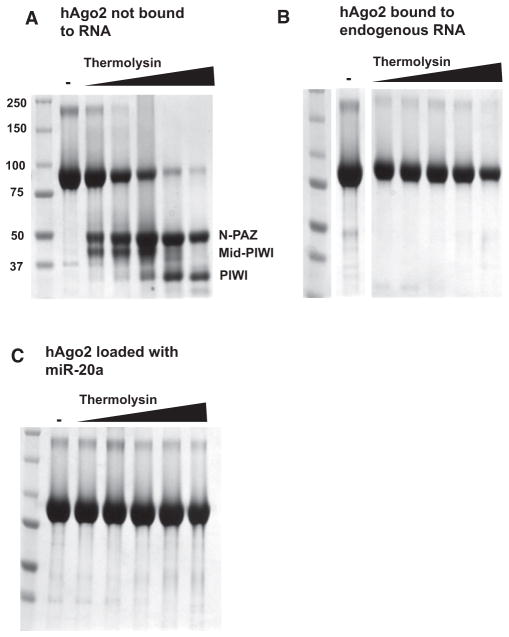
The complex of hAgo2 and endogenous small RNAs copurified with the nonloaded hAgo2 protein through several chromatographic steps. However, we were able to separate these species by cation exchange chromatography (Figure S2A). We investigated the domain structure of hAgo2 in the loaded and non-loaded state by limited proteolytic digestion with thermolysin. The hAgo2 that was not loaded with a small RNA showed a digestion pattern typical of multidomain proteins. Moderate concentrations of protease released portions of hAgo2 that correspond to structured domains. In this case, thermolysin liberated a fragment corresponding to the N-terminal and PAZ domains (N-PAZ), a fragment corresponding to the Mid and PIWI domains (Mid-PIWI), and a fragment comprising the PIWI domain alone (PIWI; Figure 1A). In contrast, hAgo2 that was loaded with a small RNA resisted digestion by thermolysin at concentrations of protease that reduced nearly all of the nonloaded protein to its constituent domains (Figure 1B). Structural differences between the two hAgo2 populations could be conferred by RNA binding or could reflect another difference between these protein fractions, for example a post-translational modification of hAgo2 that occurred in vivo following small RNA loading.
Figure 1. Small RNAs Stabilize hAgo2.
(A) Digestion of hAgo2, purified without bound, endogenous small RNAs (Figure 1, lane “not bound to RNA”) with thermolysin resulted in a digestion pattern of typical multidomain proteins. Increasing concentrations of enzyme (indicated by the wedge) released stable protein fragments corresponding to known domains (indicated), as determined by N-terminal sequencing or mass spectrometry. In contrast, treatment of hAgo2 purified along with endogenous SF9 small RNAs was remarkably protease resistant (B).
(C) Addition of synthetic miR-20a (1–20) to the fraction shown in (A) conferred stability to protease. See also Figures S1 and S2.
We previously found that we could add synthetic guide sequences of various lengths to hAgo2 purified from bacteria (Rivas et al., 2005). Similarly, we could efficiently incorporate RNAs ranging from 12 to 21 nucleotides in length into the hAgo2 that was purified from insect cells without a native small RNA (data not shown). hAgo2 loaded in this manner showed the same unusual resistance to protease digestion as did hAgo2 purified along with SF9-derived guides, strongly indicating that RNA binding is sufficient to lock the protein into a highly stable, protease-resistant structure (Figure 1C). Given its tendency to select 20 nucleotide guides in vivo, we elected to load hAgo2 with a 20 nucleotide small RNA in vitro for crystallographic studies. For this purpose, we synthesized a small RNA corresponding to the first 20 nucleotides of human miR-20a (Figure S2B).
miR-20a is a component of the miR-17-92 cluster, which has been ascribed a variety of context-dependent biological functions. This cluster is important for heart and lung development and for proper lymphoid differentiation (Kent and Mendell, 2006). It can also act as an oncogene or a tumor suppressor depending upon the cell type in which it is expressed (Rizzo et al., 2010). miR-20a itself is highly abundant in ES cells, where it regulates the cell cycle and promotes proliferation (Wang et al., 2008a).
Overall Structure of hAgo2 in the hAgo2-miR-20a Complex
We obtained crystals of full-length hAgo2 in complex with miR-20a (1–20). These diffracted to 2.2 Å resolution. We solved the structure of the complex by Single Anomalous Dispersion (SAD) by using a selenomethionine-substituted protein and refined it to 2.2 Å resolution with Rwork/Rfree of 20.6%/25.4%. For details of the structure solution, data collection, and refinement statistics, please refer to Experimental Procedures and to Table 1.
Table 1.
Data Collection and Refinement Statistics
| Data collection | Native | SeMet |
|---|---|---|
| Cell dimensions | ||
| a, b, c (Å) | 63.4, 107.7, 68.7 | 63.3, 108.2, 68.6 |
| α, β, γ (°) | 90.0, 106.7, 90.0 | 90.0, 107.2, 90.0 |
| Wavelength (Å) | 1.075 | 0.979 |
| Resolution (Å)a | 40.0–2.25 (2.29–2.25) | 40.0–2.62 (2.67–2.62) |
| Rmerge (%)a | 4.3 (30.3) | 4.1 (29.4) |
| I/σ(I)a | 28.8 (2.2) | 38.2 (4.6) |
| Completeness (%)a | 97.6 (92.6) | 100 (99.0) |
| Redundancya | 2.7 (2.5) | 3.9 (3.8) |
| Refinement | ||
| Resolution (Å) | 47.4–2.25 | |
| No. reflections | 41,069 | |
| Rwork/Rfree (%) | 20.6/25.4 | |
| No. atoms (nonhydrogen) | 6,884 | |
| Protein | 6,436 | |
| RNA | 298 | |
| Water | 136 | |
| Phenol | 14 | |
| B-factors (Å2) | ||
| Protein | 57.3 | |
| RNA | 78.0 | |
| Water | 46.8 | |
| Phenol | 50.2 | |
| Rmsds | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.674 | |
| MolProbity statistics | ||
| All atom clashscore | 4.82 | |
| Poor rotamers (%) | 0.98 | |
| Ramachandran outliers (%) | 0.0 | |
| Ramachandran favored (%) | 98.1 | |
| MolProbity score | 1.42 | |
The space group for both native and SeMet protein is P21.
Numbers in parenthesis refer to the highest resolution shell.
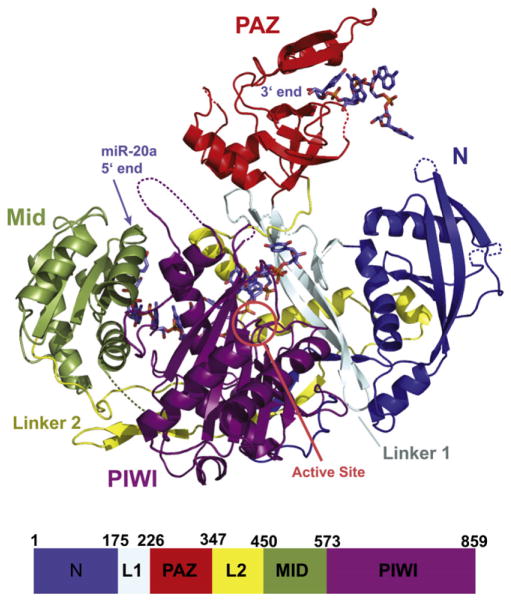
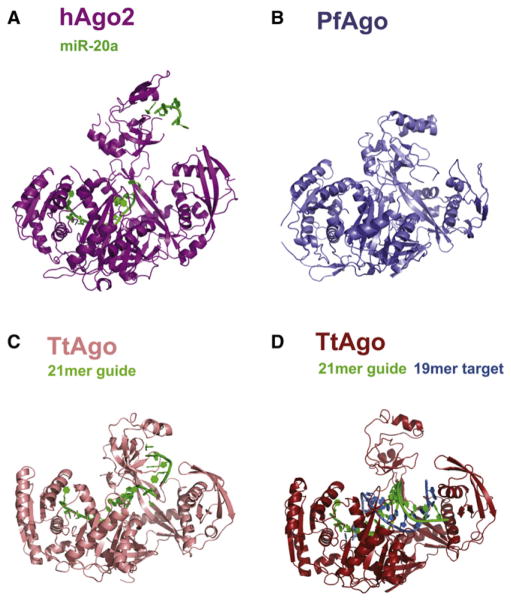
In its overall structure, hAgo2 in complex with miR-20a is very similar to the complex obtained with a mixed population of RNA (Schirle and Macrae, 2012). These structures are remarkably similar to the archaeal and bacterial Argonautes, whose structures were previously determined (Figures 2 and S3). The Mid and PIWI domains form an almost continuous lobe with the N domain completing a crescent or cradle shape. Above this, the PAZ domain wraps over the RNA binding groove. We previously likened the structure of Argonaute to a duck. The crescent formed by the Mid and PIWI domains forms the body, and the PAZ domain serves as the head, with the 3′ end of the RNA guide being held in the bill. The N domain could be thought of as a wing folding over the RNA guide and mRNA substrate. Using this analogy, we found that the neck holding the PAZ domain appears to be elongated in comparison to prokaryotic Argonautes (Figure 3). This makes hAgo2 the most open Argonaute structure to date with the PAZ domain lying further away from the other domains and the “bill” pointing upwards. hAgo2 is even more open than the thermophilus protein (TtAgo) that was crystallized with both the guide and target strands (Figure 3). In the crystal, there is a packing interaction between the PAZ domain and a neighboring molecule in the form of a salt bridge, but it is unlikely that this interaction influences the position of the PAZ domain relative to the body of the protein in our structure.
Figure 2. Structure of hAgo2 in Complex with miR-20a.
A schematic representation of the hAgo2-miR-20a complex is displayed with each domain and interdomain linker labeled. The N domain is shown in blue, the PAZ domain in red, the Mid domain in green and the PIWI domain in purple. The miRNA is shown in stick representation. The 5′ end of miR-20a is bound to the Mid domain. Nucleotides 2 to 10 track along the RNA binding groove, and nucleotides 17 to 20 are bound to the PAZ domain. The location of the active site is circled. For reference, a bar diagram is given below, color-coded as described, indicating the positions of boundaries between structural domains. Other views of the complex are shown in Figure S2.
Figure 3. Structural Comparison of hAgo2, PfAgo, and Tt Ago.
Argonaute proteins across the three kingdoms of life show striking similarities in their overall structures. All structures were superimposed based on their respective PIWI domains and are oriented as in Figure 2. (A) hAgo2-miR-20a complex adopts the most open conformation of all full-length Argonaute structures determined to date. This is compared to the archaeal PfAgo (B), eubacterial TtAgo bound to a 21-mer DNA guide (C), and TtAgo in complex with a 21-mer DNA guide and a 19-mer RNA target (D). See also Figure S4.
Similarity was particularly striking when we aligned prokaryotic and eukaryotic Argonaute structures based upon their PIWI domains. This was true despite the fact that some of these structures were determined in the presence of the guide and/or target strands, whereas others reflected nonloaded proteins. All showed a remarkably similar position and orientation of the N and Mid domains. Thus, our findings and those of Schirle and Macrae (2012) reveal that the overall structure and organization of Argonaute proteins from evolutionarily distant organisms and from all three kingdoms of life has been superbly maintained.
Relative to their prokaryotic counterparts, eukaryotic Argonautes contain a C-terminal insertion, whose structure was revealed by crystallization of a fragment comprising the Mid and PIWI domains of Neurospora QDE2 (Boland et al., 2011). The insertion forms a pair of antiparallel helices that extend from the PIWI domain and lead into the C terminus that nestles between the Mid and PIWI domains. This insertion is only partially visible in our structure of the hAgo2-miR-20a complex. In a superposition of hAgo2 and QDE2, the insertion appears to partially cover the small RNA binding groove of hAgo2; however, the base edges would still be available for base pairing. Thus, these remain available for pairing to the target strand (Figure S4). The insertion also appears to come fairly close to the PAZ domain, leaving open the possibility that the two could potentially interact. Given that the insertion is not resolved in our structure, it might be sufficiently flexible to shift away from the RNA binding groove, perhaps upon loading or target recognition. Overall, the QDE-2 Mid-PIWI lobe aligns closely with the same portion of the hAgo2-miR-20a complex, though the hAgo2 Mid domain appears to be tilted a bit closer toward its PIWI domain.
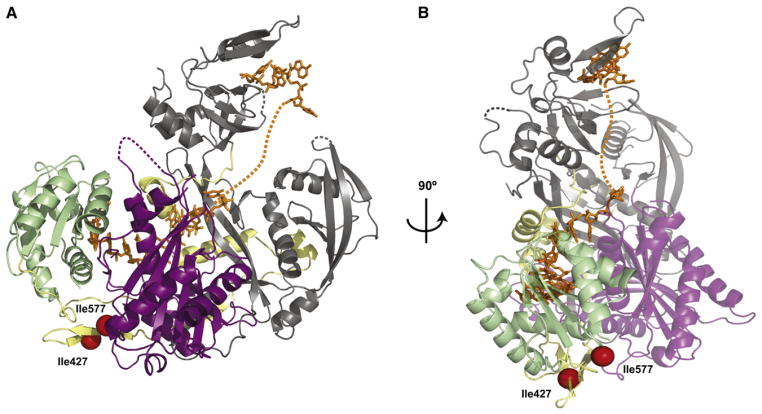
Our biochemical studies demonstrated that small RNA binding conferred striking stability to hAgo2. Limited proteolysis of the nonloaded protein by thermolysin produced two stable fragments, with cuts at I427 and I577. In the hAgo2-miR-20a complex, these residues are close neighbors, lying just about 10 Å apart. Spatially, they are located between the Mid and PIWI domains, even though one sits in the primary sequence on the linker between the PAZ and Mid domains (red spheres; Figure 4). The overall structure suggests an explanation for small RNA-induced resistance to proteolysis, namely that RNA binding essentially “glues” the two domains together from the opposite side. Given the proximity of these residues, thermolysin would likely require some flexibility in the protein to access these loops and cleave. This would seem possible only when the bound small RNA is absent. In fact, the miRNA interacts with all four domains of hAgo2 as well as with the linkers, likely providing cohesion and rigidity to the whole structure.
Figure 4. Structural Basis of RNA-Induced Stability of hAgo2.
(A) hAgo2 is shown in the canonical view as in Figure 2 to highlight interactions of the RNA across all constituent domains of the protein. The two thermolysin cleavage sites at I427 and I577 are shown as red spheres, which are only 10 Å apart and likely require flexibility in the protein for access by the protease. Extensive interactions between the protein and RNA are proposed to “glue” the structure into place, preventing the needed flexibility.
(B) An additional view from the MID domain is shown, clarifying the location of the cleavage sites and the path of the RNA.
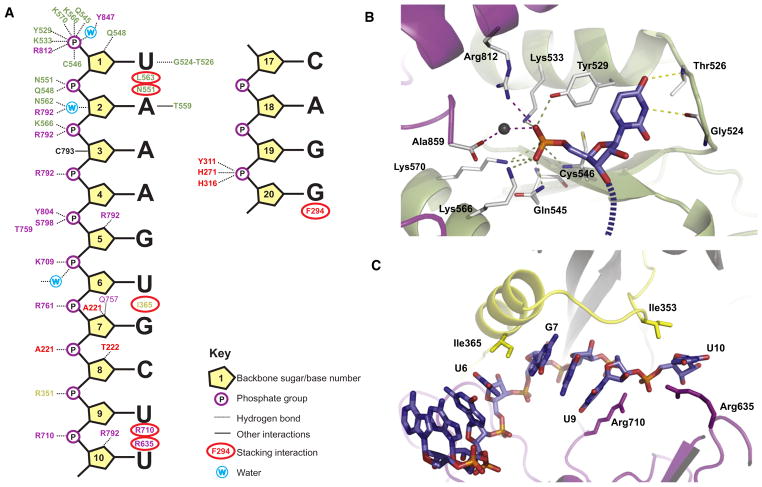
In the complex, we can clearly resolve the positions of 14 nucleotides of miR-20a. These include the first 10 bases and the final 4 (Figure S5). The electron density reflecting the central six bases is less clear but is sufficient to serve as a guide for model building (see below). The miRNA is anchored at both ends to the protein, binding to the Mid and PAZ domains as expected (Figure 5A).
Figure 5. miR-20a Interactions with hAgo2.
(A) A guide to the interactions between miR-20a (the first ten and last four bases that appear unambiguously in the structure) and hAgo2 is shown schematically. Interactions with backbone phosphates, sugars, and bases are indicated according to the key. Interacting residues are color-coded with respect to their domain location as in Figure 3.
(B) The site in the Mid domain (light green), which binds the 5′ end of miR-20a is shown. The first miR-20a base (U1) and the terminal monophosphate are displayed along with the interactions they make with hAgo2. Interacting protein side chains are shown in stick representation in atom colors with carbons in white, nitrogens in blue and oxygens in red. The 5′U is shown in atom colors as above, but with carbons in blue and phosphorous in orange. The direction of the RNA chain is shown as a dashed line. The portion of the PIWI domain, which caps the pocket, is shown in purple.
(C) The U6-G7 kink and the U9-U10 kink are shown in cartoon form. The L2 Linker is displayed in yellow, the PIWI domain in purple, and the miRNA in stick as in (B).
See text for details. See also Figures S5 and S6.
The 5′-End Binding Pocket of hAgo2
The 5′ end of miR-20a is tethered to Argonaute through a multitude of interactions to form a very tight binding pocket composed of residues mostly from the Mid domain, as previously observed (Boland et al., 2011; Boland et al., 2010; Frank et al., 2010; Ma et al., 2005; Parker et al., 2005). In the full-length structure, it is clear that this pocket is also capped on one side by PIWI domain residues (Figure 5B). All three nonbridging oxygen atoms of the 5′ phosphate interact with several residues. The first interacts with the hydroxyl of Y529 and the amine of K533 from the Mid domain and the R812 side chain from the PIWI domain. This oxygen also forms a water-mediated interaction with the C-terminal carboxylate of the protein. The second oxygen interacts with the K570, K566, and Q545 side chains. The main-chain amide of C546 and the side chain of K570 bind the third oxygen. Many of these residues are firmly positioned by interactions with each other and with additional residues. In particular, T544 stabilizes K533, the C-terminal carboxylate of the protein stabilizes K566 and K570, and the water, which mediates interaction between the C terminus and the 5′ phosphate of the RNA, is stabilized by the main-chain carbonyl of Y857 and the side chain of K533, among others.
We posit that the strong and extensive interactions between the miRNA 5′ phosphate and hAgo2 exquisitely define the position of the RNA guide with respect to the active site to ensure that cleavage of targets occurs at a well-defined and predictable position. The other interactions that occur between hAgo2 and the 5′ end of the guide are neither as strong nor as specific as for the 5′-terminal phosphate. Thus, in the absence of a 5′ phosphate, the RNA guide might adopt a variety of positions with the RNA binding groove of hAgo2, resulting in a loss of fidelity in the definition of the target cleavage site. In fact, this is precisely what was observed when hAgo2, purified from bacteria, was programmed with guides that lacked a terminal monophosphate (Rivas et al., 2005).
The 5′ uridine in the hAgo2-miR-20a complex occupies a position in the Mid domain that is essentially identical to that of the UMP in the reported structure of the isolated hAgo2 Mid domain (Frank et al., 2010). All previously identified residues that interact with the base are in identical positions (Figure 5B). The 2′OH of the first sugar interacts with the Q548 side chain, and the base itself stacks upon the aromatic ring of Y529. The Watson-Crick base edge interacts with the peptide backbone of G524–T525. Overall the structure of the isolated Mid domain superimposes almost perfectly on the Mid domain analyzed in the context of the full-length protein, with an RMSD of 0.69 Å between α-carbons.
Placement of the 5′ nucleotide into its binding pocket appears to be favored by additional structural constraints. The second miRNA base (A2) abuts T563. L553 wedges itself near the sugar, and N551 bounds the phosphate, practically directing nucleotide 1 to insert into its binding site (Figure S6). Occupancy of its binding pocket causes the first miRNA base (U1) to be flipped away from the following nucleotides, making it unavailable for interaction with the target. This is consistent with a wealth of prior structural, biochemical, and genetic data (Bartel, 2009).
The miR-20a Seed Sequence and Kinky Turns
The seed sequence of miR-20a (A2-C8) sits in a rather narrow groove. Every backbone phosphate of the seed and the adjacent two nucleotides interacts with the protein (Figure 5A). The backbone phosphate of A2 interacts both with the main and side chains of N551 and Q548. A3 interacts with K566 and R792, A4 with R792, and G5 with Y804 and with both the main chain and side chain of S798. U6 phosphate binds K709, G7 binds R761, and C8 sees the main-chain amine of A221. The RNA is held also by interactions with the sugars, particularly with RNA-specific 2′ OH groups of G5, G7, C8, and U10. The base edges all face directly outward, leaving them available for pairing with a target sequence. Because seed sequences are highly variable, it is not surprising that we see only limited interactions between hAgo2 and seed bases. The N3 of base A2 interacts with the protein, but this atom does not contribute to Watson-Crick base pairing. N2 and N3 on the minor groove side of the G5 base are nicely coordinated by a bidentate interaction with the side chain of Q757.
From the second (A2) to the sixth (U6) nucleotides, the bases are continuously stacked, but there is a noticeable destacking of bases 6 and 7 of the miRNA. This is a result of the insertion or intercalation of the side chain of I365 between these two bases (Figure 6C). This isoleucine emanates from the linker that connects the PAZ domain with the Mid domain and lies on a helix on the back of the RNA binding groove. In the TtAgo-21-mer complex, destacking also occurs, in this case between nucleotides 7 and 8 as a result of insertion of L267. Neither is this change in conformation as pronounced nor is the side chain intercalated as deeply as is seen in the miR-20a-hAgo2 complex.
Figure 6. Theoretical Modeling of the Complete miRNA.
(A) Nucleotides 11–16 of miR-20a were modeled with guidance from residual electron density (see text). Color coding is as in Figure 3. See also Figure S6.
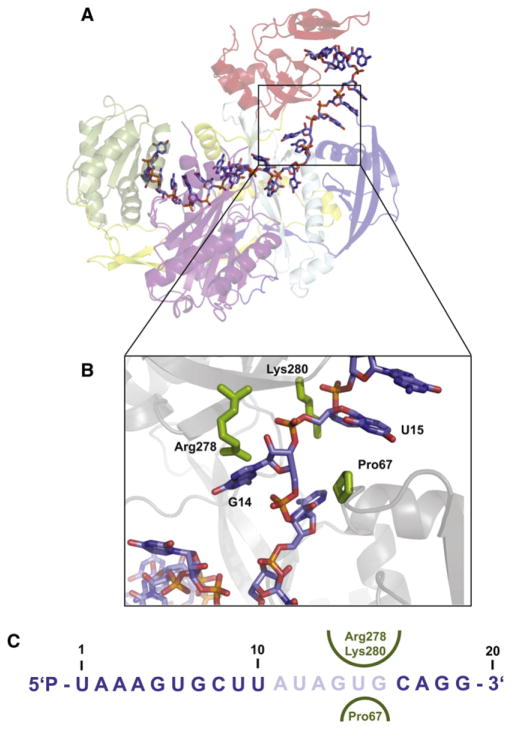
(B) A close-up of the G14-U15 turn is displayed, showing the “proline knuckle” emanating from the N domain on one side and R278 and K280 emanating from the PAZ domain on the other.
(C) The sequence of the 20-mer miR-20a is given. Ordered nucleotides are in blue, modeled ones in pale blue. The amino acids in the proline knuckle constriction are shown on both sides of the miRNA in green.
Another major kink and change in the direction of the miRNA occurs beyond the seed sequence between nucleotides U9 and U10 (Figure 5C). Here, two arginines play an important role. R710 inserts itself into miR-20a, stacking on the U9 base and interacting with the U10 phosphate. R635 stacks upon the U10 base. Importantly, residues preceding one of these arginines were shown to be important for hAgo2 function. Mutations in Q633R, H634P, and H634A did not impact siRNA binding but negated target cleavage (Liu et al., 2004). A third arginine, R351, interacts with the backbone of U9. I353 also facilitates the formation of the kink through van der Waals interactions.
The miRNA 3′ End
The 3′ end of miR-20a is tucked into the PAZ domain in a fashion similar to that previously seen for the complex of the hAgo1 PAZ domain with RNA (Ma et al., 2004). The last two nucleotides sit in a cleft of the PAZ domain, being held by the bill of our proverbial duck. The last base (A20) is stacked against F294, and the backbone phosphate that joins the last nucleotides makes a variety of contacts, as previously observed. One nonbridging oxygen is coordinated by two histidine side chains, those of H271 and H316. A tyrosine, Y311, interacts both with the other nonbridging oxygen through a hydroxyl group and with the bridging oxygen. Of the final four bases, only the middle two, A18 and G19, are stacked. G19 is destacked from the terminal base and C17 does not stack on G18 because the RNA turns sharply toward the RNA binding groove of the protein at that point.
Modeling the Missing Links—Implications for Target-Dependent RNA Movements that Enable Substrate Cleavage
Although we could only unambiguously determine the structure of 14 nucleotides, we saw clear electron density that almost certainly corresponds to the backbone of miR-20a nucleotides A11, A13, G14, and U15. We used this density as a guide for theoretical modeling of the remainder of the miRNA (Figures 6A and 6C). A11 appears to stack on top of R179 and H-bond with the side chain of S180. Interestingly, the backbone around A13 and G14 makes a turn around an N-domain residue, P67. This protrudes into the RNA binding groove like a knuckle at a relatively restricted part of the RNA binding groove that is bounded by the N domain on one side and the PAZ domain on the other. This proline knuckle might play a role in N-domain-assisted wedging of a loaded duplex strand as recently suggested (Kwak and Tomari, 2012). There are several PAZ domain residues in this area that likely provide interactions with the miRNA, most notably K278 and R280 (Figure 6B).
The active site of hAgo2 is very similar in structure, positioning and composition to Argonaute from P. furiosus (Rivas et al., 2005; Song et al., 2004). It is composed of the two canonical acidic residues, aspartates in this case (D597 and D669), projecting from the slight opening in the central β sheet of the PIWI domain. This arrangement is common to all enzymes of the RNase H family. The active site is completed by a histidine, H807, located on an adjacent α-helix. These are equivalent to D558, D628, and H745 of the archaeal enzyme. hAgo2 is a magnesium-dependent enzyme (Liu et al., 2004), and this metal is very likely coordinated by active-site residues. We did not identify any Mg2+ ions at the active site; however, Mg2+ was not included in the purification or crystallization buffers.
When examining the orientation of the miRNA with respect to the active site, we expected nucleotides 10 and 11 to lie opposite the active site. Instead, the active site sits opposite nucleotides 9 and 10. We therefore expect the domain movements associated with target binding to be a bit more complex than simple opening of the binding groove and for these to place the scissile phosphate in position to be cleaved by the active site.
DISCUSSION
It is estimated that eukaryotic Argonautes have been diverging from their archaeal counterparts for more than three billion years. Yet, structural analyses in this work and by others of full-length hAgo2 reveals remarkable similarity to both its archaeal and eubacterial counterparts (Schirle and Macrae, 2012). The active site is nearly identical between these enzymes, and the position and orientation of the remaining structural domains with respect to the catalytic center is highly conserved. This defines a shared positioning of the two ends of the nucleic acid guides used by these enzymes to identify their targets. Though we still do not know the biological functions of prokaryotic Argonautes, nor the true nature of their guides and targets in vivo—whether DNA or RNA—the deep conservation of Argonaute structure implies that all kingdoms of life most likely use this enzyme in basically the same way. This conclusion is particularly striking when one considers the diversity of known biological roles for Argonaute proteins, which seem to impact nearly every type of biological process, and the diversity of mechanisms through which Argonautes act. Not all Argonaute functions rely on their shared catalytic mechanism, but some instead depend upon the recruitment of accessory factors that enable, for example, the control of protein synthesis or modification of chromatin structure. Perhaps the evolutionary constraints implied by the high degree of structural similarity among Argonautes are simply defined by the overarching function of these proteins as programmable instruments for target recognition by complementary base pairing, but they may also reflect additional pressures.
The structure of hAgo2 in complex with miR-20a beautifully illustrates how the guide is threaded through the entire protein structure, interacting with every domain and even the linkers that join them. Nearly all of the interactions between the guide and the protein engage the RNA backbone, consistent with the ability of Argonautes to accept small RNA cofactors of essentially any sequence. The trajectory of the RNA is directed by the protein through many kinks and turns but always in a fashion that leaves most base edges accessible to be read by potential target molecules. The logic underlying the path followed by the RNA is not clear. As previously suggested, much of the seed is positioned in a way to maximally favor target binding (Wang et al., 2008b; Wang et al., 2009). Yet the circuitous route of the remainder of the RNA likely also serves a function. It is possible that this simply permits additional interactions between the protein and RNA that optimize affinity for the guide. However, the particular configuration of the RNA might also play as yet unrealized roles.
Given that RNA binding confers remarkable stability upon Argonaute proteins, the path of the guide might have evolved, in part, to maximize mutual stability, making such complexes long-lived. This is clearly critical in some settings. For example in Drosophila, Piwi protein-piRNA complexes transmit epigenetic information and likely must remain intact throughout a substantial fraction of the fly life cycle (Brennecke et al., 2008). The extensive interactions between hAgo2 and miR-20a likely restrict domain movement, as implied by the observed protease resistance of linker residues, but they might also serve to functionally couple Argonaute protein domains. Depending upon the degree of base pairing to the target, the guide RNA might induce different movements of domains relative to each other. This would permit allosteric changes in the complex that are sensitive to the mode of target interaction and that might favor one set of protein-protein interactions or one effector mode over another.
As mentioned, the structure of hAgo2 in complex with a heterogeneous mixture of bound RNAs of different lengths was reported recently (Schirle and Macrae, 2012). The structures are very similar with an RMSD of 0.336 Å over 785 α-carbon atoms. The main difference between the two structures is that the one presented here is of a complex with a homogeneous, loaded 20-mer miRNA, whereas the other contains a heterogeneous RNA. The clearer view of the bound mRNA in our structure allows us to resolve the four nucleotides at the 3′ end of miRNA-20a bound to and extending from the PAZ domain, as well as three additional nucleotides closer to the active site area with U9 and U10 forming an interesting kink. However, the path the RNA takes at nucleotides 1–7 is almost identical between the two structures, deviating a bit at nucleotides 6 and 7. Though we were able to build a few more protein residues, even the loops of the two structures are close to identical, a nice commendation of macromolecular crystallography. Schirle and MacRae (2012) also present a structure of a complex of hAgo2 with two molecules of tryptophan, which appear to bind to sites in the PIWI domain that may represent GW182 binding sites and thus a potential connection to the biochemical pathways unique to eukaryotes.
We have yet to understand the structural nuances that define different clades of Argonaute proteins, though emerging full-length structures can begin to provoke theories of how alterations in binding pockets might permit the binding of different terminal structures by Wago clade proteins or underlie preference for different terminal bases by Arabidopsis Argonautes. The more variable positioning of the PAZ domain with respect to the remainder of the protein might enable Piwi clade proteins to selectively harbor longer RNA guides than members of the Ago clade typically do.
Having the high-resolution 3D structure of the human version of the most versatile of Argonautes in complex with a miRNA provides an important framework to further understand its role and mechanism of action in essential processes in human development and disease.
EXPERIMENTAL PROCEDURES
Preparation of Small RNA Libraries
RNA was prepared by using TRIZOL (Invitrogen) from SF9 cell pellets and from a crystallization-grade hAgo2 preparation that retained endogenous small RNAs; 19–29 nt long RNAs were PAGE purified from total SF9 RNA. Small RNAs associated with hAgo2 were labeled with 32P-gamma ATP and separated by Urea-PAGE. A size range of 19 to 25 nucleotides was selected, and small RNAs were eluted from the gel. Small RNAs from SF9 cells and hAgo2 were both cloned according to (Brennecke et al., 2007), and sequenced on an Illumina GA2 platform.
Bioinformatics Analysis of Small RNA Libraries
Illumina reads were stripped of the 3′ linker and collapsed to a single representative of each sequence with multiplicity information retained. Reads that matched to synthetic cloning markers as well as low complexity reads were discarded. Only reads between 19 and 29 nt in length and that had a multiplicity of at least ten in each sample were subjected to further analyses. For sequence comparison between small RNAs associated with hAgo2 and endogenous small RNAs from SF9 cells, sequences were compared by BLAT with tile Size 11, minimum score 19 nt, and minimum identity of 90%.
Protein Expression and Purification
The full-length wild-type hAgo2 gene was cloned into the pFL vector of the MultiBac baculovirus expression system (Bieniossek et al., 2008) with a Two-Strep-SumoStar (TSS) tag (based on the One-Strep-SumoStar [OSS] tag [Schalch et al., 2011]) followed by a TEV cleavage site. SF9 insect cells, grown in Hyclone CCM3 media (Thermo Scientific), were infected with a baculovirus expressing hAgo2 for 72 hr. Initial purification was done by using Strep-Tactin superflow resin (IBA bioTAGnology) followed by tag removal with TEV protease. Protein was then loaded onto a Mono S 5/50 column (GE Healthcare) equilibrated with 20 mM Tris pH = 8.0, 50 mM KCl, and 5 mM DTT. Elution with the same buffer at 150 mM KCl and 375 mM KCl concentrations resulted in two separated fractions of protein, one that was bound to endogenous RNA originating from the insect cells and a second that was RNA free as determined by the ratio of 260/280 nm UV absorption and RNA extractions from the two samples.
Limited Proteolysis
The protocol used was adapted from Phan and colleagues (Phan et al., 2004); 1 mg/ml thermolysin (EMD Biosciences) stock solution was prepared in thermolysin reaction buffer (10 mM Tris-HCl [pH 8.0], 2 mM CaCl2, and 5% glycerol). Ten micrograms of hAgo2 were incubated with a 1:10, 1:40, 1:640, and 1:2,560 serial dilutions of the thermolysin protease for 30 min at 37°C. The reactions were quenched by the addition of SDS loading dye supplemented with 25 mM EDTA and boiling for 5 min. Samples were analyzed by SDS-PAGE, N-terminal sequencing, and mass spectrometry.
Complex Preparation
Human miR-20a-derived 20-mer of the sequence 5′ p-UAAAGUGCUUAUA GUGCAGG-3′ (Dharmacon RNAi technologies) was added to the RNA-free protein fraction at a 1:2 (protein:miRNA) molar ratio. Excess miR-20a was separated from the hAgo2-miR-20a complex by size exclusion chromatography that used a Superdex 200 10/300 column equilibrated with 10 mM Tris pH = 8.0, 100 mM NaCl, and 5 mM DTT.
Selenomethionine Protein Expression and Purification
High-five cells were infected with a baculovirus that directs expression of full-length, wild-type hAgo2 in ESF921 media (Expression Systems) for 6 hr, followed by media exchange to a methionine-free media and further growth for an additional 4 hr. Selenomethionine (SeMet) was then added to a final concentration of 198 mg/L. The cells were harvested after 48 hr. Incorporation was assessed by using mass spectrometry. SeMet protein purification and crystallization were done in the same manner as with the native protein.
Protein Crystallization and Data Collection
Crystals were initially obtained by hanging-drop vapor diffusion after mixing the protein-RNA complex at 5 mg/ml with an equal volume of 100 mM Tris [pH = 9.0], 14% PEG 3350 (w/v), 11% 2-propanol (v/v), and 0.12 M phenol. The inclusion of phenol was based on the early identification by the MacRae lab of phenol as a critical component of the crystallization conditions, as subsequently reported in Schirle and MacRae (2012). Further optimization of crystal growth was done by microseeding by mixing 1 μl of 100 mM Tris [pH = 9.0], 10%–12% PEG 3350 (w/v), 8%–10% 2-propanol (v/v), and 0.12 M phenol (v/v) with 1–3 μl of the protein. For data collection, crystals were cryoprotected by transferring the crystals briefly into the reservoir solution supplemented with 25% (v/v) ethylene glycol and flash freezing in liquid nitrogen.
X-ray diffraction data of native and SeMet-derivatized hAgo2 were collected to 2.2 Å and 2.6 Å resolution at beamline X29 at the National Synchrotron Light Source (NSLS) at Brookhaven National Laboratory (BNL). Diffraction data were indexed, integrated, and scaled with HKL2000 (Otwinowski and Minor, 1997). The structure of the SeMet crystal was determined by SAD at a wavelength optimized for the f″ signal of selenium. Eighteen selenium sites were found by using the HKL2MAP (Pape and Schneider, 2004) interface of SHELXD (Sheldrick, 2008). Heavy atom phasing was performed with SHARP (Bricogne, 1997). Density modification and phase extension to 2.2 Å, starting from the initial SHARP phases, was performed with the native data by solvent flattening as implemented in SOLOMON (Abrahams and Leslie, 1996). Automated model building with PHENIX (Adams et al., 2010) resulted in a partial model that was completed with manual correction with COOT (Emsley et al., 2010). After initial cycles of refinement with PHENIX and Refmac (Murshudov et al., 1997) automatically detected TLS groups were included as implemented in PHENIX (Version 1.7.2.869). Geometrical restraints were tightly monitored, and the inclusion of riding hydrogens resulted in significantly improved geometry of the refined model. The final structure could be refined to an Rwork/Rfree of 20.6%/25.4%.
The final model is missing residues 1–22, 120–126, 186–188, 245–247, 273–275, 603–606, 821–836 (58 of 859 residues). It contains two phenols retained from the crystallization condition and comprises nucleotides 1–10 and 17–20 of miR-20a. Nucleotides 11–16 were not included in refinement; however, fragmented density for nucleotides 13–15 allowed the modeling of the complete miRNA.
Figures were generated by using PyMol (The PyMOL Molecular Graphics System, Version 1.3 Schrödinger.).
ACCESSION NUMBERS
Coordinates and structure factors reported have been deposited in the Protein Data Bank under ID code 4F3T. The GEO accession number for the Illumina sequencing libraries reported in this study (libraries GSM930152 and GSM930153) is GSE37931.
Supplementary Material
Acknowledgments
We thank Deanna Bahel for technical support; Chris Faehnle, Thomas Schalch, and members of the Joshua-Tor and Hannon laboratories for discussions and advice; Howard Robinson and Neil Whelan for help at the National Synchrotron Light Source, which is supported by the Department of Energy, Office of Basic Energy Sciences. C.-D.K. was supported by a Jane Coffin Childs Memorial Fund for Medical Research Fellowship. This work was supported by the Louis Morin Charitable Trust (to L.J.) and by GM062534 to G.J.H. L.J. and G.J.H. are Howard Hughes Medical Institute Investigators.
Footnotes
Supplemental Information includes six figures and can be found with this article online at doi:10.1016/j.cell.2012.05.017.
Note Added in Proof
After this paper was accepted, a related paper was published online by Nakanishi et al., reporting the crystal structure of the yeast Kluyveromyces polysporus Argonaute loaded with guide RNA. Nakanishi, K., Weinberg, D.E., Bartel, D.P., and Patel, D.J. (2012). Structure of yeast Argonaute with guide RNA. Nature. Published online May 18, 2012. 10.1038/nature11211.
References
- Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Richmond TJ, Berger I. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr Protoc Protein Sci. 2008;Chapter 5(Unit 5.20) doi: 10.1002/0471140864.ps0520s51. [DOI] [PubMed] [Google Scholar]
- Boland A, Tritschler F, Heimstädt S, Izaurralde E, Weichenrieder O. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010;11:522–527. doi: 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci USA. 2011;108:10466–10471. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G. Ab initio macromolecular phasing: blueprint for an expert system based on structure factor statistics with built-in stereochemistry. Methods Enzymol. 1997;277:14–18. doi: 10.1016/s0076-6879(97)77004-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: an Ago-centric perspective. Cold Spring Harb Perspect Biol. 2011;3:a003772. doi: 10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent OA, Mendell JT. A small piece in the cancer puzzle: micro-RNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- Kwak PB, Tomari Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol. 2012;19:145–151. doi: 10.1038/nsmb.2232. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. In: In Processing of X-ray Diffraction Data Collected in Oscillation Mode. Carter CW, Sweet RM, editors. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pape T, Schneider TR. HKL2MAP: a graphical user interface for phasing with SHELX programs. J Appl Cryst. 2004;37:843–844. [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J, Tropea JE, Waugh DS. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr D Biol Crystallogr. 2004;60:1591–1599. doi: 10.1107/S0907444904017597. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Rizzo M, Mariani L, Pitto L, Rainaldi G, Simili M. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. J Cell Mol Med. 2010;14:2633–2640. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Shanker S, Partridge JF, Joshua-Tor L. The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS. Nat Struct Mol Biol. 2011;18:1351–1357. doi: 10.1038/nsmb.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Macrae IJ. The crystal structure of human Argonaute2. Science. 2012;336:1037–1040. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet. 2008a;40:1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008b;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008c;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.