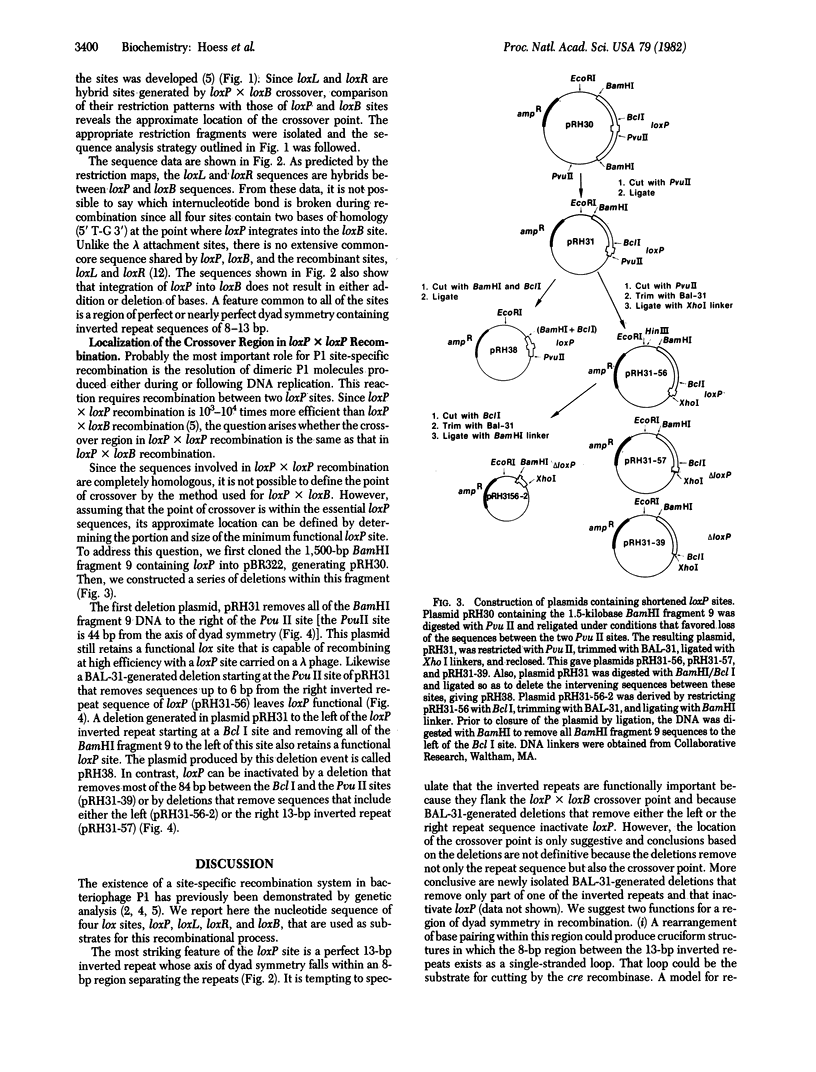
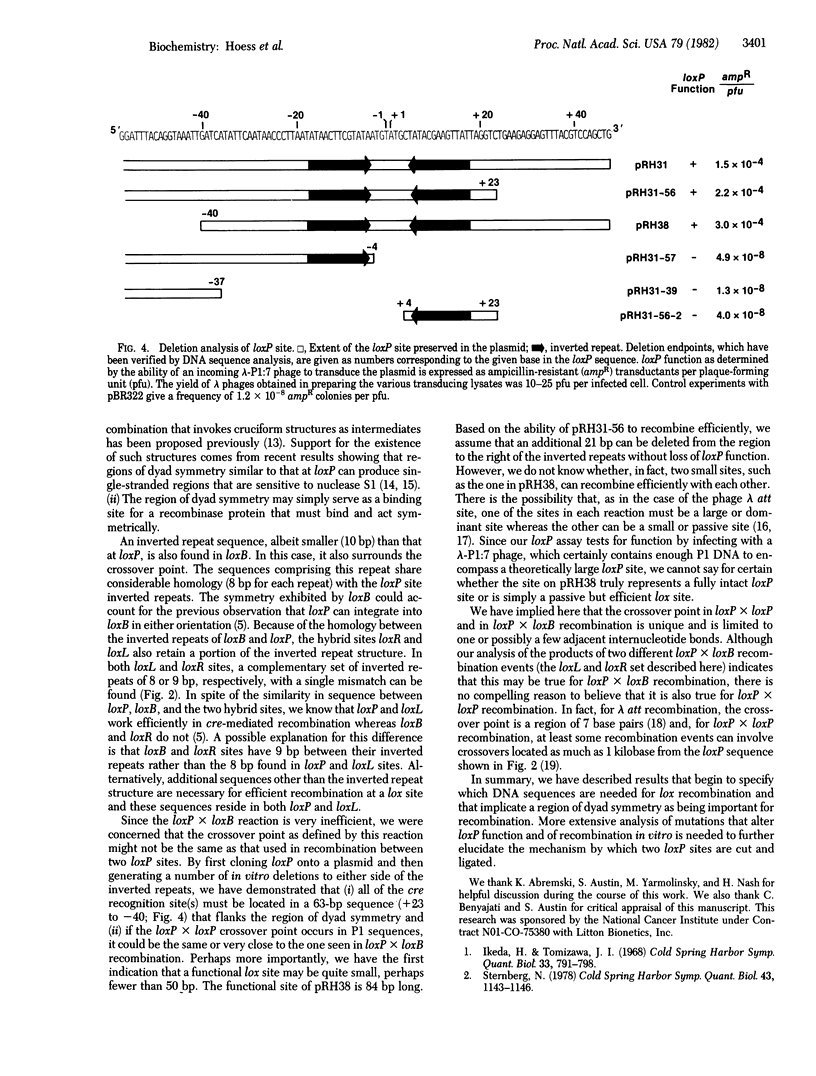
Abstract
Site-specific recombination between molecules of bacteriophage P1 DNA occurs at sites called loxP and requires the action of a protein that is the product of the P1 cre gene. Although recombination between two loxP sites is very efficient, recombination between loxP and a unique site in the bacterial chromosome (loxB) is inefficient and generates two hybrid lox sites called loxR and loxL. We present here the nucleotide sequences of all four lox sites. Analysis of these sequences indicates that (i) a region of extensive homology is not present at the loxP X loxB crossover point, in contrast to the 15-base pair common-core sequence in the bacteriophage lambda att sites, and (ii) the sites contain a region of dyad symmetry with 8- to 13-base pair inverted repeats separated by an 8- to 9-base pair sequence. The loxP X loxB crossover point falls in the sequence that separates the inverted repeats, and deletions that remove either the left or the right inverted repeat of loxP inactivate the site. These two observations are consistent with the conclusion that the region of dyad symmetry is important in los recombination. We have shown further that the loxP X loxP crossover point occurs in a 63-base pair sequence containing the loxP X loxB crossover point, suggesting that, despite the great difference in efficiencies of the two reactions, the crossover points may occur at the same place in both. Explanations for the different recombination properties of the various lox sites are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981 Sep;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Tomizawa J. Prophage P1, and extrachromosomal replication unit. Cold Spring Harb Symp Quant Biol. 1968;33:791–798. doi: 10.1101/sqb.1968.033.01.091. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legerski R. J., Hodnett J. L., Gray H. B., Jr Extracellular nucleases of pseudomonas BAL 31. III. Use of the double-strand deoxyriboexonuclease activity as the basis of a convenient method for the mapping of fragments of DNA produced by cleavage with restriction enzymes. Nucleic Acids Res. 1978 May;5(5):1445–1464. doi: 10.1093/nar/5.5.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M. Integrative recombination of bacteriophage lambda: in vitro study of the intermolecular reaction. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1111–1114. doi: 10.1101/sqb.1979.043.01.123. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Weisberg R., Enquist L., Mizuuchi M., Buraczynska M., Foeller C., Hsu P. L., Ross W., Landy A. Structure and function of the phage lambda att site: size, int-binding sites, and location of the crossover point. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):429–437. doi: 10.1101/sqb.1981.045.01.057. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. Integrative recombination of bacteriophage lambda: extent of the DNA sequence involved in attachment site function. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3220–3224. doi: 10.1073/pnas.77.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Sobell H. M. Molecular mechanism for genetic recombination. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2483–2487. doi: 10.1073/pnas.69.9.2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. Bacteriophage P1 site-specific recombination. III. Strand exchange during recombination at lox sites. J Mol Biol. 1981 Aug 25;150(4):603–608. doi: 10.1016/0022-2836(81)90384-3. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Demonstration and analysis of P1 site-specific recombination using lambda-P1 hybrid phages constructed in vitro. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1143–1146. doi: 10.1101/sqb.1979.043.01.128. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981 Aug 25;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- Sternberg N., Hamilton D., Hoess R. Bacteriophage P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol. 1981 Aug 25;150(4):487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]



