Abstract
Apolipoproteins are able to associate with lipopolysaccharides (LPS), potentially providing protection against septic shock. To gain insight in the molecular details of this binding interaction, apolipophorin III (apoLp-III) from Galleria mellonella was used as a model. The binding of apoLp-III to LPS was optimal around 37–40 °C, close to the LPS phase transition temperature. ApoLp-III formed complexes with LPS from E. coli (serotype O55:B5) with a diameter of 24 nm, a molecular weight of ~390 kDa, containing four molecules of apoLp-III and 24 molecules of LPS. The LPS-bound form of the protein was substantially more resistant to guanidine-induced denaturation compared to unbound protein. The denaturation profile displayed a multiphase character with a steep drop in secondary structure between 0–1 M guanidine, and a slower decrease above 1 M guanidine HCl. In contrast, apoLp-III bound to detoxified LPS was only slightly more resistant to guanidine HCl induced denaturation compared to unbound protein. Analysis of size-exclusion FPLC elution profiles of mixtures of apoLp-III with LPS or detoxified LPS indicated a much weaker binding interaction with detoxified LPS compared to intact LPS. These results indicate that apoLp-III initially interacts with exposed carbohydrate regions, but that the lipid A region is required for a more stable LPS binding interaction.
Exchangeable apolipoproteins have been well documented for their role in lipid transport processes. They exist in a lipid-free form as helix bundle proteins, and associate to lipoprotein surfaces facilitated by the presence of amphipathic α-helices. Bound to lipoproteins, they function to maintain lipoprotein integrity, direct lipoproteins to specific receptors or provide enzymatic activity needed for lipid exchange (1–3). A prototype exchangeable apolipoprotein that shares strong similarities in structure and function with human apolipoproteins is found in the hemolymph of certain insects such as locusts and moths (4). This protein, apolipophorin III (apoLp-III), is predominantly present in a lipid-free form but can quickly associate to lipoprotein surfaces following diacylglycerol loading, thereby providing additional stabilization for the growing lipoprotein particle. This process results in an increased flux of diacylglycerol transport to flight muscles to meet the high metabolic energy demands of insect flight (5–6). ApoLp-III is a relatively small apolipoprotein (18 kDa) and has been well characterized; NMR and X-ray structures of the protein in the lipid-free form show a bundle of five amphipathic α-helices (7–8).
Apolipoproteins are also known to play roles in other processes, which includes a potentially important role in innate immunity (9, 10). Human apolipoproteins such as apoA-I, apoE, and apoC have been shown to bind to lipopolysaccharides (LPS), providing protection against gram-negative sepsis (11–13). In order to investigate the molecular details of the binding interaction of apolipoproteins with LPS, we used apoLp-III as a model apolipoprotein (14). ApoLp-III isolated from the greater wax moth Galleria mellonella has been shown to display immune stimulating and antimicrobial properties (15–17). We have previously demonstrated that this apoLp-III is able to associate with various forms of LPS, including lipid A and the carbohydrate region (18). However, the molecular details of the apoLp-III LPS binding interaction are still not fully understood. Therefore, we have characterized the complex formed between G. mellonella apoLp-III and LPS in detail, and present a model for the binding interaction of apoLp-III with LPS.
Materials and Methods
Recombinant protein expression and purification
To produce recombinant apoLp-III, a plasmid (pET22b+) containing the coding sequence of G. mellonella apoLp-III was transformed to E. coli BL21 cells (19). One colony was used to inoculate 50 mL of LB broth culture containing 55 μg/mL of ampicillin and grown for 16 hours at 37 °C. Half of the overnight culture was then transferred into 500 mL of minimal media (47.75 mM Na2HPO4, 22 mM KH2PO4, 8.56 mM NaCl, 18.7 mM NH4Cl, pH 7.4) containing 50 μg/mL ampicillin. The media was supplemented with 2 mM MgSO4, 0.1 mM CaCl2, and 13.3 mM glucose (final concentrations). The cells were grown at 37 °C in a shaking incubator (300 rpm). When the optical density at 600 nm reached 0.6, protein expression was induced by adding isopropyl-β-D-thiogalactopyranoside (final concentration 2 mM) after which the cells were incubated for four more hours. The cells were removed by centrifugation (5,500 g for 15 minutes at 4 °C). The supernatant was concentrated by tangential flow filtration (10 K membrane, Pall Corp., Ann Arbor, MI) and a Stirred-Cell ultra filtration unit. ApoLp-III was isolated by size-exclusion chromatography (Sephadex G-75, GE Healthcare, Waukesha WI) using phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4). Further purification was achieved by reversed-phase HPLC (Beckman Coulter, Fullerton, CA) using a Zorbax 300 SB-C8 column (Agilent, Santa Clara, CA). A gradient of water and acetonitrile containing 0.05% trifluoroacetic acid was used to elute apoLp-III, which was then collected and lyophilized (Labconco freeze-dryer) for 48 hours. The dried protein was stored at −20 °C until use.
Protein and carbohydrate analysis
The concentration of apoLp-III was determined by the bicinchoninic acid (BCA) assay using BSA as a standard. The concentrations obtained were corrected based on amino acid analysis of G. mellonella apoLp-III (UC Davis Molecular Structure Facility). The 2-keto-3-deoxy-manno-octonic (KDO) assay was used to determine LPS concentrations (20). To 50 μL of KDO standards with concentrations from 0 to 0.5 mM and unknown LPS samples, 60 μl of 0.018 N H2SO4 was added and incubated at 100°C for 20 min. The samples were cooled to room temperature and oxidized by the addition of 25 μL periodic acid (HIO4.2H2O, 9.1 mg/mL in 0.125 N H2SO4) and incubated in the dark for 20 min. The samples were removed from the dark, and 50 μl of NaAsO2 (2.6% in 0.5 N HCl) was added and mixed until the yellow color disappeared. Two hundred fifty μL of 0.3% thiobarbituric acid was added, and the samples were boiled for 10 min. Then 125 μL dimethyl sulfoxide was added while still hot. The samples were cooled to room temperature and the absorbance was measured at 550 nm using a Fluoroskan Multiskan Ascent plate reader (Labsystems). The calculated KDO concentration of the unknown LPS samples were then converted into LPS concentration by assuming two KDO residues in E. coli O55:B5 LPS since KDO residues in the C-4 and C-5 positions cannot be detected using this assay (20).
LPS purification
LPS and detoxified LPS from E. coli serotype O55:B5 and K. pneumoniae were purchased from Sigma Aldrich. LPS was further purified by size-exclusion chromatography using a XK 16/70 column (GE Healthcare, USA) packed with Sepharose CL-6B. LPS (10 mg) was dissolved in 0.2 M NaCl and loaded on the column and eluted with a flow rate of 0.5 mL/min. NaCl (0.2 M) with 0.0005% sodium azide was used for the elution of LPS. Fractions (4 mL) were collected and the absorbance was monitored at 206 nm (21). LPS eluted in the void volume which was confirmed by sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis (PAGE) analysis. Fractions corresponding to the LPS peak were pooled and concentrated by cold ethanol precipitation as follows. The final NaCl concentration of the pooled sample was increased to 0.5 M (from 0.2 M) by adding the appropriate amount of NaCl. The sample was then placed on ice and cold 100% ethanol, chilled for one hour at −20 °C, was added drop by drop to a final volume of 4 x the sample volume. The mixture was incubated at −20 °C for 24 hours followed by centrifugation at 4,300 g using a Sorvall RC 5C Plus centrifuge with a FiberLite F13 rotor for 20 min. The pellet was suspended in distilled water and the concentration of LPS was determined by the KDO assay. Typically, the yield was 6–7 mg of purified LPS.
Cross-linking of apoLp-III/LPS complexes
Dimethyl suberimidate•2 HCl (DMS) is a water soluble cross-linking reagent which reacts with amine groups at alkaline pH. A solution of DMS was prepared by dissolving 20 mg of DMS in 1 mL of 1 M triethanolamine pH 9.8. LPS (500 μg) was incubated in the presence of apoLp-III (150 μg) and 20 μl of DMS (20 mg/mL) for 2 hours at 37 °C. The mixture (100 μl) was fractionated by FPLC with a Superdex 200 10/300 GL Tricorn size-exclusion column; fractions corresponding to the complex were pooled and dialyzed overnight at 4 °C in 25 mM sodium phosphate, pH 7.4. The sample was then lyophilized overnight and dissolved in 10 μl Milli-Q water. This was mixed with 10 μl SDS-sample loading buffer and analyzed by SDS-PAGE using 4–20% Tris-glycine pre-cast gels (Invitrogen, Carlsbad, CA) followed by amido black staining (0.1 % in 45 % methanol, 45% water, 7% glacial acetic acid (v/v/v)).
Circular Dichroism and Fluorescence analysis
ApoLp-III (0.4 mg/ml) was incubated with increasing concentrations of guanidine HCl (from 0 to 6.0 M) in the presence or absence of LPS for 16 hours at 24 °C. LPS was added in excess to ensure that no free apoLp-III was left in the solution. Mass ratios of LPS to apoLp-III of 5:1 were used for E. coli and K. pneumoniae LPS. These ratios were determined by binding saturation experiments performed with non-denaturing PAGE and size exclusion chromatography. ApoLp-III and LPS were incubated for 30 min at 37 °C before the addition of guanidine HCl followed by incubation for 16 hours at 24 °C. The ellipticity was measured at 222 nm using, a Jasco 810 spectropolarimeter and a 1 mm circular quartz cuvette. The midpoint of guanidine-induced denaturation of apoLp-III was calculated by sigmoidal curve fitting using Sigma Plot software (Systat, USA).
For tyrosine fluorescence studies, apoLp-III (20 μg/mL) was incubated in the presence or absence of LPS (100 μg/mL) in PBS for 30 min at 37 °C followed by incubation at 24 °C for 1 hour. Fluorescence was measured by Perkin Elmer LS-55 fluorescence spectrometer (Perkin Elmer, Massachusetts, USA). A quartz fluorescence cuvet with a 10 mm path length and 1 mL volume was used. Samples were excited at 278 nm and the emission spectrum was monitored between 285 and 400 nm with an excitation/emission slit width of 5 nm and scan rate of 50 nm/min. Fluorescence intensity of LPS-free apoLp-III was corrected for PBS. The fluorescence intensity of LPS-bound apoLp-III samples was obtained by subtracting the fluorescence intensity of LPS in PBS. To determine the temperature dependence of LPS binding, samples containing 100 μg/mL LPS and 20 μg/ml apoLp-III were prepared and incubated for 30 min at the following temperatures: 24 °C, 28 °C, 37 °C and 42 °C. The samples were then cooled to room temperature and the fluorescence intensity was measured.
Chromatography
ApoLp-III in the presence or absence of LPS, heparin (sodium salt, porcine intestinal mucosa, Calbiochem, USA), N-acetyl-D-glucosamine, D-(+)-mannose (USB, Cleveland, OH, USA), and D-(+)-galactose (Sigma-Aldrich) was analyzed and or purified using a Superdex 200 10/300 GL Tricorn (1 × 30 cm) size-exclusion column connected to an FPLC instrument (GE healtcare, AKTA). LPS (500 μg) was incubated with apoLp-III at 37 °C for 30 min in PBS in a final volume of 200 μL. Samples were then loaded to the column which was equilibrated with PBS, which was used as the elution buffer with a flow rate of 0.5 mL/min. The absorbance was monitored at 210 and 280 nm. The fractions corresponding to the apoLp-III/LPS peak were collected and pooled. The molecular mass of the complex was calculated based on the elution of reference proteins thyroglobulin (669 k), ferritin (440 k), catalase (232 k), lactate dehydrogenase (140 k) and bovine serum albumin (66 k) (GE healthcare, USA). The ratio of the retention volume of the calibration proteins and the void volume were plotted versus the logarithm of the molecular weight of the proteins, and linear regression was used to calculate the molecular weight of the complexes.
Dynamic light scatter
The hydrodynamic radius of apoLp-III/LPS complexes was determined by dynamic light scattering (DLS), using a Zetasizer Nano ZS Malvern instrument. The samples were prepared in PBS at concentrations of 0.5 mg/mL LPS and 0.5 mg/mL apoLp-III. The solutions were filtered through a 0.22 μm Millipore Millex sterile syringe filters prior to the illumination of the sample in a quartz cuvette of 45 μL volume (Hellma, Germany). The viscosity of PBS (0.8882 cP) used for the calculations was provided by the instrument software. Each sample was measured three times to obtain the average radius of the particles.
Electron microscopy
LPS samples in the presence or absence of apoLp-III were visualized by JEOL 1200EX-II transmission electron microscope (TEM) using Formvar/carbon-coated 300-mesh copper grids (Electron Microscopy Sciences) which were subjected to glow discharge before sample adsorption. LPS samples (0.5 mg/mL) were prepared and 10 μL of each sample was applied to the sample grids. The grids were allowed to dry for 2 min followed by negative staining by 10 μL of a 2% solution of phosphotungstic acid in water, pH 7.4, for 1 min. The grids were allowed to dry for another 2 min and visualized at a voltage of 80 kV.
Results
Temperature dependence of LPS binding
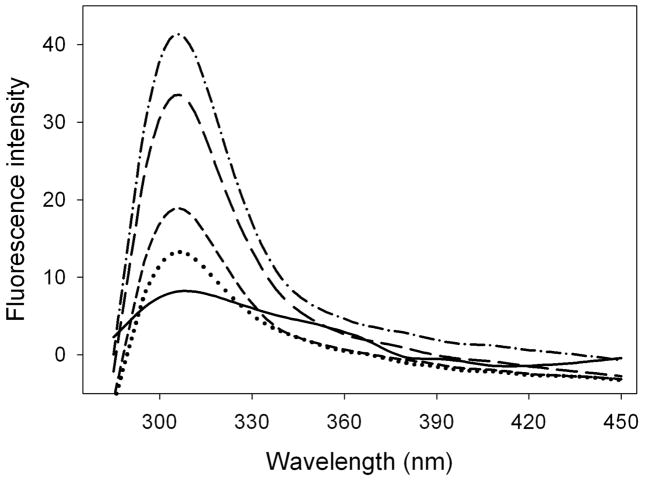
ApoLp-III from G. mellonella lacks tryptophan but contains a single tyrosine residue positioned in the 5th helix (residue 142), and the intrinsic fluorescence properties of this residue can be exploited to measure changes in the local environment. Tyr-142 is in a quenched state in the unbound protein, while binding to LPS results in a significant increase in the fluorescence emission intensity and thus can be used to monitor the LPS binding interaction (18). To determine the temperature optimum for LPS binding, apoLp-III (20 μg/mL) was incubated in the presence of 100 μg/mL E. coli LPS (serotype O55:B5) at 22, 28, 37 and 42 °C. The fluorescence emission intensity of the lone Tyr residue, with a maximum emission wavelength of 304 nm, increased substantially with higher temperatures (Figure 1). An incubation temperature of 37 or 42 °C elicited the strongest fluorescence emission intensity of the apoLp-III LPS mixtures. The gel-to-liquid phase transition temperature for enterobacterial LPS is in the range of 30 to 37 °C (22), thus the optimum binding around 37 °C may suggest that the binding interaction of apoLp-III to LPS is dependent on the LPS phase transition. Therefore, all subsequent LPS binding studies were carried out at 37 °C.
Figure 1.
Temperature dependence of LPS binding. ApoLp-III binding to LPS was measured by the increase in tyrosine fluorescence emission at 308 nm. Solid line: apoLp-III tyrosine fluorescence in the absence of LPS. ApoLp-III was incubated in the presence of LPS at the temperatures indicated, and the spectra were recorded at 22 °C (dotted line), 28°C (short dashed line), 37 °C (long dashed line), and 42 °C (dashed-dotted line).
Characterization of apoLp-III/LPS complexes
Previous studies have indicated the formation of a complex between LPS and apoLp-III upon mixing (18, 23). To understand the binding interaction in more detail, the complex formed between LPS and apoLp-III was characterized. LPS/apoLp-III complexes were isolated by size exclusion FPLC, analyzed for size, apoLp-III and LPS content, and the stoichiometry was subsequently determined. FPLC analysis showed that incubation of apoLp-III with LPS increased the elution volume of LPS, which is present as large aggregates at the concentrations used, and now contained significant amounts of apoLp-III. This indicated that when apoLp-III associated with LPS, the overall size of the LPS aggregates decreased, which is in good agreement with previous analysis based on non-denaturing PAGE (18). Based on the elution profile of a set of reference proteins ranging from 66 to 669 kDa, the elution peak of the newly formed apoLp-III/LPS complexes corresponded to a molecular mass of approximately 390 kDa (not shown). The size of the apoLp-III/LPS complex was then measured by dynamic light scattering. Figure 2 shows the DLS profile, indicating that the diameter of the LPS aggregates was reduced from 52.6 to 18.7 nm upon incubation with apoLp-III. Thus both DLS and FPLC analysis of the apoLp-III/LPS complexes indicate that the large LPS aggregates were transformed into smaller apoLp-III/LPS complexes. This result was confirmed by electron microscopy. LPS appeared as ribbon-like structures on TEM micrographs (Figure 3A). This image resembles a typical structure observed for smooth LPS of gram-negative bacteria (24, 25). Figure 3B shows the TEM image of LPS incubated in the presence of apoLp-III. The ribbon-like structure of LPS disappeared; instead small spheres were visible in the apoLp-III/LPS mixtures indicating that the LPS micelles were disaggregated.
Figure 2.
DLS analysis of LPS in the presence or absence of apoLp-III. The size distribution of LPS is shown as a solid line which peaked with a size of 52.6 nm, while after incubation with apoLp-III the size decreased to 18.7 nm (dashed line).
Figure 3.
TEM image of LPS of E. coli O55:B5. Panel A: LPS alone; panel B: LPS after addition of apoLp-III. The bar shown at the bottom of panel A represents 100 nm.
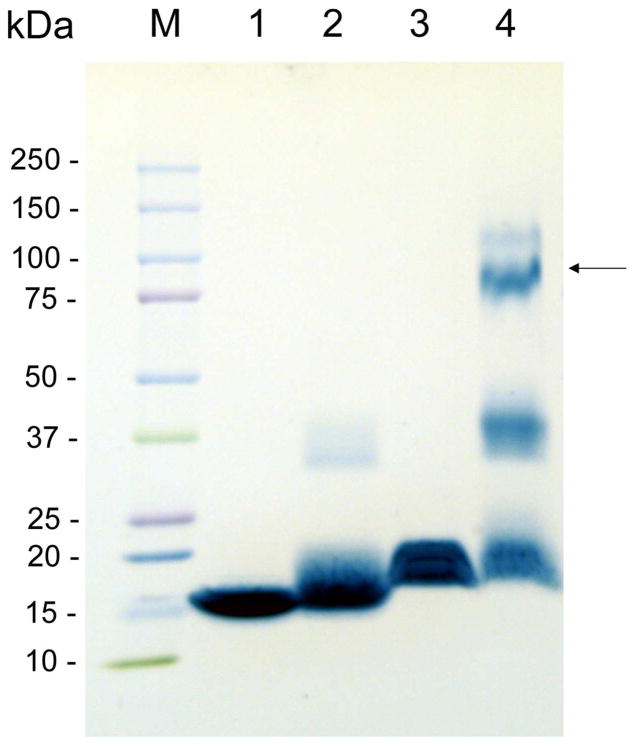
The FPLC purified apoLp-III/LPS complex was analyzed for their apoLp-III and LPS content with protein and KDO assays, respectively. This analysis provided a molar ratio of apoLp-III to LPS of 1: 6.15 ± 0.30 (n=5). The large size of the complexes indicated the presence of multiple copies of apoLp-III. To determine the number of apoLp-III molecules, the complex was treated with DMS to cross-link closely spaced apoLp-IIIs. The DMS-treated complexes were isolated by size exclusion FPLC and the peak fractions were subjected to SDS-PAGE analysis (Figure 4). This showed that apoLp-III, upon LPS association, was primarily present as a tetramer (apparent molecular weight of 87 kDa). Monomers and dimers were also observed, which could be due to incomplete cross-linking. This result implies that the majority of the complexes contained four molecules of apoLp-III, and 24 molecules of LPS based on the 1:6 molar ratio. Assuming an average size of 15 kDa for LPS and 18 kDa for apoLp-III, this translated into an overall molecular mass of 432 kDa for the apoLp-III/LPS complex, which closely resemblances the molecular mass based on the FPLC elution profile.
Figure 4.
SDS-PAGE of apoLp-III cross-linking. Lane 1: apoLp-III; lane 2: apoLp-III + DMS; lane 3: apoLp-III/LPS complex; lane 4: apoLp-III/LPS complex + DMS. Marker proteins (M) and there sizes are shown on the left; the arrow indicates the position of the main cross-linked band in lane 4 with a size of 87 kDa.
Guanidine-HCl denaturation
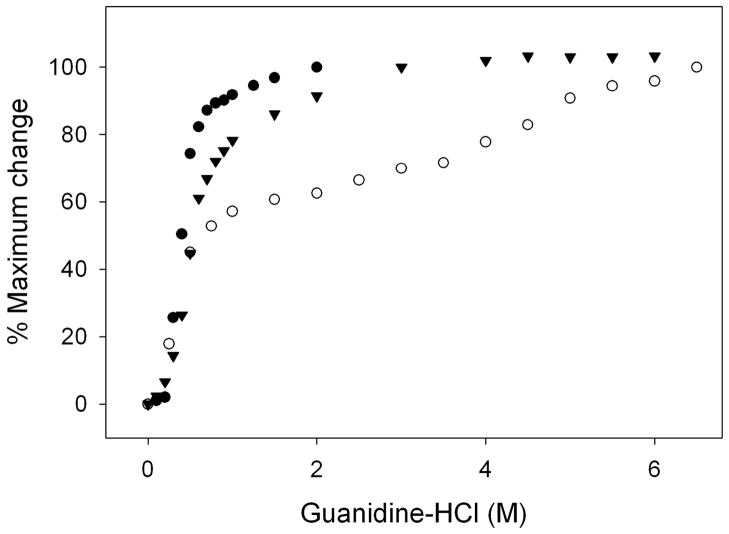
The stability of apoLp-III in complex with LPS was determined by measuring the resistance to guanidine-HCl induced denaturation. Figure 5 shows that the molar concentration of guanidine HCl at which the ellipticity at 222 nm was decreased by 50 % (midpoint of guanidine-HCl denaturation, [Gdn-HCl]1/2) was 0.40 M, similar to previously reported values for this apoLp-III (26). The denaturation profile of LPS-bound apoLp-III displayed a multiphase behavior. A steep drop in ellipticity at 222 nm was observed between 0 and 1 M guanidine-HCl, which was followed by a more gradual decrease between 1 and 7 M guanidine-HCl. The second part of the denaturation profile looks similar to the denaturation profile of apoLp-III in the lipid-bound state in which the protein was associated with dimyristoylphosphatidylcholine in the form of discoidal protein-lipid complexes (26–28). Since carbohydrates constitute a large part of LPS, the importance of LPS carbohydrates for the binding interaction was analyzed using detoxified LPS (D-LPS), which is composed of the inner and outer core carbohydrates as well as the O-antigen carbohydrate repeats, but lacks the lipid A portion (29). Previous analysis employing tyrosine fluorescence and by non-denaturing PAGE showed that apoLp-III is able to interact with D-LPS (18). The denaturation profile of apoLp-III in the presence of D-LPS looked similar to unbound protein, except that the presence of D-LPS caused a modest increase in the [Gdn-HCl]1/2 of apoLp-III to 0.56 M. Therefore, the steep decrease in negative ellipticity at low denaturant concentrations in the apoLp-III/LPS complexes may be attributed to weaker apolipoprotein-carbohydrate interactions. The more gradual decrease in ellipticity and increased resistance to denaturation may result from the interaction of apoLp-III with the lipid A portion of LPS.
Figure 5.
Gdn-HCl denaturation profile of apoLp-III in the presence or absence of LPS and D-LPS. The denaturation of apoLp-III was followed by measuring the ellipticity at 222 nm. Shown are apoLp-III alone (closed circles), apoLp-III + D-LPS (closed triangles), and apoLp-III + LPS (closed circles).
FPLC analysis of apoLp-III and LPS in diluted mixtures
The binding interaction of apoLp-III to the LPS carbohydrates was further analyzed by comparing the size-exclusion FPLC elution profile at various concentrations. At a concentration of 0.05 mg/mL apoLp-III and 0.25 mg mL D-LPS, the apoLp-III peak in the absence of D-LPS eluted at ~18 mL and overlapped completely with apoLp-III incubated with D-LPS, indicating the absence of a measurable binding interaction (Figure 6, panel A). However, when the concentration of both apoLp-III and D-LPS was increased, while maintaining the 1:5 mass ratio, the protein peak started to shift to lower elution volumes (panels A–E). At a 10 fold higher concentration of 0.5 mg/ml apoLp-III and 2.5 mg/mL D-LPS, apoLp-III eluted at ~14 mL (panel E). This result indicated that the equilibrium of apoLp-III shifted from the unbound state towards a D-LPS bound state in the more concentrated mixtures. A control experiment was carried out if the change in elution observed for D-LPS was a result of non-specific binding. In this case, apoLp-III (0.5 mg/mL) was incubated with a mixture of D-mannose, D-galactose, and N-acetyl-D-glucosamine (2.5 mg/mL). The monosaccharides had no measurable effect on the elution profile of apoLp-III. In addition, heparin, which is known to bind apoE, also had no effect on the elution profile (Panel F). This indicates a lack of binding interaction of heparin or the monosaccharides with apoLp-III. This result suggests that the carbohydrates in D-LPS specifically interact with apoLp-III, and is not a result of non-specific binding.
Figure 6.
FPLC of Superdex-200 elution profiles of apoLp-III incubations with LPS. Panel A–H: apoLp-III incubated with D-LPS (at a constant protein-LPS mass ratio of 1:5) at the following protein concentrations: 0.05 mg/mL (A), 0.1 mg/mL (B), 0.2 mg/mL (C), 0.3 mg/mL (D), and 0.5 mg/mL (E). Solid line: apoLp-III only; dotted line: LPS only, dashed line: apoLp-III in the presence of LPS. Panel F: apoLp-III (0.5 mg/mL) in the presence of heparin (2.5 mg/mL). Panels G and H: apoLp-III in the presence of LPS at a 1:5 mass ratio using a protein concentration of 0.05 mg/mL (G), and 0.5 mg/mL (H).
In contrast to D-LPS, the elution of the apoLp-III/LPS complex was not affected by the concentration of LPS and protein. The complex present in the mixtures eluted at 10 mL at all concentrations tested, shown are the incubations with the lowest concentration (panel G: 0.05 mg/mL protein and 0.25 mg/mL LPS) and highest concentration (panel H: 0.5 mg/mL protein and 2.5 mg/mL LPS). This result indicates that while apoLp-III is able to associate with D-LPS, this binding interaction is significantly weaker compared to LPS with the lipid A region present.
Klebsiella pneumonia LPS
A recent study showed that apoLp-III was able to inhibit growth of cultures of K. pneumoniae, while it was less potent to inhibit the growth of E. coli cultures (30). Therefore, binding of apoLp-III to LPS of a known pathogen, K. pneumoniae was also characterized. Another recent study showed significant changes to the membrane surface of this pathogen after incubation with apoLp-III from G. mellonella (16). Similar to E. coli, apoLp-III was able to disaggregate K. pneumoniae LPS (140 nm) by forming small protein-LPS complexes with a diameter of 16.3 nm (Figure S1, Supporting information). Calculations based on the elution profile of size-exclusion FPLC provided a molecular mass of 200 kDa for the complexes. Based on the protein and LPS content in the complexes and cross-linking analysis (Figure S2, Supporting information), the complexes contained three apoLp-III and nine LPS molecules. Similar to E. coli LPS, apoLp-III showed an increased resistance to guanidine-HCl induced denaturation and displayed a multiphase profile when associated with LPS (Figure S3, Supporting information). FPLC analysis of retention profiles of various amounts of LPS showed that the retention volume of LPS/apoLp-III complexes was not dependent on the concentration, similar to the results obtained for E. coli LPS (Figure S4, Supporting information).
Discussion
In the present study we demonstrated that apoLp-III is able to bind to LPS aggregates, leading to the formation of smaller LPS apoLp-III complexes. This binding interaction was dependent on the temperature, being most strongly at the LPS phase transition temperature. DLS measurements showed that the size of the E. coli LPS aggregates decreased from 53 to 19 nm upon incubation with apoLp-III. TEM analysis of LPS showed the presence of ribbon-like structures, which disappeared after addition of apoLp-III, and instead small spheres with a radius of 10–15 nm were found. While this size is somewhat different from the DLS analysis, both experiments indicate that apoLp-III is able to break up the large LPS aggregates. This is also in agreement with previous data in which the formation of apoLp-III/LPS complexes was confirmed by the ability of apoLp-III to increase the electrophoretic mobility of LPS (18). The disaggregation of LPS may be a key-step in neutralizing LPS toxicity. Aggregates of E. coli LPS form at a concentration of >10 μg/mL in PBS, and similar values have been reported for LPS from other species (31–32). It was shown that LPS aggregates were able to activate mononuclear cells and induce toxicity whereas monomeric LPS did not, indicating that LPS aggregates are the biologically active form (33). LPS disaggregation has been observed for hemoglobin (34–35), apoA-II (36), lysozyme (37), high density lipoproteins (38), lactoferrin (39) and polymyxin B (25). These proteins were reported to inhibit the cell stimulating activity of LPS, suggesting that LPS is ineffective to induce toxicity upon disaggregation.
The E. coli LPS/apoLp-III complexes characterized in the present study contained 4 molecules of apoLp-III and 24 molecules of LPS. The complex size, determined by the protein and carbohydrate content and the degree of apoLp-III cross-linking, produced a size of 432 kDa, which was close to the value obtained by size-exclusion FPLC (390 kDa). The complexes formed with K. pneumoniae LPS were significantly smaller. Analysis by FPLC provided insight into the relative strength of the binding of apoLp-III to LPS and D-LPS. Since dilution of apoLp-III and LPS resulted in increased elution volumes of the complex, this indicates that the binding equilibrium changed in favor of the protein in the unbound state. This concentration dependent distribution was not observed for incubations of apoLp-III with complete LPS, and suggests that binding of apoLp-III was stronger compared to D-LPS. Since the lipid A portion is absent in D-LPS, this result suggests that lipid A is a major contributor to the apoLp-III binding interaction. We have previously shown the binding of apoLp-III to lipid A and also carbohydrate-truncated variants of LPS (18). While binding of apoLp-III to the carbohydrate portion of LPS may be weaker compared to complete LPS, the carbohydrate binding interaction was evidently demonstrated. Addition of D-LPS to apoLp-III resulted in a significant increase in tyrosine fluorescence intensity, and changes the electrophoretic mobility of apoLp-III (18). In addition, the current study showed that the FPLC elution volume of D-LPS in the presence of apoLp-III increased, albeit dependent on the concentration, while no changes in elution profiles were observed with heparin, D-mannose, D-galactose, and N-acetyl-D-glucosamine.
The stability analysis of LPS/apoLp-III complexes also provided evidence for a weaker carbohydrate interaction. The amount of guanidine-HCl required to denature LPS/apoLp-III complexes was substantially higher compared to unbound apoLp-III. The denaturation plot shows a 1st phase in which the protein denatures at a low denaturant concentration. The second phase, in which substantially more denaturant is required to unfold the protein, resembles the denaturation profile of lipid-bound apoLp-III (26–28). In contrast, detoxified LPS/apoLp-III complexes unfolded similar to the free protein, with only a small increase in the amount of guanidine-HCl needed to reach the midpoint of denaturation. Thus, the initial unfolding observed in LPS/apoLp-III complexes may be explained by the unfolding of apoLp-III bound to the LPS carbohydrates, while for the 2nd phase much more denaturant was required to unfold apoLp-III bound to lipid A.
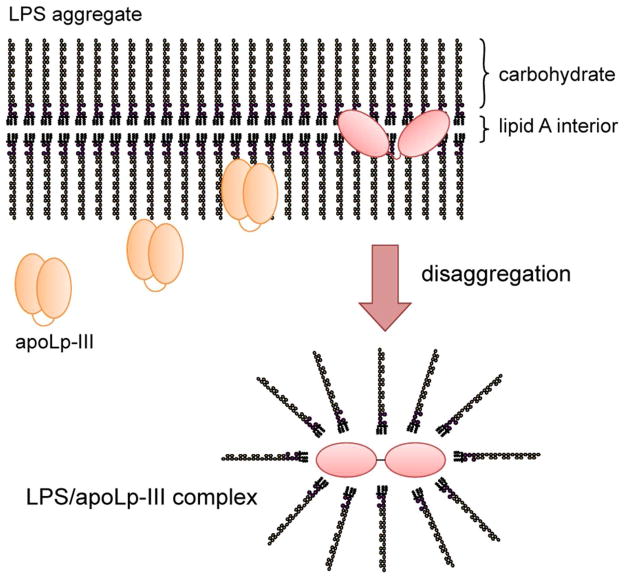
Based on these results the following model is proposed to describe the binding of apoLp-III to LPS (Figure 7). Our data suggest a mode of binding interaction in which the carbohydrates on the surface of the aggregates and facing the aqueous environment may trigger an initial moderately weak binding. Secondly, this is followed by a considerably stronger binding interaction in which apoLp-III makes direct contact with the lipid A portion of LPS. For this interaction to occur, the protein needs to gain access to lipid A and thus traverse the carbohydrate O-antigen repeat and core oligosaccharide regions. Binding is facilitated by a temperature close to the LPS phase transition, allowing access to the hydrophobic interior of LPS. Previous data have shown that apoLp-III needs to rearrange its helices upon binding to LPS, similar to binding to lipid surfaces upon binding to lipoprotein complexes (40). Thus it is conceivable that the second step requires helix bundle opening, resulting in a more stable LPS/apoLp-III complex. While ionic interactions may be important for the initial carbohydrate binding, the second step is likely driven by hydrophobic interactions. The non-polar face of the amphipathic α-helix may come in direct contact with the lipid A surface, similar as observed for the binding of exchangeable apolipoproteins to lipid or lipoprotein surfaces (41). This is further supported by previous data which have shown that apoLp-III variants lacking any two terminal helices, either on the N- or C-terminus, retained the ability to associate with LPS (26). Thus it is conceivable that the amphipathic nature of the α-helices is the critical structural feature required for LPS binding.
Figure 7.
Proposed model of the binding of apoLp-III to LPS. ApoLp-III first associates superficially to the LPS carbohydrates which protrude into the aqueous environment. Penetration into the interior of the LPS micelles provide access to the hydrophobic Lipid A region, which is then followed by LPS disaggregation. A conformational change of the protein allows direct interaction of the hydrophobic protein interior with the lipid A region, leading to the formation of a stable apoLp-III/LPS complex. The resulting product is an assembly of 4 apoLp-III and 24 LPS molecules with a size of ~ 400 kDa.
This proposed mode of action resembles the mechanism proposed for the binding of a number of antimicrobial peptides, which are potential therapeutic drugs for LPS mediated infections. Antimicrobial peptides disaggregate LPS micelles into smaller particles similar to apoLp-III (25). In addition, it has been suggested that the peptides initially bind to the carbohydrates of LPS predominantly through ionic interactions, and then traverse into the lipid core of LPS leading to disaggregation (25, 42–43). Some bacterial strains displayed resistance to some of the peptides depending on the variable O-antigen repeats (42). It has been suggested that the O-antigen acts as a barrier preventing the passage of antimicrobial peptides to lipid A. The antimicrobial peptides are amphipathic in nature and rich in lysine and arginine residues, similar to G. mellonella apoLp-III (43). This supports the view that the amphipathic helix is the key feature of LPS binding of apoLp-III, and probably also other vertebrate apolipoproteins such as apoE and apoA-I.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health, award number SC3GM089564 from the National Institute of General Medical Sciences.
Abbreviations
- apoLp-III
apolipophorin III
- D-LPS
detoxified lipopolysaccharides
- DMS
dimethyl suberimidate
- DLS
dynamic light scattering
- KDO
2-keto-3-deoxy-manno-octonic
- LPS
lipopolysaccharides
- PAGE
polyacrylamide gel electrophoresis
- [Gdn-HCl]1/2
midpoint of guanidine-HCl denaturation
- SDS
sodium dodecyl sulfate
- TEM
transmission electron microscope
Footnotes
LPS binding analysis of K. pneumonia with apoLp-III by DLS, cross-linking SDS-PAGE, Gdn-HCl denaturation, and size-exclusion FPLC elution profile. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Narayanaswami V, Ryan RO. Molecular basis of exchangeable apolipoprotein function. Biochim Biophys Acta. 2000;1483:15–36. doi: 10.1016/s1388-1981(99)00176-6. [DOI] [PubMed] [Google Scholar]
- 2.Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Narayanaswami V, Kiss RS, Weers PMM. The helix bundle: a reversible lipid binding motif. Comp Biochem Physiol A Mol Integr Physiol. 2010;155:123–133. doi: 10.1016/j.cbpa.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van der Horst DJ, Van Hoof D, Van Marrewijk WJA, Rodenburg KW. Alternative lipid mobilization: the insect shuttle system. Mol Cell Biochem. 2002;239:113–119. [PubMed] [Google Scholar]
- 6.Van der Horst DJ, Rodenburg KW. Locust flight activity as a model for hormonal regulation of lipid mobilization and transport. J Insect Physiol. 2010;56:844–853. doi: 10.1016/j.jinsphys.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Breiter DR, Kanost MR, Benning MM, Wesenberg G, Law JH, Wells MA, Rayment I, Holden HM. Molecular structure of an apolipoprotein determined at 2.5-Å resolution. Biochemistry. 1991;30:603–608. doi: 10.1021/bi00217a002. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Sykes BD, Ryan RO. Structural basis for the conformational adaptability of apolipophorin III, a helix bundle exchangeable apolipoprotein. Proc Natl Acad Sci. 2002;99:1188–1193. doi: 10.1073/pnas.032565999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Amersfoort ES, Van Berkel TJC, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16:379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berbee JF, Havekes LM, Rensen PC. Apolipoproteins modulate the inflammatory response to lipopolysaccharide. J Endotoxin Res. 2005;11:97–103. doi: 10.1179/096805105X35215. [DOI] [PubMed] [Google Scholar]
- 11.Van Oosten M, Rensen PCN, Van Amersfoort ES, Van Eck M, Van Dam A-M, Breve JJP, Vogel T, Panet A, Van Berkel TJC. Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat gram-negative sepsis. J Biol Chem. 2001;276:8820–8824. doi: 10.1074/jbc.M009915200. [DOI] [PubMed] [Google Scholar]
- 12.Berbee JF, van der Hoogt CC, Kleemann R, Schippers EF, Kitchens RL, Van Dissel JT, Bakker-Woudenberg IA, Havekes LM, Rensen PC. Apolipoprotein CI stimulates the response to lipopolysaccharide and reduces mortality in Gram-negative sepsis. FASEB J. 2006;20:2162–2164. doi: 10.1096/fj.05-5639fje. [DOI] [PubMed] [Google Scholar]
- 13.Biedzka-Sarek M, Metso J, Kateifides A, Meri T, Jokiranta TS, Muszyński A, Radziejewska-Lebrecht J, Zannis V, Skurnik M, Jauhiainen M. Apolipoprotein A-I exerts bactericidal activity against Yersinia enterocolitica serotype O:3. J Biol Chem. 2011;286:38211–38219. doi: 10.1074/jbc.M111.249482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weers PMM, Ryan RO. Apolipophorin III: role model apolipoprotein. Insect Biochem Mol Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Wiesner A, Losen S, Kopacek P, Weise C, Gotz P. Isolated apolipophorin III from Galleria mellonella stimulates the immune reactions of this insect. J Insect Physiol. 1997;43:383–391. doi: 10.1016/s0022-1910(96)00113-8. [DOI] [PubMed] [Google Scholar]
- 16.Zdybicka-Barabas A, Januszanis B, Mak P, Cytryńska M. An atomic force microscopy study of Galleria mellonella apolipophorin III effect on bacteria. Biochim Biophys Acta. 2011;1808:1896–906. doi: 10.1016/j.bbamem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Halwani AE, Niven DF, Dunphy GB. Apolipophorin-III and the interactions of lipoteichoic acids with the immediate immune responses of Galleria mellonella. J Invertebr Pathol. 2000;76:233–241. doi: 10.1006/jipa.2000.4978. [DOI] [PubMed] [Google Scholar]
- 18.Leon LJ, Pratt CC, Vasquez LJ, Weers PMM. Tyrosine fluorescence analysis of apolipophorin III-lipopolysaccharide interaction. Arch Biochem Biophys. 2006;452:38–45. doi: 10.1016/j.abb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Niere M, Meisslitzer C, Dettloff M, Weise C, Ziegler M, Wiesner A. Insect immune activation by recombinant Galleria mellonella apolipophorin III. Biochim Biophys Acta. 1999;1433:16–26. doi: 10.1016/s0167-4838(99)00148-x. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, Tsai CM. Quantification of bacterial lipopolysaccharides by the purpald assay: measuring formaldehyde generated from 2-keto-3-deoxyoctonate and heptose at the inner core by periodate oxidation. Anal Biochem. 1999;267:161–168. doi: 10.1006/abio.1998.2961. [DOI] [PubMed] [Google Scholar]
- 21.Rolando P, Vivian M. Purification of E. coli O55:B5 lipopolysaccharides by size exclusion chromatography. Biotecnologia Aplicada. 2006;23:124–129. [Google Scholar]
- 22.Brandenburg K, Seydel U. Investigation into the fluidity of lipopolysaccharide and free lipid A membrane systems by Fourier-transform infrared spectroscopy and differential scanning calorimetry. Eur J Biochem. 1990;191:229–236. doi: 10.1111/j.1432-1033.1990.tb19114.x. [DOI] [PubMed] [Google Scholar]
- 23.Pratt CC, Weers PMM. Lipopolysaccharide binding of an exchangeable apolipoprotein, apolipophorin III, from Galleria mellonella. Biol Chem. 2004;385:1113–1119. doi: 10.1515/BC.2004.145. [DOI] [PubMed] [Google Scholar]
- 24.Shands JW, Jr, Graham JA, Nath K. The morphologic structure of isolated bacterial lipopolysaccharide. J Mol Biol. 1967;25:15–21. doi: 10.1016/0022-2836(67)90275-6. [DOI] [PubMed] [Google Scholar]
- 25.Lopes J, Inniss WE. Electron microscopy of effect of polymyxin on Escherichia coli lipopolysaccharide. J Bacteriol. 1969;100:1128–1130. doi: 10.1128/jb.100.2.1128-1130.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dettloff M, Niere M, Ryan RO, Luty R, Kay CM, Wiesner A, Weers PMM. Differential lipid binding of truncation mutants of Galleria mellonella apolipophorin III. Biochemistry. 2002;41:9688–9695. doi: 10.1021/bi0200108. [DOI] [PubMed] [Google Scholar]
- 27.Wientzek M, Kay CM, Oikawa K, Ryan RO. Binding of insect apolipophorin III to dimyristoylphosphatidylcholine vesicles. Evidence for a conformational change. J Biol Chem. 1994;269:4605–4612. [PubMed] [Google Scholar]
- 28.Weers PMM, Kay CM, Oikawa K, Wientzek M, Van der Horst DJ, Ryan RO. Factors affecting the stability and conformation of Locusta migratoria apolipophorin III. Biochemistry. 1994;33:3617–3624. doi: 10.1021/bi00178a019. [DOI] [PubMed] [Google Scholar]
- 29.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zdybicka-Barabas A, Cytryńska M. Involvement of apolipophorin III in antibacterial defense of Galleria mellonella larvae. Comp Biochem Physiol B. 2011;158:90–98. doi: 10.1016/j.cbpb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Bergstrand A, Svanberg C, Langton M, Nyden M. Aggregation behavior and size of lipopolysaccharide from Escherichia coli O55:B5. Colloids Surf B Biointerfaces. 2006;53:9–14. doi: 10.1016/j.colsurfb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Aurell CA, Wistrom AO. Critical aggregation concentrations of gram-negative bacterial lipopolysaccharides (LPS) Biochem Biophys Res Commun. 1998;253:119–123. doi: 10.1006/bbrc.1998.9773. [DOI] [PubMed] [Google Scholar]
- 33.Mueller M, Lindner B, Kusumoto S, Fukase K, Schromm AB, Seydel U. Aggregates are the biologically active units of endotoxins. J Biol Chem. 2004;279:26307–26313. doi: 10.1074/jbc.M401231200. [DOI] [PubMed] [Google Scholar]
- 34.Brandenburg K, Garidel P, Andra J, Jurgens G, Muller M, Blume A, Koch MH, Levin J. Crosslinked hemoglobin converts endotoxically inactive pentaacyl endotoxins into a physiologically active conformation. J Biol Chem. 2003;278:47660–47669. doi: 10.1074/jbc.M304743200. [DOI] [PubMed] [Google Scholar]
- 35.Kaca W, Roth RI, Ziolkowski A, Levin J. Human hemoglobin increases the biological activity of bacterial lipopolysaccharides in activation of Limulus amebocyte lysate and stimulation of tissue factor production by endothelial cells in vitro. J Endotoxin Res. 1994;1:243–252. [Google Scholar]
- 36.Thompson PA, Berbee JF, Rensen PC, Kitchens RL. Apolipoprotein A-II augments monocyte responses to LPS by suppressing the inhibitory activity of LPS-binding protein. Innate Immun. 2008;14:365–374. doi: 10.1177/1753425908099171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brandenburg K, Koch MH, Seydel U. Biophysical characterization of lysozyme binding to LPS Re and lipid A. Eur J Biochem. 1998;258:686–695. doi: 10.1046/j.1432-1327.1998.2580686.x. [DOI] [PubMed] [Google Scholar]
- 38.Brandenburg K, Jurgens G, Andra J, Lindner B, Koch MH, Blume A, Garidel P. Biophysical characterization of the interaction of high-density lipoprotein (HDL) with endotoxins. Eur J Biochem. 2002;269:5972–5981. doi: 10.1046/j.1432-1033.2002.03333.x. [DOI] [PubMed] [Google Scholar]
- 39.Brandenburg K, Jurgens G, Muller M, Fukuoka S, Koch MH. Biophysical characterization of lipopolysaccharide and lipid A inactivation by lactoferrin. Biol Chem. 2001;382:15–25. doi: 10.1515/BC.2001.152. [DOI] [PubMed] [Google Scholar]
- 40.Leon LJ, Idangodage H, Wan CP, Weers PMM. Apolipophorin III: Lipopolysaccharide binding requires helix bundle opening. Biochem Biophys Res Commun. 2006;348:1328–1333. doi: 10.1016/j.bbrc.2006.07.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segrest JP, Jones MK, De Loof H, Brouillette CG, Venkatachalapathi YV, Anantharamaiah GM. The amphipathic helix in the exchangeable apolipoproteins: a review of secondary structure and function. J Lipid Res. 1992;33:141–166. [PubMed] [Google Scholar]
- 42.Rosenfeld Y, Shai Y. Lipopolysaccharide (Endotoxin) - host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochim Biophys Acta. 2006;1758:1513–1522. doi: 10.1016/j.bbamem.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 43.Tossi A, Sandri L, Giangaspero A. Amphipathic, α-helical, antimicrobial peptides. Biopolymers. 2000;55:4–30. doi: 10.1002/1097-0282(2000)55:1<4::AID-BIP30>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.