Abstract
Alterations in minisatellite DNA repeat tracts in humans have been correlated with a number of serious disorders, including cancer. Despite their importance for human health, the genetic factors that influence minisatellite stability are not well understood. Previously, we identified mutations in the Saccharomyces cerevisiae zinc homeostasis genes ZRT1 and ZAP1 that significantly increase the frequency of minisatellite alteration specifically during stationary phase. In this work, we identified mutants of END3, PKC1, and RAD27 that increase minisatellite instability during stationary phase. Genetic analysis reveals that these genes, along with ZRT1 and ZAP1, comprise multiple pathways regulating minisatellite stability during stationary phase. Minisatellite alterations generated by perturbation of any of these pathways occur via homologous recombination. We present evidence that suggests formation of ssDNA or ssDNA breaks may play a primary role in stationary phase instability. Finally, we examined the roles of these pathways in the stability of a human minisatellite tract associated with the HRAS1 oncogene and found that loss of RAD27, but not END3 or PKC1, destabilizes the HRAS1 minisatellite in stationary phase yeast. This result indicates that the genetic control of stationary phase minisatellite stability is dependent on the sequence composition of the minisatellite itself.
Keywords: DNA stability, stationary phase, G0, quiescence
Changes in DNA repeat tracts have long been associated with a number of serious human disorders (Mirkin 2007; Richard et al. 2008). Expansions in trinucleotide repeats can cause Huntington’s disease, myotonic dystrophy, and spinocerebellar ataxia (Mirkin 2007). Altered alleles of minisatellites (a class of tandem DNA repeats with repeat units 16–100 nucleotides in length) have also been correlated with several cancer subtypes (Jeong et al. 2007; Krontiris et al. 1993; Wang et al. 2003), progressive myoclonus epilepsy (Lafreniere et al. 1997), insulin-dependent diabetes mellitus (Kennedy et al. 1995), attention-deficit hyperactivity disorder (Yang et al. 2007), asthma (Kirkbride et al. 2001), and ulcerative colitis (Kyo et al. 1999). Unlike trinucleotide repeat stability, which has been well studied (Mirkin 2007; Richard et al. 2008), minisatellite repeat tract stability is only beginning to be understood.
Minisatellites are found throughout eukaryotic genomes (Richard et al. 2008). Some bind transcription factors to regulate the expression of nearby genes (Green and Krontiris 1993; Trepicchio and Krontiris 1992) or regulate splicing of the transcript (Jeong et al. 2007; Kirkbride et al. 2001; Kyo et al. 1999). In humans, minisatellites undergo frequent tract length alterations and repeat rearrangements during germline formation, as well as relatively rare somatic tract alterations (Buard et al. 2000; Jeffreys and Neumann 1997; Jeffreys et al. 1994). We previously showed that human HRAS1 minisatellite alleles integrated into the genome of Saccharomyces cerevisiae recapitulates this pattern of stability (Jauert et al. 2002), as have other groups (Debrauwere et al. 1999), thus allowing yeast to serve as a model organism for studying minisatellite stability.
The yeast genome has a number of native minisatellite sequences, with many clustered near the telomeres [reviewed in Richard and Dujon (2006)], but the stability of most of these repetitive DNA tracts has not been examined systematically. However, some native and a number of introduced minisatellite sequences have been shown to alter during meiosis. Meiotic minisatellite alterations require the recombination-initiated endonuclease Spo11, the large loop repair endonuclease Rad1 (Jauert et al. 2002), and the RAD50 recombination protein (Debrauwere et al. 1999). During mitotic growth, minisatellites are relatively stable but can be destabilized by loss of the flap endonucleases Rad27 and Dna2, yeast PCNA Pol30, DNA polymerase Pol3, or DNA helicase Pif1 (Kokoska et al. 1999; Lopes et al. 2002; Maleki et al. 2002; Ribeyre et al. 2009).
We recently demonstrated that minisatellite stability is controlled in stationary phase cells. We used a colony color assay system for assessing minisatellite stability (Kelly et al. 2007) that is unique in its ability to distinguish mitotic alterations, which are seen as sectored colonies, from minisatellite alterations that occur during stationary phase, which are seen as white microcolonies forming on the surface of the main colony; we call this novel color segregation phenotype “blebbing.” Using this system, we found that mutations in the zinc homeostasis genes ZRT1 and ZAP1 lead to increased minisatellite alterations during stationary phase (Kelly et al. 2007), specifically in the truly quiescent subset of G0 cells in stationary phase (Kelly et al. 2011). Further, these alterations require homologous recombination and also occur in a human-derived minisatellite associated with the HRAS1 oncogene (Kelly et al. 2011).
In this study, we describe additional mutants that show increased minisatellite alterations during stationary phase. Mutations in the endocytosis gene END3, the essential protein kinase encoded by PKC1, or the flap endonuclease gene RAD27 all lead to an increase in stationary phase minisatellite tract expansions or contractions, and these tract alterations are dependent on recombination factors. Genetic analysis indicates that multiple pathways regulate minisatellite stability during stationary phase. A common factor affecting minisatellite stability in these mutants may be an effect on ssDNA formation. Finally, we examine the stability of a human disease-associated minisatellite in these mutants.
Materials and Methods
Media, plasmids, and strains
Standard media (Guthrie and Fink 1991) was used, except for YPD + G418, which was made by the addition of 200 mg/L of G418 sulfate (geneticin) to standard YPD solid media. Sporulation and tetrad dissection protocols used in this study have been previously reported (Jauert et al. 2002).
All S. cerevisiae strains examined in this study (Table 1) are derived from EAS28 (Sia et al. 2001), a W303 derivative closely related to S288c (Schacherer et al. 2007). Strains whose construction is not reported here have been previously described (Kelly et al. 2007). Two minisatellite alleles were used in this study: ade2-min3 (Kelly et al. 2007), which is an artificial minisatellite initially used to examine mismatch repair in yeast (Sia et al. 1997), and ade2-h7.5, (Kelly et al. 2011), which was derived from a minisatellite associated with the human HRAS1 gene which was inserted into the HIS4 locus on chromosome III as described (Jauert et al. 2002). Strains DTK1088 and DTK1266, bearing deletions of END3, were constructed by PCR of the end3Δ::KAN cassette using DNA from the END3 deletion Saccharomyces Deletion Consortium (SDC) strain and primers 28278222 and 28278223. The irc10Δ::KAN strain DTK1091 was constructed in similar fashion, using primers 28234947 and 28234948 and DNA from the corresponding SDC strain. Strains DTK1012 and DTK1225, bearing deletions of RAD27, were also generated by PCR with the DNA of a rad27Δ::KAN SDC strain using primers 23094593 and 23094594. DTK1379, bearing a deletion of PTC1, was constructed by PCR with DNA from the appropriate SDC strain using primers 44775228 and 44775229. DTK1360, bearing a deletion of ETR1, and DTK1361, bearing a deletion of POR1, were constructed transformation of DTK271 with PCR products as above using primers 37616572 and 37616573 and primers 37616574 and 37616575, respectively. All PCRs generated a product containing the KANMX4 geneticin resistance gene flanked with 5′ and 3′ homology to the targeted gene. These cassettes were transformed into the parental strains and integration events were selected on YPD + G418. All transformants were verified by PCR.
Table 1. Yeast strains used in this study.
| Strain | Relevant Genotype | Construction Details (Reference) |
|---|---|---|
| EAS28 | Wild-type | MATa his7-2 trp1-289 ura3-52 (Sia et al. 2001) |
| DTK260 | leu2::HisG | EAS28 with pNKY85 (Kelly et al. 2007) |
| DTK264 | ade2-min3 | DTK260 with pDTK123 (Kelly et al. 2007) |
| DTK271 | ade2-min3, MATα | DTK264 with pGal-HO (Herskowitz and Jensen 1991) |
| DTK284 | ade2-min3, arg8::HisG | DTK264 with pDS27 |
| DTK904 | ade2-min3, zrt1::LEU2 | DTK284 with zrt1::LEU2a (Kelly et al. 2007) |
| DTK1012 | ade2-min3, zrt1::LEU2, rad27::KAN | DTK904 with rad27::KANa |
| DTK1056 | ade2-min3, rad50::KAN | DTK271 with rad50::KAN (Kelly et al. 2007) |
| DTK1074 | ade2-min3, rad51::KAN | DTK271 with rad51::KANa |
| DTK1088 | ade2-min3, end3::KAN | DTK271 with end3::KANa |
| DTK1091 | ade2-min3, irc10::KAN | DTK271 with irc10::KANa |
| DTK1174 | ade2-min3, zrt1::LEU, end3::KAN | DTK904 x DTK1088, isolated spore |
| DTK1185 | ade2-min3, end3-1, ras2::KAN | Y797 with ras2::KANa |
| DTK1186 | ade2-min3, ras2::LEU2 | DTK271 with ras2::LEU2a |
| DTK1187 | ade2-min3, end3::KAN, ras2::KAN | DTK1088 x DTK1186, isolated spore |
| DTK1188 | ade2-h7.5 | DTK260 with pKK055, FOAR isolate |
| DNY101 | rad52::URA3 | (Nag and Petes 1993) |
| DTK1199 | ade2-min3, rad27::KAN | DTK271 x DTK1012, isolated spore |
| DTK1205 | jnm1::KAN | Spore isolated from Yeast Deletion Consortium strain dissection |
| DTK1218 | ade2-min3, end3-1, rad27::KAN | Y797 x DTK1199, isolated spore |
| DTK1224 | ade2-min3, rad27::KAN, rad52::URA3 | DTK1199 x DTK1253, isolated spore |
| DTK1225 | ade2-h7.5, rad27::KAN | DTK1188 with rad27::KANa |
| DTK1227 | ade2-min3, end3-1, rad52::URA3 | Y797 x DTK1191, isolated spore |
| DTK1247 | ade2-min3, jnm1::KAN | DTK271 x DTK1205, isolated spore |
| DTK1253 | ade2-min3, rad52::URA3 | DTK1191 x DTK284, isolated spore |
| DTK1266 | ade2-h7.5, end3::KAN | DTK1188 with end3::KANa |
| YKH27 | pkc1-4 | (Huang and Symington 1995) |
| DTK1279 | ade2-min3, pkc1-4 | DTK271 x YKH27, isolated spore |
| DTK1288 | ade2-min3, zrt1::LEU2, pkc1-4 | DTK904 x DTK1279, isolated spore |
| DTK1289 | ade2-min3, rad50::KAN, rad52::URA3 | DTK1268 x DTK284, isolated spore |
| DTK1290 | ade2-min3, rad51::KAN, rad52::URA3 | DTK1269 x DTK284, isolated spore |
| DTK1293 | ade2-min3, end3::KAN, pkc1-4 | DTK1088 x DTK1279, isolated spore |
| DTK1294 | ade2-min3, rad27::KAN, pkc1-4 | DTK1199 x DTK1279, isolated spore |
| DTK1316 | ade2-min3, dnl4::KAN | DTK271 with dnl4::KANa |
| DTK1346 | ade2-min3, pkc1-4, rad52::URA3 | DTK1191 x DTK1279, isolated spore |
| DTK1357 | ade2-min3, pkc1-4, rad50::KAN, rad52::URA3 | DTK1056 x DTK1346, isolated spore |
| DTK1358 | ade2-min3, pkc1-4, rad51::KAN, rad52::URA3 | DTK1074 x DTK1346, isolated spore |
| DTK1360 | ade2-min3, etr1::KAN | DTK271 with etr1::KANa |
| DTK1361 | ade2-min3, por1::KAN | DTK271 with por1::KANa |
| DTK1362 | ade2-min3, pkc1-4, rad50::KAN | DTK271 x DTK1357, isolated spore |
| DTK1363 | ade2-min3, pkc1-4, rad51::KAN | DTK271 x DTK1358, isolated spore |
| DTK1364 | ade2-min3, rad27::KAN, etr1::KAN | DTK1199 x DTK1360, isolated spore |
| DTK1367 | ade2-min3, pkc1-4, etr1::KAN | DTK1279 x DTK1360, isolated spore |
| DTK1368 | ade2-min3, pkc1-4, por1::KAN | DTK1279 x DTK1361, isolated spore |
| DTK1370 | ade2-min3, rad27::KAN, por1::KAN | DTK1199 x DTK1361, isolated spore |
| DTK1371 | ade2-min3, end3::KAN, etr1::KAN | DTK1088 x DTK1360, isolated spore |
| DTK1372 | ade2-min3, end3::KAN, por1::KAN | DTK1088 x DTK1361, isolated spore |
| DTK1373 | ade2-h7.5, end3::KAN | DTK1188 with end3::KANa |
| DTK1375 | ade2-h7.5, pkc1-4 | DTK1188 x DTK1279, isolated spore |
| DTK1379 | ade2-min3, zrt1::LEU, ptc1::KAN | DTK904 with ptc1::KANa |
| DTK1386 | ade2-min3, pkc1-4, ptc1::KAN | DTK1279 x DTK1379, isolated spore |
| DTK1408 | ade2-min3, pkc1-4, dnl4::KAN | DTK1279 x DTK1316, isolated spore |
Strain was made using a PCR-generated construct.
Strain DTK1247, bearing a deletion of JNM1, was constructed by mating. The jnm1Δ::KAN diploid SDC strain was sporulated and dissected, and a haploid spore (DTK1205) of the desired mating type was isolated. DTK1205 was mated to DTK271, and the resulting diploid was sporulated and dissected. An ade2-min3 jnm1Δ::KAN spore was isolated by color and ability to survive on YPD + G418 media. This spore isolate was backcrossed twice to DTK271, and each time an ade2-min3 jnm1Δ::KAN spore isolate was identified as described above. DTK1247 is the spore isolate of the final backcross. DTK1279, bearing the temperature-sensitive point mutation pkc1-4, was also generated by mating. The pkc1-4 strain YKH27 (Huang and Symington 1994) was crossed to DTK271, and the resulting diploid was sporulated and dissected. An ade2-min3 pkc1-4 spore was isolated by color and temperature sensitivity at 37°. This spore isolate was backcrossed to DTK271 twice to generate DTK1279, the final ade2-min3 pkc1-4 spore isolate.
Whole-genome hybridization
Whole-genome hybridization of DTK271 and Y797 DNA and analysis of the resulting profiles were conducted as previously described (Gresham et al. 2006).
Minisatellite tract length analysis by PCR
White cells from independent blebs were picked with sterile toothpicks, patched on YPD, and incubated at 30° overnight. Whole-cell PCR across the ade2-min3 minisatellite tract was conducted for each independent bleb isolate, plus the wild-type ade2-min3 strain using primers 43901571 and 43901572. Five random PCR products were sequenced using primers 17339862 and 17339863 to confirm that changes in size compared with wild-type ade2-min3 were due to changes in the minisatellite repeat tract.
Flow cytometry
Flow cytometry was conducted as previously reported (Gourlay and Ayscough 2006), with minor alterations. The wild-type parent (DTK271), end3Δ (DTK1088), and end3Δ ras2Δ (DTK1187) strains were grown at 30° for 48 hr in 5ml of YPD in the presence of 5μg/ml 2′,7′-dichlorodihydrofluorescein (H2DCF-DA; Molecular Probes). Cells were sonicated briefly prior to analysis, and fluorescence was analyzed on a FACSCalibur benchtop cytometer (BD Biosciences). Data were analyzed using CellQuest Pro (BD Biosciences).
Quantification of blebbing
The frequency of bleb formation on individual colonies was determined using previously described protocols (Kelly et al. 2011). Colonies were grown at 30° for 3 days and then incubated at room temperature for 4 days. Colonies were photographed, and the number of blebs on the colony surface were counted. At least 100 colonies were examined for each strain, and each strain was assayed three times independently. Subsequently, the mean number of blebs per colony was calculated and the 95% confidence interval for the mean was determined.
Primers
The following primers were used in this study:
Primer 28278222 (End3F): GAGTTAGTGGGTATTGGAAAGGC
Primer 28278223 (End3R): CCACACCGTTACTGGATAGA
Primer 28234947 (Irc10F): TGAGTGGACACAGAAAACGC
Primer 28234948 (Irc10R): CAGTACAGTTTCGCTAAGTAAGG
Primer 23094593 (Rad27F): GCGTCCCATCGCGCAAATGAAG
Primer 23094594 (Rad27R): TCCACGTTCAAGTTCCCAGAAA
Primer 44775228 (Ptc1F): ACAGACCCCAAACACAACAAG
Primer 44775229 (Ptc1R): CCTCATTCGTCATGTGAGAGATGC
Primer 43901571 (ade2-min3F): GGTGCGTAAAATCGTTGGATCTC
Primer 43901572 (ade2-min3R): GCTCAATCTCAATCGTTAGCAC
Primer 17339862 (ade2-min3 seqF): CGGACAAAACAATCAAGTATGG
Primer 17339863 (ade2-min3 seqR): ATGTTGAGCCTGTTTGCTG
Primer 37616573 (Etr1F): TGTACCCAGGGGTGGTTTCCAT
Primer 37616572 (Etr1R): TTGAAGGGTCGACGTCCCCTTTTA
Primer 37616575 (Por1F): CCAATCAAACACCGCCATTTCG
Primer 37616574 (Por1R): TTCTCACTGCCAAGCAACCA
Results
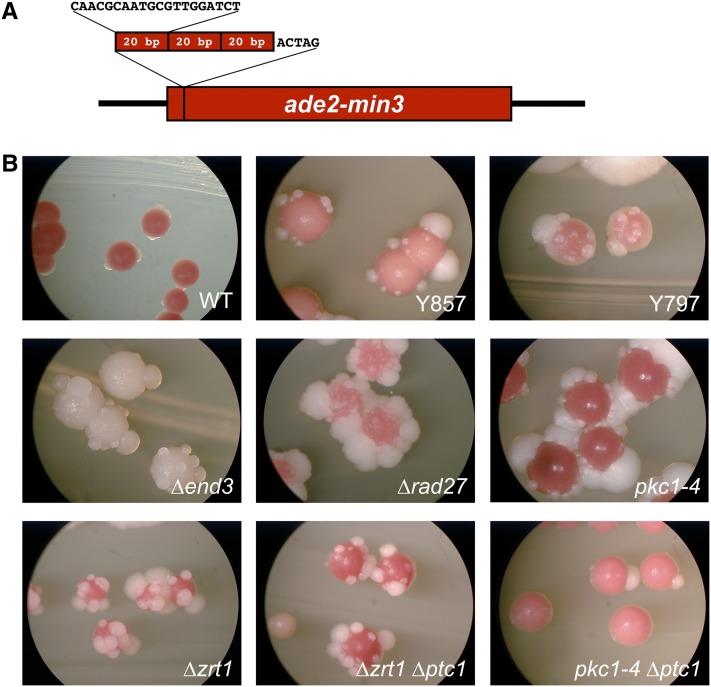
We previously described the ade2-min3 allele, a color-based reporter of minisatellite stability in S. cerevisiae (Kelly et al. 2007). This allele is composed of three tandem 20 base pair minisatellite repeats, plus 1 nucleotide, integrated into the ADE2 gene at an XbaI site (Figure 1A). With duplication of the 4 nt XbaI overhangs, this insertion shifts the reading frame of ADE2, disrupts adenine biosynthesis, and results in a red colony color. However, loss of one 20 bp minisatellite repeat unit restores the correct ADE2 reading frame, adenine production, and white color. If such minisatellite alterations occur during growth of the colony, a white sector forms within the red colony. If minisatellite alterations occur after growth of the colony has arrested, white papillations will form on the surface of the red colony. We designated this novel color segregation phenotype “blebbing.”.
Figure 1.
The color-based ade2-min3 reporter was used to identify factors that regulate minisatellite stability. (A) The ade2-min3 allele. Three 20 bp minisatellite repeats plus one additional bp were inserted into the ADE2 gene at the XbaI site. Duplication of the 4 nt XbaI overhang yielded a 65 bp insertion, resulting in a frameshift that disrupts ADE2. Loss of one 20 bp repeat unit, or gain of two repeat units, restores ADE2 to the correct reading frame. (B) Red/white color segregation in ade2-min3 strains. Strains were grown at 30° for 3 days, and then at room temperature for 4 days. The pkc1-4 mutant was grown at the semi-permissive temperature of 35° for 7 days. The wild-type ade2-min3 parent is DTK271. Y857 and Y797 are UV-generated point mutants of JNM1 and END3, respectively. Construction of the remaining strains is described above: end3Δ (DTK1088), rad27Δ (DTK1199), pkc1-4 (DTK1279), pkc1-4 ptc1Δ (DTK1386), zrt1Δ (DTK904), and zrt1Δ ptc1Δ (DTK1379).
Identification and characterization of blebbing mutants
We utilized the ade2-min3 reporter in a screen for mutants that increased the frequency of minisatellite alterations (Kelly et al. 2007). Four complementation groups with a blebbing phenotype were identified; the frequency of alleles in each group indicates that the screen was not saturated and that there are likely to be other genes that may be mutated to give a blebbing phenotype. The genes mutated in two of the four complementation groups were ZRT1 and ZAP1. While we previously described the cloning and characterization of these genes and their role in stationary phase minisatellite stability (Kelly et al. 2007, 2011), the mutations in the complementation groups represented by strains Y797 and Y857 (Figure 1B), each composed of one allele, remained to be cloned. These mutant strains were crossed to the parental DTK271 strain; the resulting diploids did not bleb, indicating that the mutations in Y797 and Y857 are recessive. We sporulated the diploids from these backcrosses, dissected tetrads, and observed the segregation of the blebbing phenotype. The resulting tetrads exhibited 2:2 segregation of the blebbing phenotype, indicating that a single mutation is responsible for blebbing in Y797 and Y857. Y797 is temperature sensitive at 37°, and the temperature sensitivity is tightly linked to the Y797 blebbing phenotype.
To clone the mutation responsible for blebbing in Y857, we used our previously described yeast genomic library on a geneticin-resistance plasmid vector (Jauert et al. 2005). Library plasmids were rescued from Y857 transformants that exhibited the parental (non-blebbing) phenotype, and the genomic insert was sequenced to identify candidate genes. The complementing plasmid 2a1b carries nucleotides 846000 to 863501 of chromosome XII; one of the genes in this interval, JNM1, encodes a dynactin subunit. Sequencing of JNM1 in Y857 revealed a C to T substitution that changes amino acid 133 from a glutamine to a stop codon. Deletion of JNM1 in the ade2-min3 background resulted in a blebbing phenotype, but the phenotype was highly variable and prevented further analysis.
Identification of the mutated gene in the Y797 complementation group was difficult. Initially we attempted to clone the mutation by complementation using the same protocols we used with Y857. However, Y797 did not survive the heat shock step of the transformation protocol, and it was also recalcitrant to electroporation or spheroplast transformation, presumably due to the temperature sensitivity that is linked to the blebbing phenotype. Attempts at cloning by complementation using low-temperature variations of the transformation protocols were also unproductive. Finally, we were unsuccessful in attempts to map the location of the mutation using mapping strains (Wakem and Sherman 1990). Therefore, we utilized whole-genome hybridization (Gresham et al. 2006) to identify the mutation responsible for blebbing in Y797. Briefly, Y797 was backcrossed to the parental strain DTK271 three times to limit the amount of polymorphism between these strains. Genomic DNA was then isolated from Y797 and DTK271 and hybridized to two separate yeast genomic tiling arrays. These microarrays are specially designed with multiple overlapping probes to detect single nucleotide polymorphisms (SNP) between the reference strain used to construct the microarray and the query strain. The SNP profiles of DTK271 and Y797 were then compared with highlight differences between the strains. Two such polymorphisms were identified in Y797, and the genes at the indicated locations were sequenced. A single nucleotide deletion in the END3 gene changed amino acid 174 from a leucine to a stop codon, and a point mutation in the IRC10 gene changed amino acid 138 from a lysine to a stop codon. Deletion of IRC10 in the ade2-min3 background did not result in a blebbing phenotype, but an ade2-min3 end3Δ mutant blebbed and was temperature sensitive at 37° (Figure 1B and data not shown). The diploid product of a cross between Y797 and an ade2-min3 end3Δ strain exhibits a blebbing phenotype. Finally, we monitored formation of white cells in liquid culture using a time course protocol previously performed with the ade2-min3 zrt1Δ strain (Kelly et al. 2007). Culture growth was monitored by OD600 at intervals, and aliquots were concurrently diluted and plated on rich media to determine the frequency of minisatellite alterations by relative number of white Ade+ colony-forming units (CFU) for the parental strain (DTK271), Y797, and an end3Δ mutant (DTK1088). Strains reached stationary phase after 96 hr at room temperature; at 120 hr we observed the first significant increase in the percentage of white CFUs in both of the end3 mutants compared with the parental strain (P < 0.02 for both, using Student t-test). These results confirm that, as with ZRT1, blebbing in an end3 mutant is caused by minisatellite alterations occurring specifically during stationary phase.
We examined mutants that were previously shown to affect minisatellite stability in actively growing cells, and we identified two additional genes that affect minisatellite stability in stationary phase. Deletion of RAD27, which encodes the yeast FEN-1 flap endonuclease (Liu et al. 2004), has been linked to minisatellite instability in actively growing cells (Lopes et al. 2002; Maleki et al. 2002). Loss of RAD27 results in a blebbing phenotype in the ade2-min3 strain background (Figure 1B), demonstrating a role for RAD27 in stationary phase cells. Sectors can also be observed in some ade2-min3 rad27Δ colonies, in agreement with prior reports that RAD27 regulates minisatellite stability during mitotic growth (Lopes et al. 2002; Maleki et al. 2002). A temperature-sensitive allele of PKC1, an essential protein kinase, previously was shown to have a hyper-recombination phenotype using a direct-repeat recombination assay (Huang and Symington 1994). Lorraine Symington provided us with the pkc1-4 temperature-sensitive point mutant, as the colony morphology of the strain was reminiscent of the blebbing phenotype of our zrt1Δ mutants. We crossed this mutation into our ade2-min3 background, using the temperature sensitivity to track the pkc1-4 allele, and we found that the pkc1-4 ade2-min3 strain blebs at 35° (Figure 1B).
To characterize the minisatellite alterations that result in white Ade+ blebs in each of our blebbing mutants, we conducted a PCR analysis of the ade2-min3 minisatellite tract length from white cells isolated from individual blebs. At least 100 independent PCR products were examined from each of the end3Δ, rad27Δ, and pkc1-4 mutant strains. Bleb formation in both the end3Δ and pkc1-4 mutants was exclusively caused by loss of one ade2-min3 minisatellite repeat unit, as described for the zrt1Δ mutant (Kelly et al. 2007). In contrast, loss of one minisatellite repeat was responsible for 51% (52/102) of blebs formed in the rad27Δ mutant, while gain of two minisatellite repeats accounted for the remaining 49% (50/102) of blebs.
Stationary phase S. cerevisiae cells can be in a truly quiescent state or in a nonquiescent, very slowly dividing, state (Allen et al. 2006). These states can be distinguished genetically: loss of ETR1, encoding a thiolester reductase, prevents quiescent cells from reentering the cell cycle, whereas loss of POR1, encoding a mitochondrial porin, has a similar effect on nonquiescent cells. We previously demonstrated that a zrt1Δ etr1Δ mutant does not bleb [0.09 blebs/colony; Kelly et al. 2011)], indicating that the minisatellite alterations occurring in the zrt1Δ mutant arise in quiescent cells. We performed a similar analysis on rad27Δ, end3Δ, and pkc1-4 mutants. Alterations in the rad27Δ mutant (Table 2, 25.4 blebs/colony) occur primarily in quiescent cells: loss of ETR1 has a significant effect (0.5 blebs/colony), whereas loss of POR1 has only a small effect (20.0 blebs/colony). Similarly, in end3Δ cells (4.8 blebs/colony), loss of ETR1 (0.4 blebs/colony) has a much greater effect than does loss of POR1 (2.3 blebs/colony). The pkc1-4 mutant (11.7 blebs/colony) was harder to evaluate; loss of ETR1 significantly reduced blebbing (to 0.03 blebs/colony), but loss of POR1 also had a strong effect (to 1.4 blebs/colony).
Table 2. Quantitative analysis of blebbing in double mutant strains.
| Second Relevant Genotype |
|||||
|---|---|---|---|---|---|
| WT | rad27Δ | end3Δ | pkc1-4a | ||
| First Relevant Genotype | WT | 3.7 ± 0.4b | 25.4 ± 1.0 | 4.8 ± 0.4 | 11.7 ± 0.7 |
| zrt1Δ | 20.6 ± 0.8 | 32.0 ± 1.2 | 3.4 ± 0.3 | 16.3 ± 1.0 | |
| pkc1-4a | 11.7 ± 0.7 | 28.5 ± 1.1 | 2.9 ± 0.3 | ND | |
| end3-1 | 8.5 ± 0.6 | 29.4 ± 1.9 | ND | ND | |
ND, no data.
Denotes colonies grown at 35°.
Mean blebs per colony ± 95% confidence interval.
Genetic analysis of blebbing mutants
The blebbing phenotypes of the various mutants differed significantly (Figure 1B). We quantified the amount of blebbing in each strain (Table 2) to compare the relative level of minisatellite instability. The rad27 and zrt1 mutants exhibited the highest level of blebbing (25.4 and 20.6 blebs/colony, respectively), both significantly above the parental DTK271 strain (3.7 blebs). The pkc1-4 strain had 11.7 blebs/colony at the 35° restrictive temperature. The two END3 mutants we evaluated exhibited significant differences: the end3-1 allele isolated from Y797 had 8.5 blebs/colony, whereas the deletion of END3 had 4.8. In addition, colonies from the deletion mutant were significantly less red than was the point mutant (Figure 1B). These data indicate that the Y797 end3-1 mutation is likely a hypomorphic allele rather than a complete loss-of-function allele.
To determine how many independent pathways monitor minisatellite stability during stationary phase, we constructed double mutant strains containing pairwise combinations of ZRT1, END3, RAD27, and PKC1 mutant alleles (Table 2). Where blebbing was higher in the double mutant than in either of the parental single mutants (as determined by non-overlap of the 95% confidence intervals for the mean), the two genes were considered to potentially participate in separable pathways regulating stationary phase minisatellite stability, whereas if blebbing was lower or not significantly different, the two genes were considered to participate in similar pathways.
Quantification of blebbing revealed that the rad27Δ zrt1Δ mutant had an average of 32.0 blebs per colony (Table 2), higher than the rad27Δ single mutant (25.4 blebs/colony) or the zrt1Δ single mutant (20.6 blebs/colony). This result indicates that RAD27 and ZRT1 have at least partially independent roles in monitoring minisatellite stability during stationary phase. Similar results were seen for rad27Δ with END3 or PKC1 mutants. A rad27Δ end3-1 strain had 29.6 blebs/colony, higher than the rad27Δ (25.4 blebs/colony) or end3-1 (8.5 blebs) single mutants. Likewise, a rad27Δ pkc1-4 strain had 28.5 blebs/colony, whereas a pkc1-4 mutant had 11.7 blebs/colony. Therefore, the RAD27 protein acts in pathways that are partially, but not fully, distinct from the END3 and PKC1 protein pathways. This classification is supported by the alteration types seen in the rad27Δ mutant (described above); only the rad27Δ mutant exhibited ade2-min3 tract length increases. No other double mutants exhibited higher blebbing than the parental single mutants, indicating that END3, PKC1, and ZRT1 could potentially function in overlapping pathways regulating minisatellite stability during stationary phase.
Minisatellite alterations in a zrt1Δ mutant require recombination factors (Kelly et al. 2007, 2011). We examined END3 and PKC1 mutants to determine the influence recombination factors have in those backgrounds. The majority of homologous recombination in S. cerevisiae requires RAD52 (Coic et al. 2008). Deletion of RAD52 in an end3-1 strain reduces blebbing to 0.6 blebs/colony from 8.5 blebs/colony (Table 3). Because blebbing in the end3-1 rad52Δ double mutant is not significantly different from spontaneous blebbing in the rad52Δ single mutant (1.4 blebs/colony) as determined by overlap of the 95% confidence intervals, we conclude that all minisatellite alterations in END3 mutants occur via RAD52-dependent recombination.
Table 3. Quantitative analysis of blebbing in strains with deletions of recombination factors.
| Second Relevant Genotype |
||||||||
|---|---|---|---|---|---|---|---|---|
| WT | rad50Δ | rad51Δ | rad52Δ | rad51Δ rad52Δ | rad50Δ rad52Δ | dnl4Δ | ||
| First Relevant Genotype | WT | 3.7 ± 0.4a | 0.9 ± 0.2 | 2.1 ± 0.3 | 1.4 ± 0.2 | 1.7 ± 0.2 | 0.6 ± 0.2 | 1.8 ± 0.3 |
| pkc1-4b | 11.7 ± 0.7 | 3.1 ± 0.5 | 3.9 ± 0.4 | 5.7 ± 0.6 | 2.1 ± 0.5 | 0.4 ± 0.2 | 9.0 ± 1.0 | |
| end3-1 | 8.5 ± 0.6 | ND | ND | 0.6 ± 0.2 | ND | ND | ND | |
ND, no data.
Mean blebs per colony ± 95% confidence interval.
Denotes strains grown at 35°.
In a PKC1 mutant strain, loss of RAD52 only partially reduces blebbing, to 5.7 blebs/colony compared with 11.7 blebs/colony in the parental pkc1-4 strain (Table 3). Therefore, approximately 50% of minisatellite alterations in a pkc1-4 mutant occur by RAD52-dependent recombination. In S. cerevisiae, RAD52-independent homologous recombination requires RAD50 and/or RAD51. A pkc1-4 rad51Δ rad52Δ triple mutant had 2.1 blebs/colony, a greater reduction in blebbing than the pkc1-4 rad52Δ double mutant displayed, indicating that some RAD52-independent recombination requires RAD51 in the pkc1-4 mutant. The pkc1-4 rad50Δ rad52Δ strain exhibited 0.5 blebs/colony, which was not significantly different from the rad50Δ rad52Δ double mutant (0.6 blebs/colony), showing that all RAD52-independent minisatellite alterations in the pkc1-4 mutant require RAD50. As RAD50 is required for non-homologous end-joining (NHEJ) as well as recombination, we deleted DNL4, which encodes a DNA ligase required for NHEJ, in the pkc1-4 strain background. The pkc1-4 dnl4Δ double mutant displayed an average of 9.0 blebs/colony, not substantially different from the pkc1-4 parent strain (11.7 blebs/colony).
The results of our recombination mutant analysis provide further differentiation between pathways regulating minisatellite stability during stationary phase. Because mutations in ZRT1 or PKC1 generate both RAD52-dependent and RAD52-independent minisatellite alterations (Kelly et al. 2011), their gene products clearly act differently than the END3 protein, as disruption of END3 results in only RAD52-dependent minisatellite alterations. We could not perform a similar analysis with RAD27, as rad27Δ rad52Δ mutants are inviable (Symington 1998).
We were next interested in determining whether ZRT1 and PKC1 function in the same pathway. It has been shown that pkc1-4 displays a hyper-recombination phenotype that can be suppressed by loss of the protein phosphatase encoded by PTC1 (Huang and Symington 1995). PTC1 is required for TOR signaling in yeast (Gonzalez et al. 2009). Deletion of PTC1 in our ade2-min3 pkc1-4 mutant suppressed blebbing (Figure 1B). However, loss of PTC1 in a zrt1Δ mutant did not suppress blebbing. The differential effect of the PTC1 mutation indicates that ZRT1 and PKC1 regulate minisatellite stability via differing pathways during stationary phase.
Reactive oxygen species production in the end3Δ mutant
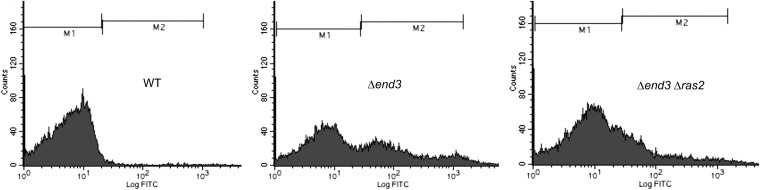
The RAD27 and PKC1 proteins have roles that influence genome maintenance (Ayyagari et al. 2003; Huang and Symington 1994; Wu and Wang 1999), but END3, which encodes a protein involved in endocytosis (Benedetti et al. 1994), has no obvious role. However, it has been reported that during stationary phase END3 mutants produce high levels of reactive oxygen species (ROS) via RAS2 hyperactivation (Gourlay and Ayscough 2006). ROS can produce a variety of DNA lesions, including DNA single- and double-strand breaks (Jackson and Loeb 2001). To investigate whether elevated ROS may be responsible for blebbing in our END3 mutant strain, we first assessed ROS production in our strains by staining with H2DCF-DA as described (Gourlay and Ayscough 2006). Wild-type, end3Δ, and end3Δ ras2Δ strains were incubated in rich liquid media in the presence of H2DCF-DA for 48 hr, then evaluated by flow cytometry. The end3Δ strain displayed elevated levels of ROS staining compared with the wild-type parent (Figure 2). Loss of RAS2 in the END3 mutant background reduced but did not completely abolish ROS staining. Similar results were observed when this experiment was repeated with end3-1 and end3-1 ras2Δ strains (data not shown). An end3-1 mutant displays an average of 8.5 blebs per colony. Blebbing in the end3-1 ras2Δ double mutant is reduced to 3.5 blebs/colony. The concurrent drop in ROS staining levels and blebbing in end3-1 ras2Δ double mutants suggests that ROS production is possibly linked to minisatellite instability in END3 mutants.
Figure 2.
END3 mutants display RAS2-dependent ROS accumulation during stationary phase. ROS accumulation in wild-type, end3Δ, and end3Δ ras2Δ stationary phase cells was assayed using H2DCF-DA by flow cytometry (see Materials and Methods). The data from one of three independent assays are displayed on histograms and divided into M1 (low ROS) and M2 (high ROS) populations.
Stability of the human HRAS1 minisatellite in RAD27, END3, and PKC1 mutants
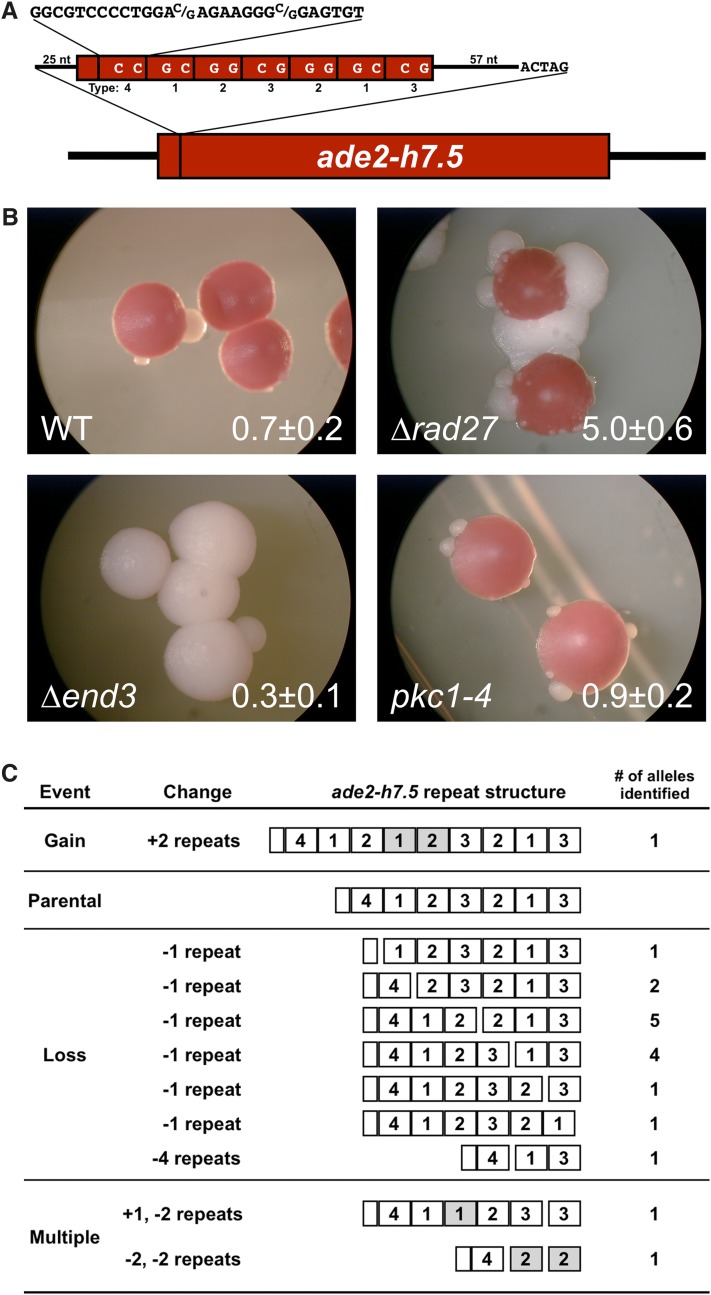
The ade2-min3 minisatellite tract consists of three identical tandem repeats. However, many human minisatellites, including the cancer-associated HRAS1 minisatellite (Green and Krontiris 1993; Krontiris et al. 1993), contain repeat units with some sequence variation. Because sequence variation could limit homologous recombination between minisatellite repeats, different mechanisms could affect the stability of direct repeat minisatellite tract and variable repeat minisatellite tracts. To determine whether RAD27, END3, and PKC1 regulate the stability of a variable repeat minisatellite tract, we utilized the previously described ade2-h7.5 allele (Kelly et al. 2011), which contains seven and one-half repeats of 28 bp derived from the human HRAS1 minisatellite inserted into the ADE2 gene at the XbaI site (Figure 3A). The primary repeat sequence is 5′ GGCGTCCCCTGGAG/CAGAAGGGG/CGAGTGT 3′, with either a G or C at the 14th and 22nd positions, as indicated in italics.
Figure 3.
The color-based ade2-h7.5 reporter was used to identify factors that regulate the stability of the human HRAS1 minisatellite. (A) The ade2-h7.5 allele. Seven and one-half repeats of 28 bp derived from the human HRAS1 minisatellite were inserted into ADE2 at the XbaI site. With unique flanking DNA and a 6 bp duplication of the XbaI site, the insert is 301 bp long, resulting in a frameshift that disrupts ADE2. As shown, repeats vary at the 14th and 22nd nucleotide. Type 1 repeats contain a G at position 14 and a C at position 22, Type 2 repeats contain G at both, Type 3 are C G, and Type 4 contain C at both. (B) Red/white color segregation in ade2-h.75 strains. Strains were grown at 30° for 3 days, and then at room temperature for 4 days. The wild-type ade2-min3 parent is DTK1188. Construction of the following strains is described above: rad27Δ (DTK1225), end3Δ (DTK1373), and pkc1-4 (DTK1375). Note the presence in the rad27Δ strain of both blebs and sectors in the upper-right of the top colony. (C) Altered alleles of ade2-h7.5 from 18 independent bleb isolates of DTK1225 (ade2-h7.5 rad27Δ) were sequenced. Altered allele structures are shown using the repeat number designations from the parental allele in Figure 3A. Repeats shown in gray have been added or modified. The location of deletions is illustrated as a gap in the repeat tract. For consistency, added repeats are shown to the right of the repeats they duplicate, although it is not possible to distinguish added and original repeats in the sequence.
We previously showed that loss of ZRT1 destabilizes the ade2-h7.5 minisatellite tract (Kelly et al. 2011). Similarly, the ade2-h7.5 rad27Δ strain exhibits a blebbing phenotype (Figure 3B). However, end3Δ and pkc1-4 mutants do not display a blebbing phenotype in the ade2-h7.5 background, raising the possibility that they might only regulate the stability of direct repeat minisatellite tracts. Whole-cell PCR across the minisatellite repeat tract was used to examine the nature of the tract alterations in independent bleb isolates of the ade2-h.7 rad27Δ strain. Of 112 alleles examined by PCR, 12 exhibited gain of two repeats, 7 exhibited loss of four repeats, and the majority, 92, exhibited loss of one repeat. A single allele was the same size as the unaltered ade2-h7.5 tract; sequencing of this allele showed that it had a single nucleotide deletion, which reduces the size of the HRAS1 minisatellite insert in ADE2 from 301 bp to 300 bp and restores the correct reading frame (data not shown). Sequencing of 18 of the ade2-h7.5 rad27Δ minisatellite alleles examined by PCR revealed a wide range of events (Figure 3C). Of the 14 alleles that had lost a single repeat, 9 showed deletion of the fourth or fifth repeat. However, we saw examples of deletion of nearly every individual repeat in the seven and one-half repeat ade2-h7.5 tract. We also obtained alleles exhibiting a loss of four repeats and a gain of two repeats. Finally, two strains had complex rearrangements indicative of multiple events. The first had a duplication of the second repeat coupled with a deletion of the fifth and sixth repeats. The second strain suffered two deletions of two repeats each. Both of these deletion events likely occurred between the two variable nucleotides in the repeat, leading to the formation of a novel repeat (indicated by gray repeats in Figure 3C, last row). The first deletion occurred between the G and C nucleotides of repeat 2 (at nucleotides 14 and 22, respectively) and the C and G nucleotides of repeat 4, forming a G G repeat (type 2), while the second deletion occurred between the G and G nucleotides of repeat 5 and the C and G nucleotides of repeat 7, forming a new type 2 repeat. The sequencing data indicate that very precise recombination events are occurring between minisatellite repeats in stationary phase cells.
Discussion
Mutations in END3, RAD27, or PKC1 stimulate alterations in a minisatellite tract while cells are in stationary phase; these alterations manifest as a blebbing phenotype in our assay system, allowing us to investigate aspects of post-mitotic genome maintenance using minisatellite tract alterations as an indicator of genome instability. Genetic analysis revealed that END3, RAD27, and PKC1, plus ZRT1 and ZAP1 (whose roles in minisatellite maintenance have been previously reported (Kelly et al. 2007), act in multiple independent pathways monitoring minisatellite stability during stationary phase. When the function of any of these pathways is disrupted, minisatellite alterations occur, with alterations being dependent on homologous recombination. Also, the human cancer-associated HRAS1 minisatellite tract is destabilized during stationary phase by mutations in RAD27 and ZRT1, but not END3 or PKC1.
This is the first report of roles for RAD27, PKC1, and END3 in post-mitotic genome maintenance. Rad27, a flap endonuclease, has a well-known role in Okazaki fragment processing and flap excision during long-patch base excision repair (BER) (Ayyagari et al. 2003; Wu and Wang 1999), and loss of RAD27 has been shown to increase minisatellite alterations in actively dividing cells (Lopes et al. 2002; Maleki et al. 2002). In agreement with these data, we have observed a sectoring phenotype in ade2-min3 rad27Δ colonies, indicative of minisatellite repeat tract alterations in actively dividing cells, in addition to the strong stationary phase blebbing phenotype (Figure 1B). It is likely that a role for RAD27 in post-mitotic genome stability has not been previously reported because other assay systems cannot easily distinguish between mitotic and post-mitotic events. Pkc1 is an essential protein kinase involved in signal transduction (Nishizuka 1992). PKC1 mutants have a previously described hyper-recombination phenotype, which was interpreted as due to mitotic events (Huang and Symington 1994), but similarities between color segregation in the hyper-recombinant PKC1 mutant (Huang and Symington 1994) and our previously reported blebbing strains (Kelly et al. 2007) prompted further investigation. We found that the pkc1-4 mutation stimulates minisatellite alterations during stationary phase (Figure 1B); the lack of sectoring in the ade2-min3 pkc1-4 strain indicates that PKC1 does not regulate minisatellite stability during mitotic growth. The END3 protein is involved in endocytosis (Benedetti et al. 1994) and has not previously been implicated in genome maintenance.
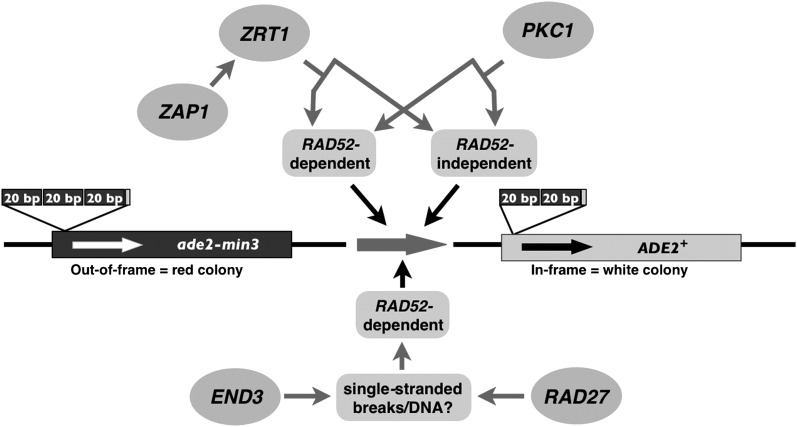
Genetic analysis with blebbing mutants identified in this study and previous work (Kelly et al. 2007) demonstrate that up to four pathways monitor minisatellite stability during stationary phase (Figure 4). One interpretation of our data is that each acts in an independent pathway: the first pathway is represented by ZRT1 and ZAP1, the second by RAD27, the third by END3, and the fourth by PKC1. While these pathways are at least partially independent, one common factor may be the involvement of single-stranded DNA (as shown in Figure 4). A second possibility is that END3 may be acting in the ZRT1 and the PKC1 pathways as a component of the RAD52-dependent repair activity, rather than representing an independent pathway.
Figure 4.
Model for pathways regulating ade2-min3 minisatellite stability during stationary phase. Loss of components in multiple pathways can lead to ade2-min3 minisatellite alterations. In the first pathway, loss of ZRT1 or ZAP1 stimulates loss of one ade2-min3 repeat unit via both RAD52-dependent and RAD52-independent recombination (Kelly et al. 2007). In a second pathway, mutation of PKC1 stimulates loss of one ade2-min3 repeat unit via both RAD52-dependent and RAD52-independent recombination. A third independent pathway is represented by END3, loss of which stimulates deletion of one ade2-min3 repeat unit via RAD52-dependent recombination. This deletion may be due to formation of ssDNA as a consequence of oxidative DNA damage. Alternatively, END3 could be acting in the RAD52-dependent portions of the ZRT1 and PKC1 pathways (not shown). In the final pathway, loss of RAD27 stimulates both loss of one and gain of two ade2-min3 repeats. These minisatellite alterations may be due to formation of ssDNA when RAD27-dependent DNA flap removal does not occur properly. RAD27-dependent minisatellite alterations during mitotic growth require RAD52, so it is possible that stationary phase minisatellite alterations in this pathway also occur via RAD52-dependent recombination.
The ZRT1/ZAP1 pathway has been characterized in detail elsewhere (Kelly et al. 2007, 2011). Loss of the RAD27 pathway causes an increase in stationary phase minisatellite alterations, which include both gain and loss of repeat units. Rad27 processes DNA flaps during Okazaki fragment maturation and long-patch BER in actively growing cells (Ayyagari et al. 2003; Wu and Wang 1999), but faulty Okazaki fragment processing is not a likely cause of minisatellite alterations during stationary phase, as bulk DNA synthesis does not occur in post-mitotic cells. However, replication during DNA repair events, such as long-patch BER, does occur in post-mitotic cells (Barzilai et al. 2008), and in the absence of RAD27, unprocessed DNA flaps could be resolved by homologous recombination. If such an event occurred within the minisatellite tract, misalignment of repeat units during recombination could account for the stationary phase minisatellite alterations seen in the RAD27 mutant. In addition, absence of Rad27 may cause an increase in ssDNA due to flap processing failure; this ssDNA may accumulate breaks or suffer repeat misalignment during repair synthesis, leading to changes in repeat number.
Loss of the PKC1-dependent pathway results in a significant increase in minisatellite alterations that occur via both RAD52-dependent and RAD52-independent recombination, but not NHEJ. PKC1 is known to have a role in stationary phase entry, downstream of TOR inactivation (Gray et al. 2004). Mutants defective for PKC1 show a substantial decrease in viability when starved (Krause and Gray 2002). Some PKC1 mutant cells fail to enter stationary phase when stressed, continuing to grow and replicate their DNA in spite of severely limited resources. DNA synthesis under these circumstances is likely to lead to ssDNA formation, replication fork stalling, and collapse, which can serve as a substrate for homologous recombination (Branzei and Foiani 2005). If pkc1-4 ade2-min3 cells that do enter stationary phase also exhibit DNA replication control abnormalities, ssDNA formation followed by fork stalling might occur within the minisatellite tract. Subsequent misalignment of the repeat units during recombination might explain the stationary phase blebbing phenotype and minisatellite alterations seen in the PKC1 mutant. However, few of the downstream effectors of the Pkc1 kinase have been identified, so it is possible that this protein may play an as yet unknown role in minisatellite stability during stationary phase.
Stationary phase minisatellite alterations are also increased in cells with a deletion of END3. Minisatellite alterations in an end3Δ mutant arise through RAD52-dependent recombination, unlike the ZRT1 and PKC1 pathways. Although END3 has no previously described role in genome maintenance, end3Δ mutants display a stationary phase–specific increase in ROS production (Gourlay and Ayscough 2006). Consistent with this result, our ade2-min3 end3Δ and end3-1 strains display an elevated level of ROS production during stationary phase (Figure 2). ROS production is reduced in an END3 mutant with a RAS2 deletion, a change that is concomitant with a decrease in blebbing. ROS can cause many types of DNA damage, including ssDNA formation and DNA breaks (Jackson and Loeb 2001). If ROS-triggered DNA breaks occur within the minisatellite, misalignment of repeat units during repair by homologous recombination could account for RAD52-dependent minisatellite alterations in stationary phase END3 mutants.
Mutations in RAD27, END3, and PKC1 all destabilize the ade2-min3 minisatellite tract, but only loss of RAD27 destabilizes the HRAS1 minisatellite repeats in ade2-h7.5 (Figure 3B). The ade2-min3 tract is composed of identical repeat units (Figure 1A), but ade2-h7.5 is composed of repeat units whose sequence varies at two nucleotides (Figure 3A). While minisatellite alterations are more frequent in the ade2-min3 rad27Δ strain (32.0 blebs/colony; Table 2) than in the ade2-h7.5 rad27Δ strain (5.0 blebs/colony; Figure 3B), both mutant strains show a similar fold increase compared with the parental wild-type strain (9-fold for ade2-min3 and 7-fold for ade2-h7.5). Thus, the HRAS1 minisatellite tract is more stable than the ade2-min3 tract, but the effect of RAD27 loss is approximately the same for both minisatellites. In contrast, END3 and PKC1 mutations affect the stability of the ade2-min3 repeat tract but not the ade2-h7.5 tract. This result indicates that pathways monitoring ade2-h7.5 stability may differ from those monitoring ade2-min3 stability, with the influence of END3 and PKC1 being limited to direct repeat minisatellites.
Almost all of the altered alleles in an ade2-h7.5 rad27Δ strain exhibit tract expansions or contractions; less than 1% of minisatellite tracts examined by PCR were similar in size to the parental ade2-h7.5 allele. Due to the sequence variation between repeats in ade2-h7.5 strains, we are able to determine the exact nature of the alterations that give rise to blebs. We sequenced 18 altered alleles (Figure 3C). Most alleles are the result of one deletion or duplication event in the original repeat tract, likely occurring via homologous recombination, with a strong bias toward alteration of the center repeats. However, two alleles (5′-411233-3′ and 5′-422-3′) are more complex, indicating that some tract alterations in the ade2-h7.5 rad27Δ mutant may derive from more than one event.
Many mechanisms for genome maintenance are conserved between yeast and humans (Taylor and Lehmann 1998). Therefore, it is likely that our results will be applicable to post-mitotic genome maintenance in human cells, especially as both RAD27 and PKC1 have human homologs (Liu et al. 2004; Nishizuka 1992). We have shown that RAD27 regulates the stability of the human cancer-associated HRAS1 minisatellite. Our results indicate PKC1 and END3 may regulate the stability of direct repeat minisatellites only. As some minisatellites that are correlated with human disease are composed of direct repeats, such as the minisatellite associated with progressive myoclonus epilepsy (Lafreniere et al. 1997), our results establish a mechanistic link between factors controlling direct repeat stability and human disease.
Acknowledgments
We thank Cheryl Tucker for technical assistance in cloning END3. This work was supported in part by National Institutes of Health grant 5RO1-GM-072598 to D.T.K. M.J.D. was supported in part by National Institutes of Health grant P50-GM-071508 to the Lewis Sigler Institute at Princeton University.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Allen C., Buttner S., Aragon A. D., Thomas J. A., Meirelles O., et al. , 2006. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 174: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyagari R., Gomes X. V., Gordenin D. A., Burgers P. M., 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278: 1618–1625 [DOI] [PubMed] [Google Scholar]
- Barzilai A., Biton S., Shiloh Y., 2008. The role of the DNA damage response in neuronal development, organization and maintenance. DNA Repair (Amst.) 7: 1010–1027 [DOI] [PubMed] [Google Scholar]
- Benedetti H., Raths S., Crausaz F., Riezman H., 1994. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol. Biol. Cell 5: 1023–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M., 2005. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 17: 568–575 [DOI] [PubMed] [Google Scholar]
- Buard J., Collick A., Brown J., Jeffreys A. J., 2000. Somatic vs. germline mutation processes at minisatellite CEB1 (D2S90) in humans and transgenic mice. Genomics 65: 95–103 [DOI] [PubMed] [Google Scholar]
- Coic E., Feldman T., Landman A. S., Haber J. E., 2008. Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Genetics 179: 199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere H., Buard J., Tessier J., Aubert D., Vergnaud G., et al. , 1999. Meiotic instability of human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nat. Genet. 23: 367–371 [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Ruiz A., Casamayor A., Arino J., 2009. Normal function of the yeast TOR pathway requires the type 2C protein phosphatase Ptc1. Mol. Cell. Biol. 29: 2876–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay C. W., Ayscough K. R., 2006. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 6487–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. V., Petsko G. A., Johnston G. C., Ringe D., Singer R. A., et al. , 2004. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 68: 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M., Krontiris T. G., 1993. Allelic variation of reporter gene activation by the HRAS1 minisatellite. Genomics 17: 429–434 [DOI] [PubMed] [Google Scholar]
- Gresham D., Ruderfer D. M., Pratt S. C., Schacherer J., Dunham M. J., et al. , 2006. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science 311: 1932–1936 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R., 1991. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc., San Diego [Google Scholar]
- Herskowitz I., Jensen R. E., 1991. Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol. 194: 132–146 [DOI] [PubMed] [Google Scholar]
- Huang K. N., Symington L. S., 1994. Mutation of the gene encoding protein kinase C 1 stimulates mitotic recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 6039–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. N., Symington L. S., 1995. Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics 141: 1275–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L., Loeb L. A., 2001. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat. Res. 477: 7–21 [DOI] [PubMed] [Google Scholar]
- Jauert P. A., Edmiston S. N., Conway K., Kirkpatrick D. T., 2002. RAD1 controls the meiotic expansion of the human HRAS1 minisatellite in Saccharomyces cerevisiae. Mol. Cell. Biol. 22: 953–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauert P. A., Jensen L. E., Kirkpatrick D. T., 2005. A novel yeast genomic DNA library on a geneticin-resistance vector. Yeast 22: 653–657 [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Neumann R., 1997. Somatic mutation processes at a human minisatellite. Hum. Mol. Genet. 6: 129–132; 134–136. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Tamaki K., MacLeod A., Monckton D. G., Neil D. L., et al. , 1994. Complex gene conversion events in germline mutation at human minisatellites. Nat. Genet. 6: 136–145 [DOI] [PubMed] [Google Scholar]
- Jeong Y. H., Kim M. C., Ahn E. K., Seol S. Y., Do E. J., et al. , 2007. Rare exonic minisatellite alleles in MUC2 influence susceptibility to gastric carcinoma. PLoS ONE 2: e1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. K., Jauert P. A., Jensen L. E., Chan C. L., Truong C. S., et al. , 2007. Zinc regulates the stability of repetitive minisatellite DNA tracts during stationary phase. Genetics 177: 2469–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. K., Alver B., Kirkpatrick D. T., 2011. Minisatellite alterations in ZRT1 mutants occur via RAD52-dependent and RAD52-independent mechanisms in quiescent stationary phase yeast cells. DNA Repair (Amst.) 10: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy G. C., German M. S., Rutter W. J., 1995. The minisatellite in the diabetes susceptibility locus IDDM2 regulates insulin transcription. Nat. Genet. 9: 293–298 [DOI] [PubMed] [Google Scholar]
- Kirkbride H. J., Bolscher J. G., Nazmi K., Vinall L. E., Nash M. W., et al. , 2001. Genetic polymorphism of MUC7: allele frequencies and association with asthma. Eur. J. Hum. Genet. 9: 347–354 [DOI] [PubMed] [Google Scholar]
- Kokoska R. J., Stefanovic L., Buermeyer A. B., Liskay R. M., Petes T. D., 1999. A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics 151: 511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause S. A., Gray J. V., 2002. The protein kinase C pathway is required for viability in quiescence in Saccharomyces cerevisiae. Curr. Biol. 12: 588–593 [DOI] [PubMed] [Google Scholar]
- Krontiris T. G., Devlin B., Karp D. D., Robert N. J., Risch N., 1993. An association between the risk of cancer and mutations in the HRAS1 minisatellite locus. N. Engl. J. Med. 329: 517–523 [DOI] [PubMed] [Google Scholar]
- Kyo K., Parkes M., Takei Y., Nishimori H., Vyas P., et al. , 1999. Association of ulcerative colitis with rare VNTR alleles of the human intestinal mucin gene, MUC3. Hum. Mol. Genet. 8: 307–311 [DOI] [PubMed] [Google Scholar]
- Lafreniere R. G., Rochefort D. L., Chretien N., Rommens J. M., Cochius J. I., et al. , 1997. Unstable insertion in the 5′ flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet. 15: 298–302 [DOI] [PubMed] [Google Scholar]
- Liu Y., Kao H. I., Bambara R. A., 2004. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 73: 589–615 [DOI] [PubMed] [Google Scholar]
- Lopes J., Debrauwere H., Buard J., Nicolas A., 2002. Instability of the human minisatellite CEB1 in rad27Delta and dna2–1 replication-deficient yeast cells. EMBO J. 21: 3201–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleki S., Cederberg H., Rannug U., 2002. The human minisatellites MS1, MS32, MS205 and CEB1 integrated into the yeast genome exhibit different degrees of mitotic instability but are all stabilised by RAD27. Curr. Genet. 41: 333–341 [DOI] [PubMed] [Google Scholar]
- Mirkin S. M., 2007. Expandable DNA repeats and human disease. Nature 447: 932–940 [DOI] [PubMed] [Google Scholar]
- Nag D. K., Petes T. D., 1993. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 13: 2324–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka Y., 1992. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science 258: 607–614 [DOI] [PubMed] [Google Scholar]
- Ribeyre C., Lopes J., Boule J. B., Piazza A., Guedin A., et al. , 2009. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 5: e1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard G. F., Dujon B., 2006. Molecular evolution of minisatellites in hemiascomycetous yeasts. Mol. Biol. Evol. 23: 189–202 [DOI] [PubMed] [Google Scholar]
- Richard G. F., Kerrest A., Dujon B., 2008. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol. Mol. Biol. Rev. 72: 686–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schacherer J., Ruderfer D. M., Gresham D., Dolinski K., Botstein D., et al. , 2007. Genome-wide analysis of nucleotide-level variation in commonly used Saccharomyces cerevisiae strains. PLoS ONE 2: e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Kokoska R. J., Dominska M., Greenwell P., Petes T. D., 1997. Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol. 17: 2851–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia E. A., Dominska M., Stefanovic L., Petes T. D., 2001. Isolation and characterization of point mutations in mismatch repair genes that destabilize microsatellites in yeast. Mol. Cell. Biol. 21: 8157–8167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L. S., 1998. Homologous recombination is required for the viability of rad27 mutants. Nucleic Acids Res. 26: 5589–5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. M., Lehmann A. R., 1998. Conservation of eukaryotic DNA repair mechanisms. Int. J. Radiat. Biol. 74: 277–286 [DOI] [PubMed] [Google Scholar]
- Trepicchio W. L., Krontiris T. G., 1992. Members of the rel/NF-kappa B family of transcriptional regulatory proteins bind the HRAS1 minisatellite DNA sequence. Nucleic Acids Res. 20: 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakem L. P., Sherman F., 1990. Chromosomal assignment of mutations by specific chromosome loss in the yeast Saccharomyces cerevisiae. Genetics 125: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Soria J. C., Chang Y. S., Lee H. Y., Wei Q., et al. , 2003. Association of a functional tandem repeats in the downstream of human telomerase gene and lung cancer. Oncogene 22: 7123–7129 [DOI] [PubMed] [Google Scholar]
- Wu X., Wang Z., 1999. Relationships between yeast Rad27 and Apn1 in response to apurinic/apyrimidinic (AP) sites in DNA. Nucleic Acids Res. 27: 956–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Chan R. C., Jing J., Li T., Sham P., et al. , 2007. A meta-analysis of association studies between the 10-repeat allele of a VNTR polymorphism in the 3′-UTR of dopamine transporter gene and attention deficit hyperactivity disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B: 541–550 [DOI] [PubMed] [Google Scholar]