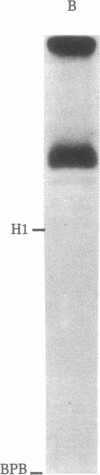
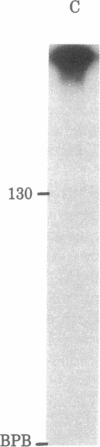
Abstract
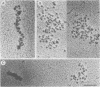
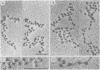
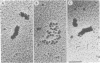
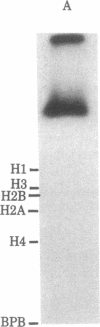
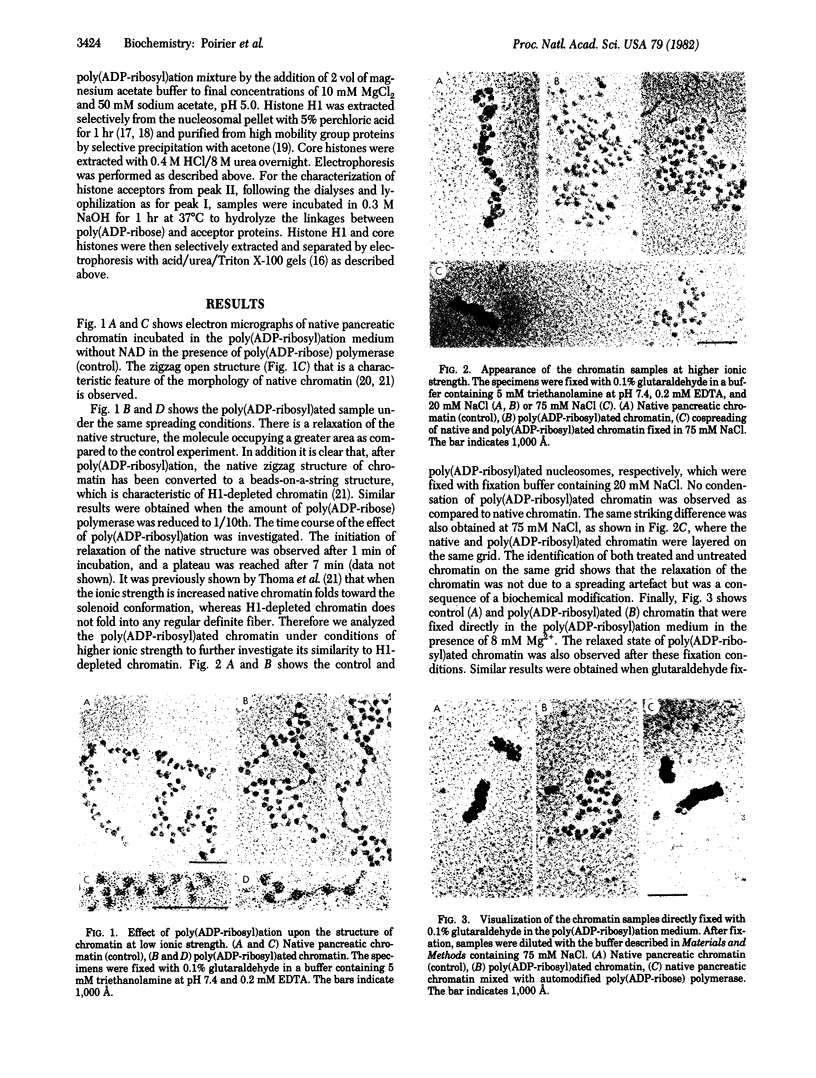
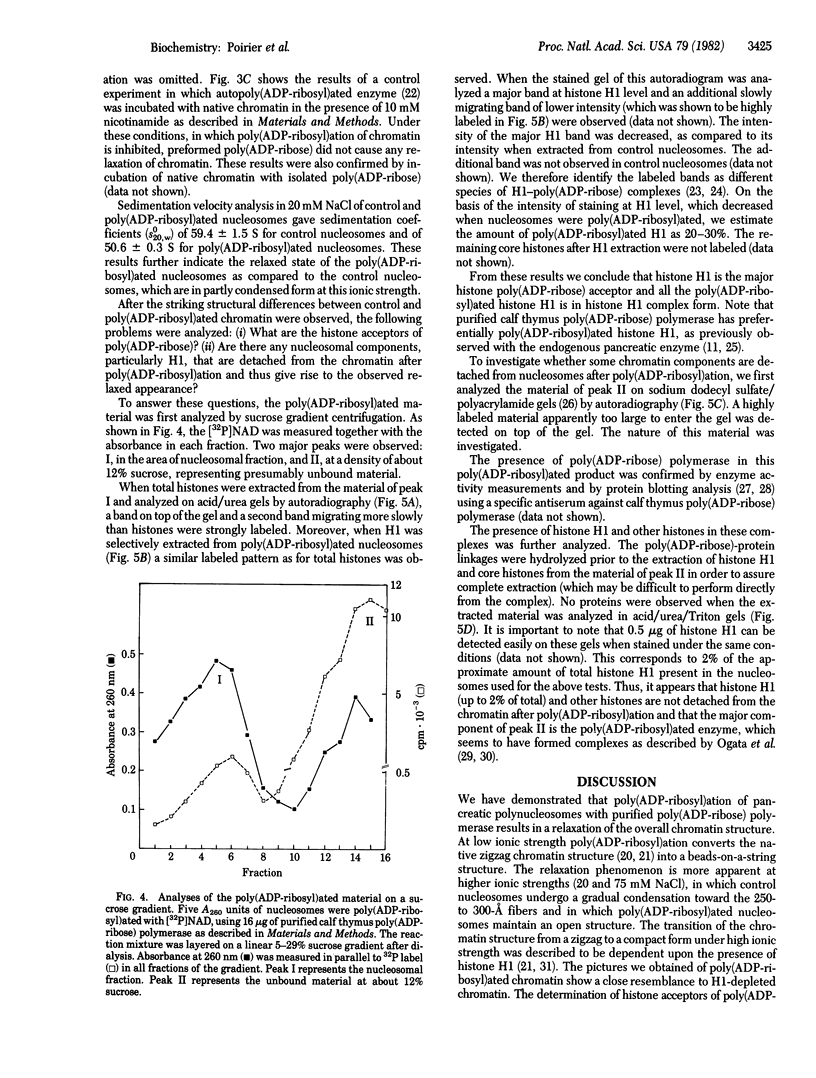
When rat pancreatic polynucleosomes were poly(ADP-ribosyl)ated with purified calf thymus poly(ADP-ribose) polymerase and examined by electron microscopy, a relaxation of their native zigzag structure was observed. At high ionic strengths control nucleosomes condensed into 250-A-thick fibers, but poly(ADP-ribosyl)ated polynucleosomes did not; they showed a close resemblance to chromatin depleted of histone H1. The relaxed state of poly(ADP-ribosyl)ated polynucleosomes was also confirmed by sedimentation velocity analysis. Histone H1 was found to be the major histone acceptor of poly(ADP-ribose). Poly(ADP-ribose) linked to histone H1 did not seem to cause its dissociation from the chromatin, but it impaired significantly its effect on chromatin condensation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger N. A., Sikorski G. W., Petzold S. J., Kurohara K. K. Defective poly(adenosine diphosphoribose) synthesis in xeroderma pigmentosum. Biochemistry. 1980 Jan 22;19(2):289–293. doi: 10.1021/bi00543a006. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., West M. H., Stedman J. D. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980 Aug;109(1):17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- Braeuer H. C., Adamietz P., Nellessen U., Hilz H. ADP-ribosylated histone H1. Isolation from Ehrlich-ascites-tumor-cell nuclei and partial characterization. Eur J Biochem. 1981;114(1):63–68. [PubMed] [Google Scholar]
- Butt T. R., Brothers J. F., Giri C. P., Smulson M. E. A nuclear protein-modifying enzyme is responsive to ordered chromatin structure. Nucleic Acids Res. 1978 Aug;5(8):2775–2788. doi: 10.1093/nar/5.8.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt T. R., DeCoste B., Jump D. B., Nolan N., Smulson M. Characterization of a putative poly(adenosine diphosphate ribose)-chromatin complex. Biochemistry. 1980 Nov 11;19(23):5243–5249. doi: 10.1021/bi00564a014. [DOI] [PubMed] [Google Scholar]
- Butt T. R., Smulson M. Relationship between nicotinamide adenine dinucleotide concentration and in vitro synthesis of poly(adenosine diphosphate ribose) on purified nucleosomes. Biochemistry. 1980 Nov 11;19(23):5235–5242. doi: 10.1021/bi00564a013. [DOI] [PubMed] [Google Scholar]
- Dam V. T., Faribault G., Poirier G. G. Separation of poly(ADP-ribosylated) nuclear proteins by polyacrylamide gel electrophoresis at acidic pH and low temperature. Anal Biochem. 1981 Jul 1;114(2):330–335. doi: 10.1016/0003-2697(81)90489-9. [DOI] [PubMed] [Google Scholar]
- Durkacz B. W., Omidiji O., Gray D. A., Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980 Feb 7;283(5747):593–596. doi: 10.1038/283593a0. [DOI] [PubMed] [Google Scholar]
- Giri C. P., West M. H., Ramirez M. L., Smulson M. Nuclear protein modification and chromatin substructure. 2. Internucleosomal localization of poly(adenosine diphosphate-ribose) polymerase. Biochemistry. 1978 Aug 22;17(17):3501–3504. doi: 10.1021/bi00610a012. [DOI] [PubMed] [Google Scholar]
- Giri C. P., West M. H., Smulson M. Nuclear protein modification and chromatin substructure. 1. Differential poly(adenosine diphosphate) ribosylation of chromosomal proteins in nuclei versus isolated nucleosomes. Biochemistry. 1978 Aug 22;17(17):3495–3500. doi: 10.1021/bi00610a011. [DOI] [PubMed] [Google Scholar]
- Johns E. W. Studies on histones. 7. Preparative methods for histone fractions from calf thymus. Biochem J. 1964 Jul;92(1):55–59. doi: 10.1042/bj0920055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra-Bilen J., Ittel M. E., Jongstra J., Mandel P. Similarities in the molecular weight of poly(ADPR) polymerase from different tissues and species. Biochem Biophys Res Commun. 1981 Nov 30;103(2):383–390. doi: 10.1016/0006-291x(81)90464-2. [DOI] [PubMed] [Google Scholar]
- Juarez-Salinas H., Sims J. L., Jacobson M. K. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979 Dec 13;282(5740):740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- Jump D. B., Butt T. R., Smulson M. Nuclear protein modification and chromatin substructure. 3. Relationship between poly(adenosine diphosphate) ribosylation and different functional forms of chromatin. Biochemistry. 1979 Mar 20;18(6):983–990. doi: 10.1021/bi00573a008. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mandel P., Okazaki H., Niedergang C. Poly(adenosine diphosphate ribose). Prog Nucleic Acid Res Mol Biol. 1982;27:1–51. doi: 10.1016/s0079-6603(08)60596-6. [DOI] [PubMed] [Google Scholar]
- Mandel P., Okazaki H., Niedergang C. Purification and properties of calf thymus poly adenosine diphosphate ribose polymerase. FEBS Lett. 1977 Dec 15;84(2):331–336. doi: 10.1016/0014-5793(77)80719-9. [DOI] [PubMed] [Google Scholar]
- Mathis D. J., Oudet P., Wasylyk B., Chambon P. Effect of histone acetylation on structure and in vitro transcription of chromatin. Nucleic Acids Res. 1978 Oct;5(10):3523–3547. doi: 10.1093/nar/5.10.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Mullins D. W., Jr, Giri C. P., Smulson M. Poly(adenosine diphosphate-ribose) polymerase: the distribution of a chromosome-associated enzyme within the chromatin substructure. Biochemistry. 1977 Feb 8;16(3):506–513. doi: 10.1021/bi00622a026. [DOI] [PubMed] [Google Scholar]
- Niedergang C., Okazaki H., Mandel P. Properties of purified calf thymus poly(adenosine diphosphate ribose) polymerase. Comparison of the DNA-independent and the DNA-dependent enzyme. Eur J Biochem. 1979 Dec;102(1):43–57. doi: 10.1111/j.1432-1033.1979.tb06261.x. [DOI] [PubMed] [Google Scholar]
- Nolan N. L., Butt T. R., Wong M., Lambrianidou A., Smulson M. E. Characterization of poly(ADP-ribose)--histone H1 complex formation in purified polynucleosomes and chromatin. Eur J Biochem. 1980 Dec;113(1):15–25. doi: 10.1111/j.1432-1033.1980.tb06133.x. [DOI] [PubMed] [Google Scholar]
- Ogata N., Ueda K., Kawaichi M., Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981 May 10;256(9):4135–4137. [PubMed] [Google Scholar]
- Okazaki H., Niedergang C., Mandel P. Adenosine diphosphate ribosylation of histone H1 by purified calf thymus polyadenosine diphosphate ribose polymerase. Biochimie. 1980;62(2-3):147–157. doi: 10.1016/s0300-9084(80)80190-8. [DOI] [PubMed] [Google Scholar]
- Oudet P., Gross-Bellard M., Chambon P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell. 1975 Apr;4(4):281–300. doi: 10.1016/0092-8674(75)90149-x. [DOI] [PubMed] [Google Scholar]
- Purnell M. R., Stone P. R., Whish W. J. ADP-ribosylation of nuclear proteins. Biochem Soc Trans. 1980 Apr;8(2):215–227. doi: 10.1042/bst0080215. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978 Apr;13(4):691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- Stone P. R., Lorimer W. S., 3rd, Kidwell W. R. Properties of the complex between histone H1 and poly(ADP-ribose synthesised in HeLa cell nuclei. Eur J Biochem. 1977 Nov 15;81(1):9–18. doi: 10.1111/j.1432-1033.1977.tb11921.x. [DOI] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Nedospasov S. A., Schmatchenko V. V., Bakayev V. V., Chumackov P. M., Georgiev G. P. Compact form of SV40 viral minichromosome is resistant to nuclease: possible implications for chromatin structure. Nucleic Acids Res. 1977 Oct;4(10):3303–3325. doi: 10.1093/nar/4.10.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia G., Mazen A., Erard M., Pouyet J., Champagne M. Isolation and physical characterization of a stable core particle. Nucleic Acids Res. 1980 Feb 25;8(4):767–779. [PMC free article] [PubMed] [Google Scholar]