Abstract
Expression of sterol carrier protein-2 (SCP-2) in Aedes aegypti shows a distinct temporal/spatial pattern throughout the life cycle. In order to identify the transcription factors responsible for the larval temporal/spatial regulation of AeSCP-2 transcription, AeSCP-2 promoter activities were studied in vivo via transient transfection of promoter/reporter gene assays. Regulatory sequences upstream −1.3 kb of the transcription start site of AeSCP-2 were found to be critical for the in vivo temporal/spatial promoter activity. Interestingly, the −1.6 kb promoter sequence efficiently drove the larval midgut-specific siRNA expression, indicating that the −1.6 kb upstream sequence is sufficient for temporal/spatial AeSCP-2 transcriptional activity. Four transcription factors were identified in the midgut nuclear extract from feeding larvae via labeled −1.6/−1.3 kb DNA probe pull-down and proteomic analysis. Co-transfection of the promoter/reporter gene with inducible siRNA expression of each transcription factor was performed to confirm the regulatory function of individual transcription factor on AeSCP-2 transcriptional activities in the larval midgut. The results indicate that two of the identified transcription factors, Thanatos-associated protein (THAP) and activating transcription factor-2 (ATF-2), antagonistically control AeSCP-2 transcriptional activity in the midgut of feeding larvae via the regulatory sequences between −1.6 to −1.3 kb 5′ upstream of the transcription start site. In vivo expression knockdown of THAP and ATF-2 resulted in significant changes in developmental progression, which may be partially due to their effects on AeSCP-2 expression.
Introduction
Cholesterol is an important component of all animal cellular membranes and is the precursor for steroid hormone biosynthesis. Insects, unlike the vertebrates, are not able to synthesize cholesterol de novo due to the lack of key enzymes in the cholesterol synthesis pathway [1]. Therefore, insects must depend on dietary and/or symbiotic microbes to meet their physiological requirements for cholesterol [2], [3], [4], [5], [6]. Sterol carrier protein-2 (SCP-2) is a small intracellular protein involved in cholesterol and lipid intracellular trafficking in insects [7], [8]. AeSCP-2, the first insect SCP-2 identified from the yellow fever mosquito, Aedes aegypti, was found to bind to cholesterol [7] and fatty acids [9]. Moreover, functional studies of AeSCP-2 in Aedes aegypti revealed that AeSCP-2 plays an important role in cholesterol uptake, mosquito development and reproduction [10], [11].
It has been confirmed that the midgut and possibly foregut are the sites of cholesterol uptake and absorption in insects [12], [13], [14], [15]. In Aedes aegypti, AeSCP-2 was expressed strongly throughout feeding larval stages and decreased to low levels after pupation [7]. Interestingly, high levels of AeSCP-2 expression are found in the larval midgut tissue [7]. Knockdown of AeSCP-2 gene expression in vivo effectively interferes with AeSCP-2 gene expression in the larval midgut, leading to the observed decreases in cholesterol uptake in larvae [10]. Results from earlier studies have indicated that AeSCP-2 gene expression is stage and tissue specific [7]. However, the transcriptional regulatory mechanism is unknown.
Results from the vertebrate SCP-2 gene transcriptional regulation studies have demonstrated that the vertebrate SCP-2 gene expression appears to be under the control of factors such as adrenocorticotropic hormone and gonadotropins via cyclic adenosine monophosphate (cAMP) activation [16]. In insects, 20-hydroxyecdysone (20E) up-regulates the transcription of AeSCP-2 by 2-fold in cultured gut tissues [7], [17]. Promoter/reporter gene transfection assays in cultured mosquito Aag-2 cells demonstrated that 20E-induced up-regulation of AeSCP-2 transcription requires HR3, an ecdysone-inducible transcription factor; and Ftz-F1, a 20E-responsive-late gene, may be involved in the down regulation of the AeSCP-2 gene [17]. There are significant increases in ecdysteroid levels in 24 hour-old larvae [18], [19], [20], which is sufficient to induce the expression of some 20E-inducible genes such as HR3 and E75, but, not Ftz-F1 [19]. Therefore, it is unlikely that Ftz-F1 is involved in AeSCP-2 transcriptional regulation in Day 1 4th instar larvae. Factors that control the temporal/spatial AeSCP-2 expression in vivo in feeding larvae are unknown. Considering its critical role in mosquito’s cholesterol metabolism and development [10], [11], it is important to further investigate the mechanism of AeSCP-2 gene expression regulation. Based on the newly developed gene delivery method in Aedes aegypti [11], the AeSCP-2 promoter regulatory sequence and potential promoter regulatory proteins were studied in vivo in this study.
Results
Identification of the Spatial/temporal Regulatory Sequences in AeSCP-2 Promoter
AeSCP-2 expression during the 4th instar is 5-fold higher in 24 hour-old larvae (feeding) than that of post feeding cessation at 62 hours post 3rd molt [21]. In order to identify the temporal/spatial expression regulatory sequence in AeSCP-2 promoter, six serial truncated promoter constructs [17] containing the CAT reporter gene were mixed with polyethylenimine (PEI) and microinjected to blood fed female mosquitoes at 16-18 hours the post blood meal (PBM). The CAT expression from the promoter/CAT constructs were measured in F0 4th instar larval samples. The -0.06 kb construct showed significantly higher CAT expression levels than that of in the non-transfected larvae (non-transfected control vs. −0.06 kb, p<0.05, data not shown), suggesting that the −0.06 kb 5′ flanking sequence may contain the basal promoter that drove the basal level reporter gene expression.
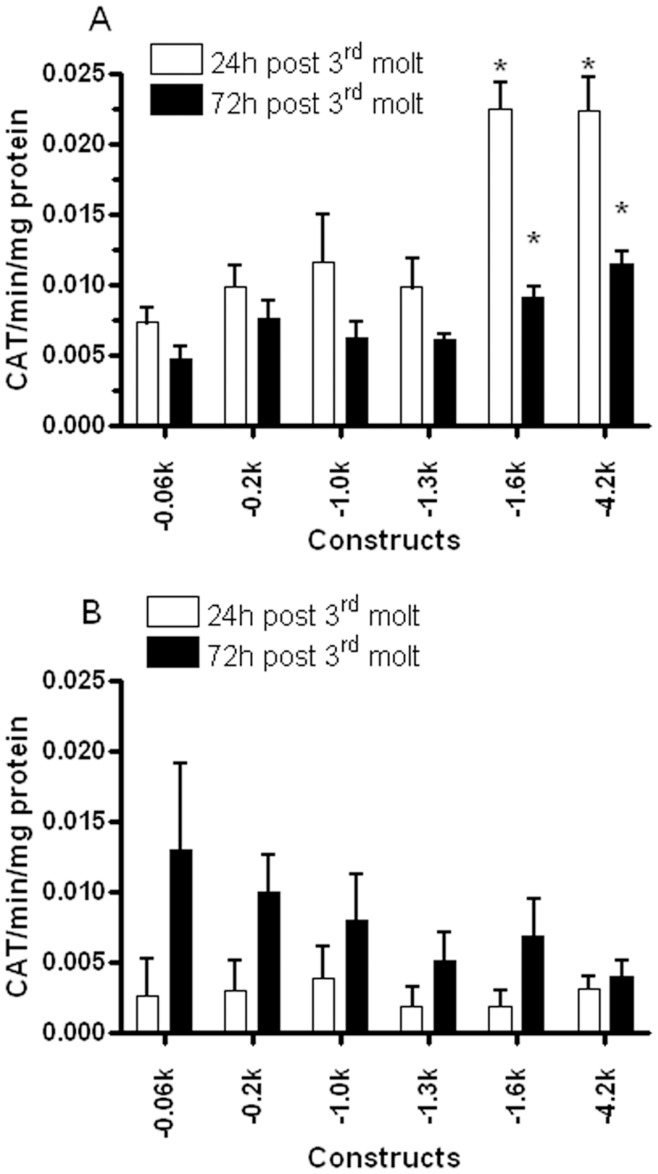
In the larval midgut, there were no significant differences in levels of CAT expression driven by the −0.06, −0.2, −1.0, and −1.3 kb 5′ flanking sequence at both 24 hour-old (24 h) and 72 hour-old (72 h) 4th instar larval stages (Fig. 1A, −0.06 to −1.3 kb), suggesting that temporal regulatory elements are not located within the −1.3 kb 5′ flanking sequence. The −4.2 kb and −1.6 kb 5′ flanking sequences showed 3.1- to 3.0-fold significantly higher CAT reporter gene expression (p<0.05) than that of the −0.06 kb construct in the midgut at 24 h time point, respectively(Fig. 1A). There was significantly (p<0.002) lower promoter activities driven by −1.3 kb than that of −1.6 kb in 24 hour-old samples (Fig. 1A), indicating that regulatory sequences between −1.6 and −1.3 kb is important for sustaining the high level of promoter activity in the feeding 4th instar larvae. In the larval midgut, the promoter activities of the −4.2 and −1.6 kb constructs at 72 h decreased by 59% and 49% compared to 24 h, respectively (p<0.01, Fig. 1A). The results suggest that sequence upstream of −1.3 kb is important for temporal promoter activity in the midgut in feeding 4th instar larvae. At the 72 h time point (feeding ceased), promoter activities of −4.2 and −1.6 kb were significantly lower than that of at 24 h, but, the levels were still significantly higher (p<0.05) than that of −0.06 kb (Fig. 1A), indicating that regulatory sequences upstream of −1.3 kb are responsible for the continued above basal level transcriptional activity of AeSCP-2 in the midgut.
Figure 1. Quantitative analysis of in vivo CAT expression from promoter/CAT constructs.
(A) The midgut samples from synchronized 4th instar larvae (30 larvae/sample) were taken at 24 and 72 hours post 3rd molt. (B) The carcass samples were from the same larvae as the midgut sampling. CAT quantity in each sample was defined as CAT unit/min of the reaction/µg protein. Mean and standard deviation are shown (N = 5−6). * Indicating significantly different (p<0.05) from the −0.06 kb construct at the same developmental time point.
In the larval carcass samples, there were no significant differences in levels of CAT expression among the six truncated promoter constructs at 24 h time point (Fig. 1B). The results were in sharp contrast to increased promoter activities of −4.2 and −1.6 kb in the midgut at 24 h (Fig. 1A vs. B, 24 h), indicating that regulatory sequences upstream of −1.3 kb are important for the spatial promoter activity of AeSCP-2. Promoter activities of −0.06 kb was significantly increased at 72 h compared to 24 h (p = 0.043, Fig. 1B), whereas upstream −0.2 kb promoter activities did not change significantly in the carcass tissues overtime (Fig. 1B).
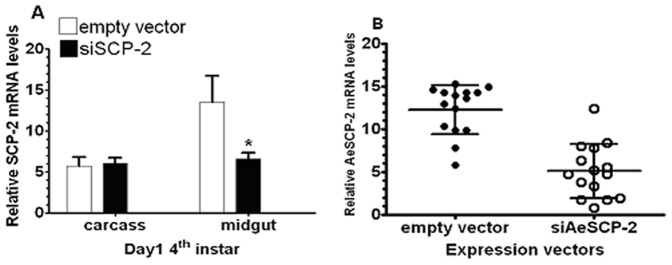
To confirm that the −1.6 kb AeSCP-2 promoter sequence would be sufficient to regulate tissue specific transcriptional activities, a siAeSCP-2 expression vector under the control of the −1.6 kb AeSCP-2 promoter was constructed. Larvae transfected with the −1.6 kb-siAeSCP-2 construct via DNA/PEI injection of vitellogenic females were reared to 4th instar. Larval tissues from synchronized 4th instar F0 larvae were collected. To estimate the efficiency of delivery −1.6 kb-siAeSCP-2 construct into F0 larvae, RNA sample was also extracted individually from 15 randomly selected 24 h larvae from one batch of the transfection. Relative AeSCP-2 mRNA levels (vs. Actin-1) were determined via RT-qPCR analysis. The −1.6 kb 5′ flanking sequence drove siAeSCP-2 expression that knocked down AeSCP-2 expression by 51% in the midgut, whereas the level of AeSCP-2 mRNA in the carcass was unchanged (Fig. 2A). The efficiency of the −1.6 kb AeSCP-2 promoter driven siRNA was 67% in randomly selected larvae that showed significantly reduced the targeted mRNA levels below the 50% average of the vector control (p = 0.02, Fig. 2B), which is consistent with previously reported in vivo transfection efficiency [11]. The lack of −1.6 kb driven siRNA expression knockdown in carcasses would not have been due to the lack of transfection in the larval carcass because heat shock inducible β-gal activities have been detected in 4th instar larval carcass in previous studies using the same DNA/PEI delivery method [11]. The results of −1.6 kb/siRNA targeted tissue specific expression knockdown (Fig. 2A) is consistent with the −1.6 kb/CAT reporter gene assays in 24 h 4th instar larvae (Fig. 1, −1.6 kb/CAT at 24 h, midgut vs. carcass ). The −1.6 kb 5′ flanking sequence of AeSCP-2 is the first confirmed larval midgut specific promoter in A. aegypti.
Figure 2. Tissue-specific transcriptional activities of the −1.6 kb AeSCP-2 construct and efficiency for driving siRNA expression in transfected larvae.
(A) Expression knockdown of AeSCP-2 via the −1.6 kb AeSCP-2 promoter driven siRNA expression. Tissues were collected from the same larvae (10 larvae/sample). Mean and standard deviation are shown (N = 3). (B) Fifteen synchronized 24 hour-old 4th instar larvae were randomly selected and the total RNA was extracted from each individual larva. Mean and standard deviation are shown (N = 15). Relative (vs. Actin-2) AeSCP-2 mRNA levels from each sample were determined via RT-qPCR. * Indicating significant difference (p<0.05) from that of the empty vector construct.
Potential Transcription Factors Bound to AeSCP-2 Promoter Regulatory Region
Based on the temporal/spatial regulatory activities of the sequences between −1.6 to −1.3 kb 5′ upstream of AeSCP-2 transcription start site (Fig. 1A and B), we focused on the 305 bp regulatory sequence between −1.6 to −1.3 kb 5′ flanking region of AeSCP-2 in search for potential regulatory proteins. Biotin-labeled 305 bp DNA fragment of the −1.6 to −1.3 kb 5′ upstream sequence of AeSCP-2 was amplified via PCR and used as probes for the biotin-streptavidin pull-down assay. Due to the promoter activities varied significantly in a temporal fashion between 24 hour-old and 72 hour-old 4th instar larval midgut samples (Fig. 1A); midgut nuclear extracts from these two stages were used to search for the potential binding proteins that interact with −1.6 kb/−1.3 kb sequence of the promoter. Following the DNA probe/protein pull-down experiment, protein mixtures that bound to the labeled DNA probe were purified and the proteins were identified via LC-MS spectrometry (see M&M). There were six proteins bound to the 306 bp regulatory sequence in the midgut nuclear extract of 24 hour-old 4th instar and six bound proteins were identified in the midgut nuclear extract of 72 hour-old 4th instar (Table 1). AAEL002827, the ATP synthase beta subunit, was one common protein bound to the AeSCP-2 −1.6/−1.3 kb promoter sequence in both 24 h and 72 h larval midgut nuclear extracts (Table 1). We considered AAEL002827 as a non-specific contaminant in the pull-down assays.
Table 1. Nuclear proteins bound to the AeSCP-2 −1.6/−1.3 kb regulatory sequence.
| 24 h 4th instar larval stage | 72 h 4th instar larval stage |
| AAEL002827 (novel gene)ATP synthase beta subunit | AAEL002827 (novel gene)ATP synthase beta subunit |
| AAEL010577 (novel gene)Unknown protein containing THAP domain, homology of human THAP3 | AAEL000494 (novel gene)histone H2A |
| AAEL011794 (novel gene)Unknown protein containing IBR domain | AAEL015674 (novel gene)histone H2B |
| AAEL013261 (VectorBase gene: AAEL801171)Protein containing C2H2-type zinc fingers, homolog of Drosophila ATF-2 (CG30420) | AAEL011795 (novel gene)allergen, putative |
| AAEL005286 (novel gene)Unknown protein containing CCCH-type Zinc finger domain | AAEL006857 (novel gene)Unkwon protein containing RRM domain |
| AAEL017566 (novel gene)Homolog of Drosophila DNA ligase 1 (CG5602) | AAEL001950 (novel gene)Unknown protein containing UBA domain |
Of the five unique proteins bound to the −1.6/−1.3 kb probe in the 24 hour-old 4th instar larval midgut nuclear extract, AAEL017566, is the homolog of Drosophila DNA ligase 1 (Table 1, 24 h) that is involved in DNA replication, repair, and recombination [22]. DNA ligase 1 is known to bind oligonucleotides [22], and it is likely that the labeled DNA probe functioned as an oligonucleotide substrate to pull DNA ligase 1 down. In the 24 h 4th instar larval midgut nuclear extract, four of the −1.6/−1.3 kb pulled-down proteins are zinc finger transcription factors with unknown function in mosquitoes (Table 1, 24 h). Analysis of putative functional domains of these proteins revealed that the hypothetical protein AAEL010577 contains a Thanatos-associated protein (THAP) domain (DNA-binding domain) that is homologous to the human THAP3 and Drosophila CG14965 with 26.86% and 26.13% identity (54.23% and 50.81% similarity), respectively. Proteins containing THAP domain are involved in diverse biological processes [23]. AAEL013261 is the homolog of Drosophila activating transcription factor-2 (ATF-2) with 30.33% identity (63.12% similarity). The Drosophila ATF-2 has been shown to be involved in lipid metabolism [24]. AAEL011794 and AAEL005286 have orthologs only in mosquito species based on similarity search in the Blastp database (Blast database, NCBI).
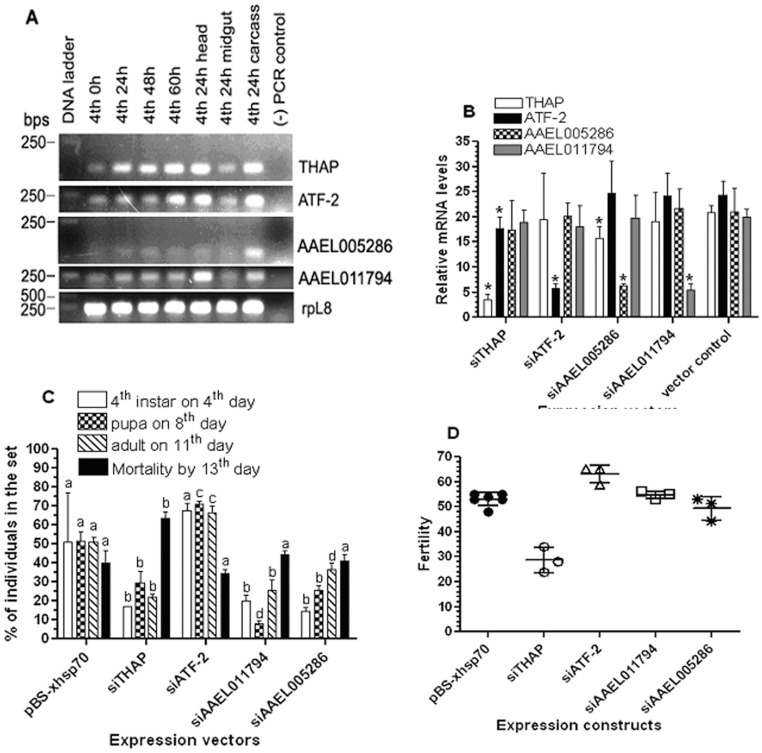
Endogenous expression of the transcription factors (Table 1, 24 h) in 4th instar larvae were confirmed via semi-quantitative RT-PCR analysis. The mRNA levels of THAP were high between 24–60 h in the 4th stadium and were much lower in the midgut than that of in the head and the carcass in 24 hour-old 4th instar larvae (Fig. 3A, THAP). Increased ATF-2 transcription was detected only in 60 hour-old 4th instar larvae when feeding had ceased and the expression in the midgut was much lower than that of in other tissues in 24 hour-old 4th instar larvae (Fig. 3A, ATF-2). The levels of AAEL005286 and AAEL011794 transcripts did not show detectable change throughout 4th instar and the mRNA levels in the larval midgut were lower than that of in the other tissues (Fig. 3A). The results indicate that all four transcription factor genes were actively transcribed in 4th instar larvae and detectable levels of transcripts were found in the 24 h 4th instar larval midgut, where AeSCP-2 expression is high [7]. Therefore, it is possible that those transcription factors may regulate AeSCP-2 transcription in the larval midgut.
Figure 3. Effects of expression knockdown of the transcription factors (Table 1, 24 h) on the progression of development, mortality, and female fertility.
The siRNA vector is driven by the Drosophila hsp70 short promoter. Thirty larvae were synchronized on Day 1 2nd instar, heat shock at 37°C started on Day 1 of 2nd instar through pupal stage and adults were returned to 26°C. (A) Temporal/spatial transcription profiles of the transcription factors in 4th instar larvae via semi-quantitative RT-PCR (30 cycles). (B) RNA sample of pooled 24 h 4th instar larval midguts (10 larvae/sample) was taken from randomly selected larvae in each respective group (triplicate batches). Relative quantity of mRNA (vs. rpL8) was determined via RT-qPCR analysis. Mean ± standard deviation (N = 3). * Indicate significantly different (p<0.05, paired t-test) from the vector control. (C) Developmental progress and mortality was recorded daily. The same letter as the vector control above the bars represents that the mean values did not differ from the control significantly in paired t-test (p>0.05). Different letters above each construct represent that the mean values were significantly different between constructs (p<0.05). (D) Female fertility was measured as producing viable 2nd instar larvae per blood-fed female (after the 1st bloodmeal) in 3–6 batches (>5 female/batch) of each mating group. Each group in panel “A” had ≥5 surviving pairs per batch. Mean and standard deviation are shown (N = 3−6).
Those transcription factors (Table 1, 24 h) were expressed in the larval midgut at approximately similar levels (Fig. 3B, vector control). To determine whether those transcription factors have essential functions in vivo, expression knockdown via siRNA vectors was carried out for each of the transcription factor. Each perspective siRNA effectively knocked down its own gene expression by at least 70% in the 24 h 4th instar larval midgut (Fig. 3B). To determine the effect of siRNA of each perspective transcription factor (Fig. 3B) on each other’s expression, mRNA levels of each transcription factor in the midgut was measured via RT-qPCR in samples of siRNA-treated larvae. Interestingly, siTHAP also reduced ATF-2 expression by 27% (Fig. 3B, siTHAP and ATF-2), whereas siAAEL005286 led to a 24% decrease in THAP expression (Fig. 3B, siAAEL005286 and THAP). The expression knockdown effects of siTHAP and siAAEL005286 on ATF-2 and THAP transcription were not due to the hairpin sequences of the perspective gene since there was no sequence similarity between the siTHAP and siAAEL005286 and ATF-2 and THAP mRNA, respectively (see M&M).
Thirty F0 larvae transfected with a siRNA expression vector were synchronized on Day 1 2nd instar and heat shocked at 37°C as described in M&M. Mortality and larval development was recorded daily, surviving adults were allowed to mate within each group and the female fertility was recorded after the 1st bloodmeal on adult day 4th. Heat-shock at 37°C throughout the 2nd -pupal stages resulted in a high mortality rate at 40% in control groups (Fig. 3C, total death by 13th day, pBS-xhsp70). However, siTHAP groups had significantly higher mortality rate (a 23% increase) than that of in the vector control (Fig. 3C, pBS-xhsp70 vs. siTHAP, p = 0.0004, t = 6.944 df = 6). Most of the increased mortality in the siTHAP group occurred in the pupal stage. On the other hand, the mortality rates in siATF-2, siAAEL011794, and siAAEL005286 groups were similar to the vector control (Fig. 3C, pBS-xhsp70 vs. siATF-2, siAAEL011794, and siAAEL005286). The results suggest that heat shock alone was not the only cause of higher mortality in siTHAP-treated groups. It is likely that THAP plays some roles in survivorship. In vivo expression knockdown of THAP, AAEL011794, and AAEL005286 significantly delayed larval development, pupation and adult emergence compared to the vector control (Fig. 3C, pBS-xhsp70 vs. siTHAP, siAAEL011794, and siAAEL005286, F 1,21 = 28.02, 39.21, and 23.34, respectively, p<0.0001). Interestingly, siATF-2-treatment led to significantly accelerated developmental pace compared to the vector control (Fig. 3C, pBS-xhsp-70 vs. siATF-2, F 1.21 = 10.76, p = 0.0036). Female fertility in the vector control groups (Fig. 3B) was similar to previously reported [11]. There was a 52% decrease in fertility in siTHAP-treated groups compared to the control (Fig. 3D, pBS-xhsp70 vs. siTHAP, p = 0.0158, t = 7.863 df = 2), whereas there was a 22% increase in fertility in siATF-2-treated groups (Fig. 3D, pBS-xhsp70 vs. siATF-2, p = 0.0216, t = 4.414 df = 3). Treatments with siAAEL011794 or siAAEL005286 did not significantly alter the female fertility compared to the control group (Fig. 3D, pBS-xhsp70 vs. siAAEL011794 and siAAEL005286). The results showed that all four transcription factors may be involved in the control of development progression in Aedes aegypti, but only THAP and ATF-2 were likely contributing to female fertility.
In the 72 h 4th instar larval midgut nuclear extract, two histone proteins, the histone H2A and H2B, was found to bind to the −1.6/−1.3 kb regulatory sequence (Table 1, 72 h). Both histone H2A and H2B are in the core structure of nucleosome that involves in chromatin remodeling machinery [25], suggesting that the down-regulation of AeSCP-2 expression in the larval midgut post larval feeding stage might be due to chromosome condensation. Three other proteins from the 72 h nuclear extract, AAEL006857, AAEL001950, and AAEL011795, have no DNA-binding motifs (Table 1, 72 h) and are functionally unknown proteins in mosquitoes. AAEL001950 has an Ubiquitin Associated domain (UBA) close to the C-terminal and an Ubiquitin-like domain close to the N-terminal. AAEL011795 has a CAP domain that is found mostly in extracellular proteins [26]. AAEL006857 has a RNA recognition motif (RRM) that is involved in RNA and protein interactions [27], [28].
In vivo Regulatory Functional Study of Transcription Factors on AeSCP-2 Expression
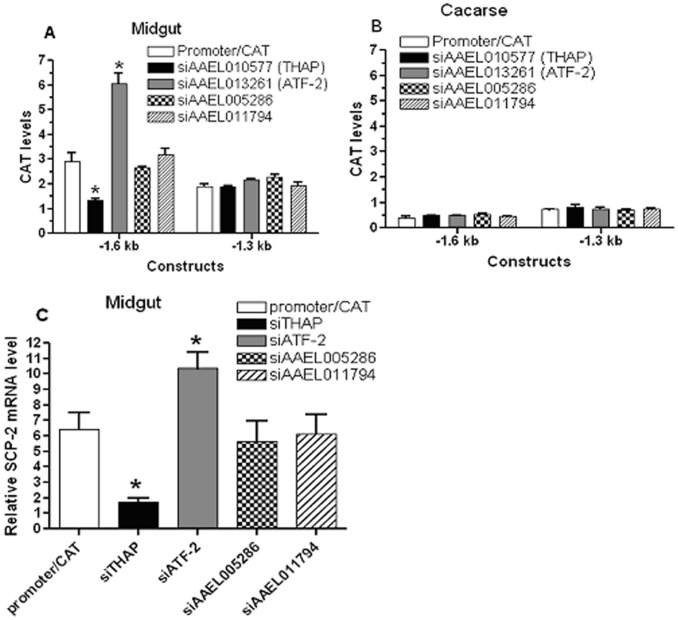
To confirm that the transcription factors identified above control AeSCP-2 expression via the −1.6/−1.3 kb promoter sequence, we performed in vivo promoter/reporter gene assays under the condition of expression knockdown of each transcription factor. F0 larvae were hatched and synchronized on Day 1 2nd instar. Selected Day 1 2nd instar larvae were heat shocked for 24 hours at 37°C and returned to 26°C, a second heat shock at 37°C for 24 hours was given to Day 1 4th instar to ensure continuous high levels of expression knockdown throughout the life cycle (Fig. S2). Quantities of CAT reporter gene product in 24 h 4th instar larval midgut and carcass were determined. The levels of expression knockdown of each transcription factor were determined via RT-qPCR analysis for triplicates of 10 pooled larvae from the same batch for each of the promoter/reporter gene assays (Fig. 3B). Three experimental repetitions were performed for each promoter/reporter gene construct. In the promoter/CAT control groups, the −1.6 kb construct drove significantly higher levels of reporter gene expression than that of −1.3 kb construct in 24 h 4th instar larvae (p = 0.047, Fig. 4A), which is consistent with the results in figure 1A (Fig. 1A, −1.3 kb vs. −1.6 kb in 24 h samples). The absolute CAT levels in each corresponding tissue sample were lowered in the reporter gene construct/siRNA co-transfected larvae than that of the reporter gene construct alone (Fig. 1 vs. Fig 4), this might be due to the amount of reporter gene constructs injected in the co-transfection experiments being only ½ of the amount of reporter gene construct alone assays (see M&M).
Figure 4. Quantitative analysis of in vivo CAT expression from co-transfection of transcription factor siRNA vector and AeSCP-2 promoter/CAT constructs (1∶1 ratio).
In the promoter/CAT group, hsp70 short promoter vector plasmid was added in at 1∶1 ratio to normalize the total amount of promoter/CAT each group received. Larvae were synchronized on Day 1 2nd instar, heat shocked at 37°C for 24 hours on Day 1 of 2nd and 4th instar, respectively. Day 1 4th instar larval samples were taken after the 2nd heat shock-treatment. (A) The midgut samples from 4th instar larvae (30 larvae/sample) were taken at 24 hour post 3rd molt. (B) The carcass samples were from the same larvae as the midgut sampling. (C) Larval midgut endogenous transcription of AeSCP-2 in −1.3 kb/CAT and siRNA co-transfected larvae. Ten synchronized 24 h 4th instar larvae were pooled from each batch of the co-transfection experiment (in panel A, −1.3 kb). Relative AeSCP-2 mRNA levels (vs. Actin-1) from each sample were determined via RT-qPCR. Mean and standard deviation are shown (N = 3). * Indicating significantly different (p<0.05) from the promoter/CAT control.
Under the condition of expression knockdown of THAP, the −1.6 kb/CAT transcriptional activities in the midgut decreased to 45% of the control (p<0.05, Fig. 4A), whereas under the same conditions there was no significant change in reporter gene expression in the carcass samples (Fig. 4B). On the other hand, siTHAP expression did not affect the −1.3 kb promoter activity in the midgut (Fig. 4A). The results suggest that THAP may be required for maintaining high levels of AeSCP-2 transcription in the 24 h 4th instar larval midgut. It is highly likely that the THAP regulatory element resides in the −1.6/−1.3 kb 5′ flanking sequence. Under the condition of ATF-2 expression knockdown, a 2-fold increase in −1.6 kb transcriptional activities was detected only in the midgut compared to the vector plasmid control (p<0.05, Fig. 4A and B). Expression knockdown of ATF-2 had no effect on the promoter activity of −1.3 kb in either the midgut or the carcass tissues (Fig. 4A and B). The results suggest that ATF-2 may attenuate AeSCP-2 expression in the larval midgut via the regulatory sequence in the −1.6/−1.3 kb region. However, neither AAEL005286 nor AAEL011794 expression knockdown had significant effects on the in vivo −1.6/−1.3 kb transcriptional activity in 24 h 4th instar larvae (Fig. 4A and B). The results showed that THAP and ATF-2 antagonistically regulated AeSCP-2 promoter activities via the −1.6/−1.3 kb regulatory sequence.
To verify the quantitative effect of expression knockdown of the transcription factors on the endogenous AeSCP-2 expression in vivo, we took larval midgut RNA samples from 24 h 4th instar larvae (10 larvae/sample) in the −1.3 kb/CAT and siRNA co-transfection batches and measured the endogenous AeSCP-2 mRNA levels via RT-qPCR analysis. AeSCP-2 mRNA levels in the midgut decreased by 73.2% in siTHAP-treated larvae (p<0.05, Fig. 4C, siTHAP vs. promoter/CAT). When ATF-2 expression was knocked down by siATF-2, the AeSCP-2 mRNA level in the 4th instar larval midgut increased to 161% of the control (p<0.05, Fig. 4C, siATP vs. promoter/CAT). Expression knockdown of AAEL005286 or AAEL011794 had no effect on AeSCP-2 transcription in the larval midgut (Fig. 4C). The effects of transcription factor siRNA treatment on AeSCP-2 expression in larvae showed that THAP and ATF-2 played a regulatory role in the endogenous AeSCP-2 transcription.
Effects of Expression Knockdown of THAP and ATF-2 on Development and Growth
It has been shown that in vivo expression knockdown of AeSCP-2 results in delayed development, higher mortality, and lower female fertility [10], [11]. Since THAP is implicated in maintaining high levels of AeSCP-2 expression in feeding larvae in vivo (Fig. 4), we expected that the lowered AeSCP-2 expression via siTHAP treatment would result in similar phenotypic responses as that of siAeSCP-2 treatment. We speculated that ATF-2 is an AeSCP-2 suppressor; therefore, increased AeSCP-2 expression via siATF-2 treatment would rescue siAeSCP-2-induced effects on development, growth and fertility. It is likely that both THAP and ATF-2 have more than one targeted gene, however, manipulation of in vivo AeSCP-2 expression via siRNA or over-expression vectors may shed light on the functional role of THAP and ATF-2 in AeSCP-2 transcriptional regulation that impacts growth and development. The constitutive AeSCP-2EGFP fusion protein expression vector [29] was co-transfected with siTHAP to “rescue” the negative effect of THAP expression knockdown on AeSCP-2 expression (Fig. S1, AeSCP-2, lanes 2 and 3 vs. 4 and 5). The AeSCP-2EGFP fusion protein shows similar cellular function as that of AeSCP-2 in cultured mosquito cells [29] and is easily distinguishable from the wild type AeSCP-2 via western blotting analysis (Fig. S1, AeSCP-2EGFP, lanes 2–4). The siSCP-2 vector was co-transfected with siATF-2 to overcome the positive effect of ATF-2 expression knockdown on AeSCP-2 transcription.
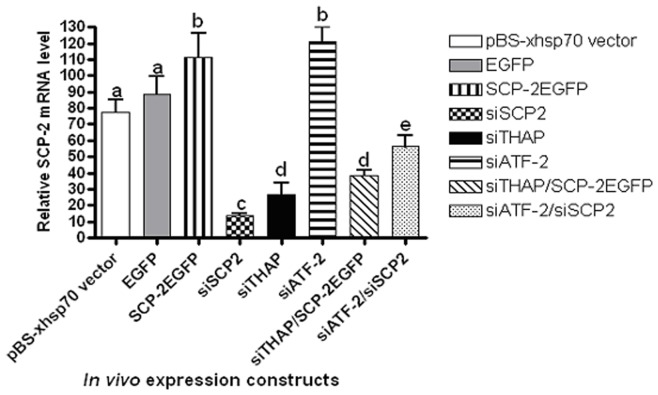
Transfected larvae were synchronized on Day 1 2nd instar and heat shocked at 37°C for 24 hours on Day 1 of 2nd and 4th instar, respectively. Midgut samples were taken from 24 h 4th instar larvae. There was no significant difference in AeSCP-2 expression in larvae transfected with either the siRNA expression vector or over-expression vector (Fig. 5, pBS-xhsp70 vector vs. EGFP). In siSCP-2-treated larvae, AeSCP-2 mRNA level decreased by 82.5% (p<0.05, Fig. 5), showing that the in vivo heat shock-induced siRNA expression functioned well. Expression knockdown of THAP resulted in a 65.3% decline in AeSCP-2 transcript (p<0.05, Fig. 5), which were consistent with the endogenous AeSCP-2 expression in larvae from the promoter/CAT reporter in assays (Fig. 4C, siTHAP). It is noted that regardless which internal control genes were used in RT-qPCR analyses (Fig. 4C using Actin-1 as the internal control vs. Fig. 5 using rpL8 as the internal control) there were no significant differences in the percentage of changes in levels of AeSCP-2 expression in siTHAP- and siATF-2-treated groups vs. vector controls, i.e. the % changes in SCP-2 mRNA levels in figure. 4C compared to figure 5 (p = 0.25 and 0.66 for siTHAP and siATF-2, respectively). Over-expression of AeSCP-2EGFP driven by the Baculovirus immediate early gene promoter (IE [17]) did not significantly increase AeSCP-2 mRNA levels in the larval midgut (Fig. 5, EGFP vs. SCP-2EGFP, p = 0.10), which did not significantly rescued the negative effect of siTHAP treatment on AeSCP-2 transcription compared to the pBS-xhsp70 vector control (Fig. 5, siTHAP vs. siTHAP/SCP-2EGFP, p = 0.07). The results indicate that the Baculovirus immediate early gene promoter was not sufficiently active in the larval midgut to drive high levels of target gene expression. On the other hand, siAeSCP-2 expression was able to effectively damp the positive effect of siATF-2 on AeSCP-2 transcription from 156% to 63.7% of the vector controls (Fig. 5, siATF-2 vs. siATF-2/SCP-2EGFP, p<0.05). The results showed that varied levels of in vivo AeSCP-2 expression were achieved effectively in most cases via combination of expression vectors.
Figure 5. In vivo AeSCP-2 transcription in the larval midgut under the influence of transfected expression vectors.
The siRNA vector is driven by the Drosophila hsp70 short promoter. Constitutive over expression is drive by the Baculovirus immediate early gene promoter [17]. In the co-transfected groups, two expression vectors were added at 1∶1 ratio. In the single expression vector groups, pBS vector plasmid was added in at 1∶1 ratio to normalized the total amount of expression vector each group received. Larvae were synchronized on Day 1 2nd instar; siRNA expression was induced via heat shocked at 37°C for 24 hours on Day 1 of 2nd and 4th instar, respectively. Heat shock-treatment was not applied to the EGFP and SCP-2EGFP groups. Day 1 4th instar larvae (10 larval midguts/sample) were taken after the 2nd heat shock-treatment. Relative (vs. rpL8) AeSCP-2 mRNA levels from each sample were determined via RT-qPCR. Mean and standard deviation are shown (N = 3). The same letters above the bars in each construct represent that the mean values did not differ from other constructs significantly (p>0.05) in paired t-tests with the vector control.
Thirty larvae from each batch were synchronized on Day 1 2nd instar and siRNA expression was induced via heat-shock at 37°C for 24 hours on Day 1 of 2nd and 4th instar, respectively. Larval growth and development were recorded daily until the 13th day when the adults were 3–5 days post emergence. The time that took to reach developmental milestones such as molting to 4th instar, pupation, and adult emergence in each batch of transfection was compared to the control (transfected with empty expression vector). For the batches that transfected only with over-expression vector, no heat shock treatment was applied. Heat shock on day 1 2nd instar for 24 hours did not significantly alter the pace of development to 4th instar (Fig. 6A, 4th instar on 4th day, pBS-xhsp70 vector vs. EGFP). After the second 24 hour-heat shock on day 1 4th larvae in the heat shocked groups reached to pupation and adult emergence at an accelerated pace compared to non-heat shocked control (Fig. 6A, pupa on 8th day and adults on 11th day, pBS-xhsp70 vector vs. EGFP, F 1,12 = 316.5, p<0.0001), although there was no significant difference in total mortality between groups of heat shock vs. non-heat shock controls (Fig. 6A, total death by 13th day, pBS-xhsp70 vector vs. EGFP). Heat-shock treatments at 37°C induced siAeSCP-2 expression (Fig. 5, siSCP-2) and resulted in delayed development compared to the vector control (Fig. 6A, pBS-xhsp70 vector vs. siSCP-2, F 1,12 = 884.1, p<0.0001), which was consistent with previous report in larvae with 3-hour 42°C heat shock-treatment (Peng et al., 2011). Co-transfection of siATF-2 with siSCP-2 vectors led to significant recovery of siSCP-2-induced developmental delay (Fig. 6A, siATF-2/siSCP-2 vs. siSCP-2, F 1,12 = 912.1, p<0.0001). The results seemed to correlate to the rescued levels of AeSCP-2 transcription mediated by siATF-2 treatment (Fig. 5, siSCP-2 vs. siATF-2/siSCP-2). Over-expression of AeSCP-2EGFP resulted in a fast developmental pace in treated larvae (Fig. 6A, EGFP vs. SCP-2EGFP, F 1,12 = 514.3, p<0.0001), however, the moderate effect of SCP-2EGFP on AeSCP-2 expression in siTHAP treated group (Fig. 5, siTHAP vs. siTHAP/SCP-2EGFP) did not overcome the siTHAP-induced developmental delay (Fig. 6A, SCP-2EGFP vs. SCP-2EGFP/siTHAP). In fact, the delay in development was more severe in siTHAP/SCP-2EGFP-treatment compared to siAeSCP-2 alone (Fig. 6A, siSCP-2 vs. siTHAP/SCP-2EGFP, F 1,12 = 8.333, p = 0.0137), suggesting that THAP may have additional target gene(s) that impact growth and development.
Figure 6. Effects of THAP and ATF-2 expression knockdown on development and fertility.
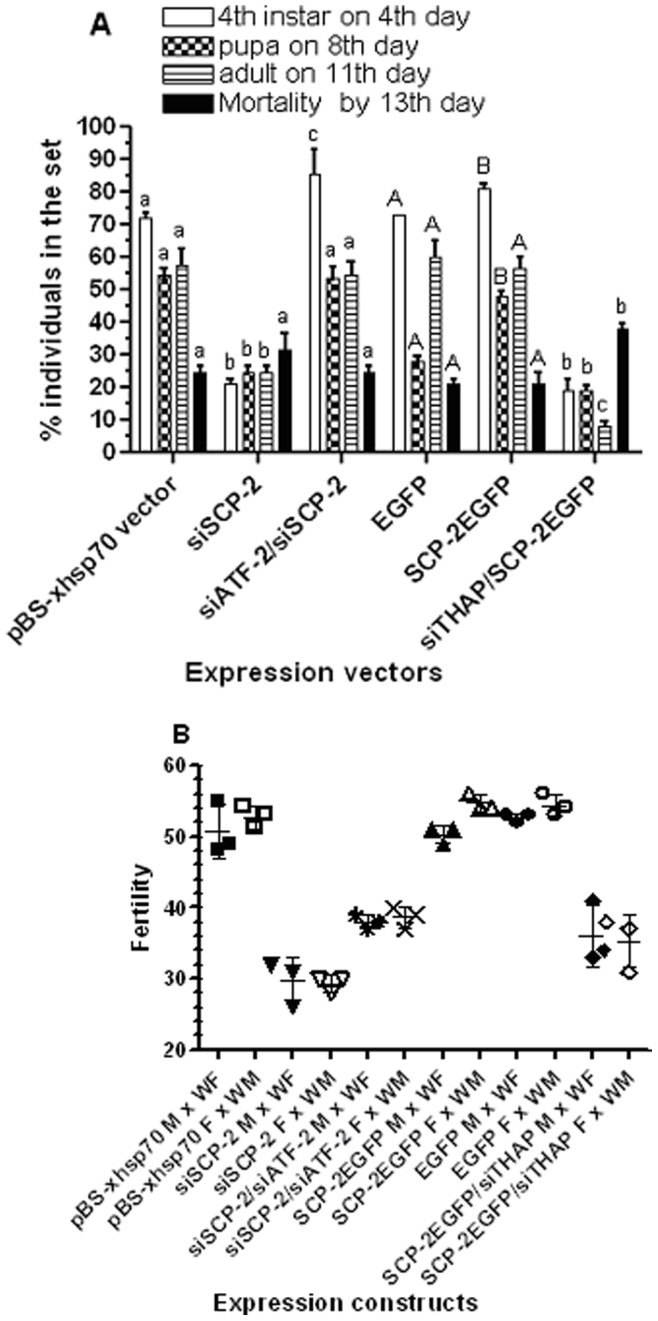
The siRNA vector is driven by the Drosophila hsp70 short promoter (see M&M). Constitutive over expression is drive by the immediate early gene promoter (Vyazunova and Lan, 2010). In the co-transfected groups, two expression vectors were added at 1∶1 ratio. In the single expression vector groups, pBS plasmid was added in at 1∶1 ratio to normalized the total amount of expression vector each group received. Thirty larvae were synchronized on Day 1 2nd instar; siRNA expression was induced via heat shocked at 37°C for 24 hours on Day 1 of 2nd and 4th instar, respectively. Heat shock-treatment was not applied to the EGFP and SCP-2EGFP groups. (A) Developmental progression and mortality. The same letters above the bars in each construct represent that the mean values did not differ from other constructs significantly (p>0.05) in paired t-tests within each observation. Lower case letter represents heat shock treated group, capital letter represents non-heat shocked group. (B) Female Fertility (after the 1st bloodmeal). Fertility is defined as viable 2nd instar larvae/female. Survived pupae in each group from (Fig. 6A) were separated by sex and adults emerged in separated cages. Adults from each group (8–12) were mated with 10 wild type opposite sex (WF = wild type female or WM = wild type male). Eggs of blood-fed females were hatch 5–6 days after egg deposition. The average fertility/female in each mating group is presented. Mean and standard deviation are shown (N = 3). The same letters above the bars in each construct represent that the mean values did not differ from other constructs significantly (p>0.05) in paired t-tests within each observation. Capital letters above the bar represent non-heat shock groups; lower case letters above the bar represent heat shock-treated groups.
Heat shock- or osmotic stress-induced phosphorylation of dATF-2 results in its release from heterochromatin [30], which leads to changes in target gene expression [30], [31]. We used heat shock-treatment to induce siATF-2 expression; therefore, it is possible that heat-shock might activate the transcriptional function of endogenous ATF-2, which may in turn lead to down-regulation of AeSCP-2 expression. However, we did not detect heat-shock-mediated changes in AeSCP-2 expression (Fig. 5, pBS-xhsp70 vector vs. EGFP), indicating that heat shock alone (presumable stress-induced activation of endogenous ATF-2) was not sufficient to regulate AeSCP-2 transcription. We showed that it was the heat-shock-induced siATF-2 expression that led to the up-regulation of AeSCP-2 transcription in the midgut via the −1.6/−1.3 kb 5′ flanking sequence (Fig. 4A and C). Moreover, the siATF-2-induced accelerated growth and development (Fig. 2A, pBS-xhsp70 vs. siATF-2) was significantly reduced by the co-expression of siAeSCP-2 (Fig. 6A, pBSxhsp70 vector vs. siSCP-2/siATF-2). It is possible that expression knockdown of ATF-2 under the condition of heat-shock exacerbated the effect of ATF-2 on AeSCP-2 regulation; the results did suggest that ATF-2 is involved in AeSCP-2 expression regulation.
Males and female adults from above batches were mated with wild type (non-transfected) and eggs/blood-fed female (the average of eggs/female in 3 batches of 5–7 females/sample), % of egg hatching, and number of surviving 2nd instar from each mating group were recorded. Female fertility was defined as viable 2nd instar/blood-fed female produced in each mating group. It should be noted that the data from egg/female were not the life time egg production of the female; instead, they were only the egg production after the 1st bloodmeal. We previously reported that siAeSCP-2-treated pairs have significantly lower fertility than that of the control pairs [11]. However, the backcross to non-transfected sexes separately also had significantly lowered fertility compared to controls (Fig. 6B, pBS-xhsp70 M × WF vs. siSCP-2 M × WF and pBS-xhsp70 F × WM vs. siSCP-2 F × WM), the egg hatching rate in “siSCP-2 M × WF” and “siSCP-2 F × WM” were similar to reported “siSCP-2 pairs” [11]. The results suggest that the function of AeSCP-2 in males affected mated female’s fertility by unknown mechanisms. Co-transfection of siATF-2 with siSCP-2 led to significantly recovery of fertility (31% increase) in the backcross of both sexes compared to siSCP-2-treatment alone (Fig. 6B, siSCP-2/siATF-2 vs siSCP-2, p<0.05), which seemed to correlate to the significantly siATF-2-rescued AeSCP-2 expression in the co-transfected larvae (Fig. 5, siATF-2/siSCP-2 vs. siSCP-2). Over-expression of SCP-2EGFP did not affect the fertility compared to controls (Fig. 6B, EGFP vs. SCP-2EGFP). However, co-transfection of SCP-2EGFP with siTHAP did not significantly affect siTHAP-mediated reduced fertility (Fig. 6B, SCP-2EGFP vs. SCP-2EGFP/siTHAP). The results suggest that either over-expression of SCP-2EGFP was not sufficient to overcome the siTHAP-mediated down-regulation of endogenous AeSCP-2 expression (Fig. 5, SCP-2EGFP vs. SCP-2EGFP/siTHAP) or THAP targets more genes than just AeSCP-2 that affect fertility.
It is unlikely that AeSCP-2 gene is the only target of THAP and ATF-2 based on the effects of siTHAP and siATF-2 on development progression and fertility. The siTHAP-treatment was significantly less effective in knocking down AeSCP-2 expression than that of siAeSCP-2 (Fig. 5), although siTHAP/SCP-2EGFP-treated larvae experienced a significantly more severe delayed developmental progression (Fig. 6A, siSCP-2 vs. siTHAP/SCP-2EGFP, F 1,12 = 8.333, p = 0.0137), noting that there were no significant differences in AeSCP-2 transcription between siTHAP and siTHAP/SCP-2EGFP treatments (Fig. 5). The results suggest that THAP may control AeSCP-2 and other gene(s) involved in growth and development. Over-expression of SCP-2EGFP resulted similar levels of AeSCP-2 transcripts as that of in siATF-2-treated larvae (Fig. 5, siATF-2 vs. SCP-2EGFP). However, over-expression of AeSCP-2EGFP did not lead to higher female fertility (Fig. 6B, SCP-2EGFPxW vs. EGFPxW) as that observed in siATF-2-treatment (Fig. 3D, siATF-2 vs. pBS-xhsp70 vector), implying that ATF-2 targets other genes involving in fertility. It is noted that THAP and AFT-2 expression are not tissue-specific and the higher levels of mRNA was detected in the carcass (Fig. 3A), therefore, expression knockdown in the whole body including the midgut (Fig. 3B) may lead to biological effects beyond the midgut physiology. The larval midgut specific target expression knockdown using the AeSCP-2′s −1.6 kb 5′ flanking sequence would enable us to test the in vivo function of THAP and ATF-2 in the larval midgut in future studies.
Discussion
High levels of AeSCP-2 expression are found in the larval midgut tissues [7] and AeSCP-2 transcription maintains at high levels throughout the feeding stage and decrease significantly 60 hours post 3rd molt in developing pharate pupae [21]. Results from earlier in vitro studies have indicated that 20E/HR3 up-regulates the −1.0 kb AeSCP-2 promoter activity, whereas 20E/Ftz-F1 down-regulates −1.6 kb AeSCP-2 promoter activity in cultured mosquito cells [17]. Using the newly developed extrachromosomal in vivo expression method in Aedes aegypti [11], we evaluated the in vivo AeSCP-2 promoter activities of six truncation constructs in 4th instar larvae. A regulatory element between −1.6 and −1.3 kb 5′ flanking sequence interacts with in an endogenous factor in culture mosquito cells and that 20E/Ftz-F1 significantly weakens this interaction [17]. However, there is no detectable level of Ftz-F1 expression in Day 1 4th instar larvae [19]. Therefore, it is unlikely that Ftz-F1 is involved in AeSCP-2 transcriptional regulation in Day 1 4th instar larvae. Factors that control the temporal/spatial AeSCP-2 expression in vivo in feeding 4th instar larvae are unknown. To search for regulatory factors that control AeSCP-2 transcription in vivo, it is necessary to define the 5′ flanking region that determines the temporal/spatial expression pattern. Using microinjection of promoter/reporter gene constructs to define temporal/spatial specific regulatory sequence in vivo has only been reported in Aedes aegypti adults [32], to study promoter activities in mosquito larvae via direct promoter/reporter DNA vector microinjection would be impossible due to high mortality rate post microinjection in 4th larvae [10]. Using the newly developed extrachromosomal in vivo expression method in Aedes aegypti [11], we evaluated the in vivo AeSCP-2 promoter activities of six truncation constructs in 4th instar larvae.
To investigate transcription factors that regulate high levels of AeSCP-2 expression in the larval midgut during feeding, we focused our attention to the transcription factors identified in the 24 h sample. THAP and ATF-2 were found to antagonistically regulate the AeSCP-2 promoter activities via the −1.6/−1.3 kb 5′ flanking sequence in the larval midgut (Fig. 4A and B). THAP proteins are known to bind to DNA regulatory elements [33], [34] as well as interact with other proteins, both THAP/DNA and THAP/protein interactions lead to changes in the target gene expression [35], [36]. Similarly, ATF-2 is known to bind to regulatory sequence as well as to interaction with other regulatory proteins in vertebrates [37]. Whether AAEL011794 and AAEL005286 were pulled down by the −1.6/−1.3 kb regulatory sequence of AeSCP-2 through the interaction with THAP and ATF-2 is unknown. AAEL011794- and AAEL005286-controlled development progression (Fig. 3C, pBS-xhsp70 vs. siAAEL011794 and siAAEL005286) is unlikely through the direct regulation AeSCP-2 expression (Fig. 4), but through other unidentified target gene (s). Interestingly, the −1.6/−1.3 kb temporal/spatial regulatory sequence did not pull down any known 20E-regulated gene product (Table 1). There is no predicted ecdysone receptor regulatory element (EcRE) within the −1.6/−1.3 kb region [17]. The results suggest that 20E-regulated AeSCP-2 expression in the larval midgut [7], [17] may be via further upstream sequences or via indirect regulations. Whether ATF-2 and THAP expression is under the influence of 20E in vivo needs further investigation.
THAP belongs to a family of proteins with a N-terminal C2CH zinc finger DNA-binding domain [23] that shares similarity with the DNA-binding domain of Drosophila P element transposase [34], [38]. The mammalian THAP proteins show diverse function such as cell proliferation regulation [39], [40], cell cycle progression control [41], and transcription repression via inhibition of histone acetylation [35]. In C. elegans, there are five proteins containing the THAP domain that are involved in chromatin-modifying [42], larval growth [43], cell cycle regulation [44], and gene expression regulation via the recruitment of another transcription factor [45]. In insects, P element transposase contains an N-terminal THAP domain and is known to bind to the consensus THAP binding site [34]. However, functions of other Drosophila THAP domain proteins are unknown [23]. We showed that Aedes THAP is involved in maintaining high levels of AeSCP-2 transcription in the larval midgut (Fig. 4). In the 5′ flanking sequence of AeSCP-2 gene (FJ554568) from −1573 to −1566 bps upstream of the transcription site, the 5′-TACGGGTA-3′ sequence matches 100% to the consensus THAP binding sequence of 5′-TXXGGGX(A/T)-3′ [34]. Whether AeTHAP binds to the −1.6/−1.3 kb AeSCP-2 5′ flanking sequence via the putative THAP binding site (5′-TACGGGTA-3′) needs further investigation.
ATF-2 is a C2H2 zinc finger transcription factor. ATF-2 has diverse functions depending on the developmental time and tissue. In mammalians, ATF-2 is important for normal cellular development and survival through its phosphorylation by JNK/p38 or ATM/ATR respectively [37], [46]. Mammalian ATF-2 is implicated in the control of CNS development in embryos [47]. It has been shown that Drosophila ATF-2 (dATF-2) positively regulates the transcription of dPEPCK (a key enzyme for both gluconeogenesis and glyceroneogenesis) via several CRE half-sites in the PEPCK promoter [24]. There is no sequence within 305 bps of the −1.6/−1.3 kb region in the AeSCP-2 promoter (FJ554568) matching 100% to the ATF/CRE core binding site (5′-TGACGTCA-3′) of the ATF-2 responsive element. The ATF-2 responsive element in the −1.6/−1.3 kb region in the AeSCP-2 promoter needs further investigation. SCP-2 is not a known ATF-2 target gene [24], [31]. This is the first report of ATF-2′s regulatory role in regulating an intracellular lipid carrier protein gene. In Drosophila, expression knockdown of dATF-2 leads to smaller triglyceride reserves [24], [48] via dATF-2 regulated expression of PEPCK (dPEPCK) gene and decreased survival under starvation conditions [24]. We did not test siATF-2-treated larvae under starvation conditions. Therefore, whether AeATF-2 has similar in vivo function as that of dATF-2 is unknown.
The antagonistic role of THAP and ATF-2 in AeSCP-2 expression regulation in the larval midgut is intriguing. However, the regulation of AeSCP-2 expression may be more complex considering that other factor(s) may control AeSCP-2 expression indirectly. Expression knockdown of AAEL005286 led to a 24% reduction in THAP expression that affected neither ATF-2 nor AeSCP-2 mRNA levels (Fig. 3B and Fig. 4C), although an 83% expression knockdown of THAP (Fig. 3B) did result in a 27% and 73% decrease in ATF-2 and AeSCP-2 expression, respectively (Fig. 3B and Fig. 4C). The results suggest that there may be a threshold level of THAP transcription below which would lead to reduction in ATF-2 and AeSCP-2 expression in the midgut. On the other hand, a 77% reduction in ATF-2 expression in the larval midgut (Fig. 3B) resulted in a 1.56-fold increase in AeSCP-2 expression in the larval midgut (Fig. 5). Therefore a forward negative modulating mechanism may exist between THAP and ATF-2 in AeSCP-2 expression regulation in the larval midgut. High levels of THAP could potentially lead to higher levels of AeSCP-2 and ATF-2 transcription (a positive modulating effect); the THAP-mediated up-regulation of ATF-2 expression in turn may attenuate AeSCP-2 transcription (Fig. 7). Expression knockdown of THAP led to 65% and 73% decreased in AeSCP-2 transcript in the midgut, respectively (Fig. 5 and Fig. 4C), which is significantly less effective than the 83% reduction in AeSCP-2 expression mediated by siAeSCP-2 (Fig. 7). It is unclear whether siTHAP-mediated 27% reduction in ATF-2 expression (Fig. 3B) led to less efficiency in the siTHAP-mediated decreasing AeSCP-2 transcription compare to siSCP-2 alone (Fig. 5) through the forward-negative modulation mechanism (Fig. 7). Any factor that influences the level of THAP or ATF-2 in the larval midgut would ultimately affect the overall levels of AeSCP-2 expression (Fig. 7).
Figure 7. Schematic diagram showing THAP- and ATF-2-regulated AeSCP-2 expression in the midgut of feeding 4th instar larvae.
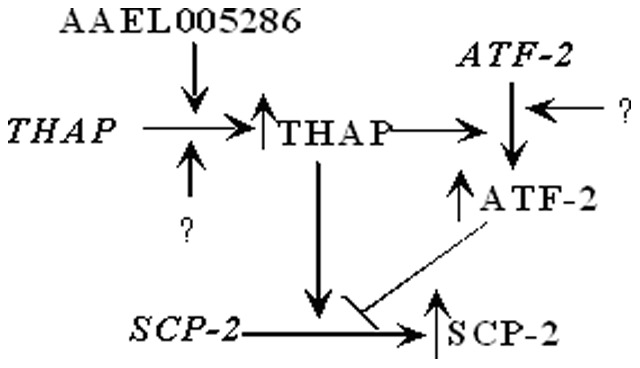
Arrows indicate up regulation, bar represents down regulation, “?” denotes unknown factors.
The endogenous THAP and AFT-2 expression in 24 h 4th instar is ubiquitous with higher levels detected in the head and carcass (Fig. 3A). It is speculated that THAP and ATF-2 have multiple target genes in different tissues at different developmental time points. Clearly, THAP and ATF-2 have the opposite effects on growth and development of Aedes aegypti (Fig. 3C, F 1,12 = 840.5, p<0.0001). In this study, we focused on the THAP and ATF-2 in vivo function in the larval midgut regarding AeSCP-2 promoter activity (Fig. 4), the biological effects of global expression knockdown of THAP and ATF-2 on development, mortality, fertility (Fig. 3C, 3D, 6A, and 6B) may only be partially due to regulation of AeSCP-2 in the larval midgut. A tissue-specific approach would help teasing out the specific THAP and ATF-2 function and the biological importance in the midgut tissue. Identification of the first larval midgut specific promoter (Fig. 1 and Fig. 4) should facility future studied.
In summary, results from the promoter/reporter gene assays indicate that −1.6 kb 5′ flanking sequence was sufficient for temporal/spatial regulation of AeSCP-2 transcription in the larval midgut (Fig. 1 and 4). AeSCP-2 is the first reported target gene of ATF-2 and THAP in mosquitoes. The results also validated the potential of using the vertical DNA delivery method for in vivo promoter activity studies in mosquito larvae.
Materials and Methods
Chemicals and Reagents
Chemicals and reagents were purchased from Sigma (St. Louis, MO), Thermo Fisher Scientific (Pittsburgh, PA) and ICN (Costa Mesa, CA) if their origins are not mentioned in the text. Enzymes for manipulating DNA during cloning processes were purchased from New England Biolabs (NEB, Ipswich, MA) or Promega (Madison, WI). Molecular biological reagents were purchased from Invitrogen (Grand Island, NY), BioRad (Hercules, CA), Qiagen (Valencia, CA).
Mosquitoes
The yellow fever mosquito, Aedes aegypti, is an inbred laboratory strain (Rockefeller) that was reared at 26°C in 16 h day light/8 h night cycle in 70–80% humidity. Larvae were fed with fish food (TretraMin, Tetra Holding, Inc., Blacksburg, VA, USA). Female adults were blood fed with defibrinated rabbit blood (Hemostat Laboratories, Dixon, CA) using a glass feeder and circulation of water heated to 37°C.
Plasmids
To construct the hsp70 short promoter expression vector, the pBS-hsp70-SV40 poly(A) expression vector [11] was digested with XbaI/XhoI, made to be blunt-ended using Klenow-exo (NEB) in the presence of 2 mM dNTPs, and then self-ligated using T4 DNA ligase (NEB), which resulted in the pBS-xhsp70-SV40 poly(A) expression vector (−194 bp 5′ flanking), the Drosophila hsp70 short promoter. To select hairpin siRNA sequences for targeted genes, we use the online free tool from Ambion (http://www.ambion.com/techlib/misc/siRNA_finder.html). The potential target sequences was blast-searched in the EST database in VectorBase of Aedes aegypti, if the sequence only match perfectly to ESTs of the targeted gene without partial match to ESTs of other genes, then, we identified the siRNA as target specific sequence. DNA oligo of sense and antisense nucleotides for the small hairpin RNA targeting THAP, ATF2, AAEL005286, and AAEL011794 (Table 2) was synthesized (IDT, Coralville, IA). One hundred microliters ddH2O was added to dissolve each primer (∼0.60 mg), 5 µl sense and antisense primer were mixed, heated at 95°C for 5 min, returned to room temperature to cool for 5 min. The annealed DNA oligo was cloned into the EcoRV site in MCS in pBS-xhsp70-SV40 plasmid. DNA miniprep of cloned plasmid was prepared using the Spin column kit (Qiagen). Two micrograms of plasmid DNA were digested with a restriction enzyme (recognition site inserted in the loop of the siRNA, Table 2) at 37°C for 2 h and was cleaned up with the spin column (Qiagen) before sequencing using the M13 reverse primer. DNA sequencing confirmed the hairpin insert for each gene. The plasmid of pBS-hsp70-siAeSCP2 [11] was digested with XbaI and self-ligated to produce the pBS-xhsp70-AeSCP2siRNA plasmid with a shortened hsp70 promoter (−254 bp 5′ flanking hsp70 sequence). The efficiency of heat shock induced expression knockdown was that more than 90% F0 larvae showed a significant reduction in which the target gene mRNA level was lower than the mean-S.D. in the control (Fig. S2A). Larvae were heat shocked at 37°C for 24 h on Day 1 2nd and Day 4th induced continuous siRNA expression through the 2nd instar-pupal-adult stages (Fig. S2C).
Table 2. Nucleotide sequences of primers for PCR and oligos for siRNA.
| Gene | Oligo* | Tm | Amplicon | Efficiency |
| Rpl8 | F: 5′-TACCTGAAGGGAACCGTCAAGCAA-3′ | 60°C | 221 bp | 99.3% |
| R: 5′-ACAATGGTACCTTCGGGCATCAGA-3′ | 60°C | |||
| AeAct-1 | F: 5′-CCCTGAAGTACCCCAATGAGC-3′ | 59°C | 51 bp | 99.7% |
| R: 5′-CCATGTCATCCCAGTTGGTG-3′ | 58°C | |||
| AeSCP-2 | F: 5′-GCTGGTCGAGTCCGACGATGC-3′ | 60°C | 82 bp | 105.7% |
| R: 5′-CAGGGCACCGGTTCCGATGG-3′ | 60°C | |||
| −1.6/−1.3 kb AeSCP-2 Probe | F: 5′-biotin-AGCTTACCTTGAATAATTAGGTACGG-3′ | 59°C | 305 bp | |
| R: 5′-biotin-AAAGGCTAAATTACCAAAAATGTAAT-3′ | 57°C | |||
| THAP | F: 5′-ATGAGGTCAGCGAACGGGAAGAAT-3′ | 60°C | 148 bp | 95% |
| R: 5′-ACGACCGGCGTTATGTTGAACTCT-3′ | 60°C | |||
| ATF-2 | F: 5′-ATGTTCTCTCGCTGCACAAGGACT-3′ | 60°C | 191 bp | 95% |
| R: 5′-AGCAGCGTTAACAACAGGATTCGC-3′ | 60°C | |||
| AAEL005286 | F:5′-CAGCGATTTCCCTGCTTTCCCAAT-3′ | 60°C | 92 bp | 95% |
| R:5′-AGGAGCCCTTCCTGTTCCATCAAT-3′ | 60°C | |||
| AAEL011794 | F:5′-TCGATAAGTGCTTCCGGTTCGGTT-3′ | 60°C | 198 bp | 95% |
| R:5′-ACTGTTCGGTGTGGTAAGGTGGAA-3′ | 60°C | |||
| −1.6 kb AeSCP-2 promoter | F: 5′-AGCTTACCTTGAATAATTAGGTACGG-3′ | 55°C | 1612 bp | |
| R: 5′-GTCGAAACTCGAAACTGATGG-3′ | 54°C | |||
| THAP siRNA | Sense: 5'-gatccgCGTGGAAACGAATGAGGTCtttgaattcAGACCTCATTCGTTTCCACGttttttggaaa-3' | |||
| Antisense: 5'-tttccaaaaaaCGTGGAAACGAATGAGGTCTgaattcaaaGACCTCATTCGTTTCCACGcggatc-3' | ||||
| ATF-2 siRNA | Sense: 5'-gatccgCTGCCCAACAAGTCGAATCttgaattcGATTCGACTTGTTGGGCAGttttttggaaa-3 | |||
| Antisense: 5′-tttccaaaaaaCTGCCCAACAAGTCGAATCgaattcaaGATTCGACTTGTTGGGCAGcggatc-3′ | ||||
| AAEL005286 siRNA | Sense: 5'-gatccgATTTGTCGATACAAGACCttgaattcGGTCTTGTATCGACAAATCttttttggaaa-3' | |||
| Antisense: 5′-tttccaaaaaaGATTTGTCGATACAAGACCgaattcaaGGTCTTGTATCGACAAATcggatc-3′ | ||||
| AAEL011794 siRNA | Sense: 5'-gatccACGCAAGCTCGGAGAATTCtttggtaccGAATTCTCCGAGCTTGCGTttttttggaaa-3' | |||
| Antisense: 5′-tttccaaaaaaACGCAAGCTCGGAGAATTCggtaccaaaGAATTCTCCGAGCTTGCGTggatc-3′ | ||||
Capital letters represent sequences of perspective genes; lower case letters represent added sequence for cloning purpose or the loop in the hairpin structure.
The −1.6 kb 5′ flanking sequence of AeSCP-2 was amplified using specific PCR primers (Table 2) and the −1.6 kb/CAT construct [17] as the template in Phusion HF Master Mix reaction solution (NEB) and was cloned into the pBlunt PCR cloning vector (Invitrogene), the cloned 1.6 kb promoter was removed from pBlunt-1.6 kb via BamHI/PstI digestion. The hsp70 promoter sequence was removed from pBS-hsp70-SV40 poly(A) expression vector [11] via BamHI/PstI digestion and the gel purified BamHI/PstI fragment of the 1.6 kb AeSCP-2 promoter sequence was inserted to replace the hsp70 promoter to create the pBS-1.6 kb AeSCP-2promoter-SV40 expression vector. The AeSCP-2 hairpin sequence was removed from the pBS-hsp70-siAeSCP-2 vector [11] using PstI/HindIII and inserted into PstI/HindIII sites in pBS-1.6 kbAeSCP-2promoter-SV40 vector to construct the pBS-1.6 kb-siAeSCP-2 plasmid. DNA sequencing confirmed the siAeSCP-2 hairpin insert.
AeSCP-2 promoter/CAT constructs have been previously described [17] and were used for in vivo transfections. The pIE over-expression vector [17] were used to construct constitutive over-expression constructs of ATF-2 and THAP (PCR cloning primers were listed in Table 2). The over-expression vectors were sequenced to confirm the correct orientation of the inserted coding regions of ATF-2 and THAP. All plasmids used in microinjection were purified using the EndoFree plasmid Maxi Kit (Qiagen), and then filtered through 0.22 µm MCE Syringe Filter (Fisher brand, cat #09-719A). The quantity of the DNA was determined using UV OD260 absorption on NanoDrop (NonoDropTM1000, NanoDrop Products, Wilmington, DE, USA).
Microinjection
The In vivo jetPEI (Polyplus transfection, Illkirch, France)/plasmid DNA mixture (N/P = 10) was prepared and microinjected as described previously by Peng et al. [11]. Ten female adults were microinjected with jetPEI/plasmid DNA mixture at 16–18 h post the blood meal (PBM). The amount of 0.5 µl jetPEI/plasmid DNA mixture (0.5 µg DNA plasmid/female) was injected into the hemolymph through the anterior thorax using a micromanipulator as described [11]. To obtain the consistent in vivo transfection results, technically 90% of the injected females should show uniformly high levels of ovarian uptake of injected DNA/PEI material from the hemolymph [11] and the mortality of microinjected adults must be <80% for each performance.
CAT Reporter Gene Assays
In vivo transfected larvae were synchronized at molting to 4th instar. Larval midgut and carcass was dissected in ice-cold phosphate saline buffer (PBS, pH 7.4) from 24 h 4th instar larvae. Tissues (30 larvae/sample) were put separately into 500 µl 1× lysis buffer (CAT ELISA Kit, Roche Applied Science, Mannheim, Germany) containing the cocktail of protease inhibitors (Sigma). Samples were homogenized briefly using a micropestle and centrifuged at 9300×g for 5 min at 4°C. The amount of 200 µl supernatant was used for CAT quantity assay using the CAT ELISA kit according to the manufacturer’s instructions. The supernatant was diluted 10 times and the protein concentration for the same sample was determined using the BCA protein assay kit (Thermo Scientific Pierce, IL). A concentration standard curve (0.015–1.00 ng/ml) of pure CAT (provided in the assay kit) was constructed for each batch of assay and the amount of CAT in each sample was calculated using the CAT standard curve. CAT quantity in each sample was defined as CAT unit/min of the reaction/µg protein.
Nuclear Protein Extraction and Boiotin-Streptavidin Pull-down Assay
Twenty four hour- and seventy two hour-old 4th instar larvae were dissected in ice-cold PBS containing protease inhibitors (Protease inhibitor cocktail, Sigma) to obtain midguts (500 larvae/sample). Midguts were washed once in 500 µl ice-cold PBS/protease inhibitors and homogenized in 1 ml of hypotonic lysis buffer provided in the CelLytic NuCLEAR Extraction Kit (Sigma). The 150 µl nuclear extracts from 24 h and 72 h 4th instar larval midguts were prepared according to the protocol of CelLytic NuCLEAR Extraction Kit (Sigma) and were snap-frozen using liquid nitrogen and were stored in −80°C. The protein concentration was determined using the BCA Protein Assay Kit (Thermo Scientific Pierce, IL). DNA fragment between −1.6 kb to −1.3 kb 5′ flanking sequence of the AeSCP2 promoter was used to make the biotinylated probe for pull-down assay. Biotinylated DNA fragments were amplified by PCR. The 5′ biotin-labeled primer pairs were listed in Table 2. The amplified biotin-labeled DNA probes were gel purified and the concentration was measured using NanoDrop Spectrophotometer. Prior to use, 100 ng Dynabeads were prewashed twice with 300 µl binding buffer using magnetic separation according to the protocol (Dynabeads kilobaseBINDER™ Kit, Invitorgen). Eight hundred nanograms of biotin-labeled probes were incubated with 100 ng Dynabeads in 300 µl binding buffer (12% glycerol, 20 mM Hepes pH 7.9, 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol) at 4°C for 3 h on a roller to keep the beads in suspension. The probe-captured beads were washed 3 times in 100 µl wash buffer (15% glycerol, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA). Eight hundred microgram proteins of each nuclear extract was incubated with probe-captured beads for 30 min at 4°C in 300 µl binding buffer containing 12% glycerol, 20 mM Hepes pH 7.9, 60 mM KCl, 1 mM EDTA, 1 mM dithiothreitol, and 5 µg of poly (dI-dC) non-specific competitor. The tubes were set on a roller to keep the beads in suspension. The protein-DNA-Dynabeads complex was washed 3 times with 50 µl buffer A (15% glycerol, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA) containing 75 mM KCl. Bound proteins were eluted with 200 µl of buffer A (15% glycerol, 20 mM Tris-HCl pH 8.0, 1 mM EDTA) containing 1 M KCl. The eluted protein mixtures were then dialyzed in 250 ml dialysis buffer (15% glycerol, 20 mM Tris-HCl pH 8.0, 1 mM EDTA) overnight at 4°C to remove the salt.
“In Liquid” digestion and mass spectrometric analysis was conducted at the Mass Spectrometry Facility (Biotechnology Center, University of Wisconsin-Madison). In short, proteins have been extracted by precipitation with 5 times excess 10% (w/v) Trichloroacetic acid (TCA)/Acetone, incubated on ice for 30 min, centrifuged for 10 min at 16,000×g and pellets washed twice with ice-cold acetone, followed by once with ice-cold methanol. Pelleted proteins were re-solubilized and denatured in 20 µl of 8 M Urea/100 mM NH4HCO3 for 10 minutes then diluted to 90 µl for tryptic digestion with: 5 µl of 25 mM Dithiothreitol (DTT), 5 µl Acetonitrile (ACN), 50 µl of 50 mM NH4HCO3 and 10 µl trypsin solution (100 ng/µl Trypsin Gold from Promega Corp. in 25 mM NH4HCO3). Digestion was conducted for 2 hours at 42°C then another 10 µl of trypsin solution was added and the reaction proceeded overnight at 37°C. The digestion was terminated by acidification with 2.5% Trifluoroacetic Acid (TFA) to 0.3% final and 8 µl loaded for nanoLC-MS/MS analysis.
Peptides were analyzed by nanoLC-MS/MS using the Agilent 1100 nanoflow system (Agilent Technologies, Palo Alto, CA) connected to a hybrid linear ion trap-orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Fisher Scientific, San Jose, CA) equipped with a nanoelectrospray ion source. Capillary HPLC was performed using an in-house fabricated column with integrated electrospray emitter essentially as described [49] but using 360 µm × 75 µm fused silica tubing. The column was packed with Jupiter 4 µm C12 particles (Phenomenex Inc., Torrance, CA) to approximately 12 cm. Sample loading (8 µl) and desalting were achieved using a trapping column in line with the autosampler (Zorbax 300SB-C18, 5 µM, 5×0.3 mm, Agilent Technologies). HPLC solvents were as follows: Isocratic loading: 1% (v/v) ACN, 0.1% Formic acid; Gradient elution: Buffer A: 0.1% formic acid in water, and Buffer B: 95% (v/v) ACN, 0.1% formic acid in water. Sample loading and desalting were done at 10 µl/min whereas gradient elution was performed at 200 nl/min and increasing %B from A of 1 to 40% in 195 minutes, 40 to 60% in 20 minutes, and 60 to 100% in 5 minutes. The LTQ-Orbitrap was set to acquire MS/MS spectra in data-dependent mode as follows: MS survey scans from m/z 300 to 2000 were collected in centroid mode at a resolving power of 100,000. MS/MS spectra were collected on the 5 most-abundant signals in each survey scan. Dynamic exclusion was employed to increase dynamic range and maximize peptide identifications. This feature excluded precursors up to 0.55 m/z below and 1.05 m/z above previously selected precursors. Precursors remained on the exclusion list for 40 sec. Singly-charged ions and ions for which the charge state could not be assigned were rejected from consideration for MS/MS. Raw MS/MS data was searched against user-defined Aedes aegypti amino acid sequence database (17,402 protein entries, VectorBase) using in-house Mascot search engine (Matrix Science Inc., Boston, MA, USA) with Methionine oxidation as variable modification, peptide mass tolerance was set at 20 ppm and fragment mass at 0.6 Da. Criteria to assign protein identification (annotation) was that at least two peptides with ion score greater then ∼25 or one with ion score greater than 40. Database Blast search of the predicted sequence against NCBI was performed to remove mismatches against common contaminant.
Reverse Transcription Polymerase Chain Reaction (RT-PCR)
Quantitative analysis of AeSCP2 expression was performed by real-time quantitative RT-PCR (RT-qPCR). Staged animals were washed with ddH2O, rinsed once with diethylpyrocarbonate (DEPC)-H2O and excess water was blotted off using clean Kimwipes. For the whole body samples total RNA was extracted from individual larva or pooled 30 2nd instar larvae or 10 3rd or 4th instar larvae or 10 pupae or adults/sample using Trizol reagent (Invitrogen). The animals from each experiment were randomly collected from each batch of transfection. Five micrograms of total RNA were treated twice at 37°C for 30 minutes with DNaseI using the Turbo DNA-free Kit (Applied Biosystems/Ambion, Austin, TX) to remove DNA contamination. RNA concentration was then measured using NanoDrop Spectrophotometer, and 0.5 µg of DNA-free total RNA was used for single-stranded cDNA synthesis using High-Capacity cDNA Archive Kit (Applied Biosystems, Austin, TX). Quantitative PCR (qPCR) was performed using the iQ™ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA). The PCR reaction solution contained 1 µl of cDNA from the RT reaction (equivalent to 25 ng RNA). PCR was performed under the following conditions: 94°C for 3 min, 40 cycles at 94°C for 10 s, 54°C for 30 s, and a final extension at 72°C for 2 min. Primers for PCR reactions were listed in Table 2. Critical qPCR parameters of the internal control were described (Table 2). The relative mRNA levels of AeSCP-2 in RT-qPCR were compared to Actin-2 or rpL8 mRNA levels.
Semi-quantitative RT-PCR was performed to determine the temporal/spatial expression profiles of THAP, ATF-2, AAEL005286, and AAEL011794 in 4th instar larvae. Total RNA samples were prepared from stages larvae and were made DNA-free as described above for RT-qPCR. The PCR reaction solution contained 1 µl of cDNA from the RT reaction (equivalent to 25 ng RNA) using the 2× Phusion Matster Mix (NEB). PCR was performed under the following conditions: 98°C for 3 min, 30 cycles at 98°C for 15 s, 58°C for 15 s, 72°C for 1 min, and a final extension at 72°C for 2 min. Primers for the RT-PCR were listed in Table 2. The PCR product was resolved in a 1% or 2% agarose gel in TAE buffer (40 mM Tris-acetate and 1 mM EDTA, pH 8.3) which was stained using 5 µg/ml ethidium bromide (EtBr). The gel electrophoresis image was captured using the Eagle Eye™ II Imaging System (Agilent Technologies, Santa Clara, CA).
Western Blotting Analysis
Intact pupae selected from transfected groups (siTHAP/SCP-2EGFP and SCP-2EGFP) were washed with ddH2O and the excess water was blotted off with a clean Kimwipe (KIMTECH, Dallas, TX). Western blotting analysis was performed as described [29] using SDS 4–20% gradient PAGE gel (ISC BioExpress, Kaysville, UT).
Statistical Analysis
Data were analyzed with two-way ANOVA (GLM procedure) to determine if several components of the biological parameter in the control groups and treated groups differed significantly using the GraphPad PRISM software version 4.0 (GraphPad). Unpaired t-test with Welch's correction was performed for two groups with unequal numbers of samples to determine whether the differences were significant using the GraphPad PRISM software version 4.0 (GraphPad). Student’s t-test was used in cases where a pair of treatments with equal numbers of samples was compared to determine the significance of the differences [50].
Supporting Information
Western Blotting analysis of AeSCP-2 expression in transfected pupae. Lane 1: Protein molecular weight markers; lane 2: siTHAP/SCP-2EGFP co-transfected male pupae; lane 3: siTHAP/SCP-2EGFP co-transfected female pupae; lane 4: siRNA vector/SCP-2EGFP transfected male pupae; lane 5: siRNA vector/SCP-2EGFP transfected female pupae. Total soluble proteins (20 µg/lane) were resolved on 4–20% gradient SDS PAGE gel. Affinity purified rabbit anti-AeSCP-2 antibody (1∶1000 dilution) and horse reddish peroxidase (HRP) conjugated goat anti-rabbit antibody (1∶2000 dilution) were used.
(DOC)
Effects of duration of heat shock on AeSCP-2 siRNA in vivo expression driven by the short Drosophila hsp70 promoter (M&M). Relative AeSCP-2 mRNA levels (vs. rpL8 ) were determined via RT-qPCR. (A) Day 1 F0 4th instar larvae were heat shocked for indicated durations and the total RNA from each randomly selected 15 individuals was extracted as described (M&M). Bars = mean and standard deviation (N = 15). (B) Synchronized Day 1 2nd instar larvae were heat shocked at 37°C for 24 hours and returned to 26°C for the rest of the growth till samples (10 per sample) were taken. (C) Larvae were synchronized on Day 1 2nd instar, heat shocked for 24 hours on Day 1 of 2nd returned to 26°C until Day 1 4th instar. A 2nd heat shock-treatment at 37°C for 24 hours was applied to Day 1 4th instar, and then returned to 26°C for the rest of the growth. Day 1 4th samples (10 larvae/sample) were taken after the second heat shock treatment. Pupae and adult samples were mixed sexes. Mean and standard deviation are shown (N = 3). Heat-shock for 24 hours on Day 1 of 2nd and 4th instar, respectively, led to significant expression knockdown of the target gene throughout the entire 2nd instar to adult development.
(DOC)
Acknowledgments
The authors thank Mr. David H. Dyer for comments on the manuscript.
Funding Statement
This work is supported by the United States National Institutes of Health Research Grant #5R01AI067422 and University of Wisconsin-Madison Graduate School award # MSN123516 to Q.L. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zdobnov EM, von Mering C, Letunic I, Torrents D, Suyama M, et al. (2002) Comparative genome and proteome analysis of Anopheles gambiae and Drosophila melanogaster . Science 298: 149–59. [DOI] [PubMed] [Google Scholar]
- 2. Clayton RB (1964) The utilization of sterols by insects. J Lipid Res 15: 3–19. [PubMed] [Google Scholar]
- 3. Dwivedy AK, Shukla SP (1982) Utilization of cholesterol by the adults of house fly Musca domestica . Entomon 7: 411–22. [Google Scholar]
- 4. Noda H, Wada K, Saito T (1979) Sterols in Laodelphax striatellus with special reference to the intracellular yeast-like symbiotes as a sterol source. J Insect Physiol 25: 443–7. [Google Scholar]
- 5. Ritter KS, Nes WR (1981) The effects of cholesterol on the development of Heliothis zea . J Insect Physiol 27: 175–82. [Google Scholar]
- 6. Nes WD, Lopez M, Zhou W, Guo D, Dowd PF, et al. (1997) Sterol utilization and metabolism by Heliothis zea. . Lipids 32: 1317–23. [DOI] [PubMed] [Google Scholar]
- 7. Krebs KC, Lan Q (2003) Isolation and expression of a sterol carrier protein-2 gene from the yellow fever mosquito, Aedes aegypti . Insect Mol Biol 12: 51–60. [DOI] [PubMed] [Google Scholar]
- 8. Dyer DH, Wessely V, Forest KT, Lan Q (2008) Three-dimensional structure/function analysis of SCP-2-like2 reveals differences among SCP-2 family members. J Lipid Res 49: 644–53. [DOI] [PubMed] [Google Scholar]
- 9. Dyer DH, Lovell S, Thoden JB, Holden HM, Rayment I, et al. (2003) The structural determination of an insect sterol carrier protein-2 with a ligand-bound C16 fatty acid at 1.35-A resolution. J Biol Chem 278: 39085–91. [DOI] [PubMed] [Google Scholar]
- 10. Blitzer EJ, Vyazunova I, Lan Q (2005) Functional analysis of AeSCP-2 using gene expression knockdown in the yellow fever mosquito, Aedes aegypti . Insect Mol Biol 14: 301–7. [DOI] [PubMed] [Google Scholar]
- 11. Peng R, Maklokova VI, Chandrashekhar JH, Lan Q (2011) In vivo functional genomic studies of sterol carrier protein-2 gene in the yellow fever mosquito. PLoS One 6: e18030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langley PA, Ogwal LM, Felton T, Stafford K (1987) Lipid digestion in the tsetse fly, Glossina morsitans . J Insect Physiol 33: 981–6. [Google Scholar]
- 13. Kuthiala A, Ritte KS (1988a) Uptake of cholesterol and cholestanol by the intestine, hemolymph, and fat body of Heliothis zea . Arch Insect Biochem Physiol 7: 225–36. [Google Scholar]
- 14. Komnick H, Giesa U (1994) Intestinal absorption of cholesterol, transport in the haemolymph, and incorporation into the fat body and Malpighian tubules of the larval dragonfly Aeshna cyanea . Comp Physiol 107A: 553–7. [DOI] [PubMed] [Google Scholar]
- 15. Jouni ZE, Zamora J, Wells MA (2002a) Absorption and tissue distribution of cholesterol in Manduca sexta . Arch Insect Biochem Physiol 49: 167–75. [DOI] [PubMed] [Google Scholar]
- 16. Lopez D, Niesen M, Bedi M, Hale D, McLean MP (2007) Activation of the SCPx promoter in mouse adrenocortical Y1 cells. Biochem Biophys Res Commun 357: 549–53. [DOI] [PubMed] [Google Scholar]
- 17. Vyazunova I, Lan Q (2010) Yellow fever mosquito sterol carrier protein-2 gene structure and transcriptional regulation. Insect Mol Biol 19: 205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lan Q, Grier CA (2004) Critical period for pupal commitment in the yellow fever mosquito, Aedes aegypti . J Insect Physiol 50: 667–76. [DOI] [PubMed] [Google Scholar]
- 19. Margam VM, Gelman DB, Palli SR (2006) Ecdysteroid titers and developmental expression of ecdysteroidregulated genes during metamorphosis of the yellow fever mosquito, Aedes aegypti (Diptera:Culicidae). J Insect Physiol 52: 558–68. [DOI] [PubMed] [Google Scholar]
- 20. Telang A, Frame L, Brown MR (2007) Larval feeding duration affects ecdysteroid levels and nutritional reserves regulating pupal commitment in the yellow fever mosquito Aedes aegypti (Diptera: Culicidae). J Exp Biol 210: 854–64. [DOI] [PubMed] [Google Scholar]
- 21. Lan Q, Wessely V (2004) Expression of a sterol carrier protein-x gene in the yellow fever mosquito, Aedes aegypti . Insect Mol Biol 13: 519–29. [DOI] [PubMed] [Google Scholar]
- 22. Ellenberger T, Tomkinson AE (2008) Eukaryotic DNA ligases: structural and functional insights. Annu Rev Biochem 77: 313–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roussigne M, Kossida S, Lavigne AC, Clouaire T, Ecochard V, et al. (2003) The THAP domain: a novel protein motif with similarity to the DNA-binding domain of P element transposase. Trends Biochem Sci 28: 66–9. [DOI] [PubMed] [Google Scholar]
- 24. Okamura T, Shimizu H, Nagao T, Ueda R, Ishii S (2007) ATF-2 regulates fat metabolism in Drosophila . Mol Biol Cell 18: 1519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trotter KW, Archer TK (2007) Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol 265–266: 162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibbs GM, Roelants K, O'Bryan MK (2008) The CAP superfamily: cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–roles in reproduction, cancer, and immune defense. Endocr Rev 29: 865–97. [DOI] [PubMed] [Google Scholar]
- 27. Cléry A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18: 290–8. [DOI] [PubMed] [Google Scholar]
- 28. Cassola A, Noé G, Frasch AC (2010) RNA recognition motifs involved in nuclear import of RNA-binding proteins. RNA Bio 7: 339–44. [DOI] [PubMed] [Google Scholar]
- 29. Radek JT, Dyer DH, Lan Q (2010) Effects of mutations in Aedes aegypti sterol carrier protein-2 on the biological function of the protein. Biochemistry 49: 7532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seong KH, Li D, Shimizu H, Nakamura R, Ishii S (2011) Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell 145: 1049–61. [DOI] [PubMed] [Google Scholar]
- 31. Vlahopoulos SA, Logotheti S, Mikas D, Giarika A, Gorgoulis V, et al. (2008) The role of ATF-2 in oncogenesis. Bioessays 30: 314–27. [DOI] [PubMed] [Google Scholar]
- 32. Isoe J, Kunz S, Manhart C, Wells MA, Miesfeld RL (2007) Regulated expression of microinjected DNA in adult Aedes aegypti mosquitoes. Insect Mol Bio 16: 83–92. [DOI] [PubMed] [Google Scholar]
- 33. Campagne S, Saurel O, Gervais V, Milon A (2010) Structural determinants of specific DNA-recognition by the THAP zinc finger. Nucleic Acids Res 38(10): 3466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sabogal A, Lyubimov AY, Corn JE, Berger JM, Rio DC (2010) THAP proteins target specific DNA sites through bipartite recognition of adjacent major and minor grooves. Nat Struct Mol Biol 17: 117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Macfarlan T, Parker JB, Nagata K, Chakravarti D (2006) Thanatos-associated protein 7 associates with template activating factor-Iβ and inhibits histone acetylation to repress transcription. Mol Endocrinol 20(2): 335–47. [DOI] [PubMed] [Google Scholar]
- 36. Dejosez M, Levine SS, Frampton GM, Whyte WA, Stratton SA, et al. (2010) Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev 24(14): 1479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bhoumik A, Ronai Z (2008) ATF2: a transcription factor that elicits oncogenic or tumor suppressor activities. Cell Cycle 7: 2341–5. [DOI] [PubMed] [Google Scholar]
- 38. Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, et al. (2005) The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci USA 102: 6907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cayrol C, Lacroix C, Mathe C, Ecochard V, Ceribelli M, et al. (2007) The THAP-zinc finger protein THAP1 regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes. Blood 109: 584–94. [DOI] [PubMed] [Google Scholar]
- 40. Zhu CY, Li CY, Li Y, Zhan YQ, Li YH, et al. (2009) Cell growth suppression by thanatos-associated protein 11(THAP11) is mediated by transcriptional downregulation of c-Myc. Cell Death Differ 16: 395–405. [DOI] [PubMed] [Google Scholar]
- 41. Balakrishnan MP, Cilenti L, Mashak Z, Popat P, Alnemri ES, et al. (2009) THAP5 is a human cardiac-specific inhibitor of cell cycle that is cleaved by the proapoptotic Omi/HtrA2 protease during cell death. Am J Physiol Heart Circ Physiol 297: 643–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reddy KC, Villeneuve AM (2004) C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–52. [DOI] [PubMed] [Google Scholar]
- 43. Chesney MA, Kidd AR 3rd, Kimble J (2006) gon-14 functions with class B and class C synthetic multivulva genes to control larval growth in Caenorhabditis elegans . Genetics 172: 915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boxem M, van den Heuvel S (2002) C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol 12: 906–11. [DOI] [PubMed] [Google Scholar]
- 45. Nicholas HR, Lowry JA, Wu T, Crossley M (2008) The Caenorhabditis elegans protein CTBP-1 defines a new group of THAP domain-containing CtBP corepressors. J Mol Biol 375: 1–11. [DOI] [PubMed] [Google Scholar]
- 46. Bhoumik A, Lopez-Bergami P, Ronai Z (2007) ATF2 on the double - activating transcription factor and DNA damage response protein. Pigment Cell Res 20: 498–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ackermann J, Ashton G, Lyons S, James D, Hornung JP, et al. (2011) Loss of ATF2 function leads to cranial motoneuron degeneration during embryonic mouse development. PLoS One 6: e19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sano Y, Akimaru H, Okamura T, Nagao T, Okada M, et al. (2005) Drosophila activating transcription factor-2 is involved in stress response via activation by p38, but not c-Jun NH(2)-terminal kinase. Mol Biol Cell 16: 2934–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Martin SE, Shabanowitz J, Hunt DF, Marto JA (2000) Subfemtomole MS and MS/MS peptide sequence analysis using nano-HPLC micro-ESI fourier transform ion cyclotron resonance mass spectrometry. Anal Chem 72: 4266–74. [DOI] [PubMed] [Google Scholar]
- 50.SAS (1999) User’s guide, Version 8. SAS Institute Inc., Cary, NC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Western Blotting analysis of AeSCP-2 expression in transfected pupae. Lane 1: Protein molecular weight markers; lane 2: siTHAP/SCP-2EGFP co-transfected male pupae; lane 3: siTHAP/SCP-2EGFP co-transfected female pupae; lane 4: siRNA vector/SCP-2EGFP transfected male pupae; lane 5: siRNA vector/SCP-2EGFP transfected female pupae. Total soluble proteins (20 µg/lane) were resolved on 4–20% gradient SDS PAGE gel. Affinity purified rabbit anti-AeSCP-2 antibody (1∶1000 dilution) and horse reddish peroxidase (HRP) conjugated goat anti-rabbit antibody (1∶2000 dilution) were used.
(DOC)
Effects of duration of heat shock on AeSCP-2 siRNA in vivo expression driven by the short Drosophila hsp70 promoter (M&M). Relative AeSCP-2 mRNA levels (vs. rpL8 ) were determined via RT-qPCR. (A) Day 1 F0 4th instar larvae were heat shocked for indicated durations and the total RNA from each randomly selected 15 individuals was extracted as described (M&M). Bars = mean and standard deviation (N = 15). (B) Synchronized Day 1 2nd instar larvae were heat shocked at 37°C for 24 hours and returned to 26°C for the rest of the growth till samples (10 per sample) were taken. (C) Larvae were synchronized on Day 1 2nd instar, heat shocked for 24 hours on Day 1 of 2nd returned to 26°C until Day 1 4th instar. A 2nd heat shock-treatment at 37°C for 24 hours was applied to Day 1 4th instar, and then returned to 26°C for the rest of the growth. Day 1 4th samples (10 larvae/sample) were taken after the second heat shock treatment. Pupae and adult samples were mixed sexes. Mean and standard deviation are shown (N = 3). Heat-shock for 24 hours on Day 1 of 2nd and 4th instar, respectively, led to significant expression knockdown of the target gene throughout the entire 2nd instar to adult development.
(DOC)