Abstract
Late-onset Alzheimer's disease (AD) is 50–70% heritable with complex genetic underpinnings. In addition to Apoliprotein E (APOE) ε4, the major genetic risk factor, recent genome-wide association studies (GWAS) have identified a growing list of sequence variations associated with the disease. Building on a prior large-scale AD GWAS, we used a recently developed analytic method to compute a polygenic score that involves up to 26 independent common sequence variants and is associated with AD dementia, above and beyond APOE. We then examined the associations between the polygenic score and the magnetic resonance imaging–derived thickness measurements across AD-vulnerable cortex in clinically normal (CN) human subjects (N = 104). AD-specific cortical thickness was correlated with the polygenic risk score, even after controlling for APOE genotype and cerebrospinal fluid (CSF) levels of β-amyloid (Aβ1–42). Furthermore, the association remained significant in CN subjects with levels of CSF Aβ1–42 in the normal range and in APOE ε3 homozygotes. The observation that genetic risk variants are associated with thickness across AD-vulnerable regions of interest in CN older individuals, suggests that the combination of polygenic risk profile, neuroimaging, and CSF biomarkers may hold synergistic potential to aid in the prediction of future cognitive decline.
Keywords: Alzheimer's disease, imaging genetics, polygenic score
Introduction
Late-onset Alzheimer's disease (AD) is a complex disease with heritability estimates of 50–70% (Gatz et al. 1997; Pedersen et al. 2004). While the ε4 allele of Apoliprotein E (APOE) is the best-established genetic risk factor (Saunders et al. 1993; Strittmatter et al. 1993), recent genome-wide association studies (GWAS) have identified novel common DNA variants associated with AD (Harold et al. 2009; Lambert et al. 2009; Seshadri et al. 2010; Hollingworth et al. 2011; Naj et al. 2011), and a growing number of studies have pointed to further candidate loci (Waring and Rosenberg 2008; Bertram and Tanzi 2009). An open question is whether these newly identified genetic risk variants are associated with markers of neurodegeneration and, if so, how early in the progression of the disease.
Clinical AD is preceded by a long asymptomatic phase, which is characterized by the aberrant processing and accumulation of amyloid and tau variants in the brain (Grundke-Iqbal et al. 1986; Price and Morris 1999; Selkoe 2004; Chiti and Dobson 2006). Current in vivo markers of amyloid and tau pathology include cerebrospinal fluid (CSF) concentrations of Aβ1–42, phosphorylated tau (p-tau) and total tau (t-tau) (Buerger et al. 2006; Shaw et al. 2009), and amyloid-binding positron emission tomography (PET) tracers, for example, Pittsburgh Compound B (PIB) (Klunk et al. 2004). Superthreshold levels of amyloid and/or tau burden in clinically normal (CN) individuals have been associated with a heightened risk for future dementia (De Meyer et al. 2010; Villemagne et al. 2011). Thus, preclinical AD research has increasingly relied on stratifying CN subjects based on PIB PET and/or CSF measurements (Pike et al. 2007; Villemagne et al. 2008).
The prevailing AD model (Jack et al. 2010) predicts that amyloid pathology reaches a plateau before atrophy in vulnerable brain regions (e.g., the medial temporal lobe and association cortices) can be detected in vivo, for example, via magnetic resonance imaging (MRI) (Fox, Freeborough, et al. 1996; Jack et al. 1997; Dickerson et al. 2009; Frisoni et al. 2010). Although increased rates of atrophy have been shown to precede the clinical diagnosis of familial AD (Fox, Warrington, et al. 1996; Ridha et al. 2006), accelerated atrophy is often assumed to coincide with cognitive decline during the mild cognitive impairment (MCI) or dementia phases of late-onset AD (Fox et al. 1999; Jack et al. 1999).
Several neuroimaging studies have demonstrated APOE-associated functional, metabolic, and structural variation in CN individuals (Reiman et al. 1998, 2004; Small et al. 2000; Wishart et al. 2006; Filippini et al. 2009; Sheline et al. 2010). However, how newly identified genetic risk variants influence structural variation and how they relate to the effects of APOE remain largely unknown. In the present study, we built on a prior GWAS and computed a polygenic score (Purcell et al. 2009; Biffi et al. 2010) that is associated with clinical AD above and beyond APOE. We then examined the association between the polygenic score and the structural MRI-based thickness measurements within a priori AD-vulnerable cortical regions across the heteromodal association cortex and medial temporal lobe in CN subjects. Given the known significant heterogeneity in the risk of clinical progression among normal older individuals, even after stratification by APOE genotype and amyloid burden, we hypothesized that the polygenic score would provide additional explanation to the remaining variance in cortical thickness.
Materials and Methods
ADNI Data
Our analyses focused on the Alzheimer's Disease Neuroimaging Initiative (ADNI, http://www.adni-info.org) subjects who were clinically diagnosed as normal (zero clinical dementia rating [CDR] and no objective memory loss) with available CSF measurements of tau and Aβ1–42 concentration (N = 116). We removed 12 individuals from this sample because they either failed the genotype quality control (QC) tests or were of non-European descent, as determined based on genome-wide data, resulting in a sample of 104 CN for further analyses. In the first part of our analysis, in order to assess the clinical value of the polygenic score, we compared the CN group with the ADNI AD subjects with CSF samples (N = 100). Table 1 provides summary statistics for the analysis sample. The CN and AD groups did not significantly differ in age, gender, and education (all P > 0.1). As expected, the groups differed on measures of disease severity: Mini-Mental State Examination (MMSE) and Clinical Dementia Rating Sum of Boxes (CDR-SB) (P < 0.001). CDR-SB is a refined version of the global CDR and provides increased sensitivity in tracking disease progression within and across various stages of disease severity (O'Bryant et al. 2008).
Table 1.
Summary statistics for the ADNI sample used in the analyses
| Variable | CN (N = 104) | AD (N = 100) |
| Age | 75.9 (5.1) | 75.1 (7.8) |
| Female % | 48 | 42 |
| Education (years) | 15.9 (2.7) | 15.2 (3.3) |
| CDR-SB* | 0.02 (0.1) | 4.2 (1.5) |
| MMSE* | 29.1 (1.0) | 23.6 (1.9) |
| CSF Aβ1–42 (pg/mL)* | 205.5 (55.5) | 143.6 (40.7) |
| CSF t-tau (pg/mL)* | 71.3 (31.0) | 119.0 (59.9) |
| CSF p-tau (pg/mL)* | 25.5 (15.1) | 41.4 (19.7) |
| Cortical thickness (mm)* | 2.71 (0.17) | 2.41 (0.20) |
Note: Mean values are listed with SD in parentheses. Cortical thickness: average thickness in AD-vulnerable cortical regions, AD: Alzheimer's patients.
*Indicates statistically significant group differences, all P < 0.001 (two-sampled t-test).
An independent portion of the ADNI sample (called the discovery sample) in which CSF data were not available was utilized to construct the polygenic score (N = 197).
Genotype Processing
We merged individual-level genotype data from the ADNI database (ADNI) into a single data set containing genome-wide single nucleotide polymorphism (SNP) information for 810 individuals (female ratio 41.85%). We performed the following genotype QC using PLINK version 1.07 (Purcell et al. 2007). First, we removed individuals with missing genotype rates of greater than 0.05. Subjects were additionally removed if they were outliers based on nearest neighbor-based clustering using identity-by-state measures. Next, we applied SNP-level filtering, removing SNPs with a minor allele frequency less than 0.01, missing rates greater than 0.05, and significant departure from Hardy–Weinberg Equilibrium (P < 1 × 10−6). SNPs were further removed if genotype missingness was significantly different between cases and controls (P < 1 × 10−6) or if there was nonrandom missingness based on the PLINK haplotype test. To control for population stratification, we conducted a multidimensional scaling (MDS) analysis on the ADNI data with the HapMap phase 3 reference data set (Thorisson et al. 2005; Price et al. 2006). Based on clustering with 988 HapMap subjects using the first 2 MDS components, we retained 745 ADNI individuals with European ancestry (female ratio of 40.81%). The first 3 MDS components were highly correlated with the ADNI case/control data and thus were included as covariates in all remaining analyses. Total genotyping rates for final QC'ed ADNI genotype data were 99.72% for 548 340 SNPs. Ungenotyped SNPs were imputed using Beagle v.3.3.1 (Browning BL and Browning SR 2009) with HapMap phase 3 CEU reference data. Further analysis focused on a total of 986 993 autosomal SNPs with imputation quality scores not less than 0.8.
Computation of Polygenic Score
To compute the polygenic AD-risk score, we used the “score” utility in PLINK (Purcell et al. 2009) and the detailed GWAS results reported in the supplemental material of Harold et al. (2009). First, we obtained the list of 761 SNPs that showed nominal association with AD (P ≤ 1 × 10−3) in (Harold et al. 2009) (3941 cases and 7848 controls). To account for only independent association signals from the list of AD-susceptibility SNPs, we conducted linkage disequilibrium –based clumping, implemented in PLINK, and selected the index SNPs with the most significant association P value from each clumped association region based on the (Harold et al. 2009) GWAS. The polygenic score of ADNI subjects was then calculated as the sum of the number of susceptibility alleles of the index SNPs, weighted by the logarithm of the corresponding odds ratios (ORs). Of note, for imputed SNPs in the ADNI data, expected allele counts (i.e., dosage data) were used in the scoring process in order to reflect imputation uncertainty.
The SNPs that make up the score were determined by identifying those exceeding a certain statistical threshold in the (Harold et al. 2009) GWAS. In general, a more stringent threshold yields a score consisting of a smaller number of SNPs with a higher fraction of them being truly associated with AD. On the other hand, an increasing number of truly associated SNPs (which can be obtained by relaxing the statistical threshold) can increase the effect size of the score and thus improve our statistical power for detecting associations with disease correlates on a limited sample size. Our initial goal was therefore to conduct a discovery analysis and identify a good threshold that would yield a score with as many truly associated SNPs as possible. Follow-up analyses were then conducted to demonstrate the utility and clinical relevance of this score on an independent analysis sample.
The discovery analysis to determine the statistical threshold for establishing the list of SNPs that would contribute to the polygenic score was performed on a portion of the ADNI data (discovery sample; CN and AD patients with no CSF, N = 197) that was independent from the analysis sample that consisted of the ADNI data with CSF measurements. We evaluated 5 different threshold values: 1 × 10−3, 1 × 10−4, 1 × 10−5, 1 × 10−6, and 1 × 10−7, yielding 5 different polygenic scores. These scores were entered into a logistic regression (CN vs. AD) and correlated with MMSE and CDR-SB in the ADNI discovery sample (see Supplementary Fig. 1). 1 × 10−5 yielded the highest correlation values and strongest association with AD diagnosis (P value = 1.47 × 10−7) and was therefore used to compute the polygenic score in the remaining analyses on the independent analysis sample. The list of SNPs contributing to this polygenic score and corresponding GWAS ORs are listed in Supplementary Table 1. The closest genes for these SNPs and a list of studies that report these genes as promising associations with AD are included in Supplementary Table 2.
MRI Processing
We used FreeSurfer version 4.5 to process all MRI scans automatically (FreeSurfer, http://surfer.nmr.mgh.harvard.edu/). We first utilized the independent cross-sectional The Open Access Series of Imaging Studies (OASIS) data set (Marcus et al. 2007) consisting of 94 participants to generate an exploratory map of cortical thickness differences between older controls (N = 47, 58% female, mean age ± standard deviation [SD]: 78 ± 5.6 years) and CDR 0.5 individuals clinically classified as incipient AD (N = 47, 63% female, 76.4 ± 4.7 years). Based on the resulting statistical maps, we delineated seven regions of interest (ROIs) on the average cortical surface template that demonstrated the greatest magnitude of bilateral cortical thinning in incipient AD participants relative to older controls. These regions include the entorhinal cortex, temporopolar cortex, lateral temporal cortex, inferior parietal cortex, inferior parietal sulcus, posterior cingulate cortex, and inferior frontal cortex. These ROIs and corresponding statistical maps have been published elsewhere (Sabuncu et al. 2011) and are made publicly available with FreeSurfer. We further used FreeSurfer's longitudinal stream to process a set of serial MRIs (baseline and month 12) from each study participant; this stream yields accurate and unbiased estimates of subtle changes over time (Reuter and Fischl 2011).
We transferred the OASIS-derived AD-vulnerable ROIs from the surface template onto the individual ADNI subjects’ cortical representations via surface-based registration (Fischl, Sereno, Dale 1999; Fischl, Sereno, Tootell, et al. 1999; Fischl et al. 2001; Ségonne et al. 2007). We computed average thickness values (across the cortical ROIs in both hemispheres) and used these measurements in all our analyses. Average thickness measurements from the automatically delineated cortical gyral ROIs (parcels covering the whole cortex) were used for a supplementary analysis (Desikan et al. 2006).
Statistical Analysis
All statistical analyses were conducted using the general linear model framework implemented in Matlab. Age, sex, and education (years) were included as covariates in all analyses. To control for APOE genotype, we included both ε2 and ε4 allele counts as covariates (The ADNI sample we analyzed included 17 subjects with the APOE ϵ2/ϵ4 combination. Only 2 of these were CN and 1 was included in the main analysis (i.e., had a CSF sample). Excluding this subject from all our analyses did not alter our main results.). As effect size, we reported partial correlations (ρ) between the polygenic score and the outcome of interest (such as cortical thickness) and linear coefficients (β) for the effect of APOE allele counts on the outcome.
Results
Polygenic Score Predicts Dementia Above and Beyond APOE
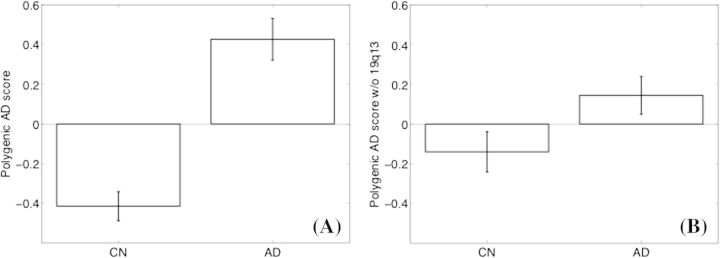
The polygenic score was significantly associated with CDR-SB (ρ = 0.40, P < 0.001), MMSE (ρ = −0.34, P < 0.001), and AD diagnosis (1 SD increase in the polygenic score was associated with an increase of 1.06 in the log-odds of AD diagnosis, P < 0.001) in the CN and AD combined analysis sample (N = 204). To explore non-APOE contributions, we excluded the APOE-linkage region 19q13 from the polygenic score computation and controlled for APOE genotype by including APOE ε4 and ε2 allele counts as covariates. The association between the non-APOE polygenic score and the CDR-SB (ρ = 0.20, P < 0.005), the MMSE (ρ = −0.14, P < 0.05), and the AD diagnosis (1 SD increase in the non-APOE polygenic score was associated with an increase of 0.44 in the log-odds of AD diagnosis, P < 0.01) remained significant. Figure 1 shows the average polygenic score values in the CN and AD groups. Supplementary Table 1 lists the individual SNPs included in the polygenic score and corresponding associations in the GWAS (Harold et al. 2009) and ADNI samples.
Figure 1.
Mean polygenic scores (z score normalized across the entire analysis sample) in CN (N = 104) and AD (N = 100) groups. Error bars show standard error of the mean. (A) The polygenic score based on all 26 linkage disequilibrium–independent SNPs, (B) excluding 19q13, the APOE-linkage region.
These results demonstrated the clinical relevance of the polygenic score using the CN and AD combined analysis sample. All remaining analyses focused on the CN analysis sample.
Polygenic Score Is Correlated with AD-Specific Cortical Thickness in CN Subjects
In CN subjects, the average thickness in AD-vulnerable cortex was significantly associated with the polygenic score (ρ = −0.195, P < 0.05). Supplementary Table 3 lists SNP-level associations with AD-specific cortical thickness, whereas Table 2 lists the partial correlations between the thickness measurements from individual ROIs and the polygenic risk score.
Table 2.
Partial correlation between polygenic score and average thickness of individual cortical ROIs in the CN group (N = 104)
| Partial correlation | |
| Inferior frontal cortex | −0.17 |
| Inferior parietal cortex | −0.10 |
| Inferior parietal sulcus | −0.06 |
| Lateral temporal | −0.08 |
| Medial temporal lobe | −0.02 |
| Retrosplenial/posterior cingulate | −0.27* |
| Temporal pole | −0.15 |
Note: Age, sex, education (years), and the first 3 principal components from the population substructure analysis were included as control variables.
*Indicates P < 0.01.
We further conducted a supplementary analysis across the entire cortex, where we correlated the average thickness within each cortical ROI of the Desikan atlas (Desikan et al. 2006) and the polygenic score (see Supplementary Fig. 2). Isthmus of the cingulate, a region that overlaps with the manually defined AD-vulnerable posterior cingulate ROI, exhibited a statistically significant association (P < 0.05, Bonferroni corrected), while inferior temporal and orbitofrontal regions, which are known to be targeted in early AD, exhibited a suggestive association. However, most other cortical regions exhibited no statistically significant association between thickness and polygenic risk. For example, across 2 regions that are known to show little, if any, atrophy during early AD (primary motor and sensory cortices) the partial correlation was +0.10 (P = 0.31, uncorrected. Note that we expect the true correlation to be negative). Furthermore, the average cortical thickness across the 2 hemispheres was not associated with the polygenic score in CN subjects (ρ = −0.006, P = 0.95, uncorrected).
The Association between AD-Specific Cortical Thickness and Polygenic Score Is Above and Beyond APOE
Next, we examined APOE and non-APOE contributions to the association between the AD-specific cortical thickness and the polygenic score. APOE ε2 allele was significantly associated with AD-specific cortical thickness in a load-dependent manner (β = 0.09, P = 0.03), but the ε4 allele was not (P = 0.83). The non-APOE polygenic score (that excludes 19q13) was significantly correlated with the thickness measurements in CN subjects (ρ = −0.26, P < 0.01; including APOE as covariate). In APOE ε3 homozygote CN subjects (N = 61, 50.8% female), the association between the non-APOE polygenic score and the AD-specific cortical thickness remained significant (ρ = −0.28, P < 0.05).
CSF Aβ1–42 Does Not Fully Explain the Association between Cortical Thickness and Polygenic Score
Of the 3 CSF measurements (CSF Aβ1–42, p-tau, and t-tau), only Aβ1–42 was significantly associated with the polygenic score in the CN group (ρ = −0.45, P < 10−5). This association remained significant (ρ = −0.29, P < 0.05) even among those CN subjects with CSF Aβ1–42 levels greater than 192 pg/mL, the threshold previously implicated in predicting progression from mild cognitive impairment to AD dementia (Shaw et al. 2009) (N = 64, 48.4% female). Henceforth, we refer to this group as CN with subthreshold amyloid burden, since high CSF Aβ1–42 is an indication of low amyloid deposition in the brain (Fagan et al. 2006).
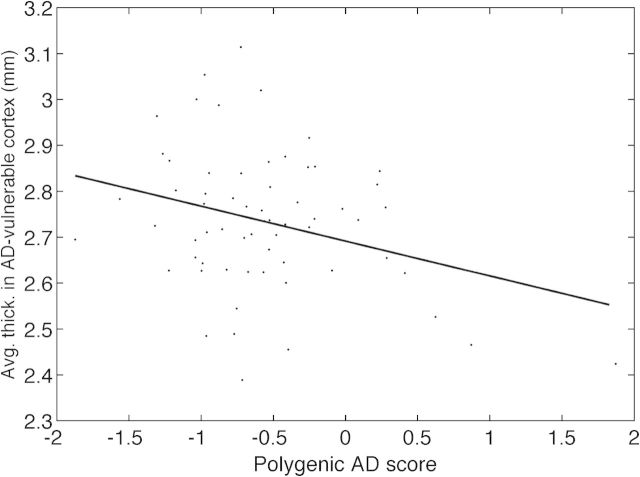
To examine whether CSF Aβ1–42 explained the correlation between the cortical thickness and the polygenic score, we first conducted an analysis where we included CSF Aβ1–42 as a covariate. In the entire CN group, AD-specific cortical thickness remained significantly correlated with the non-APOE polygenic score (ρ = −0.27, P < 0.01), whereas the association with APOE ε2 weakened to trend level (β = 0.06, P = 0.08). In CN individuals with subthreshold amyloid burden, the association between the AD-vulnerable cortical thickness and the non-APOE polygenic score remained significant (ρ = −0.29, P < 0.05; including CSF Aβ1–42 as a covariate) (see Fig. 2).
Figure 2.
Thickness across AD-vulnerable cortex versus Alzheimer-associated non-APOE polygenic score in CN subjects with subthreshold levels of amyloid burden (N = 64, ρ = −0.29, P < 0.05).
The Association between AD-specific Cortical Thickness and Polygenic Score in CN Subjects Is Possibly Driven by a Neurodegenerative Effect
Our analyses so far demonstrated polygenic AD-related variation in thickness measurements across the vulnerable cortex in CN individuals, even among those without evidence of abnormal brain Aβ deposition. There are at least 2 possible explanations for this effect: 1) the polygenic risk is associated with cortical thinning via a modulation of the neurodegenerative mechanisms or 2) the polygenic risk reflects variation in “brain reserve,” that is, the amount of brain tissue available before neurodegeneration begins.
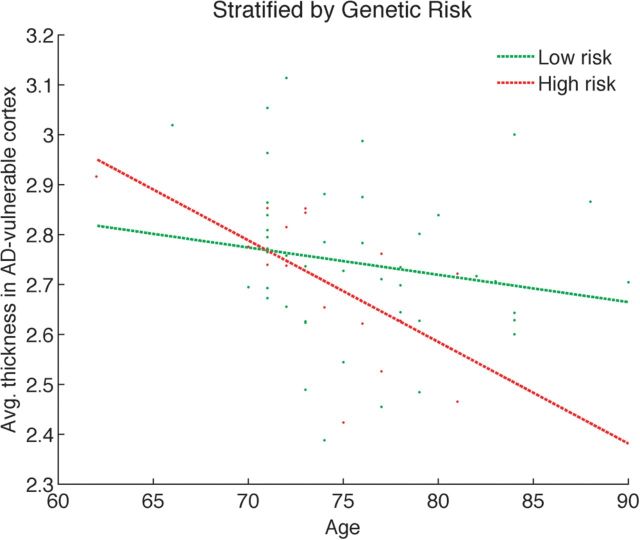
We conducted an exploratory analysis to assess which one of the 2 scenarios provides a better explanation for the association between polygenic score and AD-specific cortical thickness we observe in our data. We divided the CN group with subthreshold amyloid burden into 2 subgroups based on the non-APOE polygenic score. Those with a score greater than the average score were classified as “high risk” (N = 17), while the remaining subjects were considered “low risk” (N = 46). We then examined the relationship between AD-specific cortical thickness and subject age in these 2 groups (see Fig. 3). A stepwise regression (with sex, education, and the first 3 principal components from the population substructure analysis as control variables) revealed that the slopes of these 2 groups were statistically significantly different (P < 0.05), yet their offsets were not. These results remained robust after discarding the 2 outlier subjects who were younger than 70 years (see Supplementary Fig. 3). These results provided preliminary support for the “variable neurodegeneration” hypothesis and not the brain reserve hypothesis, which would predict different offsets for the 2 groups.
Figure 3.
Cortical thickness versus age in CN subjects with subthreshold levels of amyloid burden, stratified by polygenic score. The high risk group (N = 17) has a significantly steeper slope than the low risk group (N = 46) (P < 0.05).
Finally, to test the variable neurodegeneration hypothesis directly, we conducted a preliminary analysis on longitudinal (annual) thinning measurements in the CN subjects who received a month 12 MRI scan (N = 91, 46% female, 75.2 ± 5.0 years). Average thinning measurements were computed as baseline thickness minus month 12 thickness divided by the time difference, and the processing was done with FreeSurfer's longitudinal stream. The partial correlation between the annual thinning rate and the non-APOE polygenic score was 0.18, which was significant at a trend level (P = 0.07).
Discussion
The Clinical Associations of the Polygenic Score
Late-onset AD is a polygenic disease, with APOE contributing most to the underlying genetic susceptibility. Large-scale case–control studies are expanding the list of genetic variations associated with AD (Waring and Rosenberg 2008; Bertram and Tanzi 2009; Harold et al. 2009; Lambert et al. 2009; Seshadri et al. 2010; Hollingworth et al. 2011; Naj et al. 2011). While these newly identified genetic loci may provide valuable insights into the mechanisms that lead to AD (van Es and van den Berg 2009) and thus offer targets for intervention, their immediate clinical value is under debate (Pedersen 2010; Seshadri et al. 2010). Furthermore, the details of the associations between these genetic variants and markers of AD remain unknown.
In this study, we demonstrated that aggregating data across many genetic markers, each suspected to be associated with the disease with a small effect size, yield a score that is associated significantly with clinically defined AD dementia. This association is complementary to APOE genotype status, with a partial correlation between the CDR-SB and the polygenic score of 0.20 (N = 204, P < 0.05) after the effect of APOE genotype was removed. The polygenic score offers a way to investigate AD-related variation in biomarker measurements across healthy individuals, similar to previous studies using APOE (Small et al. 2000; Reiman et al. 2004; Filippini et al. 2009; Fleisher et al. 2009; Sheline et al. 2010).
Evidence of AD-Like Neurodegeneration in CN Individuals
Structural MRI allows for the detection of macroscopic tissue atrophy associated with neurodegeneration in AD (Jack et al. 1997; Whitwell et al. 2007; Dickerson et al. 2009; Frisoni et al. 2010). Robust markers of AD based on structural MRI include hippocampal volume (Laakso et al. 1995; Jack et al. 1999; Fischl et al. 2002) and thickness of AD-vulnerable cortex (Dickerson et al. 2009; Desikan et al. 2010). Recent evidence has suggested that cortical thickness in AD-vulnerable regions may be a sensitive marker for AD during the preclinical phase (Sabuncu et al. 2011).
It is often assumed that macroscopically detectable neurodegeneration in AD is preceded by a sequence of molecular events that involve the aberrant accumulation of the proteins Aβ and tau, (e.g., Selkoe 2004). Although there is some evidence that AD-specific atrophy patterns are detectable prior to cognitive impairment (Csernansky et al. 2005; Jagust et al. 2006; Becker et al. 2011; Sabuncu et al. 2011), most empirical observations suggest that atrophy more closely tracks with clinical decline (Jack et al. 2005; Savva et al. 2009; Frisoni et al. 2010). In the present study, evidence for an association between thickness in vulnerable cortical regions and genetic risk to AD was found in CN individuals with subthreshold amyloid burden. Our preliminary analysis suggests that this association is possibly due to a genetic modulation of neurodegeneration, which may be driven by amyloid and/or other factors. This observation agrees with results that demonstrate AD-associated accelerated atrophy rates before the onset of cognitive impairment (Mori et al. 2002; Chen et al. 2007; Schott et al. 2010). However, we need to emphasize that our interpretation is only preliminary and needs to be further elucidated in future longitudinal studies.
CSF Aβ1–42 measurements were not sufficient to explain the correlation between AD-specific cortical thickness and polygenic score in CN individuals. This result may be indicative of a disassociation between AD-like patterns of cortical thinning and fibrillar amyloid, yet, the correlation between the subthreshold levels of amyloid burden (as measured by CSF Aβ1–42) and the polygenic score is consistent with amyloid-mediated mechanisms of neurodegeneration. Another potential mechanism is mitochondrial dysfunction that may precede AD histopathology (Yao et al. 2009; Valla et al. 2010). Although we found evidence of a polygenic association with cortical thickness in AD-vulnerable regions, some of these genes may confer more general vulnerability to oxidative stress, inflammation, microvascular damage, or other processes associated with aging and neurodegenerative diseases. Conversely, some of these genes may reflect varying levels of resilience to neuronal damage due to synaptic density or plasticity mechanisms. Finally, an alternative possibility that we cannot rule out is that the genetic association is in fact due to developmental, and not degenerative, effects—a hypothesis that can be tested in a young and healthy cohort.
The SNP-level associations (Supplementary Table 3) indicate that outside of 19q13, rs7561528 is the SNP that is driving most of the observed effect. This SNP was recently confirmed to be associated with AD and is located close to BIN1 (the bridging integrator 1 gene), which is expressed abundantly in brain tissue (Wechsler-Reya et al. 1997). Amongst the several roles of BIN1 are the promotion of caspase-independent apoptosis, neuronal membrane organization, and clathrin-mediated synaptic vessel formation (Wigge et al. 1997), which is disrupted by Aβ (Kelly and Ferreira 2007). BIN1 has also been associated with schizophrenia (Carrasquillo et al. 2009).
Anatomical ROI-level analyses (Table 2) demonstrate that the associations between thickness measurements and polygenic AD score, while in the same direction, vary in effect size across different regions. This variation might reflect the spatial extent and/or magnitude of vulnerability in the different regions during the early phase of the disease. Intriguingly, the posterior cingulate cortex, a region that is particularly vulnerable to early amyloid deposition (Mintun et al. 2006) and hypometablism in very early AD (Minoshima et al. 1997), demonstrates the strongest association.
Caveats
The general philosophy behind our MRI and genetic association methodology is to build on prior independent studies for determining a priori anatomic and genomic regions the analysis can focus on. We then chose to aggregate signal across these regions in examining associations. The strength of this approach, which has been employed in neuroimaging (Kloppel et al. 2008; Wolk and Dickerson 2010; Sabuncu et al. 2011) and genetics (Purcell et al. 2009), is that statistical power can be boosted substantially and previously undetectable effects may be revealed. There are, however, 2 major weaknesses of this approach. First, the analysis does not allow for inferences outside of the a priori anatomic and genomic regions. Second, our results provide limited insights into the precise relationship between individual anatomic regions and genomic loci. A detailed characterization will require follow-up studies on independent data sets.
The a priori anatomic regions were derived from the OASIS sample and have been used in a prior study (Sabuncu et al. 2011). We restricted our analyses to these ROIs to maximize statistical power and minimize bias. In the prior study, we found that AD-specific cortical thickness was a more sensitive biomarker than hippocampal volume in asymptomatic individuals and more closely tracked CSF Aβ1–42. Therefore, we opted to focus on cortical thickness as a structural MRI-derived marker in the present study. We further note that there may be systematic discrepancies between the OASIS and the ADNI samples that may make these ROIs suboptimal. For example, gender proportion is an obvious difference between the 2 data sets. One way we attempted to control for such discrepancies was to include relevant covariates in all our analyses.
Although the lack of correlation between the polygenic score and whole-brain average cortical thickness or thickness in control regions such as the primary motor cortex would suggest otherwise, we cannot fully exclude the possibility that the reported associations are not AD-specific but due to a more general mechanism. One piece of evidence in favor of this argument is the minimal correlation between thickness in the entorhinal cortex, an ROI that exhibits neurodegeneration in early AD (Gomez-Isla et al. 1996; Du et al. 2003), and the polygenic score in CN subjects. On the other hand, the only region to exhibit an association that was strong enough to survive the correction for multiple comparisons included the posterior cingulate, a region that seems to play a central role in early AD.
The list of SNPs examined in constructing the polygenic score was derived from a single recent GWAS (Harold et al. 2009) because the complete list of nominally associated SNPs and corresponding ORs were reported in that study. The polygenic score was derived based on analyzing each candidate SNP independently and aggregating the data in an additive model (Purcell et al. 2009). Therefore, potential gene–gene interactions (epistasis) were not explicitly considered. Future research should explore more sophisticated multivariate models that will allow for the characterization of epistasis (Cordell 2009) and should incorporate additional SNPs discovered by other studies (e.g., Lambert et al. 2009; Seshadri et al. 2010; Hollingworth et al. 2011; Naj et al. 2011).
A significant weakness of the present study is due its cross-sectional nature. The characterization of Alzheimer-related variation in CN individuals was achieved by examining across subject variation of a genetic risk score. Therefore, the accuracy of the presented interpretations hinges on how predictive, at the individual-level, this score is of future clinical decline toward AD. For example, it is entirely possible that many of the studied CN subjects with “subthreshold amyloid burden,” may not eventually progress to AD, nullifying any preclinical interpretations of the presented results. Future longitudinal studies that follow healthy individuals with genetic susceptibility and/or biomarker evidence suggestive of early AD pathology are required to elucidate the precise links between genetic susceptibility, neurodegenerative/neuroprotective effects, and brain reserve. An interesting direction of research that may shed light on these links would be the further characterization of the presented effects in a younger (e.g., middle-aged) cohort.
Finally, it is important to note that our discussion of the variation in CN individuals strongly depends on our definition of “CN.” In the ADNI study, CNs were selected on the basis of the clinical dementia rating (CDR = 0) and MMSE, which had to be between 24 and 30. Additionally, CNs had to be non-MCI and thus did not suffer from an objective memory loss, as measured by education-adjusted scores on Wechsler Memory Scale-Revised (Wechsler 1987). Further criteria for the categorization of CN were: 1) no active neurological or psychiatric disorders; 2) some subjects may have had ongoing medical problems, yet the illnesses or their treatments did not interfere with cognitive function; 3) normal neurological exam; and 4) were independently functioning community dwellers. Converging evidence suggests that a combination of biomarker abnormalities and subtle cognitive decline in episodic memory and non-memory domains may portend the future onset of MCI and/or AD (Sperling et al. 2011), suggesting that more sensitive cognitive measures may be useful in characterizing preclinical AD-associated behavioral variation. Once such tests are established, future studies will examine the relationships between these test scores and imaging, genetic and CSF markers of AD, and determine the combination of markers that best predicts the likelihood of future decline.
Conclusions
We presented a polygenic score that is associated with clinical dementia, above and beyond APOE genotype and age, the 2 major risk factors for AD. The strength of the polygenic score is that it aggregates evidence from multiple weakly associated genomic loci. Similar to the use of APOE in CN subjects (Wishart et al. 2006; Morris et al. 2010; Sheline et al. 2010), we employed the polygenic score to make inferences about AD biomarkers in CN individuals, a subset of whom may be in the preclinical stages of AD.
The polygenic score was correlated with markers of amyloid but not tau in CN subjects. This is in agreement with the temporal characterization of these biomarkers (Jack et al. 2010) and extends a recent result of a similar APOE ε4/ε2 effect to other loci (Morris et al. 2010).
We further demonstrated an APOE-independent association between the AD-specific cortical thickness and the polygenic score in CN subjects, which also remained in the group of individuals with subthreshold levels of amyloid burden. Our results suggest that CSF biomarkers, structural MRI, and genotype data may provide complementary information about AD-like neurodegenerative processes in asymptomatic individuals. Future longitudinal studies will determine whether this combination of markers will prove useful in more accurately predicting prospective cognitive decline and progression to clinical stages of AD.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/
Funding
Data collection and sharing for this project was funded by the ADNI and the National Institutes of Health (NIH) (grant U01 AG024904). The ADNI is funded by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott Laboratories, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation Plc, Genentech Inc, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson Services Inc, Eli Lilly and Company, Medpace Inc, Merck and Co Inc, Novartis International AG, Pfizer Inc, F. Hoffman-La Roche Ltd, Schering-Plough Corporation, CCBR-SYNARC Inc, and Wyeth Pharmaceuticals, as well as nonprofit partners the Alzheimer's Association and Alzheimer's Drug Discovery Foundation, with participation from the US Food and Drug Administration. Private sector contributions to the ADNI are facilitated by the Foundation for the NIH. The grantee organization is the Northern California Institute for Research and Education Inc, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. This work was supported by NIH grants P01AG036694; P50AG005134; P41RR14074; R01AG021910; and R01AG027435. Further support was provided in part by the National Center for Research Resources (P41-RR14075, and the NCRR BIRN Morphometric Project BIRN002, U24 RR021382), the National Institute for Biomedical Imaging and Bioengineering (R01EB006758), the NIA (AG022381), the National Center for Alternative Medicine (RC1 AT005728-01), the National Institute for Neurological Disorders and Stroke (R01 NS052585-01, 1R21NS072652-01), and the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019, and 1S10RR023043. Additional support was provided by The Autism & Dyslexia Project funded by the Ellison Medical Foundation. Dr. Sabuncu received support from a KL2 Medical Research Investigator Training (MeRIT) grant awarded via Harvard Catalyst, The Harvard Clinical, and Translational Science Center (NIH grant #1KL2RR025757-01 and financial contributions from Harvard University and its affiliated academic health care centers), and an NIH K25 grant (NIBIB 1K25EB013649-01). Additional support was provided by the NIH Blueprint for Neuroscience Research (U01-MH093765, part of the multi-institutional Human Connectome Project).
Supplementary Material
Acknowledgments
Data used in the preparation of this article were obtained from the ADNI database. As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators is available at http://tinyurl.com/ADNI-main. Conflict of Interest: None declared.
References
- Becker JA, Hedden T, Carmasin J, Maye J, Rentz DM, Putcha D, Fischl B, Greve DN, Marshall GA, Salloway S, et al. Amyloid-β associated cortical thinning in clinically normal elderly. Ann Neurol. 2011;69:1032–1042. doi: 10.1002/ana.22333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Genome-wide association studies in Alzheimer's disease. Hum Mol Genet. 2009;18:R137–R145. doi: 10.1093/hmg/ddp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67:677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger K, Ewers M, Pirttilä T, Zinkowski R, Alafuzoff I, Teipel SJ, DeBernardis J, Kerkman D, McCulloch C, Soininen H, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer's disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD, et al. Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet. 2009;41:192–198. doi: 10.1038/ng.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Alexander GE, Caselli RJ, Gerkin R, Bandy D, Domb A, Osborne D, Fox N, Crum WR, et al. Correlations between apolipoprotein E ϵ 4 gene dose and whole brain atrophy rates. Am J Psychiatry. 2007;164:916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cordell HJ. Detecting gene–gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csernansky JG, Wang L, Swank J, Miller JP, Gado M, McKeel D, Miller MI, Morris JC. Preclinical detection of Alzheimer's disease: hippocampal shape and volume predict dementia onset in the elderly. Neuroimage. 2005;25:783–792. doi: 10.1016/j.neuroimage.2004.12.036. [DOI] [PubMed] [Google Scholar]
- De Meyer GD, Shapiro F, Vanderstichele H, Vanmechelen E, Engelborghs S, De Deyn PP, Coart E, Hansson O, Minthon L, Zetterberg H, et al. Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch Neurol. 2010;67:949–956. doi: 10.1001/archneurol.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Sabuncu MR, Schmansky NJ, Reuter M, Cabral HJ, Hess CP, Weiner MW, Biffi A, Anderson CD, Rosand J, et al. Selective disruption of the cerebral neocortex in Alzheimer's disease. PLoS One. 2010;5:e12853. doi: 10.1371/journal.pone.0012853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, et al. The cortical signature of Alzheimer's disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du AT, Schuff N, Zhu XP, Jagust WJ, Miller BL, Reed BR, Kramer JH, Mungas D, Yaffe K, Chui HC, et al. Atrophy rates of entorhinal cortex in AD and normal aging. Neurology. 2003;61:481–486. doi: 10.1212/01.wnl.0000044400.11317.ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan AM, Mintun MA, Mach RH, Lee SY, Dence CS, Shah AR, LaRossa GN, Spinner ML, Klunk WE, Mathis CA, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Aβ42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-ε4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleisher AS, Sherzai A, Taylor C, Langbaum JB, Chen K, Buxton RB. Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer's disease risk groups. Neuroimage. 2009;47:1678–1690. doi: 10.1016/j.neuroimage.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Freeborough PA, Rossor MN. Visualisation and quantification of rates of atrophy in Alzheimer's disease. Lancet. 1996;348:94–97. doi: 10.1016/s0140-6736(96)05228-2. [DOI] [PubMed] [Google Scholar]
- Fox NC, Scahill RI, Crum WR, Rossor MN. Correlation between rates of brain atrophy and cognitive decline in AD. Neurology. 1999;52:1687–1689. doi: 10.1212/wnl.52.8.1687. [DOI] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, Rossor MN. Presymptomatic hippocampal atrophy in Alzheimer's disease: a longitudinal MRI study. Brain. 1996;119:2001–2007. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- Frisoni GB, Fox NC, Jack CR, Jr, Scheltens P, Thompson PM. The clinical use of structural MRI in Alzheimer disease. Nat Rev Neurol. 2010;6:67–77. doi: 10.1038/nrneurol.2009.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer's disease: the study of dementia in Swedish twins. J Gerontol A Biol Sci Med Sci. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- Gómez-Isla T, Price JL, McKeel DW, Jr, Morris JC, Growdon JH, Hyman BT. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer's disease. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, Petersen RC, Trojanowski JQ. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurol. 2010;9:119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, Waring SC, O'Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer's disease. Neurology. 1997;49:786–794. doi: 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Jr, Shiung MM, Weigand SD, O'Brien PC, Gunter JL, Boeve BF, Knopman DS, Smith GE, Ivnik RJ, Tangalos EG, et al. Brain atrophy rates predict subsequent clinical conversion in normal elderly and amnestic MCI. Neurology. 2005;65:1227–1231. doi: 10.1212/01.wnl.0000180958.22678.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust W, Gitcho A, Sun F, Kuczynski B, Mungas D, Haan M. Brain imaging evidence of preclinical Alzheimer's disease in normal aging. Ann Neurol. 2006;59:673–681. doi: 10.1002/ana.20799. [DOI] [PubMed] [Google Scholar]
- Kelly BL, Ferreira A. Beta-amyloid disrupted synaptic vesicle endocytosis in cultured hippocampal neurons. Neuroscience. 2007;147:60–70. doi: 10.1016/j.neuroscience.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöppel S, Stonnington CM, Chu C, Draganski B, Scahill RI, Rohrer JD, Fox NC, Jack CR, Jr, Ashburner J, Frackowiak RS. Automatic classification of MR scans in Alzheimer's disease. Brain. 2008;131:681–689. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- Laakso MP, Soininen H, Partanen K, Helkala EL, Hartikainen P, Vainio P, Hallikainen M, Hänninen T, Riekkinen PJ., Sr Volumes of hippocampus, amygdala and frontal lobes in the MRI-based diagnosis of early Alzheimer's disease: correlation with memory functions. J Neural Transm Park Dis Dement Sect. 1995;9:73–86. doi: 10.1007/BF02252964. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, Klunk WE, Mathis CA, DeKosky ST, Morris JC. [11C] PIB in a nondemented population. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- Mori E, Lee K, Yasuda M, Hashimoto M, Kazui H, Hirono N, Matsui M. Accelerated hippocampal atrophy in Alzheimer's disease with apolipoprotein E ϵ 4 allele. Ann Neurol. 2002;51:209–214. doi: 10.1002/ana.10093. [DOI] [PubMed] [Google Scholar]
- Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67:122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Bryant SE, Waring SC, Cullum CM, Hall J, Lacritz L, Massman PJ, Lupo PJ, Reisch JS, Doody R, Texas Alzheimer's Research Consortium Staging dementia using Clinical Dementia Rating Scale Sum of Boxes scores: a Texas Alzheimer's research consortium study. Arch Neurol. 2008;65:1091–1095. doi: 10.1001/archneur.65.8.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NL. Reaching the limits of genome-wide significance in Alzheimer disease: back to the environment. JAMA. 2010;303:1864–1865. doi: 10.1001/jama.2010.609. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Gatz M, Berg S, Johansson B. How heritable is Alzheimer's disease late in life? Findings from Swedish twins. Ann Neurol. 2004;55:180–185. doi: 10.1002/ana.10999. [DOI] [PubMed] [Google Scholar]
- Pike KE, Savage G, Villemagne VL, Ng S, Moss SA, Maruff P, Mathis CA, Klunk WE, Masters CL, Rowe CC. Beta-amyloid imaging and memory in non-demented individuals: evidence for preclinical Alzheimer's disease. Brain. 2007;130:2837–2844. doi: 10.1093/brain/awm238. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price JL, Morris JC. Tangles and plaques in nondemented aging and preclinical Alzheimer's disease. Ann Neurol. 1999;45:358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly mJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P, Purcell SM, Stone JL, Sullivan PF, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci U S A. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Uecker A, Caselli RJ, Lewis S, Bandy D, De Leon MJ, De Santi S, Convit A, Osborne D, Weaver A, et al. Hippocampal volumes in cognitively normal persons at genetic risk for Alzheimer's disease. Ann Neurol. 1998;44:288–291. doi: 10.1002/ana.410440226. [DOI] [PubMed] [Google Scholar]
- Reuter M, Fischl B. Avoiding asymmetry-induced bias in longitudinal image processing. Neuroimage. 2011;57:19–21. doi: 10.1016/j.neuroimage.2011.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, Fox NC. Tracking atrophy progression in familial Alzheimer's disease: a serial MRI study. Lancet Neurol. 2006;5:828–834. doi: 10.1016/S1474-4422(06)70550-6. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Desikan RS, Sepulcre J, Yeo BT, Liu H, Schmansky NJ, Reuter M, Weiner MW, Buckner RL, Sperling RA, et al. The dynamics of cortical and hippocampal atrophy in Alzheimer disease. Arch Neurol. 2011;68:1040–1048. doi: 10.1001/archneurol.2011.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders AM, Strittmatter WJ, Schmechel D, St George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele ϵ 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- Savva GM, Wharton SB, Ince PG, Forster G, Matthews FE, Brayne C, Medical Research Council Cognitive Function and Ageing Study Age, neuropathology, and dementia. N Engl J Med. 2009;360:2302–2309. doi: 10.1056/NEJMoa0806142. [DOI] [PubMed] [Google Scholar]
- Schott JM, Bartlett JW, Fox NC, Barnes J, Alzheimer's Disease Neuroimaging Initiative Investigators Increased brain atrophy rates in cognitively normal older adults with low cerebrospinal fluid Aβ1-42. Ann Neurol. 2010;68:825–834. doi: 10.1002/ana.22315. [DOI] [PubMed] [Google Scholar]
- Ségonne F, Pacheco J, Fischl B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging. 2007;26:518–529. doi: 10.1109/TMI.2006.887364. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Cell biology of protein misfolding: the examples of Alzheimer's and Parkinson's diseases. Nat Cell Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, Blennow K, Soares H, Simon A, Lewczuk P, et al. Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline YI, Morris JC, Snyder AZ, Price JL, Yan Z, D'Angelo G, Liu C, Dixit S, Benzinger T, Fagan A, et al. APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF A β 42. J Neurosci. 2010;30:17035–17040. doi: 10.1523/JNEUROSCI.3987-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small GW, Ercoli LM, Silverman DH, Huang SC, Komo S, Bookheimer SY, Lavretsky H, Miller K, Siddarth P, Rasgon NL, et al. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer's disease. Proc Natl Acad Sci U S A. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Jr, Kaye J, Montine TJ, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging and the Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD. The international HapMap project web site. Genome Res. 2005;15:1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valla J, Yaari R, Wolf AB, Kusne Y, Beach TG, Roher AE, Corneveaux JJ, Huentelman MJ, Caselli RJ, Reiman EM. Reduced posterior cingulate mitochondrial activity in expired young adult carriers of the APOE ε4 allele, the major late-onset Alzheimer's susceptibility gene. J Alzheimer's Dis. 2010;22:307–313. doi: 10.3233/JAD-2010-100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Es MA, van den Berg LH. Alzheimer's disease beyond APOE. Nat Genet. 2009;41:1047–1048. doi: 10.1038/ng1009-1047. [DOI] [PubMed] [Google Scholar]
- Villemagne VL, Pike KE, Darby D, Maruff P, Savage G, Ng S, Ackermann U, Cowie TF, Currie J, Chan SG, et al. A β deposits in older non-demented individuals with cognitive decline are indicative of preclinical Alzheimer's disease. Neuropsychologia. 2008;46:1688–1697. doi: 10.1016/j.neuropsychologia.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Villemagne VL, Pike KE, Chételat G, Ellis KA, Mulligan RS, Bourgeat P, Ackermann U, Jones G, Szoeke C, Salvado O, et al. Longitudinal assessment of Aβ and cognition in aging and Alzheimer disease. Ann Neurol. 2011;69:181–192. doi: 10.1002/ana.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring SC, Rosenberg RN. Genome-wide association studies in Alzheimer disease. Arch Neurol. 2008;65:329–334. doi: 10.1001/archneur.65.3.329. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Manual for the Wechsler memory scale-revised. San Antonio (TX): Psychological Corporation; 1987. [Google Scholar]
- Wechsler-Reya R, Sakamuro D, Zhang J, Duhadaway J, Prendergast GC. Structural analysis of the human BIN1 gene. Evidence for tissue-specific transcriptional regulation and alternate RNA splicing. J Biol Chem. 1997;272:31453–31458. doi: 10.1074/jbc.272.50.31453. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Przybelski SA, Weigand SD, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr 3D maps from multiple MRI illustrate changing atrophy patterns as subjects progress from mild cognitive impairment to Alzheimer's disease. Brain. 2007;130:1777–1786. doi: 10.1093/brain/awm112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P, Köhler K, Vallis Y, Doyle CA, Owen D, Hunt SP, McMahon HT. Amphiphysin heterodimers: potential role in clathrin-mediated endocytosis. Mol Biol Cell. 1997;8:2003–2015. doi: 10.1091/mbc.8.10.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart H, Saykin A, McAllister T, Rabin L, McDonald B, Flashman L, Roth R, Mamourian A, Tsongalis G, Rhodes C. Regional brain atrophy in cognitively intact adults with a single APOE ϵ 4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- Wolk DA, Dickerson BC. Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional–executive network function in Alzheimer's disease. Proc Natl Acad Sci U S A. 2010;107:10256–10261. doi: 10.1073/pnas.1001412107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD. Mitochondrial bioenergetic deficit precedes Alzheimer's pathology in female mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2009;106:14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.