Abstract
Histone methylation performs multiple functions such as DNA replication, transcription regulation, heterochromatin formation, and chromatin condensation. How this methylation gradient is orchestrated in the centromere during chromosome segregation is not known. Here we examine the temporal dynamics of protein methylation in the centromere by SUV39H1 methyltransferase, a key mitotic regulator, using fluorescence resonance energy transfer-based sensors in living HeLa cells and immunofluorescence of native SUV39H1 substrates. A quantitative analysis of methylation dynamics, using centromere-targeted sensors, reveals a temporal change during chromosome segregation. These dynamics result in an accurate chromosome congression to and alignment at the equator as an inhibition of methylation dynamics using SUV39H1 inhibitor perturbs chromosome congression in living HeLa cells. Surprisingly, this inhibition of methylation results in a brief increase in Aurora B kinase activity and an enrichment of microtubule depolymerase MCAK in the centromere with a concomitant kinetochore–microtubule destabilization and a reduced tension across the sister kinetochores with ultimate chromosome misalignments. We reason that SUV39H1 generates a gradient of methylation marks at the kinetochore that provides spatiotemporal information essential for accurate chromosome segregation in mitosis.
Keywords: mitosis, SUV39H1, methylation, MARC, syntelin
Introduction
The kinetochore is a super-molecular complex assembled at each centromere in eukaryotes. It provides a chromosomal attachment point for the mitotic spindle, linking the chromosome to the microtubules and functions in initiating, controlling, and monitoring the movements of chromosomes during mitosis. The kinetochore of animal cells contains two functional domains; that is, the inner kinetochore, which is tightly and persistently associated with centromeric DNA sequences via the histone and its variants throughout the cell cycle, and the outer kinetochore which is composed of many dynamic protein complexes that interact with microtubules only during mitosis. The stable propagation of eukaryotic cells requires each chromosome to be accurately duplicated and faithfully segregated. During mitosis, attaching, positioning, and bi-orientating kinetochores with the spindle microtubules play critical roles in chromosome segregation and genomic stability (Rajagopalan and Lengauer, 2004; Shen, 2011). However, the molecular mechanism underlying epigenetic regulation of centromere plasticity and genomic stability has remained elusive (Cleveland et al., 2003; Lahtz and Pfeifer, 2011). Mitosis is orchestrated by signaling cascades that coordinate mitotic processes and ensure accurate chromosome segregation. Mounting evidence demonstrates that the master mitotic kinase Cdk1 together with PLK1, Aurora kinases, and the NIMA-related kinases orchestrate the chromosome segregation in mitosis (Nigg, 2001; Ke et al., 2003; Fu et al., 2007; Yang et al., 2008; Yu and Yao., 2008; Zhang et al., 2011). However, little is known about whether mitotic phosphorylation cascades cross-talk with other post-translational modifications such as methylation on the centromere during chromosome segregation in mitosis.
The methylation of histone H3 on lysine 9 (H3K9me) is a key event in the formation of heterochromatin in most eukaryotes. This modification serves as a binding site for HP1 family proteins, which play a critical role in maintaining chromatin structure and ensuring faithful chromosome segregation during cell division. SUV39H1 is an evolutionarily conserved site-specific methyltransferase responsible for H3K9 tri-methylation (Rea et al., 2000). Perturbation of SUV39h1 in mice results in cell cycle progression defects, chromosomal mis-segregation and HP1 mis-localization (Czvitkovich et al., 2001). Interestingly, it has been shown that human SUV39H1 is preferentially recruited to pericentromeric heterochromatin during entry into mitosis (Aagaard et al., 2000). Using immunofluorescent staining of H3K9me3, it was shown that the H3K9me3 level is dynamic in mitosis with the peak at metaphase and declines as sister chromatids separate (McManus et al., 2006). However, it was unclear whether and how the SUV39H1 activity gradient governs chromosome and kinetochore–microtubule dynamics in mitosis.
Here, we identified a novel regulatory mechanism underlying kinetochore–microtubule dynamics regulated by the SUV39H1 methylation gradient in mitosis. Using a fluorescence resonance energy transfer (FRET)-based methylation sensor in living HeLa cells, our quantitative analysis reveals the temporal dynamics of centromere methylation during chromosome segregation. Our study indicates that SUV39H1 activity orchestrates kinetochore–microtubule dynamics via the Aurora B-MCAK axis.
Results
Development of a novel centromere methylation sensor for real-time imaging
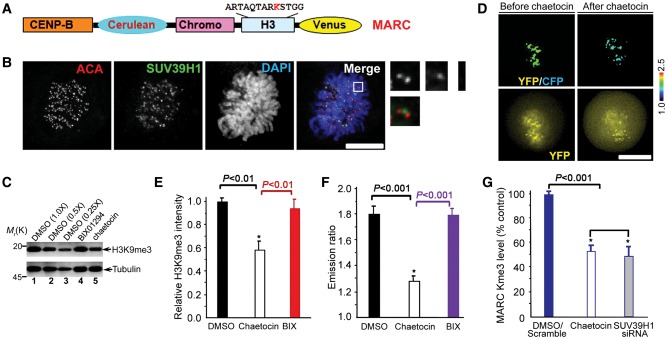
It has been shown that H3K9me3 is dynamic in mitosis using immunofluorescence staining of H3K9me3 (McManus et al., 2006). To examine if tri-methylation dynamics of H3K9 is a function of SUV39H1 methyl-transferase activity in mitosis, we sought to monitor SUV39H1 activity in the centromere using FRET-based sensors that report quantitative changes in substrate methylation in living cells. We adapted a sensor design (Lin et al., 2004), in which changes in intra-molecular FRET between cyan and yellow fluorescent proteins (CFP–YFP) depend on changes in methylation of a SUV39H1 substrate peptide such as H3K9. To mimic localizations of endogenous SUV39H1 substrates, sensors were targeted at centromeres (CENP-B fusion; Figure 1A), since SUV39H1 is primarily located at the centromere in a manner partially overlapping with the centromere mark anti-centromere antibody (ACA) staining (Figure 1B; also see magnified images). The sensor developed using the chromodomains from HP1 responds to enzymatic methylation at the H3K9 quantitatively by appropriate YFP/CFP emission ratio increases in vitro or in living cells. The sensor was named as MARC (Methylation Activity Reporter at Centromere).
Figure 1.
Development and characterization of a novel centromere methylation sensor for visualization of methylation dynamics in live cells. (A) Schematic representation of the domain structure of MARC organization. (B) Localization of SUV39H1 to centromere of mitotic cells. HeLa cells grown on a coverslip were fixed, then stained with antibodies against ACA and SUV39H1, chromosomes was stained with DAPI. Bar, 10 µm. (C) HeLa cells treated with DMSO, 10 μM BIX01294 and 1.3 μM chaetocin for 12 h were collected and processed for western blotting against H3K9me3 rabbit antibody and α-tubulin mouse antibody, respectively. Aliquots of diluted lysates were used for calibrating and quantifying the inhibition of H3K9me3 by chaetocin. (D) HeLa cells expressing MARC were imaged before and after adding chaetocin. Upper panel, color-coded images of the emission ratio; lower panel, unprocessed YFP images. Bar, 10 μm. (E) Statistic analysis of H3K9 tri-methylation western blotting in response to chaetocin and BIX01294 treatments (mean ± SE; n= 3). (F) Statistic analysis of MARC activity before and after adding chaetocin derived from experiments shown in D (mean ± SE; at least 10 cells per category). (G) Statistic analysis of MARC tri-methylation western blotting in response to SUV39H1 inhibition and knockdown (mean ± SE; n= 3).
To validate MARC response to changes in SUV39H1 activity, we first imaged mitotic cells before and after SUV39H1 inhibition with chaetocin (Greiner et al., 2005; Figure 1D). As shown in Figure 1C, western blotting analyses indicate that chaetocin specifically inhibits H3K9 tri-methylation. Aliquots of diluted lysates were run in parallel for quantifying the chaetocin-induced inhibition of H3K9 tri-methylation (Figure 1C, lanes 2 and 3). This inhibition was also observed in a decrease in the YFP:CFP emission ratio of MARC (Figure 1D), consistent with demethylation for this MARC design and previous reports (Lin et al., 2004). The MARC is specific for H3K9 tri-methylation as it does not respond to other methyltransferase inhibitors such as BIX01294, which inhibits dimethylation on H3K9 (Figure 1C and D; sensor data not shown for BIX01294). The quantitative decrease in the MARC readout in response to chaetocin (Figure 1F) is in proportion to the chaetocin-induced decrease of H3K9 tri-methylation judged by quantitative western blotting (Figure 1E). In addition, this chaetocin-induced inhibition of H3K9 tri-methylation judged by western blotting analysis was validated by SUV39H1 siRNA (Figure 1G), indicating that the measured FRET change in MARC is a faithful readout of SUV39H1 activity in the centromere.
Since phosphorylation of Ser10 was reported to interfere with H3K9 tri-methylation (Fischle et al., 2005), we sought to mutate Ser10 to non-phosphorylatable Ala so there would not be interference in the MARC readout by mitotic phosphorylation. However, mutagenesis of Ser10 to Ala resulted in an inability of MARC to respond to methylation changes. To validate the potential interference of phospho-Ser10 with H3K9 tri-methylation, we sought to inhibit Aurora B activity and measure the FRET ratio of MARC in the presence and absence of Aurora B inhibitor ZM447439. To our surprise, MARC activity readout was not altered by Aurora B activity as ZM447439 does not change the MARC FRET ratio (Supplementary Figure S1A and B) while the level of endogenous H3 Ser10 phosphorylation is minimized by ZM447439 (Supplementary Figure S1C). Thus, we conclude that MARC is an accurate reporter of H3K9 tri-methylation in mitosis.
Measuring methylation dynamics in living mitotic cells using MARC
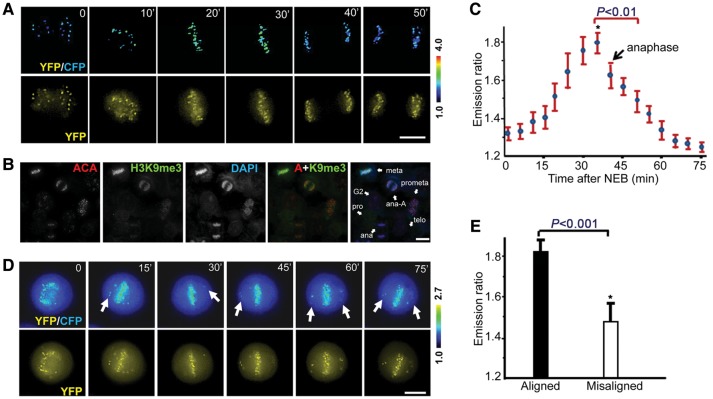
Previously envisioned methylation dynamics at the centromere are based on the still fluorescence images of H3K9me3 staining of fixed cells. To visualize methylation dynamics in living cells, HeLa cells were transiently transfected to express MARC. As shown in Figure 2A, MARC activity readout peaks in metaphase cells followed by a gradual decay at anaphase onset and reaches the bottom at telophase and cytokinesis. Trial experiments validate that H3K9me3 level can be visualized (Supplementary Figure S2A), and assessed using tri-methylation-specific antibody under a fluorescent microscopy (Supplementary Figure S2B), which suggests that centromeres of mitotic cells exhibit a higher level of H3K9 tri-methylation labeling compared with that of interphase (Supplementary Figure S2B; P< 0.05). Examination of an endogenous SUV39H1 substrate such as the tri-methylation of H3K9 of using tri-methylation-specific antibody provides a consistent profile of mitotic dynamics (Figure 2B), which indicates that tri-methylation of H3K9 peaks at metaphase-aligned chromosomes. The quantitative analyses of temporal dynamics of MARC from 15 cells were summarized in Figure 2C. Careful examination of the MARC emission ratio of congressing chromosomes revealed that methylation differences between individual centromeres depend on the centromere position as chromosomes congress to the equator during mitotic progression (Figure 2C).
Figure 2.
Quantification of centromere methylation dynamics in living mitotic cells using MARC. (A) HeLa cells expressing the MARC were imaged through the whole mitosis. Upper panel, color-coded images of the emission ratio; lower panel, unprocessed YFP images. Bar, 10 µm. (B) An aliquot of asynchronized HeLa cells was extracted, fixed and stained with ACA, H3K9me3, and DAPI. Bar, 10 µm. (C) Quantification of dynamic MARC activity during the cause of mitosis derived from the live cell imaging experiments shown in A (mean ± SE; >100 kinetochores from 15 cells). (D) HeLa cells expressing MARC were imaged for chromosome movements in the presence of syntelin, a mitotic kinesin CENP-E inhibitor. Syntelin was added to thymidine-synchronized cells before nuclear envelope breakdown. Upper panel, color-coded images of the emission ratio; lower panel, unprocessed YFP images. Bar, 10 µm. (E) Quantification of centromere methylation of aligned vs. misaligned chromosomes. MARC activity was quantified in syntelin-treated cells shown in D (mean ± SE; 15 chromosomes from 5 cells).
Our recent study has identified a small molecule inhibitor syntelin that produces misaligned chromosomes in living cells due to its specific inhibition of CENP-E motor activity (Yao et al., 2000; Ding et al., 2010). Addition of syntelin promotes the population of lagging chromosomes, which is evident by MARC readout difference seen among individual centromeres as misaligned chromosomes attempt to approach metaphase alignment at the equator (Figure 2D, arrows). Quantitative measurement of MARC readout from 15 aligned and misaligned chromosomes in living HeLa cells indicates that metaphase-aligned chromosomes exhibit a higher methylation gradient than that of misaligned chromosomes (Figure 2E), confirming that MARC provides an accurate readout of SUV39H1 methylation in space and time.
SUV39H1 activity is required for faithful mitotic progression
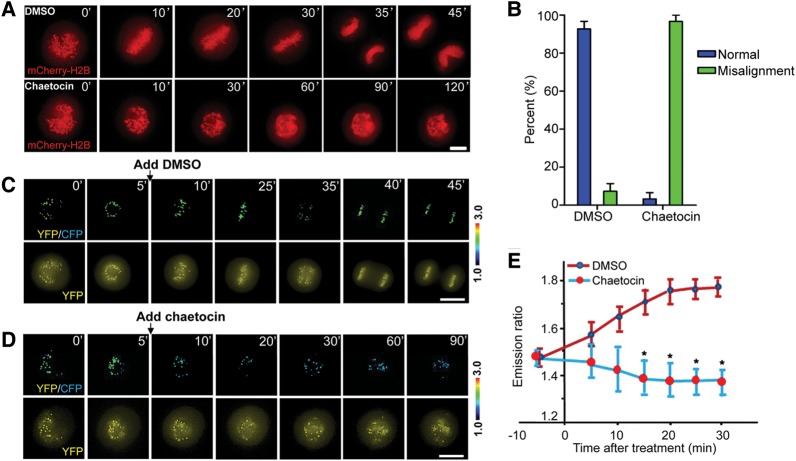
To precisely probe for the methylation dynamics in chromosome movements in mitosis, we conducted real-time imaging of HeLa cells expressing mCherry-H2B in the presence or absence of SUV39H1 inhibitor chaetocin. Chaetocin does not inhibit progression through S and G2 phases of the cell cycle but causes mitotic arrest with lagging chromosomes. If chaetocin is a specific inhibitor of SUV39H1, suppression of SUV39H1 by siRNA should produce the same phenotype. As expected, knockdown of SUV39H1 by siRNA caused mitotic arrest with many mis-aligned chromosomes (Supplementary Figure S3). However, inhibition of SUV39H1 by chaetocin did not perturb bipolar spindles, but produced misaligned chromosomes near the spindle poles (Supplementary Figure S4), and chronic arrest in mitosis (Figure 3A) with a significant increase in misaligned chromosomes (Figure 3B; P< 0.05), indicating that SUV39H1 methylation activity is essential for faithful chromosome congression.
Figure 3.
Dynamic tri-methylation of H3K9 is required for mitotic progression. (A) HeLa cells expressing mCherry-H2B were imaged in live in the presence of DMSO (upper panel) and chaetocin (lower panel). Note that chaetocin addition causes a chronic arrest of mitotic progression. Bar, 10 µm. (B) Quantification of phenotypic changes in chaetocin treatment shown in A (mean ± SE; n= 15). (C and D) HeLa cells expressing the MARC were synchronized and released. DMSO (C) or chaetocin (D) was added 5 min after the nuclear membrane breakdown. Note that the centromere methylation level is suppressed by the treatment so does the chromosome alignment. Upper panel, color-coded images of the emission ratio; lower panel, YFP images. Bar, 10 µm. (E) Chromosome alignment is a function of dynamic centromere methylation. Quantification of centromere methylation dynamics was conducted in control and SUV39H1-inhibited cells (mean ± SE; n= 10).
Small molecules that modulate specific protein functions are valuable tools for dissecting complex processes in mammalian cell division. Having demonstrated the role of methylation gradient in mitosis, we sought to test whether chaetocin inhibition of SUV39H1 activity alters the chromosome dynamics in living cells. To this end, we adopted a protocol to synchronize cells at prometaphase using monastrol and then release for progression toward the metaphase. Chaetocin is added right after the release for visualizing for real-time chromosome movements in methylation regulated living HeLa cells. We began real-time imaging of prometaphase-synchronized HeLa cells expressing MARC soon after the addition of dimethyl sulphoxide (DMSO) to visualize chromosome congression and subsequent prometaphase–metaphase transition and telophase (Figure 3C). In general, it takes an average of 35 ± 3 min (n = 20) for HeLa cells to transit from prometaphase (mono-polar) to the anaphase onset of sister chromatid separation. However, some chromosomes still failed to align at the equator in the chaetocin-treated cells even after 90 min (Figure 3D). Examination of MARC emission ratio in those living HeLa cells revealed that methylation differences between individual centromeres depend on the centromere position and a temporal dynamics of MARC as chromosomes congress to the equator. Addition of chaetocin promotes demethylation of the sensor, which is evident by the minimized MARC readout seen among individual centromeres as chromosomes failed to achieve metaphase alignment at the equator over an extended period of 90 min (Figure 3D). Quantitative analyses of 10 mitotic cells expressing MARC demonstrate that the centromere methylation level was significantly suppressed by chaetocin with aberrant chromosome alignments (Figure 3E; P< 0.001). Thus, we conclude that centromere methylation temporal dynamics are essential for accurate chromosome congression to the metaphase plate.
Suppression of SUV39H1 activity promotes Aurora B kinase activity at centromere
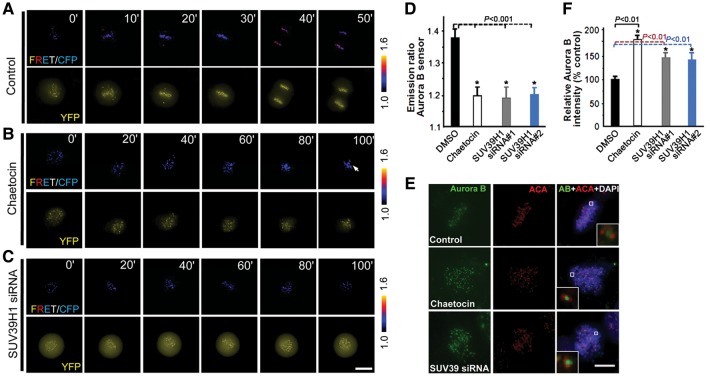
Accurate attachment of spindle microtubules to kinetochore requires Aurora B kinase (Yang et al., 2008; Hua et al., 2011). Our recent studies show that the Aurora B kinase activity is dynamically regulated by PLK1 kinase in living HeLa cells (Chu et al., 2011). To visualize if there is any Aurora B activity changes in response to chaetocin inhibition of SUV39H1 activity, we performed Aurora B kinase activity quantification in real-time mitosis using FRET-based reporter based on changes in phosphorylation of MCAK, an Aurora B substrate peptide, which results in binding to FHA2 domain and corresponding FRET changes (Fuller et al., 2008; Chu et al., 2011). Specifically, the increase in the FRET ratio is in proportion to a decrease of phosphorylation. The sensor is specific for Aurora B as it does not respond to other mitotic kinase inhibitors except for Aurora B inhibitors such as hersperadin (Chu et al., 2011). In addition, this Aurora B sensor is not a substrate of SUV39H1 and shows no response to chaetocin directly (data not shown).
An examination of the Aurora B sensor emission ratio revealed that phosphorylation differences between individual centromeres depend on the centromere position as chromosomes congress to the equator during mitotic progression in a live HeLa cells (Figure 4A). Addition of Aurora B kinase inhibitor hesperadin promotes dephosphorylation of the sensor, which is evident by minimized differences seen among individual centromeres as chromosomes approach metaphase alignment at the equator (data not shown). Surprisingly, addition of chaetocin promotes phosphorylation of the sensor as chromosomes attempt to achieve metaphase alignment visualized by a decrease of FRET ratio (Figure 4B; arrow). To validate the specific involvement of SUV39H1, we performed a parallel measurement of Aurora B activity in SUV39H1-siRNA-treated cells. Consistent with what was observed in chaetocin-treated samples, suppression of SUV39H1, by two independent siRNAs targeted at different regions of SUV39H1, resulted in an increased phosphorylation reported by the Aurora B sensor (Figure 4C), suggesting that an increase of Aurora B activity is a function of SUV39H1 suppression. Quantitative analyses of 15 mitotic cells expressing Aurora B sensor indicate that Aurora B activity is significantly elevated in misaligned chromosomes in response to chaetocin treatment (Figure 4D; P< 0.001). Suppression of SUV39H1 activity by either siRNA or chaetocin produces a persistent and significant elevation of Aurora B kinase activity at the centromere in live cells (Figure 4D; P< 0.001). To probe if there is an increase of the Aurora B protein level at the centromere, we performed immunofluorescence staining of Aurora B and the centromere marker ACA. As shown in Figure 4E, chaetocin treatment retained the inner centromere localization of Aurora B in the cells with reduced methylation (middle panel). Knockdown of SUV39H1 by siRNA also enhances the localization of Aurora B to the kinetochore (lower panel). Our quantification of normalized pixel intensities showed that chaetocin increased the kinetochore-bound Aurora B level to 181% ± 9% of its control value (Figure 4F; P< 0.01 compared with the control), while siRNA-mediated knockdown enriched Aurora B level to 143% ± 7% of its control value (P< 0.01), indicating that centromere methylation activity of SUV39H1 negatively regulates the kinetochore localization of Aurora B.
Figure 4.
Suppression of SUV39H1 activity in mitosis promotes Aurora B kinase activity. (A) HeLa cells expressing the centromere-target Aurora B kinase sensor were imaged on the duration of mitosis. Upper panel, color-coded images of the emission ratio; lower panel, YFP images; timestamps (minutes) relative to drug treatments. Bar, 10 µm. (B) Aurora B sensor is responsive to SUV39H1 activity inhibition. Chaetocin was added at time 0. Bar, 10 µm. (C) Aurora B sensor is responsive to SUV39H1 knockdown by siRNA. HeLa cells in early G1 were co-transfected with Aurora B sensor and SUV39H1 siRNA for 20 h before imaging. Bar, 10 µm. (D) Statistic analyses of the YFP:CFP emission ratio of Aurora B sensor indicate that suppression of SUV39H1 by inhibitor or siRNA promotes Aurora B kinase activity (mean ± SE; >100 kinetochores of each categories from 15 cells). Two different siRNAs (siRNA#1 and siRNA#2) targeted to two different regions of SUV39H1 were used with essential same outcome. (E) Aurora B localization in centromere is enhanced by SUV39H1 suppression. HeLa cells treated with DMSO, chaetocin, or SUV39H1 siRNA were fixed and stained for Aurora B (green), centromere (ACA; red) and DNA (DAPI). Enlarged areas (inset) show stronger staining in SUV39H1-suppressed (chaetocin-treated and siRNA-treated) cells. Bar, 10 µm. (F) Statistic analyses of Aurora B immunofluorescence intensity at kinetochore indicate that suppression of SUV39H1 promotes Aurora B protein levels in kinetochore (mean ± SE; 10 cells of each category from three different preparations).
To test if the increased level of Aurora B is a function of a misaligned chromosome, aliquots of treated cells were exposed to 20 μM nocodazole to depolymerize kinetochore–microtubule before fixation. As shown in Supplementary Figure S6A, nocodazole treatment results in a dramatic enrichment of Aurora B to kinetochore of control cells (P< 0.01), consistent with the literature (Murata-Hori and Wang, 2002). However, nocodazole treatment did not modulate the Aurora B level at the kinetochore of SUV39H1-suppressed cells compared with that of control cells (Supplementary Figure S6B). Thus, we conclude that the increased levels of Aurora B in the kinetochore of SUV39H1-suppressed cells are not due to misalignment of chromosomes.
SUV39H1 orchestrates accurate chromosome segregation by regulating kinetochore plasticity
The microtubule depolymerase MCAK is a key regulator of mitotic spindle assembly and dynamics under mitotic kinase regulation (Andrews et al., 2004; Lan et al., 2004). The elevated Aurora B activity reported by MCAK phosphorylation together with an increase of the Aurora B protein level in chaetocin-treated HeLa cells prompted us to examine the spindle microtubule plasticity. To this end, we asked whether suppression of SUV39H1 activity affects the attachment and stability of spindle microtubules to kinetochores. The control and chaetocin-treated cells were briefly chilled at 4°C before being extracted, fixed and stained with anti-tubulin antibody and ACA. While most non-kinetochore-associated microtubules were lost, images obtained by optical sectioning with deconvolution microscopy showed that centromeres labeled by ACA remained associated with spindle microtubule ends in control cells (Figure 5A, red dots, left panel). In SUV39H1-inhibited cells, many chromosomes were misaligned and scattered around the spindle pole (Figure 5A, middle panel), with kinetochore fibers relative instable when compared with those of control cells. A similar phenotype was also observed in SUV39H1-depleted cells (lower panel). Although cold-resistant attachment of kinetochores to spindle microtubules can be achieved for majority of chromosomes when SUV39H1 levels are diminished, the abundance of chromosomes without a stable bioriented kinetochore attachment even after long periods of mitotic arrest suggests that SUV39H1 must have an important function in the orchestration of dynamic kinetochore–microtubule association with chromosomes.
Figure 5.
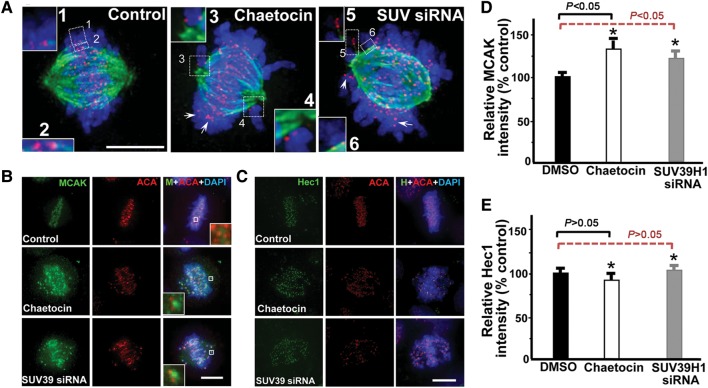
H3K9 tri-methylation dynamics is essential for accurate kinetochore–microtubule interactions. (A) Perturbation of H3K9 tri-methylation reduced stability and/or dynamics of kinetochore-associated microtubules in SUV39H1-suppressed cells. Optical sections of deconvolution images of mitotic HeLa cells 22 h after transfection with a control scramble siRNA (left) or with SUV39H1 siRNA#1 (right) are shown. A chaetocin-treated cell was also included (middle). Cells were stained with ACA (red), DAPI (blue), and anti-tubulin antibody (green). Insets 1 and 2 show bi-oriented chromosome pairs of a scramble transfected cell. Insets 3 and 4 show kinetochore pairs without stable microtubule attachment in a chaetocin-treated cell, which displays a typical syntelic attachment. Insets 5 and 6 show kinetochore pairs without stable microtubule attachment in a SUV39H1-suppressed cell. Arrows indicate kinetochores without microtubule attachment. Bar, 10 µm. (B) MCAK localization in centromere is enhanced by SUV39H1 suppression. HeLa cells treated with DMSO, chaetocin, or SUV39H1 siRNA were fixed and stained for MCAK (green), centromere (ACA; red) and DNA (DAPI). Enlarged areas (insets) show stronger staining in SUV39H1-suppressed (chaetocin and siRNA-treated) cells. Bar, 10 µm. (C) Hec1 localization in centromere is not altered by SUV39H1 suppression. HeLa cells treated with DMSO, chaetocin, or SUV39H1 siRNA were fixed and stained for Hec1 (green), centromere (ACA; red) and DNA (DAPI). Bar, 10 µm. (D) Statistic analyses of MCAK immunofluorescence intensity at kinetochore indicate that suppression of SUV39H1 promotes MCAK protein levels in kinetochore (mean ± SE; 10 cells of each category from three different preparations). (E) Statistic analyses of Hec1 immunofluorescence intensity at kinetochore indicate that suppression of SUV39H1 does not modulate Hec1 protein levels in kinetochore (mean ± SE; 10 cells of each category from three different preparations).
The shortening and unstable kinetochore–microtubule together with a rise in MCAK phosphorylation reported in Aurora B kinase sensor prompted us to examine the microtubule deploymerase MCAK level in the centromere of chaetocin-treated cells. As predicted, the level of MCAK at the centromere of chaetocin-treated or SUV39H1-siRNA-treated cells was dramatically increased, as judged by the immunofluoresence staining of MCAK (Figure 5B). On the other hand, the protein level of Hec1, a critical component linking spindle microtubule to kinetochore, was barely changed in response to the suppression and inhibition of SUV39H1 (Figure 5E; P> 0.05). Quantification of normalized pixel intensities showed that chaetocin increased the kinetochore-bound MCAK level to 133% ± 9% of its control value (Figure 5D; P< 0.05) while siRNA-mediated knockdown of SUV39H1 enriched the MCAK level to 121% ± 7% of its control value (Figure 5D; P< 0.05), indicating that the centromere methylation gradient regulates the kinetochore–microtubule plasticity by retention of MCAK. We have also evaluated if the increased level of MCAK is a function of misaligned chromosomes by depolymerizing kinetochore–microtubules with nocodazole before fixation. As shown in Supplementary Figure S7A, nocodazole treatment results in a brief but not significant enrichment of MCAK to the kinetochore of control cells (P> 0.05). In addition, nocodazole treatment did not change the MCAK level at the kinetochore in SUV39H1-suppressed cells compared with that of control cells (Supplementary Figure S7B). Thus, misalignment of chromosomes does not contribute to the enrichment of MCAK in the centromere of SUV39H1-suppressed cells.
If SUV39H1 cooperates with MCAK/Aurora B to govern the kinetochore–microtubule plasticity in vivo, inhibition of SUV39H1 should release the tension exerted on the sister kinetochores. As the inter-kinetochore distance from sister chromatids has previously been proposed as an accurate reporter for judging the tension developed across the kinetochore pair (Yao et al., 2000; Liu et al., 2007), we measured this distance in >100 kinetochore pairs in which both kinetochores were in the same focal plane, in SUV39H1-depleted, SUV39H1-inhibited, SUV39H1-and-Aurora B-inhibited, and control cells (Table 1). Control kinetochores exhibited a separation of 1.43 ± 0.18 µm, whereas the distances between misaligned kinetochores in SUV39H1-siRNA#1-treated cells and SUV39H1-inhibited cells were 1.18 ± 0.17 µm and 1.19 ± 0.18 µm, respectively. The inter-kinetochore distance of misaligned kinetochores in SUV39H1- and Aurora B-refrained cells was 1.31 ± 0.23 µm. The distance in SUV39H1-depleted and nocodazole-treated cells, in which kinetochore pairs were presumably under no tension, was 1.01 ± 0.13 µm. We conclude that SUV39H1 orchestrates mitotic chromosome dynamics by governing kinetochore–microtubule plasticity via the Aurora B-MCAK interaction axis.
Table 1.
SUV39H1 activity controls the tension between sister kinetochores.
| Treatment | Distance (μm)a |
|---|---|
| Scramble/DMSO-treated | 1.43 ± 0.18 |
| SUV39H1 siRNA#1 (mis-aligned kinetochore) | 1.18 ± 0.17 |
| SUV39H1 siRNA#2 (mis-aligned kinetochore) | 1.16 ± 0.19 |
| Chaetocin (mis-aligned kinetochore) | 1.19 ± 0.18 |
| Chaetocin+hesperadin (mis-aligned kinetochore) | 1.31 ± 0.23 |
| SUV39H1 siRNA#1 + nocodazole treatment | 1.01 ± 0.13 |
Data were obtained >150 kinetochore pairs in which kinetochores were in the same focal plane.
aDistance measured between ACA-marked sister kinetochore from same focal plane.
Discussion
The kinetochore is a complex structure that functions as a molecular machine to power chromosome movement, and as a signaling device to govern chromosome segregation and cell cycle control. We have developed a novel centromere methylation reporter MARC for spatiotemporal dynamics of SUV39H1 activity. Our studies show that perturbation of SUV39H1 temporal dynamics blocks chromosome congression to the metaphase plate in living cells. Interestingly, the temporal dynamics of centromere methylation is coupled to Aurora B activity and kinetochore–microtubule plasticity regulated by Aurora B-MCAK axis. We propose that SUV39H1 generates a gradient of methylation marks that provides spatiotemporal information essential for accurate chromosome segregation in mitosis. Our molecular dissection has suggested a regulatory mechanism for temporal regulation of methylation–phosphorylation cross-talk in the centromere during mitosis.
A central characteristic of the kinetochore–spindle interface is its ability to orchestrate stable and dynamic associations while bound microtubules are polymerizing or depolymerizing. Such properties would be best coordinated by several distinct but cooperative kinetochore–microtubule binding sites regulated by signaling cascades (Wang et al., 2004; Cheeseman et al., 2006; Zhang et al., 2011). MCAK is localized to various subcellular structures in mitotic cells, such as inner centromeres, outer kinetochores, centrosomes, spindle microtubules, kinetochore–microtubule ends and spindle midzone (Andrews et al., 2004; Lan et al., 2004; Moore et al., 2005). This diversified localization of MCAK in mitosis implies a complex regulation. In fact, our recent studies show that PLK1 phosphorylates the C-terminal domain of MCAK, and this phosphorylation promotes its depolymerization activity by releasing an intra-molecular interaction of MCAK that locks the microtubule depolymerase in a latent state in a non-phosphorylated form (Zhang et al., 2011). In addition, our studies show that Aurora B kinase activation requires PLK1-mediated phosphorylation priming events (Chu et al., 2011), which proposes a feedback mechanism in kinetochore to synergize mitotic kinase cascades for maintaining chromosome stability by orchestrating kinetochore–microtubule interactions and error correction during mitosis. A most recent study supports our working model by demonstrating that Aurora B kinase recruitment to kinetochore is regulated by PLK1 (Salimian et al., 2011). Their data support the notion that the Aurora B feedback pathway culminates in enriching the kinase at the centromeres of misaligned chromosomes, which promotes efficient destabilization of aberrant kinetochore–microtubule attachments for error correction. Upon error correction, the corresponding reduced levels of the kinase and the increased distance from its kinetochore targets switch the centromere to a mode that is stabilized until all chromosomes are properly aligned and anaphase onset begins. We reason that the kinetochore possesses a feedback mechanism in which Aurora B both regulates kinetochore–microtubule plasticity and is regulated by SUV39H1 established here and PLK1 reported previously (Chu et al., 2011; Salimian et al., 2011), which amplifies the differential phosphorylation of kinetochore substrates and increases the efficiency of error correction. Our preliminary study excluded the involvement of phosphorylation of histone 3 by haspin or Sgo1 level in promoting Aurora B localization (Supplementary Figure S5). It would be of great interest to delineate how multiple phosphorylation sites in MCAK coordinate the aberrant kinetochore–microtubule depolymerization, error correction and stabilization of accurate kinetochore–microtubule attachment. Further investigations are warranted to elucidate the structure basis underlying this phospho-regulated MCAK conformational change and the structural-functional relationship of MCAK depolymerase activity.
SUV39H1 is an essential mitotic regulator for faithful mitosis. SUV39H1 specifically accumulates at the centromere during prometaphase but dissociates from centromeric positions at the metaphase to anaphase transition by which it specifies the temporal dynamics of centromere methylation visualized by the activity sensor MARC characterized in this study. It was proposed in the binary switch hypothesis that phosphorylation of histone H3 at Ser10 negatively regulates the binding of HP1α to the adjacent H3K9me3. However, MARC activity readout was not altered by Aurora B activity, consistent with an early study in which modulation of H3 Ser10 phosphorylation did not interfere with HP1α–H3K9me3 interaction (Terada, 2006). Although our study revealed a novel mechanism underlying phospho-methylation regulation in accurate chromosome segregation, the details of this regulatory network remain for further analyses. Recent studies in budding yeast show that Set1 methyltransferase methylates microtubule stabilizing complex Dam1 by inhibiting phosphorylation of flanking serines by Aurora kinase (Zhang et al., 2005). Dam1 is critical for the correct attachment of sister chromatids to opposing spindle poles (bi-orientation) and is regulated by phosphorylation/dephosphorylation on multiple sites mediated by yeast Aurora B kinase and phosphatase 1γ, respectively (Pinsky et al., 2006). Phosphorylation of Dam1 has been proposed to reduce its affinity for microtubules or to alter its interaction with the kinetochore Ndc80 complex, thereby allowing for dissociation of faulty kinetochore–microtubule attachments. Dam1 peptides dimethylated on Lys233 could not be phosphorylated on Ser235 by Aurora B in vitro, suggesting the existence of a phospho-methyl switch. A most recent study from the same group establishes that Dam1 dimethylation occurs in the context of functional kinetochores, as deletion of either the centromeric DNA-binding protein Ndc10 or the structural kinetochore protein Ndc80 results in ablation of Dam1 methylation. Although the direct effects of Dam1 dimethylation on kinetochore structure and function are not known, it is tempting to speculate that dimethylation may stabilize the interaction of kinetochores with microtubules upon proper biorientation of sister chromatids by preventing rephosphorylation of Dam1 by Aurora B (Latham et al., 2011). Although Dam1 seems to be specific to the budding yeast as other eukaryotes lack an ortholog based on sequence homology, functional regulation of the methyl-phosphorylation regulatory cascade remains conserved in high eukaryotic kingdom. It is worth noting that SUV39H1 is phosphorylated in serines and thronines in mitosis (Aagaard et al., 2000), and the great challenge and excitement ahead are to test if Aurora B phosphorylates SUV39H1 and if this phosphorylation liberates SUV39H1 from kinetochore and therefore reduces the methylation gradient in the kinetochore.
Surprisingly, this inhibition of methylation results in a brief increase in Aurora B kinase activity and an enrichment of microtubule depolymerase MCAK in the centromere. This elevated MCAK promotes kinetochore–microtubule destabilization and reduces the tension across the sister kinetochores, which contributes to chromosome misalignment in methylation-inhibited HeLa cells. We reason that SUV39H1 generates a gradient of methylation marks that provides spatiotemporal information essential for accurate chromosome segregation in mitosis.
In summary, our findings reveal the temporal dynamics of centromere methylation in living HeLa cells during mitosis and demonstrate critical roles of SUV39H1 in mitotic congression to the metaphase plate. Our study indicates that SUV39H1 activity orchestrates kinetochore–microtubule dynamics via mitotic kinases feedback regulation and depolymerase machinery at the kinetochore. We propose that the SUV39H1 methylation constitutes a feedback regulatory network with mitotic kinases PLK1-Aurora B in the centromere, which is critical for correcting aberrant spindle microtubule attachments and chromosome stability in the cell division cycle. Further molecular delineation will uncover the underlying regulatory basis for temporal regulation of SUV39H1-MCAK axis in the centromere during mitosis.
Materials and methods
Cell culture, synchronization, and drugs
HeLa from American Type Culture Collection (Rockville) was maintained as subconfluent monolayers in Dulbecco's modified Eagle's medium (Invitrogen) with 10% fetal bovine serum (FBS; Hyclone) and 100 U/ml penicillin plus 100 μg/ml streptomycin (Invitrogen) at 37°C with 8% CO2. Cells were synchronized at G1/S with 5 mM thymidine for 16 h and then washed with phosphate-buffered saline (PBS) three times and cultured in thymidine-free medium to release. Where indicated, 200 ng/ml nocodazole (Sigma-Aldrich), 100 nM Hesperadin, 40 μM monastrol (Sigma), and/or 1.3 μM chaetocin, 10 μM BIX01294 were added.
Plasmid construction and transfection
The pcDNA 3.0 H3K9 methylation sensor was a gift from Alice Y. Ting (Lin et al., 2004). The cDNA of centromeric DNA-binding domain of CENP-B (amino acids 1–176) was cloned into H3K9 methylation sensor by HindIII and BamHI sites. We modified the sensor design to get sufficient fluorescence intensity for ratio-metric FRET measurements: CFP and YFP were replaced with mCerulean and mVenus, respectively. The centromere-targeted Aurora B sensor was constructed as previously described (Chu et al., 2011).
For immunofluorescence, cells were seeded onto sterile, acid-treated 12-mm coverslips in 24-well plates (Corning Glass Works). Thymidine-blocked and released HeLa cells were transfected with 1 μl of Lipofectamine 2000 pre-mixed with 1 μg of various plasmids as described above.
Antibodies and immunofluorescence
Immunoblots and immunofluorescence experiments were performed using the following antibodies: SUV39H1 antibody (anti-rabbit 1:500, Abcam), H3K9me3 (anti-rabbit 1:800, Cell Signaling), tubulin (anti-mouse 1:1000, Sigma), ACA (anti-human 1:400), MCAK (anti-mouse 1:1000), Hec1 (anti-mouse 1:500, Abcam), and AIM-1/Aurora B (anti-mouse 1:500, BD Biosciences).
Briefly, 24–36 h after transfection, HeLa cells were rinsed for 1 min with PHEM buffer (100 mM PIPES, 20 mM HEPES, pH 6.9, 5 mM EGTA, 2 mM MgCl2, and 4 M glycerol) and were permeabilized for 1 min with PHEM plus 0.1% Triton X-100 as previously described (Yao et al., 1997). Extracted cells were fixed in freshly prepared 3.7% paraformaldehyde in PHEM and rinsed three times in PBS. Cells on the coverslips were blocked with 0.05% Tween 20 in PBS (Tween-PBS) with 1% bovine serum albumin (Sigma). These cells were incubated with various primary antibodies in a humidified chamber for 1 h and then washed three times in TPBS. Primary antibodies were visualized using FITC-conjugated goat anti-mouse IgG or rhodamine-conjugated goat anti-rabbit IgG. DNA was stained with 4,6-diamidino-2-phenylindole (Sigma). For an analysis of cold-stable microtubules, cells were incubated in L15 media containing 20 mM Hepes (pH 7.3) on ice for 10 min and then fixed as described above.
Fluorescence intensity quantification and kinetochore distance measurement
The fluorescence intensity of kinetochore protein labeling was measured using a Zeiss LSM 510 NLO confocal microscope scan head mounted transversely on an Axiovert 200 inverted microscope with a 100×NA 1.3 PlanApo objective. The images from double labeling were collected using a dichroic filter set with Zeiss LSM 5 image processing software. The distance between sister kinetochores marked with ACA was measured as the distance between the peak fluorescence as described previously (Yao et al., 2000; Liu et al., 2007). To test whether repression of SUV39H1 activity by chaetocin or elimination of SUV39H1 protein reduces the tension across the sister kinetochore, we added 100 nM nocodazole to the aliquots of siRNA-treated cells for an additional 18 h (Yao et al., 2000). The distance between sister kinetochores marked with ACA was measured as described above.
Quantification of the level of kinetochore-associated protein was conducted as described by Liu et al. (2007). In brief, the average pixel intensities from at least 100 kinetochore pairs from 10 cells were measured, and background pixel intensities were subtracted. The pixel intensities at each kinetochore pair were then normalized against ACA pixel values to account for any variations in staining or image acquisition. The values of specific siRNA-treated cells were then plotted as a percentage of the values obtained from cells transfected with a control siRNA duplex, or DMSO-treated cells in the case of chaetocin treatment.
Live cell imaging and FRET assay
HeLa cells expressing different kinds of plasmids entered mitosis 7 h after thymidine release were observed. Transfected cells grown on glass-based dishes (IWAKI) were supplemented with CO2-independent medium (Gibco) and observed using the DeltaVision RT system (Applied Precision) at a temperature of 37°C. The mCherry images were taken at 5 min intervals with an exposure time of 0.1 sec. For live imaging of methylation sensor, CFP was excited at 470 nm, and CFP and YFP emissions were acquired simultaneously with a beam splitter (Dual-View, Optical Insights). Individual centromeres or kinetochores were defined automatically from confocal image stacks, and the YFP/CFP or FRET/CFP emission ratio was calculated at each centromere/kinetochore. For live imaging of phosphorylation sensors, the method was the same as described above.
Image acquisition and processing
Immunofluorescence images were collected on an inverted microscope (Olympus IX-70) and a 60×NA 1.4 PlanApo objective. 0.2-μm step sections were acquired using a 100×NA 1.3 PlanApo objective. Olympus Acquisition parameters, including exposure, focus, and illumination, were controlled by SoftWorks (Applied Precision). Z stack projection, subsequent analysis, and processing of images were performed using SoftWorks (Applied Precision). For the analysis of microtubule attachments, images were deconvolved using the DeltaVision software (Applied Precision). Measurements of the intensity of kinetochore localization were conducted on non-deconvolved images. All images for a specific experiment used identical exposure settings and scaling as described (Huang et al., 2012).
For live cell imaging, the medium was replaced with CO2-independent medium supplemented with 10% FBS, penicillin/streptomycin, and l-glutamine (Invitrogen) was covered with mineral oil with a 60×NA 1.4 PlanApo objective lens together with a filter wheel. Images were analyzed with DeltaVision deconvolution system (Applied Precision).
Data analyses
All distance and fluorescence intensity measurements were made using MetaMorph software. Inter-kinetochore distances were measured using the centers of the paired CENP-A dots. Kinetochore fluorescence intensities were determined by measuring the integrated fluorescence intensity within a 7 × 7-pixel square positioned over a single kinetochore and subtracting the background intensity of a 7 × 7-pixel square positioned in a region of cytoplasm lacking kinetochores. Maximal projected images were used for these measurements.
To determine the significant differences between means, unpaired t-tests assuming unequal variance were performed; differences were considered significant when P < 0.05.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by National Basic Research Program (973) (2010CB912103, 2012CB917200, 2007CB914503 and 2002CB713700), Chinese Natural Science Foundation Grants (30870990, 90508002, 90913016, 39925018 and 91129714), Chinese Academy of Science Grants (KSCX1-YW-R-65, KSCX2-YW-H-10, and KSCX2-YW-R-195), Ministry of Education (20113402130010), International Collaboration Grant (2009DFA31010) and National Institutes of Health Grants (DK-56292 and CA118948), and Anhui Province Project Grant (08040102005). The facilities were supported, in whole or in part, by NCRR Grant (G12RR03034; for use of facilities). X.Y. is a GCC Eminent Scholar.
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank Dr Alice Ting (MIT, USA) for providing pcDNA3.0-H3K9 plasmid. We thank members of our groups for insightful discussion during the course of this study.
References
- Aagaard L., Schmid M., Warburton P., et al. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- Andrews P.D., Ovechkina Y., Morrice N., et al. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Cleveland D.W., Mao Y., Sullivan K.F. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–409. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., et al. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Chu Y., Yao P.Y., Wang W., et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J. Mol. Cell Biol. 2011;3:260–267. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czvitkovich S., Sauer S., Peters A.H., et al. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech. Dev. 2001;107:141–153. doi: 10.1016/s0925-4773(01)00464-6. [DOI] [PubMed] [Google Scholar]
- Ding X., Yan F., Yao P., et al. Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 2010;20:1386–1389. doi: 10.1038/cr.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Tseng B.S., Dormann H.L., et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Fu G., Ding X., Yuan K., et al. Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 2007;17:608–618. doi: 10.1038/cr.2007.55. [DOI] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner D., Bonaldi T., Eskeland R., et al. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 2005;1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- Hua S., Wang Z., Jiang K., et al. CENP-U cooperates with Hec1 to orchestrate kinetochore–microtubule attachment. J. Biol. Chem. 2011;286:1627–1638. doi: 10.1074/jbc.M110.174946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang W., Yao P., et al. CENP-E interacts with SKAP to orchestrate chromosome segregation in mitosis. J. Biol. Chem. 2012;287:1500–1509. doi: 10.1074/jbc.M111.277194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y.W., Dou Z., Zhang J., et al. Function and regulation of Aurora/Ipl1p kinase family in cell division. Cell Res. 2003;13:69–81. doi: 10.1038/sj.cr.7290152. [DOI] [PubMed] [Google Scholar]
- Lahtz C., Pfeifer G.P. Epigenetic changes of DNA repair genes in cancer. J. Mol. Cell Biol. 2011;3:51–58. doi: 10.1093/jmcb/mjq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W., Zhang X., Kline-Smith S.L., et al. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr. Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Latham J.A., Chosed R.J., Wang S., et al. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell. 2011;146:709–719. doi: 10.1016/j.cell.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.W., Jao C.Y., Ting A.Y. Genetically encoded fluorescent reporters of histone methylation in living cells. J. Am. Chem. Soc. 2004;126:5982–5983. doi: 10.1021/ja038854h. [DOI] [PubMed] [Google Scholar]
- Liu D., Ding X., Du J., et al. Human NUF2 interacts with centromere-associated protein E and is essential for a stable spindle microtubule-kinetochore attachment. J. Biol. Chem. 2007;282:21415–21424. doi: 10.1074/jbc.M609026200. [DOI] [PubMed] [Google Scholar]
- McManus K.J., Biron V.L., Heit R., et al. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. 2006;281:8888–8897. doi: 10.1074/jbc.M505323200. [DOI] [PubMed] [Google Scholar]
- Moore A.T., Rankin K.E., von Dassow G., et al. MCAK associates with the tips of polymerizing microtubules. J. Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata-Hori M., Wang Y.L. The kinase activity of Aurora B is required for kinetochore–microtubule interactions during mitosis. Curr. Biol. 2002;12:894–899. doi: 10.1016/s0960-9822(02)00848-5. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Pinsky B.A., Kotwaliwale C.V., Tatsutani S.Y., et al. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H., Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Salimian K.J., Ballister E.R., Smoak E.M., et al. Feedback control in sensing chromosome biorientation by the Aurora B kinase. Curr. Biol. 2011;21:1158–1165. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. Genomic instability and cancer: an introduction. J. Mol. Cell Biol. 2011;3:1–3. doi: 10.1093/jmcb/mjq057. [DOI] [PubMed] [Google Scholar]
- Terada Y. Aurora-B/AIM-1 regulates the dynamic behavior of HP1α at the G2-M transition. Mol. Biol. Cell. 2006;17:3232–3241. doi: 10.1091/mbc.E05-09-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu X., Ding X., et al. Human Zwint-1 specifies localization of Zeste White 10 to kinetochores and is essential for mitotic checkpoint signaling. J. Biol. Chem. 2004;279:54590–54598. doi: 10.1074/jbc.M407588200. [DOI] [PubMed] [Google Scholar]
- Yang Y., Wu F., Ward T., et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J. Biol. Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Anderson K.L., Cleveland D.W. The microtubule-dependent motor centromere- associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J. Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., et al. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yu H., Yao X. Cyclin B1: conductor of mitotic symphony orchestra. Cell Res. 2008;18:218–220. doi: 10.1038/cr.2008.20. [DOI] [PubMed] [Google Scholar]
- Zhang K., Lin W., Latham J.A., et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Shao H., Huang Y., et al. PLK1 phosphorylates mitotic centromere-associated kinesin and promotes its depolymerase activity. J. Biol. Chem. 2011;286:3033–3046. doi: 10.1074/jbc.M110.165340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.