Abstract
We present a designed cyclic DNA motif that consists of six DNA double helices that are connected to each other at two crossover sites. DNA double helices with 10.5 nucleotide pairs per turn facilitate the programming of DNA double crossover molecules to form hexagonally symmetric arrangements when the crossover points are separated by seven or fourteen nucleotide pairs. We demonstrate by atomic force microscopy well-formed arrays of hexagonal six-helix bundle motifs both in 1D and in 2D.
Keywords: Designed DNA structures, DNA self-assembly, 1D DNA arrays, 2D DNA arrays, DNA nanotubes
DNA has been shown to be one of the most exciting molecules available for use in nanotechnology.1 The key attraction of this molecule is related to the same features that enable it to function as the genetic material of living organisms, its capacity to contain and express information through intermolecular interactions. Watson-Crick hydrogen bonding is well-known to all, and it sits at the center of DNA's role in life. A useful facet of this interaction is that relatively small sticky ends are able to direct the associations of larger DNA molecules in a sequence-specific fashion.2 In addition, it has been shown that sticky-ended cohesion leads not only to programmable affinities between molecules, but also to predictable local product structures.3 Nevertheless, the topologically linear nature of the DNA helix axis would prevent its use in many applications; central to the use of DNA in nanotechnology is the ability to extend its structure to include branched motifs.4,5 DNA molecules have been produced containing from three to six branches.6 In addition, a number of motifs have been produced from multiply-fused four-arm junctions, such as the double crossover (DX) molecule,7 the triple crossover (TX) molecule8 and the paranemic crossover (PX) molecule.9
The TX molecule described previously contains three double helical domains that are coplanar as nearly as possible, so that the dihedral angle formed by its two DX domains is about 180°. For 10.5-fold DNA,10,11 a dihedral angle of 120° ought to be an easy structure to achieve, because 7 nucleotide pairs correspond to exactly 2/3 of a turn and 14 nucleotide pairs correspond exactly to 4/3 of a turn. Here, we have tested this notion by assembling six-helix bundles of DNA. In principle, six helices, each rotated 120° out of the plane of the last pair, ought to produce an hexagonal bundle containing a hole down the middle. Here, we describe the construction of such a six-helix bundle (6HB), both as a blunt-ended molecule and as a molecule containing sticky ends on its termini. Without ligation, one sticky-ended molecule self-assembles to form one-dimensional arrays 1-15 microns long. We also describe 2D arrays that can be formed from tetrads of sticky ends. In addition to this property, another important feature of the molecule is that it is the first specifically-designed non-planar array, potentially useful for the construction of nanodevices with features that flank a central axis. Other DNA nanotubes have been described recently12,13,14 but they are a consequence of bending within double crossover (DX) tiles. By contrast, the tube described here is designed as a specific DNA motif.
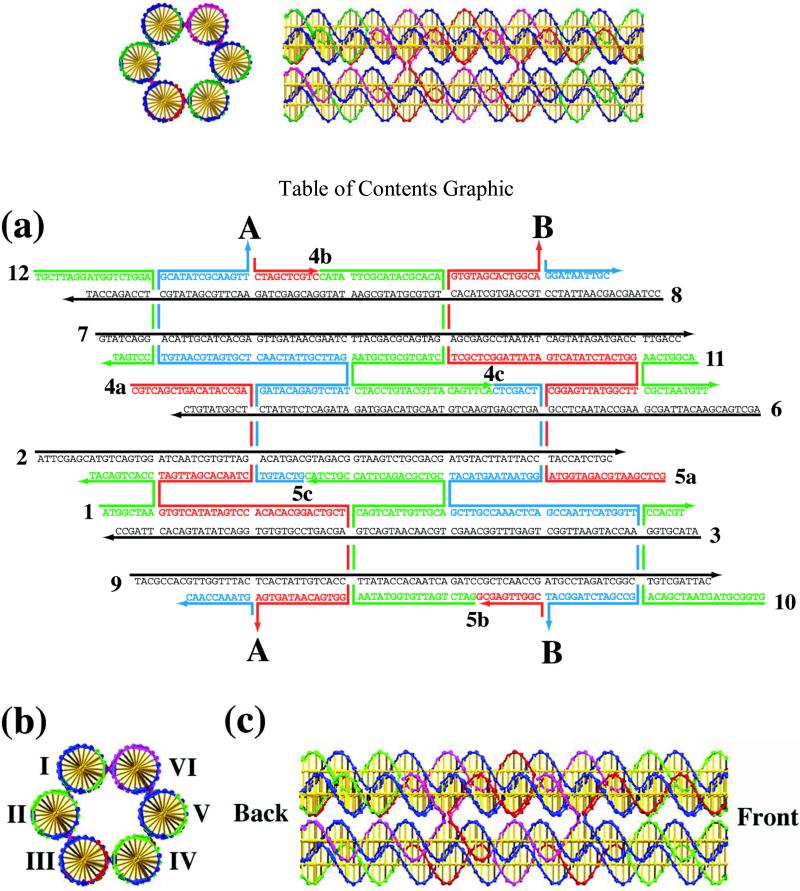
Figure 1a illustrates the sequence of a 6HB molecule in which the individual pairs of DX components are separated by fourteen nucleotide pairs. The motif shown contains sticky ends, so that it can self-assemble into a long 1-dimensional construct. Figure 1b illustrates the cross-section of a 6HB molecule, and Figure 1c shows a side view in the same style; in this case, the individual DX crossovers are separated by seven nucleotide pairs, and the ends are all blunt. From Figure 1b, it is clear that in its ideal form, the motif contains a hole down its middle that is approximately the diameter of a DNA double helix, ~2nm.
Figure 1.
Schematic Drawings of the Six-Helix Bundle Motif. (a) The strand sequences are shown for a version of this motif with 14 nucleotide pairs between crossovers. Points A and B are the places where the cyclic bundle closes. Strand numbering is indicated. There are six helical strands, shown in black, and the other strands perform the crossovers. (b) A cross-sectional geometrical view of a six helix bundle in which the helices are indicated by Roman numerals. (c) A geometrical side view of a six helix bundle in which the crossovers are separated by seven nucleotide pairs. The back and front are indicated as the ends to be used in self-assembly.
Thus, it is feasible to design a 6HB motif without stressing the polynucleotide backbone. Is it actually possible to construct such a motif? Figure 2 shows data that resolves this question. This figure shows autoradiograms of non-denaturing gels containing a blunt-ended version of the molecule shown in Figure 1a. Each lane contains all of the components of the 6HB motif, but a different strand has been labeled radioactively in each lane, as indicated by the label there. In each case, we see the same single band in the vicinity of the expected size (504 nucleotide pairs), with neither multimers nor breakdown products present. This method is an effective means of demonstrating the stability of a motif, so long as the labeled strands are present in proper stoichiometric ratios with the other strands.14 The slightly higher mobility of the motif than the linear markers of equivalent molecular weight is likely a consequence of the lowered surface area of the 6HB motif, relative to the same number of nucleotide pairs in linear duplex DNA.
Figure 2.
Autoradiograms of 5% Non-Denaturing Gels that Establish the Robustness and Proper Stoichiometry of the Six Helix Bundle Motif. Every lane contains the complete blunt-ended 6HB molecule. The lanes are labeled with the radioactive strand that is contained in that complex; strand numbering is as shown in Figure 1. The marker lanes contain a 100-base pair ladder. Note that every strand is incorporated into the motif, and that there are neither multimers nor breakdown products in this gel, demonstrating the robust character of the motif.
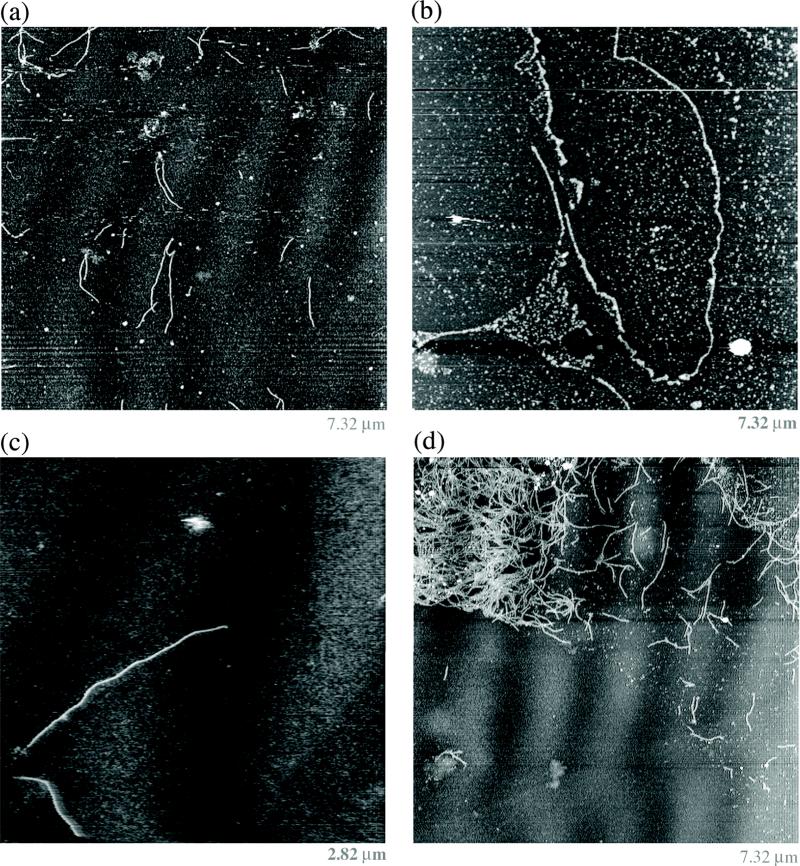
The 6HB molecule with sticky ends offers opportunities for the formation of arrays in 1D, 2D and 3D. It is straightforward to build a self-assembling 1D array of 6HB molecules, simply by using the version of the molecule shown in Figure 1a, where the sticky ends on the two ends of each helix are complementary to each other. We find that these molecules self-assemble readily into long wire-like species, as illustrated in Figure 3, which contains contact-mode atomic force microscopy (AFM) images of these assemblies. Figure 3a shows a number of the 6HB assemblies in a 7.3 micron-square field. They form discrete linear entities that do not display much curvature. Figure 3b shows the longest 1D 6HB assembly that we have been able to identify; the continuous part on the right is about 7 μm long, and it may originally have been as long as 15 μm in length before contact-mode scanning. Figure 3c illustrates a zoomed image of a typical 6HB self-assembly about 1.9 μm in length. Sometimes the assemblies form large clumps from which some individual 1D assemblies separate during spreading on the mica surface, as illustrated in Figure 3d.
Figure 3.
One-Dimensional 6HB Self-Assembled Arrays. The images are obtained by contact mode atomic force microscopy. (a) A series of 6HB wire-like images. (b) A long nanotube. (c) A zoomed view showing a large nanotube. (d) A bundle of nanotubes from which some have been separated. Note that the molecules in all panels are relatively unbent.
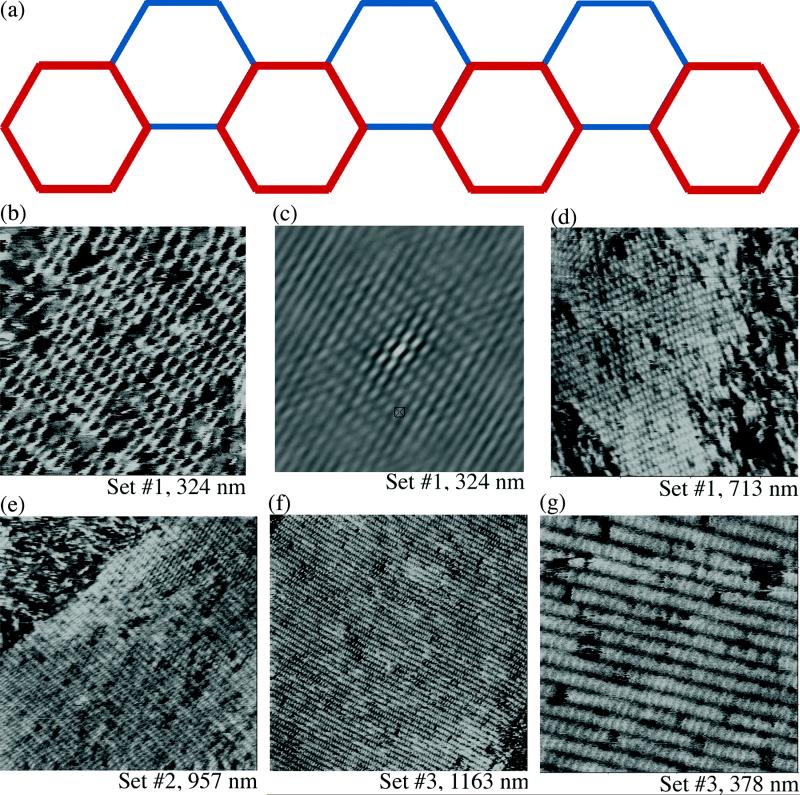
We have used the motif in Figure 1c to assemble multidimensional arrays. We have learned from our recent success in assembling hexagonal arrays16 that it is more effective to rely on cohesion between two adjacent double helices (DX cohesion) rather than on a single pair of sticky ends. Thus, the most robust way to assemble 6HB motifs in multiple dimensions appears likely to be bonding the helices in adjacent pairs. A 3D array would result if one paired sticky ends on the front ends of helices I and VI (Figure 1b) with sticky ends on the back ends of helices III and IV, the fronts of II and III with the backs of VI and V, and the fronts of IV and V with the backs of II and I. 3D arrays of 6HB motifs will be discussed elsewhere. For our purposes here, it is interesting that a well-formed 2D array of 6HB motifs will result if only two of the three pairs listed above contain sticky ends. The 2D 6HB arrangement will form a corrugated structure when viewed in projection down the helix axes, as illustrated in Figure 4a. This schematic represents the 6HB unit as hexagons whose vertices correspond to helix axes; the red hexagons are closer to the reader than the blue hexagons. This feature is evident in Figure 4b, which shows a tapping mode AFM image of a 2D array in which one of the sets of sticky ends is blunt; we use two different tiles, an A-tile and a B-tile, to form these arrays (the sequences and the specifically blunted helices are presented in the supplementary information). This is a very well-ordered array, because two directions of ordering are evident in its autocorrelation pattern, which is shown in Figure 4c. A longer view of the array is shown in Figure 4d. Figure 4e shows an array of the same 6HB components, but with a different set of blunted ends. The array looks very good, but its autocorrelation pattern only indicates one direction of distinguishable ordered components. Figure 4f shows the third set in long view, and Figure 4g shows a zoom of the same array. The ordering is somewhat better for this array, but it is clear that there are components missing from the crystal. The image in Figure 4g is reminiscent of an ear of corn from which occasional kernels are missing.
Figure 4.
Two dimensional arrays obtained from six helix bundle motifs. The three different paired sets of helical sticky ends have been blunted to produce a 2D array. A schematic where the vertices of hexagons represent helix axes is shown in (a); the red layer is closer to the reader than the blue layer. From the first set, a zoom is shown in panel (b), its autocorrelation function is shown in (c), and a long view is shown in (d). The zoom in (b) shows how the individual 6HB motifs form a corrugated arrangement. Note that the autocorrelation function indicates 2D order. A long view of each of the other two directions is shown in (e) and (f). Neither of their autocorrelation functions indicates good order in more than one direction. (g) shows that the image in (f) has clear vacancies, but that they do not appear to destroy the periodicity of the array.
The differences noted here between the three directions are seen consistently. If one assumes the absence of sequence-dependent structural effects, the three independent directions should be equivalent geometrically; the presence of differences in ordering suggests the breakdown of that assumption. The observed unit cell dimensions for the three groups (23.25 ± 0.25; 23 ± 0.3; 22.75 ± .25 nm) are close to the predicted values (20.7 nm) in the long direction. In the transverse direction, however, there is a much greater disparity between observed (14 ± 0.5; 12.75 ± 0.75; 11.5 ± 0.5 nm) and estimated (~6 nm) repeat distances. One factor leading to this discrepancy may be flattening of the tubes when in contact with the mica surface. The second factor, likely more important, is that antiparallel helices are often seen to be separated by much more than their estimated separation when observed in buffer on mica, using AFM tapping mode (ref. 14 and B. Ding and NCS, unpublished).
We have demonstrated a new motif in nucleic acid nanotechnology, the deliberately designed DNA tube. We expect it can be put to a number of uses, because it is a prototype for a surface with a designed curvature. In its current form, it appears to be useful as a potential nanomechanical strut, because it seems to be fairly stiff. In addition, there are evident applications to creating related molecules whose inner or outer surfaces are uncharged, for example by building them out of peptide nucleic acids.17 A 6HB motif containing a strategically placed neutral outer surface ought to be able to impale a cell membrane, and a 6HB motif with a neutral inner surface might serve to encapsulate neutral elongated species, such as carbon nanotubes.
The formation of a closed surface by the 6HB provides a group of helices that could be used to mount other motifs or nanodevices that could be oriented in different directions. The 6HB is only one example of deliberately designed nucleic acid-based nanotubes. It is evident that there are many different relaxed DNA nanotubes that could be built using the different sets of available dihedral angles that correspond to relaxed TX molecules. These different nanotubes could have larger radii, and even could provide a mixture of concave and convex surfaces. The design of these relaxed nucleic acid nanotubes is treated elsewhere (WB Sherman and NCS, in preparation).
Supplementary Material
ACKNOWLEDGMENT
This research has been supported by grants CTS-0103002, DMI-0210844, EIA-0086015, CCF-0432009 and DMR-01138790 from the National Science Foundation, GM-29554 from the National Institute of General Medical Sciences, N00014-98-1-0093 from the Office of Naval Research, F30602-01-2-0561 from DARPA/AFSOR and an award from Nanoscience Technologies, Inc.
Footnotes
Supporting Information Available
Strand sequences for the components of Figure 4. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 2.Cohen SN, Chang ACY, Boyer HW, Helling RB. Proc. Nat. Acad. Sci. (USA) 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiu H, Dewan JC, Seeman NC. J. Mol. Biol. 1997;267:881–898. doi: 10.1006/jmbi.1997.0918. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 5.Seeman NC. NanoLett. 2001;1:22–26. 5667–5674. [Google Scholar]
- 6.Wang Y, Mueller JE, Kemper B, Seeman NC. Biochem. 1991;30 doi: 10.1021/bi00237a005. [DOI] [PubMed] [Google Scholar]
- 7.Fu T-J, Seeman NC. Biochem. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 8.LaBean T, Yan H, Kopatsch J, Liu F, Winfree E, Reif JH, Seeman NC. J. Am. Chem. Soc. 2000;122:1848–1860. [Google Scholar]
- 9.Shen Z, Yan H, Zhang X, Seeman NC. J. Am. Chem. Soc. 2004;126:1666–1674. doi: 10.1021/ja038381e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J. Proc. Nat. Acad. Sci. (USA) 1979;76:200–203. [Google Scholar]
- 11.Rhodes D, Klug A. Nature. 1980;286:573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Park SH, Reif JH, LaBean TH. Proc. Nat. Acad. Sci. (USA) 2004;101:717–722. doi: 10.1073/pnas.0305860101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell JC, Harris R, Maio J, Bath J, Turberfield AJ. J. Am. Chem. Soc. 2004;126:16342–16343. doi: 10.1021/ja043890h. [DOI] [PubMed] [Google Scholar]
- 14.Rothemund PWK, Ekani-Nkodo A, Papadakis N, Kumar A, Fygenson DK, Winfree E. J. Am. Chem. Soc. 2004;126:16344–16352. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 15.Liao S, Seeman NC. Science. 2004;306:2072–2074. doi: 10.1126/science.1104299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding B, Sha R, Seeman NC. J. Am. Chem. Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 17.Lukeman PS, Mittal A, Seeman NC. Chem. Comm. 2004;(15):1694–1695. doi: 10.1039/b401103a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.