Abstract
Our previous studies revealed that the staphylococcal protein Gcp is essential for bacterial growth; however, the essential function of Gcp remains undefined. In this study, we demonstrated that Gcp plays an important role in the modulation of the branched-chain amino acids biosynthesis pathway. Specifically, we identified that the depletion of Gcp dramatically elevated the production of key enzymes that are encoded in the ilv-leu operon and responsible for the biosynthesis of the branched-chain amino acids isoleucine, leucine, and valine (ILV) using proteomic approaches. Using qPCR and promoter-lux reporter fusions, we established that Gcp negatively modulates the transcription of the ilv-leu operon. Gel-shift assays revealed that Gcp lacks the capacity to bind the promoter region of ilv. Moreover, we found that the depletion of Gcp did not influence the transcription level of CodY, a known repressor of the ilv-leu operon, while induced the transcription of CcpA, a known positive regulator of the ilv-leu operon. In addition, the depletion of Gcp decreased the biosynthesis of N6-threonylcarbamoyladenosine (t6A). To elucidate whether the essentiality of Gcp is attributable to its negative modulation of ILV biosynthesis, we determined the impact of the ilv-leu operon on the requirement of Gcp for growth, and revealed that the deletion of the ilv-leu operon did not affect the essentiality of Gcp. Taken together, our results indicate that the essentiality of Gcp isn’t attributable to its negative regulation of ILV biosynthesis in S. aureus. These findings provide new insights into the biological function of the staphylococcal Gcp.
Introduction
Staphylococcus aureus is an important pathogen that can cause severe human and animal infections. The prevalence of multi-drug resistant S. aureus, especially methicillin- and vancomycin-resistant S. aureus has caused serious public health concerns [1], [2]. Understanding the physiology of bacterial cells allows us to identify alternative strategies to combat S. aureus. Our previous studies have demonstrated that a Gcp is required for the viability of S. aureus during in vitro culture [3] and is involved in the modulation of bacterial autolysis [4]. However, the essential function of Gcp remains undefined.
The metabolic pathway of the branched-chain amino acids (BCAAs) isoleucine, leucine, and valine (ILV) plays an important role in bacterial physiology. These hydrophobic amino acids are crucial for maintaining protein structure and function [5]. The branched-chain alpha-keto acids, both the first catabolic and the last anabolic intermediates of ILV, are the precursors of the branched-chain fatty acids that are the major fatty acids involved in bacterial cell membrane biosynthesis [6], [7]. Moreover, the immediate precursor of valine, α-ketoisovalerate, is the precursor of leucine and involved in the synthesis of cofactors pantothenate and coenzyme A [8]. The biosynthesis of BCAAs requires several key enzymes. In Bacillus subtilis, the genes encoding these enzymes are well characterized, including the ilvBHC-leuABCD (ilv-leu) operon and ilvA, ilvD, ybgE and ywaA genes distributed around the genome [9], [10], [11]. In S. aureus, there is a similar ilv-leu gene cluster ilvDBHC-leuABCD-ilvA containing 9 genes that are involved in the ILV biosynthesis pathway. Another gene ilvE encoding an aminotransferase has a similar function with ybgE and ywaA of B. subtilis.
Several mechanisms of regulating the biosynthesis of BCAAs have been revealed and characterized in different organisms. In B. subtilis, it has been shown that a T-box anti-termination system mediates the ilv-leu operon in response to leucine availability [9], [12]; moreover, the specific cleavage sites of the full-length transcript and the stability of the cleavage product affect the transcription level of each gene in the ilv-leu operon [13]. In S. aureus, it was revealed that a global regulator CodY represses the BCAA biosynthesis [14]; whereas two global regulators of nitrogen metabolism in B. subtilis, CodY and TnrA, repress the BCAA biosynthesis through binding to an upstream regulatory region of the ilv-leu operon [14], [15], [16], [17], [18]. In addition, in B. subtilis a major regulator of carbon metabolism, CcpA, positively regulates the transcription of the ilv-leu operon by binding to the promoter region [10], [18], [19]. Although S. aureus possesses all genes necessary for biosynthesis of BCAAs, the bacterium exhibits an auxotrophic phenotype for BCAAs through an unknown mechanism [20], [21].
Recent biochemical analysis from E. coli showed that Gcp homologues are involved in the biosynthesis of N6-threonylcarbamoyladenosine (t6A) and tRNA modification [22], which is a highly conserved mechanism. In this study, we identified that the BCAA biosynthesis pathway and the biosynthesis of N6-threonylcarbamoyladenosine (t6A) are controlled by the essential protein Gcp and demonstrated that Gcp modulates the transcription of the ilv-leu operon in S. aureus. The elimination of isoleucine, leucine, and valine remarkably enhanced bacterial growth during the depletion of Gcp. Importantly, we found the essentiality of Gcp is not attributable to the negative modulation of BCAA biosynthesis. These new findings provide new insights into the biological function of the essential protein, Gcp.
Results
Identification of IlvA, IlvB and IlvD Proteins that are Elevated by the Depletion of Gcp Using Proteomic Approaches
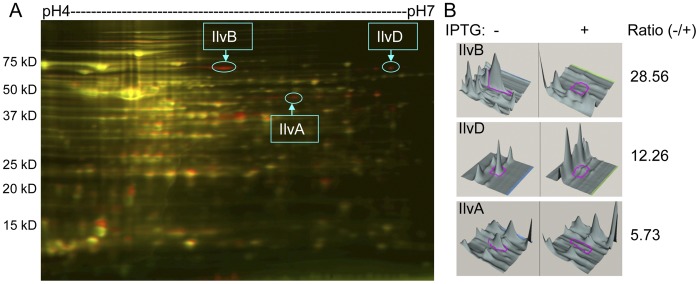
In order to explore the potential function of the essential protein Gcp, we examined the impact of Gcp on global protein production of S. aureus by using 2D-Differential In Gel Electrophoresis (DIGE) coupled with mass spectrometry. The bacterial cells were harvested from the mid-log phase cultures of the Pspac-regulated gcp expression strain with and without addition of inducer, IPTG as described [3]. The bacterial cells were lysed, and whole cell lysates was labeled with Cy2, Cy3 and Cy5 respectively as described in the Materials and Methods. The same amount of labeled proteins was loaded onto gradient SDS-PAGE following electrophoresis. Proteins with specific dyes from different samples were separated and compared. Ten differential spots were picked and followed by in-gel digestion and MALDI mass spectrometry. Based on the data obtained from the Scaffold database, three proteins were found more than once, including threonine dehydratase (IlvA, EC: 4. 3. 1. 19), acetolactate synthase (IlvB, EC: 2. 2. 1. 6), and dihydroxy-acid dehydratase (IlvD, EC: 4. 2. 1. 9) (Fig. 1A). The genes encoding the three proteins are located in an ilv-leu operon and are key enzymes for the biosynthesis of the branched-chain amino acids (BCAAs), isoleucine, leucine, and valine. Moreover, each pair of protein spots (Cy3 and Cy5-labeled) was further assessed in 3D views and statistically analyzed. We found that the expression of IlvA, IlvB and IlvD was significantly increased during the depletion of Gcp (Fig. 1B). The identity of remaining protein spots was not determined. The above results indicate that the essential staphylococcal Gcp may be involved in the modulation of the biosynthesis pathway of BCAAs.
Figure 1. Effect of the down-regulation of Gcp on global gene expression.
(A) 2D-DIGE analysis of the impact of the down-regulation of Gcp on global gene expression in the culture of the Pspac-regulated gcp expression strain (JRN0105), the arrows indicate the identity of proteins in the selected protein spots. (B) 3D review of the selected protein spots, and the changes of the selected protein spots containing IlvA, IlvB, and IlvD, respectively, during the depletion of Gcp.
The Depletion of Gcp Dramatically Increased the Transcription of the ilv-leu Operon
Bioinformatics analysis of the S. aureus genome revealed that nine genes are sequentially aligned in the ilvDBHC-leuABCD-ilvA operon, which is similar to the ilv-leu operon responsible for the biosynthesis of the branched-chain amino acids in B. subtilis [11]. Genome mapping showed that the gcp operon (20) and the ilv-leu operon were juxtaposed on the genome of S. aureus (Fig. 2). To confirm the effect of Gcp on the expression of IlvA, IlvB, and IlvD, we examined the impact of the depletion of Gcp on the transcription of ilvD and leuA located on the ilv-leu operon, and ilvE gene, which is located elsewhere in S. aureus genome and encodes an aminotransferase. The qPCR results showed that the down-regulation of Gcp had no impact on ilvE RNA levels; whereas the RNA levels of ilvD, leuA and ilvA were elevated by approximately 5- to 111- fold, suggesting that Gcp mediates the transcription of the ilv-leu operon (Table 1). Interestingly, we found that the depletion of Gcp increased the transcription of ccpA (Table 1), which encodes CcpA, a possibly positive regulator of the ilv-leu operon.
Figure 2. Illumination of the genomic context and arranges of gcp and ilv-leu operons.
Table 1. Gene transcription change due to the depletion of Gcp.
| ORF (N315) | Gene | Fold Changea |
| Sa0512 | ilvE | 0.97 |
| Sa1098 | codY | 0.78 |
| Sa1557 | ccpA | 3.24 |
| Sa1858 | ilvD | 5.45 |
| Sa1862 | leuA | 68.59 |
| Sa1866 | ilvA | 111.37 |
The fold change represents the transcription levels of genes with the depletion of Gcp compared with those during the induction of gcp transcription with IPTG (200 μM) at exponential phase of growth (OD600 nm ∼0.5).
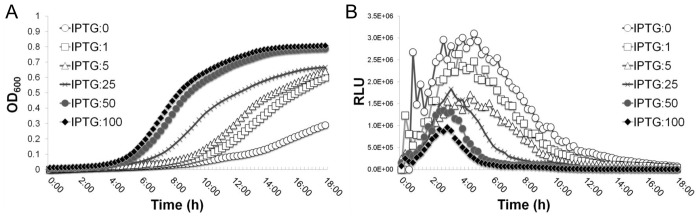
To further confirm the transcriptional regulation, we constructed an ilv promoter-lux reporter fusion in the Pspac-regulated gcp expression mutant. This system allows us to effectively down-regulate gcp expression and simultaneously to monitor the reporter gene expression by measuring the bioluminescence intensity [23]. The results showed that bacterial growth is dependent on IPTG, indicating the essentiality of Gcp (Fig. 3A); whereas the bioluminescence intensity was increased from the early log to mid-log phases of growth during the depletion of Gcp in a dose dependent manner (Fig. 3B). The Pspac-regulated gcp expression mutant carrying a promoterless lux reporter was utilized as a control. The addition of IPTG had no influence on the lux expression in the control strain (data not shown). Taken together, these data indicate that Gcp transcriptionally mediates the expression of the ilv-leu operon.
Figure 3. Effect of the down-regulation of Gcp on the ilv promoter transcription activity.
(A) The growth curves of the Pspac-regulated gcp expression mutant containing the ilv promoter-lux reporter fusion (JRN0110) at the presence of different concentrations of inducer IPTG. (B) Relative Bioluminescence intensity of JRN0110 during the culture with different concentrations of IPTG (0, 1, 5, 25, 50 and 100 µM). The optical density at OD600 nm and bioluminescence intensity of cultures were simultaneously measured every 15 min for 18 hours at 37°C using a BioTek Synergy plate reader. The experiments were repeated at least five times. These figures are one representative of three independent experiments.
The Recombinant Gcp Lacks the Capacity of Binding to the Promoter Region of ilv-leu Operon
Structural analysis of Gcp protein revealed that Gcp lacks a helix-turn-helix DNA-binding domain, suggesting that Gcp may indirectly regulate the transcription of ilv-leu operon. To further determine the mechanism of Gcp’s involvement in modulating the expression of the ilv-leu operon, we expressed and purified His-tagged Gcp protein as described [3] and performed gel-shift assays to examine whether Gcp binds to the promoter region of ilv. Negative controls included the labeled ilv promoter region without protein and the labeled ilv with BSA protein. The non-labeled ilv promoter region was used as a specific competitor, while an internal ilv fragment probe was used as a nonspecific binding control. The results showed that the addition of Gcp in the reaction mixtures containing the ilv promoter probe had no impact on the electrophoretic mobility of ilv probe compared to the controls (data not shown). This suggests that the regulation of the ilv-leu operon transcription by Gcp may not function through its binding to the ilv promoter region.
Gcp is Involved in the Biosynthesis of N6-threonyl Carbamoyladenosine (t6A)
It has been reported that the Gcp homolog YgjD in E. coli is required for the synthesis of t6A [22]. To determine whether Gcp is involved in t6A biosynthesis in S. aureus, we quantitatively measured the t6A/A ratio in tRNAs purified from the staphylococcal cells under Gcp deplete (-IPTG) or Gcp-replete (+ IPTG) conditions using LC-MS/MS. When the defined Pspac-regulated gcp expression mutant was grown in TSB with IPTG, the ratio of t6A/A was 1.3%, whereas during depletion of Gcp by growing the mutant in the absence of IPTG the ratio of t6A/A observably decreased 5-fold to 0.26% (Fig. 4). This indicates that the staphylococcal Gcp is important for t6A modification.
Figure 4. Determine the effect of depletion of Gcp on t6A content of tRNA isolated from staphylococcal cells using LC-MS/MS.
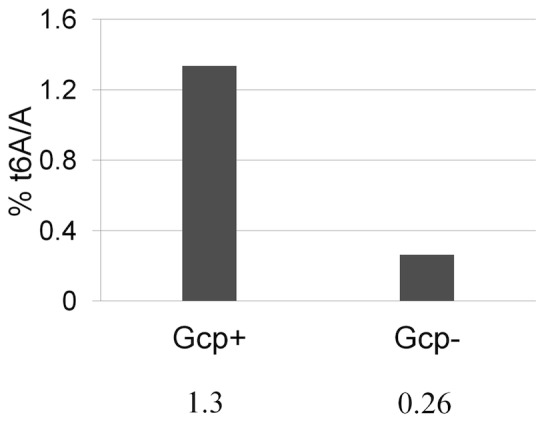
tRNA isolated and purified from the defined Pspac-regulated gcp expression mutant with (Gcp+) and without inducer IPTG (200 μM) (Gcp-) at exponential phase of growth (OD600nm ∼0.5) and processed for LC-MS/MS analysis of t6A as described in Materials and Methods. The t6A content was normalized to the respective adenosine content and represented to % t6A/A ratio (the numerical values).
Down-regulation of gcp Enhances Bacterial Growth in ILV Dropout Chemically Defined Media
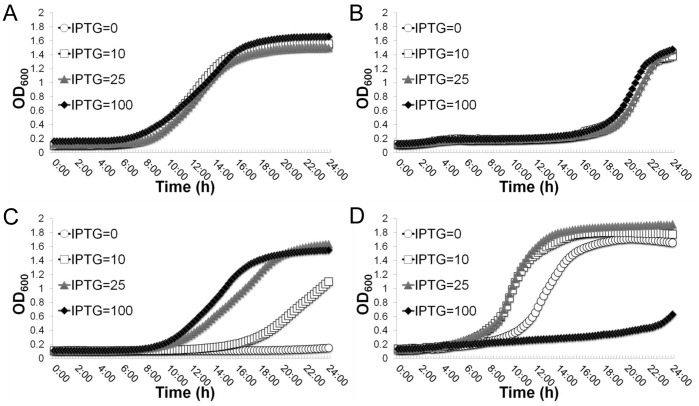
The above findings led us to hypothesize that the tight control of the branched-chain amino acids biosynthesis by Gcp may contribute to the essentiality of Gcp for growth. To test this possibility, we determined the effect of isoleucine, leucine and valine (ILV) on the requirement for Gcp by monitoring the growth of the defined Pspac-regulated gcp expression mutant in ILV dropout CDM with varying amounts of IPTG. The conditional gcp expression mutant and the wild-type control were inoculated into nutrient complete CDM and ILV dropout CDM, respectively. The wild-type control strain exhibited consistent growth curves regardless of IPTG concentrations in all growth media (Fig. 5A and 5B). A longer lag phase was seen in ILV dropout CDM (Fig. 5B) compared with nutrient complete CDM (Fig. 5A). However, in nutrient complete CDM the defined Psapc-regulated gcp expression mutant exhibited IPTG dependent growth; without IPTG induction bacterial growth was dramatically inhibited, indicating the essentiality of Gcp for growth (Fig. 5C). In contrast, in ILV dropout CDM, without IPTG induction the bacteria exhibited a robust growth phenotype; whereas in the presence of 100 µM IPTG bacterial growth was noticeably inhibited (Fig. 5D). In addition, IPTG dependent growth only occurred between 0 to 25 µM IPTG (Fig. 5D).
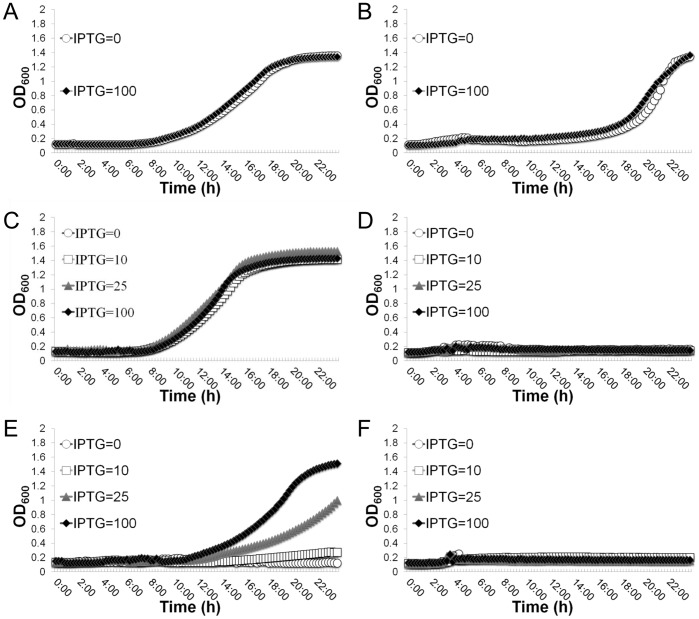
Figure 5. Effect of the depletion of the branched-chain amino acids isoleucine, leucine and valine (ILV) on the growth of the defined Pspac-regulated gcp expression strain.
The growth curves of the wild type control, WCUH29 carrying pYH4-lacI in in nutrient complete CDM (A) and ILV dropout CDM (B); the growth curves of the defined Pspac-regulated gcp expression strain (JW290111) in in nutrient complete CDM (C) and ILV dropout CDM (D). The bacterial growth in the presence of different concentrations of inducer IPTG (0, 10, 25 and 100 µM) was monitored by kinetically measuring the optical density at OD600 nm every 15 min for 24 hours at 37°C using a BioTek Synergy plate reader. These figures are one representative of three independent experiments.
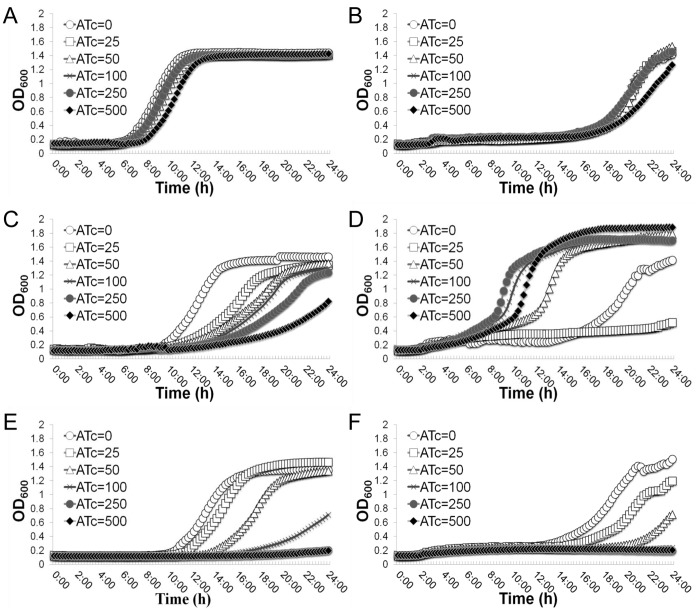
To eliminate potential effects of isoleucine, leucine and valine amino acids on the regulatory function of the Pspac-regulated gcp expression system, we confirmed the effect of the depletion of Gcp on growth in ILV dropout CDM using a TetR-regulated gcp antisense RNA expression system [4]. The wild-type strain carrying parental plasmid and a TetR-regulated eno antisense RNA expression mutant [24] were used as controls. In nutrient complete CDM, the addition of inducer, ATc, had no obvious impact on the growth of the wild-type control strain (Fig. 6A); whereas, the growth of the TetR-regulated gcp antisense RNA mutant exhibited an ATc dependent inhibition; and the growth was abolished with a higher dose inducer, 500 µM ATc, indicating the essentiality of Gcp (Fig. 6C). In ILV dropout CDM, the addition of ATc had no obvious influence on growth of the wild-type control (Fig. 6B); whereas remarkably the induced gcp antisense RNA enhanced the growth of the TetR-regulated gcp antisense RNA strain, when 50 µM or more of inducer ATc was present, indicating the stimulatory effect on growth by the down-regulation of Gcp (Fig. 6D). However, the growth of the gcp antisense RNA mutant was dramatically inhibited in the presence of 25 µM ATc compared without inducer, indicating the importance of Gcp for growth (Fig. 5D). In addition, in both nutrient complete CDM and ILV dropout CDM, the growth of the eno antisense RNA control strain was remarkably inhibited by the addition of inducer ATc in a dose dependent manner (Fig. 6E, F). These results eliminated the possibility of non-specific effect of ILV dropout on the Pspac-regulatory system and the TetR-regulated antisense RNA expression system, and demonstrated that moderate depletion of Gcp has a stimulatory effect on bacterial growth in the absence of BCAAs.
Figure 6. Effect of the depletion of the branched-chain amino acids isoleucine, leucine and valine (ILV) on the growth of the TetR-regulated gcp antisense RNA strain.
The growth curves of control strain WCUH29 carrying parental plasmid pYH4 in nutrient complete CDM (A) and in ILV dropout CDM (B); the growth curves of the TetR-regulated gcp antisense RNA strain (WCUH29/gcp-as) in nutrient complete CDM (C) and in ILV dropout CDM (D); and the growth curves of the TetR-regulated eno antisense RNA strain (WCUH29/eno-as ) in nutrient complete CDM (E) and in ILV dropout CDM (F). The growth curves are monitored by kinetically measuring the optical density at OD600 nm in the corresponding culture medium in the presence of different concentrations of inducer, anhydrotetracyclin (ATc: 0, 25, 50, 100, 250 and 500 ng/ml) every 15 min for 24 hours at 37°C using a BioTek Synergy plate reader. These figures are one representative of three independent experiments.
Modulation of ilv-leu Operon by Gcp has no Involvement in Gcp’s Essentiality
The above results suggested that the inhibitory effect on growth by the depletion of Gcp may result from the accumulation of ILV due to the over-expression of the ilv-leu operon. To test this possibility, we constructed ilv-leu operon deletion mutants in both the wild-type WCUH29 strain and the Pspac-regulated gcp expression mutant and examined the effect on the essentiality of Gcp. As expected, in ILV dropout CDM the wild-type control grew slower than that in nutrient complete CDM as expected (Fig. 7A, B). The only deletion of the ilv-leu operon had no influence on bacterial growth in nutrient complete CDM (Fig. 7C); whereas in ILV dropout CDM the growth of the ilv-leu null mutants was completely eliminated (Fig. 7D, F). These results further confirmed the null mutation of the ilv-leu operon by the deletion mutagenesis. Surprisingly, in nutrient complete CDM the deletion of the ilv-leu operon had no impact on IPTG-dependent growth of the defined Pspac-regulated gcp expression mutant (Fig. 7E). This result indicates that Gcp’s influence on ILV biosynthesis is not associated with the essential mechanism of Gcp for growth.
Figure 7. Effect of the deletion of the ilv-leu operon on the essentiality of Gcp for growth.
The growth curves of wild-type control, WCUH29 carrying pYH4-lacI, in nutrient complete CDM (A) and in ILV dropout CDM (B). The growth curves of the ilv-leu operon deletion mutant (JW290211) in nutrient complete CDM (C) and in ILV dropout CDM (D); the growth curves of the ilv-leu operon deletion and defined Pspac-regulated gcp expression strain (JW290311) in nutrient complete CDM (E) and in ILV dropout CDM (F). The growth curves are monitored by kinetically measuring the optical density at OD600 nm in the corresponding culture medium in the presence of different concentrations of inducer, IPTG (0. 10, 25 and 100 µM), in every 15 min at 37°C using a BioTek Synergy plate reader. These figures are one representative of three independent experiments.
Discussion
The staphylococcal protein Gcp is conserved, belongs to the Kae1/OSGEP/Gcp family [25], and is essential for bacterial growth in a variety species [26]; however, the essential biological function of Gcp remains largely undefined. This study provides direct evidence that the essential protein Gcp is involved in the t6A modification pathway, and negatively regulates the expression of the ilv-leu operon responsible for the biosynthesis pathway of the branched-chain amino acids (BCAAs): isoleucine, leucine and valine (ILV). In addition, our results showed that the depletion of Gcp dramatically enhanced bacterial growth in chemically defined medium lacking these branched-chain amino acids. Importantly, we demonstrated that the essentiality of Gcp for growth is not attributable to the modulation of the ilv-leu operon. These findings provide new insights into the biological function of the essential protein Gcp of S. aureus.
In this study, we identified significantly altered expression of three key enzymes of the BCAAs biosynthesis pathway, including threonine dehydratase, acetolactate synthase, and dihydroxy-acid dehydratase, during the depletion of Gcp using 2D-DIGE combined with mass spectrometry. These enzymes are encoded by ilvA, ilvB and ilvD gene, respectively, located in an ilv-leu operon. Although ilvA, ilvB, and ilvD are located in the same ilv-leu operon, we observed different expression levels of these proteins in 2D-DIGE. A possible explanation for this is that one identified protein spot on 2D-gel may contain several different proteins, although we could not determine the identities of these proteins due to limited peptides coverage and the amount of protein. Another possibility is that the protein levels for each gene are regulated at a post-translational level and could not be determined by our present experiments. Our qPCR and ilv promoter-lux reporter experiments further confirmed the 2D-DIGE and mass spectrometry results and indicated the transcriptional modulation of ilv-leu operon by Gcp in S. aureus, which is consistent with previous report that the depletion of Gcp homolog, YgjD, in E. coli increases the transcription of the thr and ilv operons [27]. Our qPCR assays revealed that Gcp has a similar influence on the transcriptional level of ilvD, leuA, and ilvA, but has no impact on ilvE, which lies outside of the ilv-leu operon in the S. aureus chromosome. These results further indicate the specific effect of Gcp on the ilv-leu operon that is important for the biosynthesis of BCAAs in S. aureus. Although our qPCR and promoter-lux reporter results indicated that the transcription of the ilv-leu operon is highly elevated during the depletion of Gcp, the protein-DNA binding experiment did not reveal any evidence of Gcp binding to the ilv promoter region, indicating indirect transcriptional regulation by Gcp. This is consistent with our structural analysis showing that Gcp is unlikely to be a DNA binding protein due to the absence of a DNA binding domain (data not shown). However, this finding is inconsistent with a previous report about a Gcp Pyrococcus abyssi homolog, Pa-Kae1, which possesses a novel iron/ATP binding site, as well as DNA binding and apurinic nuclease activity [28]. Taken together, our results indicate an important role of Gcp in the regulation of branched-chain amino acids biosynthesis in S. aureus.
Our finding that the staphylococcal Gcp is involved in the t6A modification of tRNA is consistent with recent reports regarding the function of Kae1/Ori7/YgjD in the biosynthesis of N6-threonylcarbamoyladenosine (t6A) [29], [30], which is important for the recognition of Ile tRNA synthetases and the formation of charged Ile tRNA [31]. This new finding indicates one conserved function between the staphylococcal Gcp and the E. coli homolog YjgD. It is possible that Gcp mediates the transcription of the ilv-leu operon through the t6A modification pathway, as uncharged tRNA act as a positive regulatory factor to increase gene expression during amino acid limitation [32] and the lack of t6A modification may interfere with T-box dependent regulation of the ilv operon [27]. In addition, qPCR revealed that during the depletion of Gcp the increased transcription levels of ilvD, leuA and ilvA genes in one polycistronic operon exhibited significant variations. This suggests that Gcp may affect the transcript processing of the ilv-leu operon. In addition, we cannot exclude the possibility that in S. aureus Gcp may mediates the ilv-leu operon at a posttranscriptional level, because in B. subtilis it has been revealed that posttranscriptional regulation of the ilv-leu operon by endoribonuclease and exoribonuclease proteins leads to three different of mRNA transcript lengths of the ilv-leu operon with varied half life [33].
The mechanism of mediating BCAAs biosynthesis by Gcp is different from other known regulators, including CodY, TnrA, and CcpA, that directly regulate the transcription of ilv-leu operon by specifically binding to a promoter region [18]. We found that the depletion of Gcp had no influence on codY transcription, but increased the ccpA transcription by 2-fold, suggesting that Gcp may mediate the expression of ilv-leu operon, at least in part through the modulation of the positive regulator, CcpA. We are currently working to further determine the impact of CcpA and CodY on Gcp’s modulation of ilv-leu operon. It is possible that Gcp may control the transcription of ilv-leu operon through interaction with YeaZ, as the staphylococcal Gcp binds to YeaZ, another essential protein [26]. In E. coli, it has been revealed that the Gcp homolog YgjD interacts with YeaZ, which is essential for bacteria growth [34]. Investigation is also underway to determine the impact of YeaZ on Gcp’s involvement in the modulation of BCAAs biosynthesis in S. aureus.
Although S. aureus maintains a complete set of genes for the biosynthesis of BCAAs the existence of a BCAA auxotrophic phenotype in S. aureus is still unclear [20], [35]. In this study, we revealed that moderate depletion of Gcp remarkably enhances bacterial growth in the medium lacking ILV. The ability to stimulate bacterial growth by depleting Gcp in ILV dropout CDM may result from the up-regulation of the ilv-leu operon and subsequently increase the biosynthesis necessary metabolites and ILV. The essential nature of Gcp requires a basal level expression for proper bacterial cell growth and viability, but at the same time, this expression of Gcp constitutively inhibits ILV biosynthesis. Thus, the BCAA auxotrophic phenotype is likely due to the absence of environmental ILV as well as repression of the intracellular ILV biosynthetic pathway by Gcp, which are important for protein function and bacterial physiology.
The observation that the deletion of the ilv-leu operon had no impact on the essentiality of Gcp for growth, indicating the repression of ILV biosynthesis pathway is one aspect of the biological function of Gcp, but is not involved in the essential mechanism of Gcp. However, in the absence of exogenous ILV, it seems that Gcp is not required for bacterial growth. The growth of the defined Pspac-regulated gcp expression mutant and the TetR-regulated gcp antisense RNA mutant strains in ILV dropout CDM, compared to wild-type controls, was enhanced when Gcp expression was down-regulated. The growth of these conditional gcp mutant strains in ILV dropout CDM depends on the expression of Gcp as well as the biosynthesis of ILV. In our experiments, we observed that the initial inoculum of the defined Pspac-regulated gcp mutant couldn’t be over-diluted in ILV dropout CDM (we used 1:200 inoculum of overnight culture in CDM) allowing the cells to utilize residual ILV for survival. At a 1:200 dilution, the requirement of Gcp for growth is not titratable due to the carryover of sufficiently expressed Gcp in inoculated bacterial cells compared with those in 1:105–106 diluted cells that are routinely used in down-regulated gene expression studies to determine the essentiality of Gcp. Moreover, our results indicate that the expression levels of Gcp play critical roles in bacterial growth. The lower level of Gcp is important for bacterial growth especially in the medium lacking isoleucine, leucine and valine, because the partial depletion of Gcp can activate the biosynthesis pathway of BCAAs and compensate for the growth defect due to the lack of ILV. In contrast, the high expression levels of Gcp alleviate bacterial growth due to its inhibition of the BCAA biosynthetic pathway, thereafter interfering with bacterial growth in the culture media lacking these essential metabolites.
In conclusion, we have demonstrated that the essential staphylococcal Gcp is involved in the biosynthesis of N6-threonylcarbamoyladenosine (t6A), and the repression of the branched-chain amino acids biosynthesis pathway. More importantly, our data indicate that the essentiality of Gcp is independent of its modulation of branched-chain amino acids biosynthesis. These findings provide new insights into the biological function of the staphylococcal Gcp.
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions
The strains and plasmids used in this study are listed in table 2. E. coli strain DH10B was used for plasmid constructions. Luria-Bertani (LB) liquid medium and LB-agar plates were used for the growth and maintenance of E. coli. S. aureus laboratory strain RN4220 was used as an intermediate host strain prior to introducing plasmids into wild type S. aureus strains. S. aureus WCUH29 is a clinical isolate that was used for genetic manipulation and growth characterization. Tryptic soy broth (TSB) medium and chemically defined media (CDM) were used for the cultivation of S. aureus. Glucose was used as a carbon source at a concentration of 56 mM (1%, w/v). All amino acids included in CDM were L-amino acids. When necessary, isoleucine, leucine and valine were left out of the medium during preparation resulting in ILV dropout CDM.
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference |
| Strains | ||
| RN4220 | Laboratory strain; rsbU- | [44] |
| WCUH29 | Clinical human isolate; rsbU + | [45] |
| JRN0105 | RN4220::Pspac-gcp, spac promoter regulated gcp expression mutant | [3] |
| JRN0110 | JRN0105 with plasmid plux- Pilv | This study |
| JW290111 | WCUH29 Δgcp attB:: Pspac-gcp containing plasmid pYH4-lacI | This study |
| JW290211 | WCUH29 Δilv-leu containing plasmid pYH4-lacI | This study |
| JW290311 | WCUH29 Δilv-leu Δgcp attB:: Pspac-gcp containing plasmid pYH4-lacI | This study |
| WCUH29/gcp-as | WCUH29 containing TetR regulated gcp antisense expression plasmid pYH4/gcp-as | [4] |
| WCUH29/eno-as | WCUH29 containing TetR regulated eno antisense expression plasmid pYH4/eno-as | [24] |
| Plasmids | ||
| plux-Pilv | ilv promoter fused with luxABCDE on pFF40 [38] | This study |
| pYH4-lacI | lacI gene inserted in the vector pYH4 at EcoR I site | This study |
| pLH1 | The integration vector pLL39 [39] carrying Pspac-lacI segment [46] | This study |
| pLH1/gcp | pLH1 vector carrying gcp gene under regulation of Pspac promoter | This study |
| pYH4/gcp-as | pYH4 vector carrying a gcp fragment in an antisense orientation | [3] |
| pKOR1 | E.coli/ S.aureus shuttle vector, permits lambda recombination and ccdB selection;temperature sensitive in S. aureus, | [41] |
| pKOR1/Δgcp | pKOR1 vector carrying gcp flanking sequences for gcp allelic replacement | This study |
| pKOR1/Δilv-leu | pKOR1 vector carrying operon ilv-leu flanking sequences for operon ilv-leu allelicreplacement | This study |
Two-dimensional Differential in Gel Electrophoresis (DIGE)
Sample preparation was carried out as follows: Briefly, the bacterial cells of mid-log phase culture in TSB in the present or absent IPTG were harvested by centrifugation, placed in lysis buffer (30 mM Tris (pH 8.5), 7 M urea, 2 M thiourea and 4% (w/v) CHAPS), and sonicated on ice. Samples were cleared of insoluble material by centrifugation and quantitated using the Advanced Protein Assay Reagent (Cytoskeleton Denver, CO). Proteins were then subjected to Amersham BioScience CyDye™ minimal labeling per manufacturer’s instructions. Cy2 was designated for labeling a pooled internal standard composed of equal quantities of all samples [36].
2D-DIGE comprised two treatments repeated in triplicate that were co-resolved in three DIGE gels with spots coordinated by the pooled internal standard. Cy2, Cy3 and Cy5 labeled sample was combined for each replicate to 250 µl final volume with immobilized pH gradient (IPG) running buffer (7 M urea, 2 M thiourea, trace bromophenol blue) with DTT and IPG buffer (Amersham Bioscience; pH range same as strip) added to a final concentration of 12 mM and 0.5% v/v, respectively. The samples were allowed to rehydrate into an Immobline DryStrip, pH 4–7, 13 cm strip (GE Healthcare) overnight under low current (30 V for 7 h followed by 60 V for 7 h). After overnight rehydration the samples were resolved with a voltage ramp of 500 V for 0.5 kVh, 1000 V for 0.8 kVh and 8000 V for 17.3 kVh in the Amersham Ettan™ IPGphor ™IEF until a total 19.255 kVh was reached. Strips were equilibrated for 0.5 h in SDS equilibration buffer (50 mM Tris pH 8.8, 6 M urea, 30% glycerol, 4% SDS, 1% DTT) followed by resolution on 8-16% SDS-PAGE BioRad Criterion™ gels. Gels were visualized utilizing the FujiFilm FLA-5000 with the following settings (Cy2: Ex 473 nm, Emission filter 530DF20, Cy3: Excitation 532 nm, Emission filter 570DF20, Cy5: Excitation 635 nm, Emission filter R665). Differential expression between treatments was analyzed using the DeCyderTM version 5.02 (Amersham Bioscience) software package. Spots with an average ratio change of at least 2.5 and a Student’s t-test score of at least 0.05 were deemed of interest.
Protein spots of interest were picked from 2D gels after visualizing with Deep Purple Total Protein Stain (GE Healthcare). Protein spots of interest were robotically excised from the gels using the Genomic Solution™ Investigator ProPic™, and subjected to enzymatic digestion [37] on the Genomic Solution™ ProPrep™, and followed by MALDI mass spectrometry analysis. The identity of identified proteins was revealed using Scaffold and BLAST analysis.
Construction of a Defined Pspac-regulated gcp Expression Mutant in S. aureus
The construction of a defined Pspac-regulated gcp expression mutant strain in WCUH29 was performed as described [38]. Briefly, the gcp gene was obtained by PCR with primers listed in Table 3, cloned downstream of Pspac promoter in the integration vector pLH1 (Table 2) with tetracycline for selection. The recombinant plasmid was electroporated into RN4220 and was integrated into the chromosome at the Phage 11 attB site as previously described [39]. The chromosome segment containing the Pspac-gcp cassette was then transferred from RN4220 to WCUH29 by phage transduction as described using tetracycline selection [40] and confirmed by diagnostic PCR. After the Pspac-gcp segment was transduced into the WCUH29 chromosome, the endogenous gcp gene was then deleted by homogenous recombination using the temperature sensitive plasmid pKOR1 as described [41]. Using the same strategy, we created the ilv-leu operon deletion mutants in both the defined Pspac-regulated gcp expression mutant and the parental wild-type WCUH29 strain. The ilv-leu operon is a 10 kb chromosome segment; to increase the efficiency of allelic replacement events, we deleted the gene cassettes ilvDBHC and leuABCDilvA sequentially in-frame.
Table 3. Primers used in this study.
| Name | Sequence |
| LacIForEcoRI | GCGAATTCTCATCATTTCCTTCCGAAAAAACG |
| LacIRevEcoRI | GCGAATTCTCACTGCCCGCTTTCCAGTC |
| PspacLacIForPstI | TTCTGCAGATCTGGTAATGACTCT |
| PspacLacIRev | TTAGATCTTCACTGCCCGCTTTCCAGTC |
| GcpLattBFor | BGGGGACAAGTTTGTACAAAAAAGCAGGCTATGGATTGTTATCGCTTAG |
| GcpLRev | TTCACCCACATAACCATTG |
| GcpRFor* | AATAAACGTCAGTTATTAACATG |
| GcpRattBRev | GGGGACAAGTTTGTACAAAAAAGCAGGCTTTAGGAATGTAAAATACGCC |
| IlvDLattBFor | GGGGACAAGTTTGTACAAAAAAGCAGGCTCGCATCTAATTGCTGTTTAGC |
| IlvDLRev | AGTAAATTCCCCCGTAAATTTTAATG |
| IlvCRfor* | GATAGACCTACAATGAGGAGTTG |
| IlvCRattBRev | GGGGACCACTTTGTACAAGAAAGCTGGGTTTTGTCCGCAATGGTCTTG |
| LeuALattBFor | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATAAACCTAAATAACCAATAATTTG |
| IlvARFor* | CACATAGTAAGAAAAACAGTCATAAATTG |
| IlvARattBRev | GGGGACCACTTTGTACAAGAAAGCTGGGTCTGTCCTTAGTACGAGAGGACCG |
| CodYRTFor | GCGCGCGATAAAGCTGCTATTACA |
| CodYRTRev | ATTAATAGGCCTTCCGTACCGCCA |
| CcpARTFor | GCCACAGTGTCGCGTGTTGTTAAT |
| CcpARTRev | ACCTCTAGCAACAGCATTTGGACG |
| LeuARTFor | CGGCCTTCAAAGTGCTGTTGTTGT |
| LeuARTRev | ACTTCTGCTTGGGCATCAGTACCT |
| IlvARTFor | ATTTGTGGACGGTGCATCTGTAGC |
| IlvARTRev | AAGAGCACTCACACTTAATGCGCC |
| IlvERTFor | GGCGTTGGTGCATCACATCAGTAT |
| IlvERTRev | CCACGAACAGCACGCACATATTCA |
| IlvDRTFor | CACCCGGTATGATTTAGCAG |
| IlvDRTRev | ACAAGTAGGGCAGGCATTTTG |
| IlvDproFor | ATCCATTGTTCAATCGTATC |
| IlvDproRev | GTAAATTTTAATGATTAATCATGTTTTATAG |
:5-terminal end phosphorylated.
RNA Isolation and Purification
Overnight cultures of S. aureus were inoculated at 1% in TSB medium and grown to the mid-exponential (∼4 h) phase of growth. Total RNA was purified from the above cultures, as described (11). Briefly, bacterial cells were harvested by centrifugation at 4,000×g, and the RNA was isolated using the SV total RNA isolation system (Promega). Contaminating DNA was removed with two rounds of DNase treatment (TURBO DNA-free kit,Ambion), and the RNA yield was determined spectrophotometrically at 260 nm.
Semi-quantitative Real-time RT-PCR (qPCR) Analysis
In order to determine the effect of Gcp on the expression of ilv-leu operon, we employed qPCR to compare the RNA levels as described (11, 16). The first strand cDNA was synthesized using reverse transcriptase SuperScript III and random primers (Invitrogen). For each RNA sample, we performed duplicate reactions of reverse transcription, as well as a control without reverse transcriptase, in order to determine the levels of DNA contamination. PCR reactions were set up in duplicate by using a SYBR Green PCR Master Mix (USB). Real-time sequence-specific detection and relative quantitation were performed with the Stratagene Mx3000P Real Time PCR System. Gene-specific primers were designed to yield 100∼200 bp specific product (Table 3). Relative quantification of the product was calculated using the Comparative CT method, as described for the Stratagene Mx3000P system. The housekeeping gene 16S rRNA was used as an endogenous control (16).
Construction of ilvD Promoter-lux Reporter Fusion System
To confirm whether Gcp mediates the transcription of the ilv-leu operon, we constructed an ilv promoter-lux reporter fusion system as described [23]. The ilv promoter region (25) was amplified by PCR, ligated upstream of promoterless luxABCDE in pFF40 [38] resulting in an ilv-promoter-lux fusion plasmid. The ilv promoter-lux fusion plasmid was electroporated into the Pspac-regulated gcp expression strain [3]. Both bioluminescence signals and cell growth were monitored at 37°C by measuring the bioluminescent intensity and optical density at 600 nm with a BioTek Synergy Microplate Reader. To eliminate the effect of bacterial growth, the relative light units (RLU) were calculated (light intensity/OD600) from triplicate readings at different times during growth.
Gel Mobility Shift DNA Binding Assay
To determine whether Gcp directly regulates the transcription of the ilv-leu operon, we performed gel-shift assays. The DNA fragment of the upstream region of ilv was obtained by PCR using the primers listed in Table 3. The amplified DNA fragment was purified and labeled with Digoxigenin using a DIG Gel Shift kit (Roche) according to the manufacturer’s protocol. The DNA-binding and electrophoresis were performed, as described (17, 20). Briefly, the purified PCR products were labeled with Digoxigenin using terminal transferase (Roche). The labeled DNA fragments were further purified to remove the redundant DIG-ddUTP and salts. The interaction of Gcp with DNA was conducted in a 20 μl reaction mixture containing 0.2 pmol DIG-labeled DNA, 1 μg of poly-[d(I–C)], 25 mM NaH2PO4 (pH 8.0), 50 mM NaCl, 2 mM MgCl2, 1 mM DTT, 10% glycerol, 0.1 mM EDTA, and different concentrations of Gcp protein. Unlabeled DNA fragments of the promoter region as a specific competitor were added into the reaction with 100-fold excess to labeled probe. Internal gene fragments were obtained by PCR, purified, and labeled as nonspecific controls. BSA was used as a nonspecific protein binding control. The DNA binding reaction was initiated by the addition of Gcp and incubated at room temperature for 25 min. Samples were then loaded directly onto a 5% native polyacrylamide gel [acrylamide:bisacrylamide (29:1) in 0.5×TBE buffer]. Electrophoresis was run for 2 h at 4°C with 7 V/cm, and the gels were transferred to Nylon membrane via electro-blotting in 0.5×TBE at 300 mA for 90 min. at 4°C. After cross-linking of DNA fragments using UV, the membranes were hybridized with anti-digoxigenin-AP antibody and exposed to X-ray film for 4 h to achieve the desired signal.
Purification of Bulk tRNA
The conditional S. aureus cells were grown in 100 ml TSB media at 37°C supplemented with or without 200 µM IPTG to OD600 = 0.4, harvested, and washed in cold water. The bacterial cells were broken using a Bead Beater as described [42]. Briefly, the bacterial cells were resuspended in 1.5 ml DEPC-treated water and were then transferred into two 2 ml Eppendorf tubes containing equal volume (750 µl) of 0.1 mm glass beads. The bacterial cells were broken using a Mini-Bead Beater-8 at maximum speed for 4 min at 4°C. The supernatant was transferred into a new 1.5 ml Eppendorf tube; and bulk tRNA was isolated using the mirVanaTM miRNA isolation kit (Ambion) according to the manufacturer’s instruction. The tRNA was eluted in water and stored at −80°C. The concentration of tRNA was measured by using a Nano Drop (Thermo Scientific).
LC-MS/MS Analysis of Digested tRNA
To quantify the t6A content of the tRNA, samples of each purified bulk tRNA were enzymatically hydrolyzed to nucleotides as described [43]. Briefly, 15 µg of bulk tRNA was sequentially digested with 5 units of nuclease P1 (Sigma) at 45°C for 16 h, 0.01 units of Phosphodiesterase I (Sigma) at 37°C for 2 h, and then 10 units of calf intestine alkaline phosphatase (Promega) at 37°C for 1 h in a total volume of 70 µl. The resulting nucleotides were acidified by the addition of 1–5 µl 10% formic acid and monitored using pH indicator paper, clarified by centrifugation, and then purified by passing through a C18 zip tip column (Millipore). The purified samples were dehydrated, suspended in 50 µl of 5 mM ammonium acetate, pH 6.0, and diluted (1 µl to 20 µl). The 20 µl diluted samples were then subjected to injection using an Agilent autosampler LCMSMS with an analytical Waters Symmetry C18, 3.5 µm column connected to the Applied Biosystem 4000 iontrap fitted with a turbo V electrospray source. The samples were subjected to a linear gradient of 5 mM ammonium acetate (pH 6.0) to 100 percent acetonitrile for 10 minutes at a column flow rate of 200 µl /min. Transitions monitored were the m/z 413 > m/z 281 and the m/z 413> m/z 136 for the t6A and m/z 268> m/z 136 for the adenosine. The data was analyzed using MultiQuant (ABI) providing the peak area for the transitions. A standard curve was constructed using concentrations of t6A from fentomole to nanomole in 20 µl.
Characterization of Bacteria Growth
The S. aureus strains were inoculated in TSB, CDM or ILV dropout CDM with appropriate antibiotics and different concentrations of IPTG (0, 10, 25, and 100 μM) or antisense RNA inducer, anhydrotetrocycline (ATc), at 37°C. The bacterial growth was monitored by kinetically measuring optical density at 600 nm with a BioTek Synergy Microplate reader.
Acknowledgments
We thank Mr. Jeffrey W. Hall for helpful suggestions and critically reading this manuscript.
Funding Statement
This project was supported by grant AI057451 from the National Institute of Allergy and Infectious Disease. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Appelbaum PC (2006) The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus . Clin Microbiol Infect 12 Suppl 116–23. [DOI] [PubMed] [Google Scholar]
- 2. Loomba PS, Taneja J, Mishra B (2010) Methicillin and vancomycin resistant S. aureus in hospitalized patients. J Glob Infect Dis 2: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng L, Yang J, Landwehr C, Fan F, Ji Y (2005) Identification of an essential glycoprotease in Staphylococcus aureus . FEMS Microbiol Lett 245: 279–285. [DOI] [PubMed] [Google Scholar]
- 4. Zheng L, Yu CX, Bayles K, Lasa I, Ji YD (2007) Conditional mutation of an essential putative glycoprotease eliminates autolysis in Staphylococcus aureus . Journal of Bacteriology 189: 2734–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brosnan JT, Brosnan ME (2006) Branched-chain amino acids: enzyme and substrate regulation. J Nutr 136: 207S–211S. [DOI] [PubMed] [Google Scholar]
- 6. Beck HC, Hansen AM, Lauritsen FR (2004) Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus . J Appl Microbiol 96: 1185–1193. [DOI] [PubMed] [Google Scholar]
- 7. Kaneda T (1991) Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev 55: 288–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maas WK, Vogel HJ (1953) alpha-Ketoisovaleric acid, a precursor of pantothenic acid in Escherichia coli . J Bacteriol 65: 388–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grandoni JA, Zahler SA, Calvo JM (1992) Transcriptional regulation of the ilv-leu operon of Bacillus subtilis . J Bacteriol 174: 3212–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shivers RP, Sonenshein AL (2005) Bacillus subtilis ilvB operon: an intersection of global regulons. Mol Microbiol 56: 1549–1559. [DOI] [PubMed] [Google Scholar]
- 11. Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL (2010) Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol 192: 6357–6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marta PT, Ladner RD, Grandoni JA (1996) A CUC triplet confers leucine-dependent regulation of the Bacillus subtilis ilv-leu operon. J Bacteriol 178: 2150–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mader U, Hennig S, Hecker M, Homuth G (2004) Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. J Bacteriol 186: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, et al. (2010) Direct targets of CodY in Staphylococcus aureus . J Bacteriol 192: 2861–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wray LV Jr, Ferson AE, Rohrer K, Fisher SH (1996) TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci U S A 93: 8841–8845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tojo S, Satomura T, Morisaki K, Yoshida K, Hirooka K, et al. (2004) Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA. J Bacteriol 186: 7971–7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shivers RP, Sonenshein AL (2004) Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol 53: 599–611. [DOI] [PubMed] [Google Scholar]
- 18. Tojo S, Satomura T, Morisaki K, Deutscher J, Hirooka K, et al. (2005) Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol Microbiol 56: 1560–1573. [DOI] [PubMed] [Google Scholar]
- 19. Ludwig H, Meinken C, Matin A, Stulke J (2002) Insufficient expression of the ilv-leu operon encoding enzymes of branched-chain amino acid biosynthesis limits growth of a Bacillus subtilis ccpA mutant. J Bacteriol 184: 5174–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Onoue Y, Mori M (1997) Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int J Food Microbiol 36: 77–82. [DOI] [PubMed] [Google Scholar]
- 21. Lincoln RA, Leigh JA, Jones NC (1995) The amino acid requirements of Staphylococcus aureus isolated from cases of bovine mastitis. Veterinary Microbiology 45: 275–279. [DOI] [PubMed] [Google Scholar]
- 22. Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D (2012) Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 287: 13666–13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yan M, Yu C, Yang J, Ji Y (2009) Development of shuttle vectors for evaluation of essential regulator regulated gene expression in Staphylococcus aureus . Plasmid 61: 188–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu X, Zheng L, Yang J, Lei T, Ji Y (2011) Characterization of essential enolase in Staphylococcus aureus . World Journal of Microbiology and Biotechnology 27: 897–905. [Google Scholar]
- 25. Koonin EV (2003) Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1: 127–136. [DOI] [PubMed] [Google Scholar]
- 26. Lei T, Liang X, Yang J, Yan M, Zheng L, et al. (2011) The C-terminal domain of the novel essential protein Gcp is critical for interaction with another essential protein YeaZ of Staphylococcus aureus . PLoS One 6: e20163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hashimoto C, Sakaguchi K, Taniguchi Y, Honda H, Oshima T, et al. (2011) Effects on transcription of mutations in ygjD, yeaZ, and yjeE genes, which are involved in a universal tRNA modification in Escherichia coli . J Bacteriol 193: 6075–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, et al. (2007) An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res 35: 6042–6051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, et al. (2011) A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 30: 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, et al. (2011) The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 30: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, et al. (1994) Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli . J Mol Biol 236: 710–724. [DOI] [PubMed] [Google Scholar]
- 32. Grandoni JA, Fulmer SB, Brizzio V, Zahler SA, Calvo JM (1993) Regions of the Bacillus subtilis ilv-leu operon involved in regulation by leucine. J Bacteriol 175: 7581–7593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mader U, Hennig S, Hecker M, Homuth G (2004) Transcriptional organization and posttranscriptional regulation of the Bacillus subtilis branched-chain amino acid biosynthesis genes. Journal of Bacteriology 186: 2240–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Handford JI, Ize B, Buchanan G, Butland GP, Greenblatt J, et al. (2009) Conserved network of proteins essential for bacterial viability. J Bacteriol 191: 4732–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, et al. (2001) Whole genome sequencing of meticillin-resistant Staphylococcus aureus . Lancet 357: 1225–1240. [DOI] [PubMed] [Google Scholar]
- 36. Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, et al. (2003) A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3: 36–44. [DOI] [PubMed] [Google Scholar]
- 37. Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem 68: 850–858. [DOI] [PubMed] [Google Scholar]
- 38. Fan F, Lunsford RD, Sylvester D, Fan J, Celesnik H, et al. (2001) Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus . Plasmid 46: 71–75. [DOI] [PubMed] [Google Scholar]
- 39. Luong TT, Lee CY (2007) Improved single-copy integration vectors for Staphylococcus aureus . J Microbiol Methods 70: 186–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji Y, Marra A, Rosenberg M, Woodnutt G (1999) Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J Bacteriol 181: 6585–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bae T, Schneewind O (2006) Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55: 58–63. [DOI] [PubMed] [Google Scholar]
- 42. Yu C, Sun J, Zheng L, Ji Y (2007) Genomic analysis of gene expression of Staphylococcus aureus . Methods Mol Biol 391: 169–178. [DOI] [PubMed] [Google Scholar]
- 43. Crain PF (1990) Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol 193: 782–790. [DOI] [PubMed] [Google Scholar]
- 44. Kreiswirth BN, Lofdahl S, Betley MJ, O’Reilly M, Schlievert PM, et al. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305: 709–712. [DOI] [PubMed] [Google Scholar]
- 45. Ji Y, Zhang B, Van SF, Horn, Warren P, et al. (2001) Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293: 2266–2269. [DOI] [PubMed] [Google Scholar]
- 46. Zhang L, Fan F, Palmer LM, Lonetto MA, Petit C, et al. (2000) Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255: 297–305. [DOI] [PubMed] [Google Scholar]