Abstract
Cohesion, the force that holds sister chromatids together from the time of DNA replication until separation at the metaphase to anaphase transition, is mediated by the cohesin complex. This complex is also involved in DNA damage repair, chromosomes condensation, and gene regulation. To learn more about the cellular functions of cohesin, we conducted a genetic screen in Schizosaccharomyces pombe with two different cohesin mutants (eso1-G799D and mis4-242). We found synthetic negative interactions with deletions of genes involved in DNA replication and heterochromatin formation. We also found a few gene deletions that rescued the growth of eso1-G799D at the nonpermissive temperature, and these genes partially rescue the lagging chromosome phenotype. These genes are all chromatin effectors. Overall, our screen revealed an intimate association between cohesin and chromatin.
The generation of cohesion between sister chromatids takes place during DNA replication and dissolves at the metaphase to anaphase transition. Cohesion allows sister chromatids to biorient on the mitotic spindle and segregate accurately when the cell divides. Cohesion is mediated by the cohesin complex in cooperation with additional factors. In addition to its essential role in chromosome segregation, cohesin plays roles in chromosome condensation, DNA damage repair, and gene regulation. The role in gene regulation has been proposed to occur through several different mechanisms, including cohesin promoting gene looping, barrier function, enhancer−promoter interactions, and RNA pol II elongation (Dorsett 2011).
The generation of cohesion is dependent on an acetyltransferase that acetylates the Smc3 subunit of the cohesin ring to stabilize cohesion (Rolef Ben-Shahar et al. 2008; Unal et al. 2008; Zhang et al. 2008). This acetyltransferase is known as Eco1 in budding yeast, Eso1 in S. pombe, and ESCO2 in mammals. Mutations in both copies of ESCO2 are associated with the human disease Roberts syndrome (RBS) (Vega et al. 2005). Most of the mutations are missense mutations in which case ESCO2 protein is not detected, but a mutation that affects the active site also has been identified in association with RBS (W539G) (Vega et al. 2005). One hallmark of metaphase chromosomes in RBS is that they show “heterochromatic repulsion,” which refers to regions of “puffing” at heterochromatic regions around the centromeres and nucleolar organizers (Schule et al. 2005). Heterochromatin has been shown to be important for cohesin binding at pericentric regions in Schizosaccharomyces pombe (Bernard et al. 2001; Nonaka et al. 2002). Cohesin also associates with many locations in chromosome arms (Schmidt et al. 2009).
Eso1p in S. pombe acetylates evolutionarily conserved lysine residues in Psm3, in a process that appears to be similar to that reported in S. cerevisiae and humans. Acetylation is critical for the establishment of cohesion during DNA replication in both mitosis and meiosis (Feytout et al. 2011; Kagami et al. 2011). Mutation of both lysine residues in Psm3 to the acetyl-mimicking asparagine makes eso1 dispensable, although surprisingly the nonacetylatable mutant also was viable but did have cohesion defects (Feytout et al. 2011). eso1 in S. pombe is actually a fusion of two genes that are separate in S. cerevisiae and mammals. The N-terminal two-thirds is homologous to RAD30, also known as DNA polymerase eta, which is involved in translesion synthesis during postreplication DNA repair (Tanaka et al. 2000; Madril et al. 2001). The C-terminal one-third is homologous to ECO1. The ECO1 domain is sufficient for the establishment of cohesion in S. pombe because deletion of the N-terminus increases sensitivity to ultraviolet irradiation but does not compromise cohesion (Tanaka et al. 2000). All these data suggest the importance of acetylation activity in S. pombe and the evolutionarily conserved function of eso1 in cohesion establishment.
Given the many functions of cohesin, we decided to conduct an unbiased genetic screen to identify gene deletions that would act synthetically with an allele of eso1. The results of the screen could help highlight the roles of cohesin in various aspects of chromosome metabolism. We used a query strain bearing a mutation in the acetyltransferase domain of eso1 that compromises the catalytic activity of the protein (eso1-G799D, originally eso1-H17). The eso1-G799D mutation confers sensitivity to elevated growth temperature (Tanaka et al. 2000). We chose to conduct the screen in S. pombe because 1) there is a collection of 3066 strains with deletions in the nonessential genes (Kim et al. 2010) and 2) S. pombe displays heterochromatic properties similar to higher eukaryotes. Our screen identified gene deletions that in combination with eso1-G799D had (1) negative effects on growth (synthetic sick) and (2) rescued growth at nonpermissive temperature (synthetic rescue). One of the major gene classes with negative effects were genes involved in heterochromatin function. These genes also displayed synthetic negative interactions with a second cohesin allele, mis4-242. mis4 is involved in cohesin loading (Tomonaga et al. 2000). We identified and verified three new deletions that partially rescued the growth of eso1-G799D at elevated temperatures, all of which are genes whose protein products operate on chromatin. Overall, our findings suggest an intimate relationship between cohesin and chromatin.
Materials and Methods
Strains and media
All the strains used in this study are listed in supporting information, Table S6. The culture media used for S. pombe was YES except where otherwise stated. S. pombe strains were grown at 32°, except that the temperature sensitive strains were grown at 25°. For serial dilution plating assays, 10-fold dilutions of a log-phase culture were plated on the indicated medium and grown for 3 to 4 days. Thiabendazole (10 μg/mL) was used for the sensitivity test. For silencing assays, the strains with ura4+ reporter gene inserted at outer repeat region of centromere1 (otr1::ura4+) were used. Serial dilutions of the wild-type and respective mutants were plated on YES, and YES plates containing FOA. DAPI staining were used to determine the percentage of lagging chromosomes as described previously (Gregan et al. 2007).
Mutagenesis and gene disruption
To construct the eso1 mutant strains, the C-terminus of eso1 was amplified from genomic DNA and cloned into Pclonat1 (a gift from Gregan’s laboratory). The construct was subjected to site-directed mutagenesis. Plasmids carrying mutated eso1 were linearized with MfeI and transformed into the PEM2 S. pombe strain. Positive transformations were identified by polymerase chain reaction, and point mutation was verified by sequencing. The 3066 G418-resistant, haploid single-deletion mutants were obtained from the BIONEER (V 2.0). To make gene deletion strains, each individual gene deletion cassette was amplified from the genomic DNA of BIONEER gene deletion collection. The forward and reverse primers are designed about 250 bp upstream or 150 bp downstream of the open reading frame. After transformation, proper integration of the KanMX1 cassette in positive colonies was verified by colony polymerase chain reaction. Transformation was conducted with the lithium acetate method as described previously (Gregan et al. 2006).
Genetic crosses
Genetic crosses were performed according to the PEM procedure as described previously (Roguev et al. 2007, 2008). In summary, a PEM2 strain with either the eso1-G799D or mis4-242 mutation was used as the query strain to cross against the whole gene deletion library. Taking advantage of the background of PEM2 strains, after mating and sporulation, we used cycloheximide to select against the unsporulated diploid cells and h+ haploid cells. The mutant eso1 and mis4 genes of the query strain were fused with the NatMX cassette, which confers the resistance to nourseothricin (aka clonNAT), and the test strain from deletion collection has anti-G418 background; therefore, 100 μg/mL G418 and clonNAT was used to select the double mutants after haploid selection. Images of the agar plates were analyzed and processed. The plates with and without treatment of clonNAT were set as control and test plates, respectively. The eso1 screen was performed twice, with either 4 or 12 individual spots scored for growth. The mis4 screen was performed once with four individual spots scored for growth. All primary data for this article can be found at http://srdr.stowers.org/websimr/datasetview/474/0/.
Data processing and quality assessment
Images of the agar plates were acquired and analyzed. We normalized the colony sizes to correct for differences in growth conditions(Collins et al. 2006). In summary, the colony sizes of the outermost two rows and two columns are normalized to their plate middle mean, and then the colony sizes on each plate were scaled such that the middle means for all plates were equal a fixed number, which was the median of plate middle means across all plates. We used paired t-test to compare the average colony size of double mutants to single mutants and recorded t-statistics and p-values for each test. We combined P values from two independent experiments by using the Fisher method whenever possible.
Results
Characterization of cohesin alleles in S. pombe
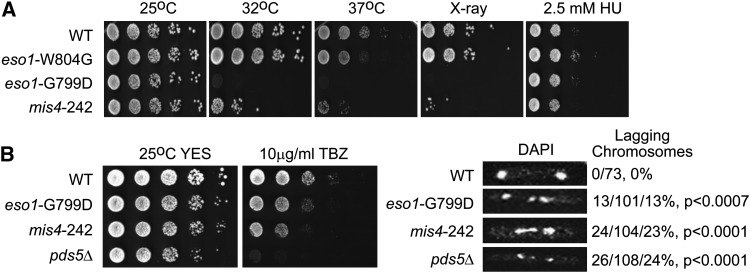
To choose alleles for genetic screening, we compared the behavior of different cohesin mutations. eso1-G799D has been previously reported to exhibit temperature-sensitive growth and defects in double-strand DNA break repair (Tanaka et al. 2000). This allele is analogous to the eco1-1 allele in budding yeast, which severely compromises acetyltransferase activity. We compared the behavior of an eso1-G799D mutant strain with one bearing eso1-W804G, which is analogous to a mutation associated with RBS and also compromises acetyltransferase activity (Vega et al. 2005). We also evaluated a mutation in Mis4/Scc2, mis4-G3965A(mis4-242), a previously reported temperature sensitive mutation (Tanaka et al. 2000; Toyoda et al. 2002). Mis4 is part of a cohesin loading complex (Furuya et al. 1998; Tomonaga et al. 2000). Mutations in mis4 are associated with Cornelia de Lange syndrome in humans (Tonkin et al. 2004). All mutant genes were expressed from the native promoter at their endogenous locus. When we grew these strains at 25°, 32°, and 37°, both eso1-G799D and mis4-242 were dead at 37° (Figure 1). A strain with eso1-W804G grew normally at 32°, and even a bit at 37°, although it was slow relative to a wild-type strain. Both the eso1-G799D and mis4-242 strains were sensitive to X-rays (Figure 1), consistent with a role for cohesin in double-strand DNA break repair (Furuya et al. 1998; Tanaka et al. 2000). The eso1-W804G strain did not show a significant growth defect upon exposure to X-rays. None of the mutants were hypersensitive to 2.5 mM hydroxyurea, a drug that will slow DNA replication (Figure 1). The eso1-W804G mutant seems to show a weaker phenotype compared with eso1-G799D, similar to what has been observed in budding yeast (Lu et al. 2010).
Figure 1 .
Basic characterization of cohesin alleles in S. pombe. (A) Cohesin mutants were grown at 25°, 32°, and 37°. eso1-G799D and mis4-242 mutants can grow at 25° but not at greater temperatures, and both of these mutants are sensitive to 75-Gy X-ray treatment. eso1-W804G, an analogous mutation to that associated with RBS, grew normally at 32° and grew only slightly slower at 37°. The eso1-W804G mutant did not show a significant growth defect upon exposure to X-rays. None of the mutants were hypersensitive to 2.5 mM hydroxyurea (HU). (B) TBZ sensitivity was tested, and lagging chromosomes were scored. P values are derived from a Fisher test using wild-type (WT) as the reference.
We further checked whether eso1-G799D showed phenotypes consistent with defects in chromosome segregation at 25° (Figure 1B). eso1-G799D and mis4-242 mutants were sensitive to thiabendazole (TBZ), a microtubule depolymerizing agent. Strains with defects in chromosome segregation are often sensitive to this agent. The pds5Δ mutant was extremely sensitive to TBZ. pds5 contributes to cohesion maintenance in S. cerevisiae (Hartman et al. 2000) but is not essential in S. pombe (Tanaka et al. 2001). Pds5, along with Rad61/Wpl1, may also function as negative regulators of cohesion (Rolef Ben-Shahar et al. 2008; Heidinger-Pauli et al. 2009; Rowland et al. 2009; Sutani et al. 2009; Feytout et al. 2011). We used DAPI staining to monitor lagging chromosomes. All the cohesin mutants had some lagging chromosomes. Overall, these results suggest that chromosome segregation is affected by the cohesin mutants at the permissive temperature.
A genetic screen with the eso1-G799D allele
To explore genetic interactions, we created a query strain with the eso1-G799D mutation in the PEM2 background (Roguev et al. 2008). Since the eso1-G799D did not grow at 37°, we could easily screen for suppressor mutations. We crossed the eso1-G799D strain against the whole gene deletion collection from BIONEER and followed published protocols to obtain double mutants (Roguev et al. 2007). The double mutants were plated at 25° to examine synthetic interactions and 37° to discover deletions that would rescue the temperature sensitivity. Colony size was used as a quantitative phenotypic readout. We used paired t-test to compare the average colony size from test plates to control plates. Genes within 500 kb of eso1 or rpl44 (a gene used as part of the selection procedure) were eliminated from the analysis as the result of linkage (Roguev et al. 2008). After elimination of linkage bias, a normal distribution of t-statistics was observed.
We found 215 genes that had a significant negative interaction with the eso1-G799D mutation by statistical analysis (Adjusted P < 0.05 and average difference of colony size >25; Table S1). Furthermore, we identified several genes whose deletion rescued growth of eso1-G799D at 37°. Many of the genes with negative interactions function in heterochromatin formation. To further confirm the negative interaction, we randomly selected several genes from these 215 genes. Serial dilution growth assay were performed, and 23 gene deletions were confirmed to be synthetically sick with the eso1-G799D mutation (Figure S1 and Table S2). Gene ontology term analysis regarding these 215 genes was conducted, and several GO categories are highly overrepresented in our synthetic sick gene list. Consistent with the fundamental role of eso1 in cohesion establishment and chromosome segregation, GO terms of chromosome segregation, and cell cycle are highly overrepresented (Table S4). Interestingly, we also found that the GO term “gene silencing” was overrepresented.
Eco1 normally establishes cohesion during DNA replication at S phase, and accumulating evidence indicates the strong connection between cohesion establishment and DNA replication (Skibbens et al. 1999; Kenna and Skibbens 2003; Skibbens 2004; Moldovan et al. 2006). Moreover, mutations that affect DNA replication also cause cohesion defects both in S. pombe and S. cerevisiae (Skibbens 2004; Ansbach et al. 2008). We found that deletion of ctf8, chl1, or swi3 showed synthetic negative effects with the eso1-G799D mutation. Ctf8 is part of an alternative replication factor C complex, whereas Chl1 is a replicative helicase; deletions were previously reported to be synthetically sick with cohesin mutants in S. cerevisiae (Costanzo et al. 2010). Swi3, a subunit of a replication fork protection complex, may normally stabilize the replication fork, which could facilitate the establishment of cohesion (Ansbach et al. 2008). We also found that deletion of the mhf1 gene, which was recently found to be important for DNA replication fork stabilization (Yan et al. 2010), showed synthetic negative growth with the eso1-G799D mutation. Thus, our data support the connection between cohesion establishment and DNA replication forks.
Deletions of genes involved in heterochromatin formation show a synthetic negative interaction with eso1 acetyltransferase mutants
In S. pombe, Swi6p, the homolog of human heterochromatin protein1, is critical for heterochromatin formation and the recruitment of cohesin to the centromeric region. swi6Δ has been shown to have synthetic growth defects with cohesin mutants (Bernard et al. 2001; Nonaka et al. 2002). Interestingly, in the cells of patients with RBS, cohesion disruption is specifically found at heterochromatin regions, further indicating a potential physical and/or genetic interaction between cohesin and heterochromatin in mammalian cells.
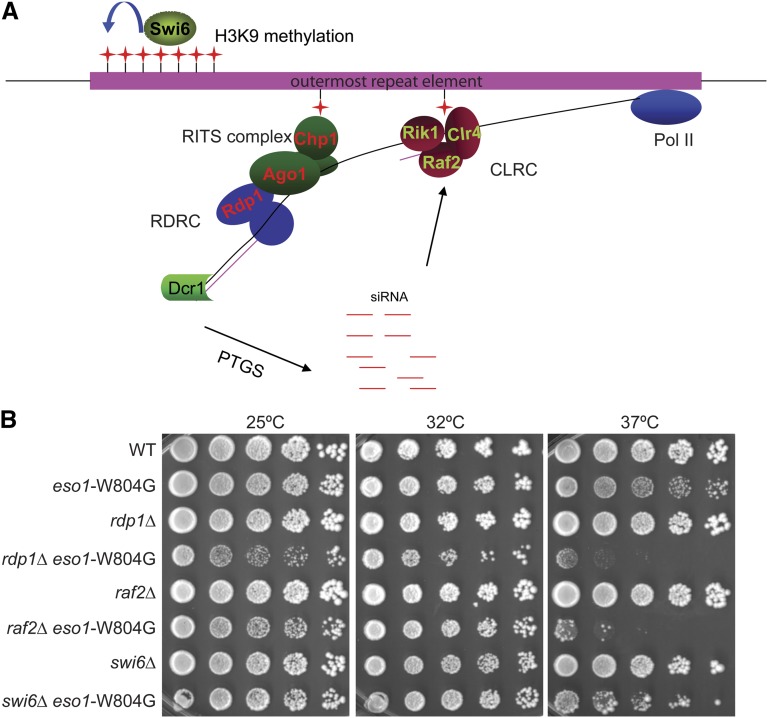
In our study, we found that the eso1-G799D mutation showed a synthetic negative effect not only with swi6Δ but several additional genes involved in heterochromatin formation. To further confirm the genetic interaction between eso1 and genes participating in heterochromatin formation, we combined the eso1-W804G mutation with rdp1Δ, raf2Δ, and swi6Δ. These genes contribute to heterochromatin formation in different ways. Rdp1p, an RNA-dependent RNA polymerase, is a component of RNA-dependent RNA polymerase complex, which facilitates the methylation of H3K9 by CLRC (containing Raf2). Rdp1 is not in our synthetic sick table (Table S2) because of the data processing to eliminate linkage biases. After H3K9 methylation, Swi6 can be recruited, and heterochromatin is established (Grewal and Jia 2007; Reddy et al. 2011). We found that all three double mutants grew poorly at 37° (Figure 2B). Interestingly, the growth defect with rdp1Δ and raf2Δ is even stronger than that observed with swi6Δ, suggesting that disrupting the function of these complexes in heterochromatin formation has a stronger effect than swi6Δ.
Figure 2 .
eso1-W804G showed negative genetic interaction with rdp1Δ, raf2Δ, and swi6Δ. (A) Illustration depicting the role of various proteins in heterochromatin formation. (B) The eso1-W804G mutant alone grew normally at 32° and was only slightly sick at 37°. However, combining the eso1-W804G mutation with rdp1, raf2, or swi6 deletion made the cells grow poorly at 37°.
Given the genetic interaction between eso1 and heterochromatin formation genes, we asked whether the eso1 mutations would affect gene silencing at heterochromatin regions. To address this question, we used strains containing ura4+ inserted at pericentric (imr1R::ura4+, otr1R::ura4+) and mating type(mat3::ura4+) heterochromatin regions. The silencing of ura4+ expression at heterochromatin region enables cell growth on plates with 5-fluoroorotic (FOA), which is toxic to cells with ura4+ expression. As a control we deleted rik1, which has previously been reported to disrupt heterochromatin in these regions. Rik1p functions at an early step in heterochromatin formation, as it is required for proper H3K9 methylation (Partridge et al. 2002) and Swi6 localization (Ekwall et al. 1996). This deletion made the cells unable to grow on FOA plates as previously reported [(Hong et al. 2005) Figure S2]. Strains with either the eso1-G799D or eso1-W804G mutation can still grow on the plates containing FOA, indicating that eso1 mutations do not have significant effects on the silencing of the ura4+ reporter gene at the outermost centromeric repeats, innermost centromeric repeats or mating type region. Thus, the acetyltransferase activity of Eso1p does not appear to contribute to silencing at these regions of heterochromatin in S. pombe.
A genetic screen with the mis4-242 allele
To distinguish whether the synthetic negative interacted genes were specific to eso1, we crossed the mis4-242 PEM2 strain against the whole-gene deletion collection from BIONEER and followed the same procedure used for eso1-G799D genetic screen. We found 92 gene deletions that showed a significant negative interaction with the mis4-242 mutant by statistical analysis (adjusted P < 0.05 and average difference of colony size >25; Table S3). Gene Ontology (GO) term analysis was conducted, and several GO categories are highly overrepresented in our synthetic sick gene list. Consistent with the fundamental role of mis4 in cohesin loading onto chromatin, GO terms such as mitotic cell cycle, sister chromatid cohesion, and response to DNA damage stimulus were highly overrepresented (Table S5). Again we found the GO category “gene silencing” was overrepresented. This result argues for a general connection between heterochromatin formation and cohesin rather than any specific connection between eso1 and heterochromatin. Interestingly, we found all checkpoint clamp complex gene deletions (rad1, hus1, rad9) showed negative synthetic interaction with mis4-242.
We found 16 gene deletions that shared negative synthetic growth interaction with both eso1-G799D and mis4-242 mutants (Figure 3). Four of these were heterochromatin genes: swi6, clr3, raf2, and epe1. In addition, genes important for DNA replication, such as ctf8 and swi3 also show synthetic negative interactions with both eso1 and mis4, which is consistent with the connection between cohesin and DNA replication. Interestingly, one of the genes encodes a protein from the 60S subunit of the ribosome. It has been recently proposed that mutations in cohesin genes may impair ribosome biogenesis by compromising nucleolar structure and function (Bose et al. 2012; Gard et al. 2009). Deletions of some ribosomal protein genes also have been shown to be synthetically sick with cohesin mutants in S. cerevisiae (Costanzo et al. 2010).
Figure 3 .
Intersection of mis4 (green) and eso1 (blue) screen and table of shared negative interactors
Deletion of chromatin effectors Spt2, Not3, and Rox3 can partially rescue the growth of the eso1-G799D mutant strain
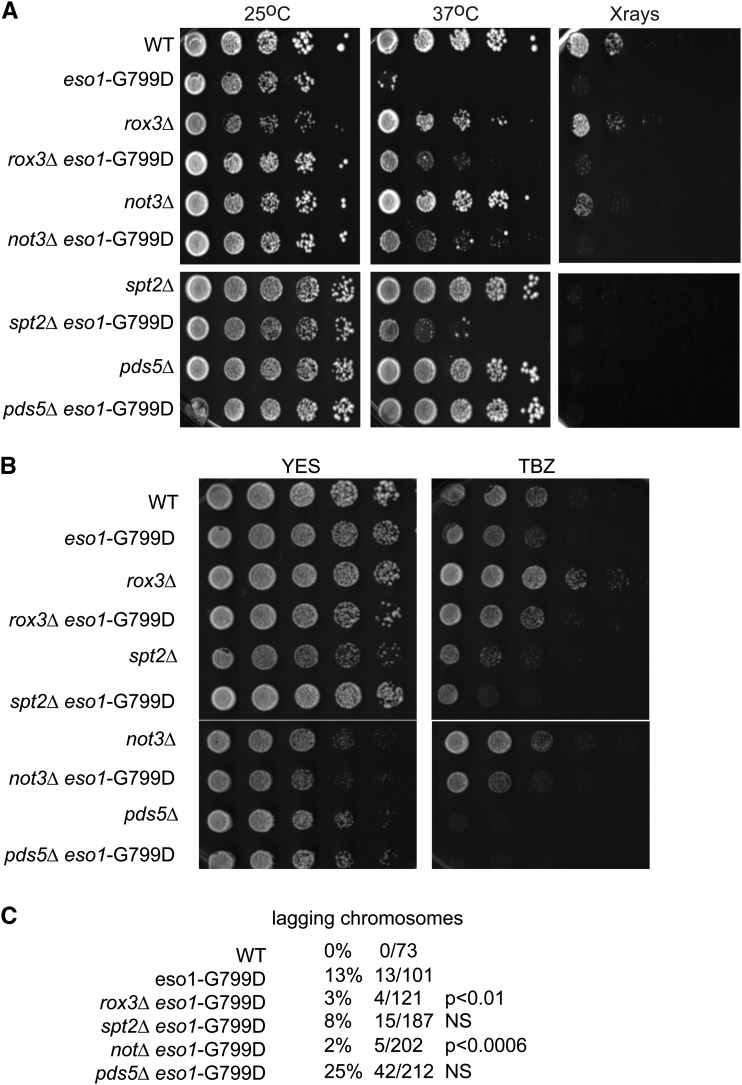
Taking advantage of the growth defect at 37° of eso1-G799D mutant, we were able to screen for suppressors of the temperature sensitive phenotype by growing the double mutants at 25° and 37°. Most of the double mutants can only grow at 25° but not 37° as predicted. Consistent with published data, we found that deletion of either pds5Δ or wpl1Δ can strongly rescue the growth defect of eso1-G799D mutant at 37°(Tanaka et al. 2001; Feytout et al. 2011). Besides pds5Δ and wpl1Δ, we also identified that spt2Δ, not3Δ, or rox3Δ/med19Δ can partially rescue the eso1-G799D mutant (Figure 4). Spt2 is an HMG-like nonhistone chromatin component, which is involved in gene regulation by affecting transcription initiation, elongation and polyadenylation (Perez-Martin and Johnson 1998; Hershkovits et al. 2006; Nourani et al. 2006). Not3 is a component of the CCR4-NOT complex, an evolutionarily conserved global transcriptional regulator (Liu et al. 1998; Chen et al. 2001). Rox3 was identified as a subunit of the evolutionarily conserved RNA pol II mediator complex (Spahr et al. 2001), which is composed of four modules (head, middle, tail, and kinase). Rox3 belongs to the head module. Deletion of rox3 in S. cerevisiae has previously been to shown to release the middle module of mediator. The net effect is that the complex can no longer function as a conduit between activators and the core transcription machinery (Baidoobonso et al. 2007).
Figure 4 .
Deletions that rescue the eso1-G799D allele at nonpermissive temperature. Deletion of rox3, not3, and spt2 can partially rescue the growth defect at 37° of eso1-G799D mutation. (A) rox3, spt2, and not3 deletion can partially rescue the growth defect at 37° caused by eso1-G799D mutation. pds5 deletion was used as the control. None of these deletions can alleviate the hypersensitivity of the eso1-G799D mutant to 75Gy X-ray treatment. Deletions that rescue the eso1-G799D allele at nonpermissive temperature do not rescue TBZ sensitivity (B), but show partial rescue of lagging chromosomes (C).
Because the eso1-G799D mutant is hypersensitive to X-ray treatment, we treated the double mutants containing both eso1-G799D mutation and rox3Δ, spt2Δ, and not3Δ deletion with X-rays. We found that none of these gene deletions could rescue the growth defect caused by eso1-G799D mutation after 75Gy X-ray treatment (Figure 4). Moreover, pds5Δ or spt2Δ alone made the cells hypersensitive to X-rays.
Next we checked rescue of TBZ sensitivity (Figure 4B). We found that rox3Δ alone made cells more resistant to TBZ, spt2Δ, or pds5Δ made cells more sensitive to TBZ, and not3Δ showed no significant effect. None of the double mutants showed increased tolerance to TBZ. We also checked the frequency of lagging chromosome in double mutants containing both eso1-G799D and spt2Δ, not3Δ, rox3Δ, or pds5Δ deletion. We found partial rescue of lagging chromosomes by deletion of rox3Δ (3%, P < 0.01), or not3Δ (2%, P < 0.0006), whereas spt2Δ was not statistically significant although it showed some rescue (8%). The P values, derived from the Fisher test, are relative to eso1-G799D (13%). The partial rescue of lagging chromosomes in the eso1-G799D background might contribute to the partial rescue of the growth defect of the double mutants at 37°. However, pds5Δ can fully rescue the growth of eso1-G799D mutant at 37° but does not rescue the lagging chromosome defect. This finding suggests that the chromatin effectors may partially rescue the cohesion defect, but interestingly, the growth rescue by pds5Δ does not include cohesion rescue.
Discussion
In the present study, we carried out a genetic screen to explore the interactions between cohesin and nonessential genes in S. pombe. We found that deletions of genes associated with heterochromatin showed significant negative effects with eso1 and mis4 mutations. In S. pombe, it has been proposed that Swi6p directly recruits cohesin (Nonaka et al. 2002). Although deletion of swi6 has been previously identified as having a synthetic negative effect with cohesin mutants, we have extended these finding to include subunits of the CLRC and RDRC complexes, which function in different aspects of heterochromatin function and have even stronger synthetic effects on growth. Thus, deletions that compromise heterochromatin in combination with a defect in cohesion establishment result in a significant synthetic negative phenotype. We speculate that the heterochromatin mutants negatively affect cohesion establishment at the pericentric regions, which can lead to defects in chromosome segregation. Because the eso1 mutants do not relieve silencing at the pericentric regions, we speculate that the heterochromatic puffing associated with chromosomes in RBS may not be associated with lack of silencing. Deletions that affect DNA replication also resulted in reduced growth when combined with the eso1 or mis4 mutation, as has been previously observed for cohesin mutations in S. cerevisiae (Costanzo et al. 2010).
Although genes involved in DNA replication and heterochromatin were common negative interacting genes with both eso1 -G799D and mis4-242, there were also some genetic interactions unique to each mutation. This finding likely reflects the different roles these genes play in cohesion, because Mis4 is a loader and Eso1 is an establishment factor. The differences in genetic interactions also may reflect additional roles of these genes. For instance, Eso1 may have acetylation targets in addition to Psm3 and Rad21 (Ghosh et al. 2012). Mis4 has been proposed to recruit acetyltransferases to specific gene promoters to influence their transcription (Jahnke et al. 2008).
Taking advantage of the temperature sensitive phenotype of the eso1-G799D mutant, we identified several gene deletions that allowed rescue at nonpermissive temperature. Although the cohesion antiestablishment factors were expected (wpl1 and pds5), deletions that affect chromatin and transcriptional processes were unexpected (not3, rox3, spt2). Consistent with this observation, deletion of SPT2 in S. cerevisiae has been reported to have a synthetic positive effect on growth of a smc3-1 cohesin mutant (Costanzo et al. 2010). The chromatin rescuers all appear to have positive effects on cohesion function. Overall, this screen indicates that cohesin and chromatin are genetically connected through many different chromatin effectors.
Supplementary Material
Acknowledgments
We thank Richard Alexander for assistance with quantification of colony size. We thank the Baumann lab, Krogan lab, Moazed lab, and Jia lab for strains. This work was supported by Stowers Institute for Medical Research.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Ansbach A. B., Noguchi C., Klansek I. W., Heidlebaugh M., Nakamura T. M., et al. , 2008. RFCCtf18 and the Swi1-Swi3 complex function in separate and redundant pathways required for the stabilization of replication forks to facilitate sister chromatid cohesion in Schizosaccharomyces pombe. Mol. Biol. Cell 19: 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baidoobonso S. M., Guidi B. W., Myers L. C., 2007. Med19(Rox3) regulates Intermodule interactions in the Saccharomyces cerevisiae mediator complex. J. Biol. Chem. 282: 5551–5559 [DOI] [PubMed] [Google Scholar]
- Bernard P., Maure J. F., Partridge J. F., Genier S., Javerzat J. P., et al. , 2001. Requirement of heterochromatin for cohesion at centromeres. Science 294: 2539–2542 [DOI] [PubMed] [Google Scholar]
- Bose T., Lee K. K., Lu S., Xu B., Harris B., et al. , 2012. Cohesin proteins promote ribosomal RNA production and protein translation in yeast and human cells. PLoS Genet. 8: e1002749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Rappsilber J., Chiang Y. C., Russell P., Mann M., et al. , 2001. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314: 683–694 [DOI] [PubMed] [Google Scholar]
- Collins S. R., Schuldiner M., Krogan N. J., Weissman J. S., 2006. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 7: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., et al. , 2010. The genetic landscape of a cell. Science 327: 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D., 2011. Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 21: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekwall K., Nimmo E. R., Javerzat J. P., Borgstrom B., Egel R., et al. , 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109(Pt 11): 2637–2648 [DOI] [PubMed] [Google Scholar]
- Feytout A., Vaur S., Genier S., Vazquez S., Javerzat J. P., 2011. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol. Cell. Biol. 31: 1771–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K., Takahashi K., Yanagida M., 1998. Faithful anaphase is ensured by Mis4, a sister chromatid cohesion molecule required in S phase and not destroyed in G1 phase. Genes Dev. 12: 3408–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard S., Light W., Xiong B., Bose T., McNairn A. J., et al. , 2009. Cohesinopathy mutations disrupt the subnuclear organization of chromatin. J. Cell Biol. 187: 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Gardner J. M., Smoyer C. J., Friederichs J. M., Unruh J. R., et al. , 2012. Acetylation of the SUN protein Mps3 by Eco1 regulates its function in nuclear organization. Mol. Biol. Cell 23: 2546–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Rabitsch P. K., Rumpf C., Novatchkova M., Schleiffer A., et al. , 2006. High-throughput knockout screen in fission yeast. Nat. Protoc. 1: 2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Riedel C. G., Pidoux A. L., Katou Y., Rumpf C., et al. , 2007. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr. Biol. 17: 1190–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S. I., Jia S., 2007. Heterochromatin revisited. Nat. Rev. Genet. 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Hartman T., Stead K., Koshland D., Guacci V., 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli J. M., Unal E., Koshland D., 2009. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol. Cell 34: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershkovits G., Bangio H., Cohen R., Katcoff D. J., 2006. Recruitment of mRNA cleavage/polyadenylation machinery by the yeast chromatin protein Sin1p/Spt2p. Proc. Natl. Acad. Sci. USA 103: 9808–9813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. J., Villen J., Gerace E. L., Gygi S. P., Moazed D., 2005. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3–K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2: 106–111 [DOI] [PubMed] [Google Scholar]
- Jahnke P., Xu W., Wulling M., Albrecht M., Gabriel H., et al. , 2008. The cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res. 36: 6450–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagami A., Sakuno T., Yamagishi Y., Ishiguro T., Tsukahara T., et al. , 2011. Acetylation regulates monopolar attachment at multiple levels during meiosis I in fission yeast. EMBO Rep. 12: 1189–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna M. A., Skibbens R. V., 2003. Mechanical link between cohesion establishment and DNA replication: Ctf7p/Eco1p, a cohesion establishment factor, associates with three different replication factor C complexes. Mol. Cell. Biol. 23: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. U., Hayles J., Kim D., Wood V., Park H. O., et al. , 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. Y., Badarinarayana V., Audino D. C., Rappsilber J., Mann M., et al. , 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17: 1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Goering M., Gard S., Xiong B., McNairn A. J., et al. , 2010. Eco1 is important for DNA damage repair in S. cerevisiae. Cell Cycle 9: 3315–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madril A. C., Johnson R. E., Washington M. T., Prakash L., Prakash S., 2001. Fidelity and damage bypass ability of Schizosaccharomyces pombe Eso1 protein, comprised of DNA polymerase eta and sister chromatid cohesion protein Ctf7. J. Biol. Chem. 276: 42857–42862 [DOI] [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S., 2006. PCNA controls establishment of sister chromatid cohesion during S phase. Mol. Cell 23: 723–732 [DOI] [PubMed] [Google Scholar]
- Nonaka N., Kitajima T., Yokobayashi S., Xiao G., Yamamoto M., et al. , 2002. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Nourani A., Robert F., Winston F., 2006. Evidence that Spt2/Sin1, an HMG-like factor, plays roles in transcription elongation, chromatin structure, and genome stability in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 1496–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge J. F., Scott K. S., Bannister A. J., Kouzarides T., Allshire R. C., 2002. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 12: 1652–1660 [DOI] [PubMed] [Google Scholar]
- Perez-Martin J., Johnson A. D., 1998. The C-terminal domain of Sin1 interacts with the SWI-SNF complex in yeast. Mol. Cell. Biol. 18: 4157–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy B. D., Wang Y., Niu L., Higuchi E. C., Marguerat S. B., et al. , 2011. Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 25: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A., Wiren M., Weissman J. S., Krogan N. J., 2007. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Methods 4: 861–866 [DOI] [PubMed] [Google Scholar]
- Roguev A., Bandyopadhyay S., Zofall M., Zhang K., Fischer T., et al. , 2008. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T., Heeger S., Lehane C., East P., Flynn H., et al. , 2008. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321: 563–566 [DOI] [PubMed] [Google Scholar]
- Rowland B. D., Roig M. B., Nishino T., Kurze A., Uluocak P., et al. , 2009. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol. Cell 33: 763–774 [DOI] [PubMed] [Google Scholar]
- Schmidt C. K., Brookes N., Uhlmann F., 2009. Conserved features of cohesin binding along fission yeast chromosomes. Genome Biol. 10: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule B., Oviedo A., Johnston K., Pai S., Francke U., 2005. Inactivating mutations in ESCO2 cause SC phocomelia and Roberts syndrome: no phenotype-genotype correlation. Am. J. Hum. Genet. 77: 1117–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 166: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens R. V., Corson L. B., Koshland D., Hieter P., 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13: 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahr H., Samuelsen C. O., Baraznenok V., Ernest I., Huylebroeck D., et al. , 2001. Analysis of Schizosaccharomyces pombe mediator reveals a set of essential subunits conserved between yeast and metazoan cells. Proc. Natl. Acad. Sci. USA 98: 11985–11990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T., Kawaguchi T., Kanno R., Itoh T., Shirahige K., 2009. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr. Biol. 19: 492–497 [DOI] [PubMed] [Google Scholar]
- Tanaka K., Yonekawa T., Kawasaki Y., Kai M., Furuya K., et al. , 2000. Fission yeast Eso1p is required for establishing sister chromatid cohesion during S phase. Mol. Cell. Biol. 20: 3459–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Hao Z., Kai M., Okayama H., 2001. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 20: 5779–5790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T., Nagao K., Kawasaki Y., Furuya K., Murakami A., et al. , 2000. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 14: 2757–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin E. T., Wang T. J., Lisgo S., Bamshad M. J., Strachan T., 2004. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 36: 636–641 [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Furuya K., Goshima G., Nagao K., Takahashi K., et al. , 2002. Requirement of chromatid cohesion proteins rad21/scc1 and mis4/scc2 for normal spindle-kinetochore interaction in fission yeast. Curr. Biol. 12: 347–358 [DOI] [PubMed] [Google Scholar]
- Unal E., Heidinger-Pauli J. M., Kim W., Guacci V., Onn I., et al. , 2008. A molecular determinant for the establishment of sister chromatid cohesion. Science 321: 566–569 [DOI] [PubMed] [Google Scholar]
- Vega H., Waisfisz Q., Gordillo M., Sakai N., Yanagihara I., et al. , 2005. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat. Genet. 37: 468–470 [DOI] [PubMed] [Google Scholar]
- Yan Z., Delannoy M., Ling C., Daee D., Osman F., et al. , 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell 37: 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Shi X., Li Y., Kim B. J., Jia J., et al. , 2008. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell 31: 143–151 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.