Abstract
Gemcitabine is a nucleoside analog that is currently the best available single-agent chemotherapeutic drug for pancreatic cancer. However, efficacy is limited by our inability to deliver sufficient active metabolite into cancer cells without toxic effects on normal tissues. Targeted delivery of gemcitabine into cancer cells could maximize effectiveness and concurrently minimize toxic side effects by reducing uptake into normal cells. Most pancreatic cancers overexpress epidermal growth factor receptor (EGFR), a trans-membrane receptor tyrosine kinase. We utilized a nuclease resistant RNA aptamer that binds and is internalized by EGFR on pancreatic cancer cells to deliver gemcitabine-containing polymers into EGFR-expressing cells and inhibit cell proliferation in vitro. This approach to cell type–specific therapy can be adapted to other targets and to other types of therapeutic cargo.
Introduction
Pancreatic cancer has a mortality rate that exceeds nearly all other cancers. In 2011, approximately 44,000 people were diagnosed with pancreatic cancer, and over 37,000 people died from the disease (Siegel et al., 2011). At the time of diagnosis, most patients have disease that is unresectable due to invasion of local structures and/or distant metastasis. Fewer than 10% of pancreatic cancer patients have tumors that are amenable to surgical resection, and the vast majority of these patients will develop recurrent disease within 5 years. Pancreatic cancer is notoriously chemoresistant, with response rates ranging from 0% to 30% in most clinical trials, even for combinations of the “best” agents (Kindler et al., 2005; Moore et al., 2007; Conroy et al., 2011). Therefore, there is an unquestionable need for more effective pancreatic cancer therapies.
Gemcitabine (Gemzar®, Eli Lilly) is a deoxycytidine analog (2′,2′-difluoro, 2′ deoxycytidine, or dFdC) (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/nat) that is the most effective single-agent therapy for pancreatic cancer. Gemcitabine (Gem) has become standard of care for patients with advanced disease based on a “clinical benefit response” of 20%, but a radiographic response rate of only 5% (Burris et al., 1997). Adjuvant Gem following potentially curative resection improved 5-year disease-free survival from 6% to 16% in the CONKO-001 study (Oettle et al., 2007), suggesting that it does eradicate microscopic disease in some patients. In vitro, all pancreatic cell lines are sensitive to Gem if you treat them with high enough concentrations. However, as with other cytotoxic agents, its effects on normal tissues limit the dose that can be applied in vivo. At the current standard dose for Gem monotherapy of 1,000 mg/m2, the most common dose-limiting toxicities are hematologic, with neutropenia occurring in approximately 25% of patients (Burris et al., 1997; Oettle et al., 2007). Additionally, tumor resistance seems to result—at least in part—from cellular mechanisms that decrease its uptake (Achiwa et al., 2004; Nakano et al., 2007). Targeted delivery of Gem into cancer cells could maximize effectiveness and concurrently minimize toxic side effects by reducing uptake into normal cells.
The majority of pancreatic cancers overexpress epidermal growth factor receptor (EGFR) (Xiong et al., 2004; Moore et al., 2007), a transmembrane receptor tyrosine kinase. Binding of EGFR to its cognate ligands causes receptor dimerization leading to autophosphorylation, internalization of the receptor, and activation of intracellular signal transduction pathways (Ullrich and Schlessinger, 1990). Since EGFR is overexpressed in pancreatic cancers and is internalized upon ligand binding, it is a good candidate for targeted therapy. In principle, any reagent that specifically binds to EGFR can be used to deliver therapeutic cargo into EGFR-expressing cells.
Patra et al. (Patra et al., 2008) have used the chimeric anti-EGFR antibody cetuximab (Erbitux®, ImClone Systems) as a targeting agent to deliver gold nanoparticles loaded with Gem to pancreatic cancer cells, demonstrating “proof of concept” that EGFR can be utilized for targeted delivery. Targeting aptamers are an alternative to antibody-based therapeutics. Aptamers are a class of oligonucleotide (RNA or DNA) molecules that share the same virtue of high affinity for their specific binders as antibodies. Aptamers are generated by an iterative screening process of complex oligonucleotide libraries (>1014 shapes per library) employing a process termed SELEX, or systemic evolution of ligands by exponential enrichment (Ellington and Szostak, 1990; Tuerk and Gold, 1990). Several recent studies have demonstrated that aptamers can be used as agents for targeted drug delivery (for reviews, refer to references Levy-Nissenbaum et al., 2008; Bunka et al., 2010; Cerchia et al., 2009; Zhang et al., 2010; Zhou and Rossi, 2011). For instance, an RNA aptamer against prostate-specific membrane antigen (PSMA), a surface protein specifically expressed in prostate cancer, is known to be internalized and has been used to selectively deliver cytotoxins (Chu et al., 2006), chemotherapeutic agents (Bagalkot et al., 2006; Farokhzad et al., 2006), and small interfering RNAs (siRNAs) (McNamara et al., 2006; Dassie et al., 2009) into PSMA-expressing cells, but not into cells that do not express PSMA.
Li et al. have generated a 2′-fluoro-modified RNA aptamer against EGFR (E07) and demonstrated that it is internalized by and inhibits proliferation of EGFR-expressing cells (Li et al., 2011). In the present work, we have utilized this EGFR aptamer to specifically deliver Gem into EGFR-expressing pancreatic cancer cells. This approach takes advantage of the fact that Gem is a nucleoside and therefore is amenable to chemical phosphorylation to its triphosphate (NTP) form with subsequent polymerization and hybridization to aptamers. We present a novel method for enzymatic polymerization of a Gem-containing oligonucleotide that can be delivered to pancreatic cancer cells by the EGFR aptamer for targeted cell death.
Materials and Methods
Cells and media
The human pancreatic cancer cell lines MiaPaCa-2 and HPAF-2 were purchased from ATCC (American Type Culture Collection) and were grown in Dulbecco's modified Eagle and Eagle's modified essential media respectively with 10% fetal bovine serum and 1% penicillin-streptomycin.
Western blot analysis
The cells were lysed in the radio-immunoprecipitation assay lysis buffer (Santa Cruz). Total protein (0.5 μg) was resolved in by 4%–20% sodium dodecyl sulfate polyacrylamide gel electrophoresis (BioRad) and transferred to a polyvinylidene difluoride membrane (BioRad). Rabbit polyclonal anti-EGFR antibody (1:1,000 dilution; BioLegend) was used as the primary antibody and horseradish peroxide (HRP)-conjugated goat anti-rabbit antibody (1:1,000 dilution; Invitrogen) was used as the secondary antibody. ß-tubulin was used as the loading control and was detected by HRP-conjugated anti-ß-tubulin antibody (1:5,000 dilution; Abcam). The signal was detected by the Pierce® ECL Western Blotting Substrate (Thermo Scientific).
Flow cytometry
The cells were grown to 70% confluence under standard growth conditions, trypsinized, and counted for the flow cytometry experiments. The EGFR aptamer (E07) and the scrambled mutant aptamer (mE07) were transcribed with a 24-nucleotide extension at the 3′ end (5′-CUG GUC AUG GCG GGC AUU UAA UUU-3′). In order to evaluate binding of aptamer–Gem polymer complexes, the aptamers were directly biotinylated at the 3′ end using RNA 3′ End Biotinylation Kit (Pierce) and annealed (1:1 ratio) to the Gem polymer. Streptavidin-phycoerythrin (SA-PE, Prozyme) was used to label the aptamer–wing Gem polymer complex. As controls, E07 and mE07 without the 24-nucleotide extension at the 3′ end were biotinylated and similarly labeled with SA-PE as described above. For experiments evaluating aptamer binding alone, the E07 and mE07 aptamers with the 24-nucleotide extension at the 3′ end were annealed (1:1 ratio) to a biotinylated DNA capture oligonucleotide “wing,” and SA-PE was used to label the aptamer–wing complex. Cells (1×106) were first incubated with the labeled aptamer in 100 μL PBS buffer for 30 minutes at 37°C. The stained cells were subsequently washed 3 times with 200 μL of PBS, resuspended in 500 μL of PBS, and analyzed using FACSCalibur (BD Biosciences).
Microscopy
The cells (1×104) were plated on round cover slips in a 24-well plate the day before the experiments. The EGFR aptamer (E07) and the scrambled mutant aptamer (mE07) were labeled with Streptavidin-Alexa-488 fluorophore using the same techniques as described above for flow cytometry. The cells were first washed in buffer E (PBS plus 5 mM MgCl2) and incubated with labeled aptamer in 300 μL buffer E for 30 minutes at 37°C. The final concentration of labeled aptamer was 100 nM. The stained cells were washed 3 times with 300 μL buffer E and fixed with 4% paraformaldehyde solution. Fixed cells were washed in buffer E and stained with DAPI (300 nM final concentration). Finally, the DAPI stained cells were washed in buffer E and mounted on the glass slides with FluorSave™ (Calbiochem) for imaging using Zeiss Axio Observer wide field fluorescence microscope.
Internalization assay (Riboshredder™ assay)
MiaPaCa-2 cells were incubated with 400 nM of the labeled (SA-PE) aptamers, annealed to the Gem polymer, in 100 μL PBS buffer for 30 minutes at 37°C or 4°C. The labeled cells were first treated with 2 μL of RiboShredder™ (Epicentre® Biotechnologies) for 10 minutes at 25°C and then washed 3 times in 200 μL PBS. Finally, the cells were resuspended in 500 μL of PBS and analyzed using FACSCalibur (BD Biosciences). The percentage of aptamer internalization was calculated using geometric means from the histogram with the formula (F2 – F0)/(F1 – F0), where F0=mE07 binding, F1=E07 binding, and F2=E07 binding after Riboshredder treatment.
RNA synthesis
The RNAs used were enzymatically synthesized using a double-stranded DNA template and a mutant T7 RNA polymerase (Y639F) (Sousa and Padilla, 1995). The DNA template (5′-AAT TTA ATA CGA CTC ACT ATA GGG AGA GAT GAT CGA TCG ATC CTG GTC ATG GCG GGC ATT TAA TTT-3′) (6 nmol) was annealed to the complementary DNA strand at an equi-molar ratio in a total volume of 90 μL of annealing buffer (10 mM Tris-HCL pH 7.5, 10 mM MgCl2). The samples were heated to 95°C for 5 minutes and allowed to cool to room temperature for 2 hours. The natural and Gem-RNA were transcribed using 5 μg of this double-stranded DNA template in a total volume of 100 μL of 1×transcription buffer (40 mM Tris-HCL pH7.5, 6 mM MgCl2, 5 mM NaCl, and 10 mM spermidine). A 2-μL aliquot of the Durascribe T7 enzyme mix (Epicentre®) containing the mutant T7 enzyme with either 5 mM or 2 mM NTP mix (ATP, UTP, GTP, and either dFdCTP or 2′F-CTP) was added to the reaction mixture. The reaction was incubated overnight at 37°C in a water bath. The DNA template was digested using RQ1 RNase-free DNase (1 U/μL) by incubation at 37°C for 15 minutes. The unincorporated NTPs were removed by G25 spin columns (GE Healthcare). The final product was ethanol precipitated, washed, resuspended in diethylpyrocarbonate (DEPC)-treated water and quantified using ultravioletc Nanodrop spectrometer. The quality and size of the RNA were analyzed on a 10% (19:1) polyacrylamide gel with 7M urea.
Cell proliferation assays
The cells (3,000 cells/well) were seeded on a 96-well plate on the day before the experiment. The cells were treated with E07–Gem polymer (400 nM); mE07–Gem polymer (400 nM), Gem polymer alone (400 nM), E07 annealed to transcript with 2′F CTP instead of Gem (400 nM), and unpolymerized Gem (1,000 nM) were used as controls. Each treatment was performed in triplicate. CellTiter-Glo assay (Promega) was performed to assess cell viability after 48 hours.
Statistical analysis
The student's t test with a 2-sided alpha of 0.05 was used to compare cell viability between treatment groups (GraphPad Prism Version 4.0b).
Results
EGFR aptamer-gemcitabine polymer (E07–Gem polymer) binds to MiaPaCa-2 cells expressing EGFR receptor
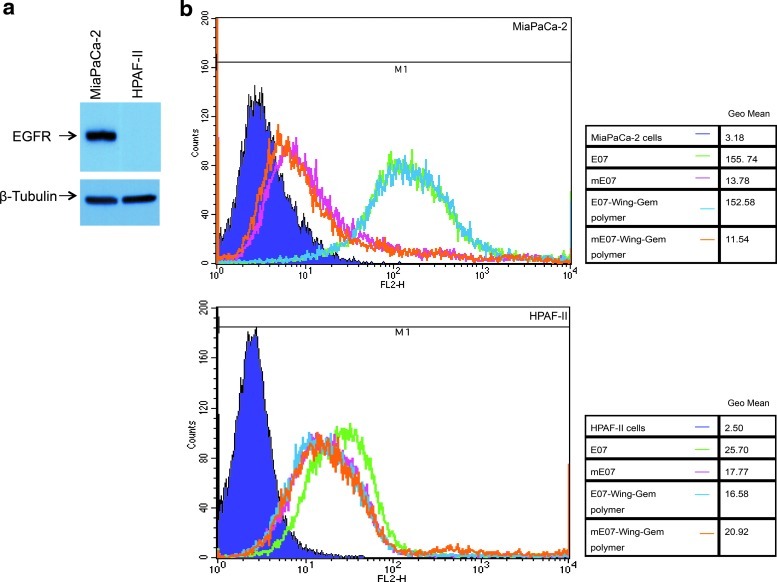
Pancreatic cancer cell lines MiaPaCa-2 and HPAF-2 were tested for EGFR expression. Western blot analysis was performed on cell-free lysate of MiaPaCa-2 and HPAF-2 cells. EGFR expression was detected in the MiaPaCa-2 but not in the HPAF-2 cell line (Fig. 1a). E07 was labeled with the fluorophore phycoerythrin (PE) and used to assess for binding to the cell surface of these 2 pancreatic cancer cell lines by flow cytometry. A scrambled aptamer (mE07) that does not bind to purified EGFR protein in vitro was used (Table 1) as a negative control to represent background (nonspecific) oligonucleotide binding to cells. At a concentration of 400 nM, there was a greater than 10-fold shift in binding of E07 to MiaPaCa-2 cells compared to background, whereas HPAF-2 cells, which do not express detectable levels of EGFR by western blot analysis, demonstrated only a minimal (less than 1-fold) shift in binding of E07 compared to background (Fig. 1b). Similar binding intensities were observed with E07 annealed to the Gem polymer, thus demonstrating that the Gem polymer does not interfere with the binding of the aptamer to the EGFR positive MiaPaCa-2 cells.
FIG. 1.
E07–gemcitabine (Gem) polymer binds to the epidermal growth factor receptor (EGFR)-expressing MiaPaCa-2 cells. (a) Western blots were done using the cell-free extracts from MiaPaCa-2 and HPAF-2 cells. EGFR was detected in the MiaPaCa-2 but not in the HPAF-2 cells. ß-tubulin was used as the loading control. (b) E07-Gem polymer (blue) and E07 (green) showed similar binding to the EFGR expressing MiaPaCa-2 cells. The controls, mE07–Gem polymer (orange) and the mE07 (pink) represent background (non-specific) binding (upper panel). There was a minimal shift in binding to HPAF-2 cells by E07 aptamer alone (green) but not E07–Gem polymer (blue) over background binding (pink and orange, lower panel). Cells were analyzed by FACSCalibur on the FL2-H channel. The number of events counted is plotted on the y-axis. Quantitation of the histograms is presented in tabular form as geometric mean (Geo Mean) for the corresponding peaks.
Table 1.
Aptamer Sequences
| GGCGCUCCGACCUUAGUCUCUG-N51-GAACCGUGUAGCACAGCAGA | |
| E07 | UGCCGCUAUAAUGCACGGAUUUAAUCGCCGUAGAAAAGCAUGUCAAAGCCG |
| mE07 | UCAGACACGGCAAUUCGUGGCGCGAGCUGAUAAAUACAUGCCCAUAUUAAG |
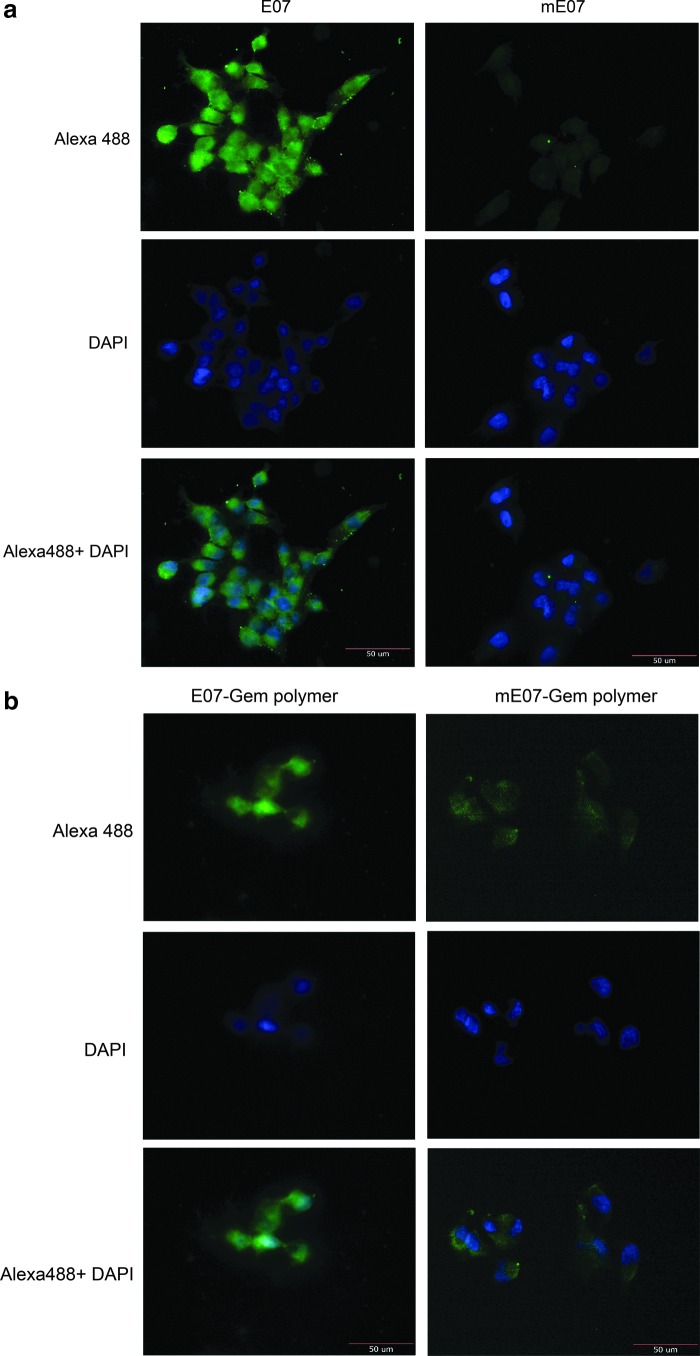
We next tested whether E07 aptamers, labeled with Alexa-488 fluorophore, could be used to image MiaPaCa-2 cells by fluorescent microscopy. MiaPaCa-2 cells were readily stained with the E07 aptamer, whereas no detectable staining above the background was visualized when the mutant aptamer (mE07) was used (Fig. 2a). Annealing of Gem polymer did not interfere with the binding of E07 to the MiaPaCa-2 cells (Fig. 2b). These data demonstrate that the aptamer E07 specifically recognizes and binds to the EGFR protein expressed on the cell surface of pancreatic cancer cell line MiaPaCa-2. Furthermore, the annealing of the Gem polymer to the E07 aptamer does not interfere with cell binding (Figs. 1b, 2), thus indicating that this might be a viable approach to deliver Gem into EGFR-expressing cells.
FIG. 2.
Imaging MiaPaCa-2 cells with E07 and E07–Gem polymer: (a) E07 and (b) E07–Gem polymer aptamers labeled with the fluorophore Alexa-488 bound to the MiaPaCa-2 cells were detected by fluorescence microscopy. No detectable staining above the background was observed when the mutant aptamer (mE07) was used. DAPI was used for the nuclear staining. DAPI, 4',6'-diamindino-2-phenylindole.
EGFR-aptamer-gemcitabine polymer (E07–Gem polymer) is internalized
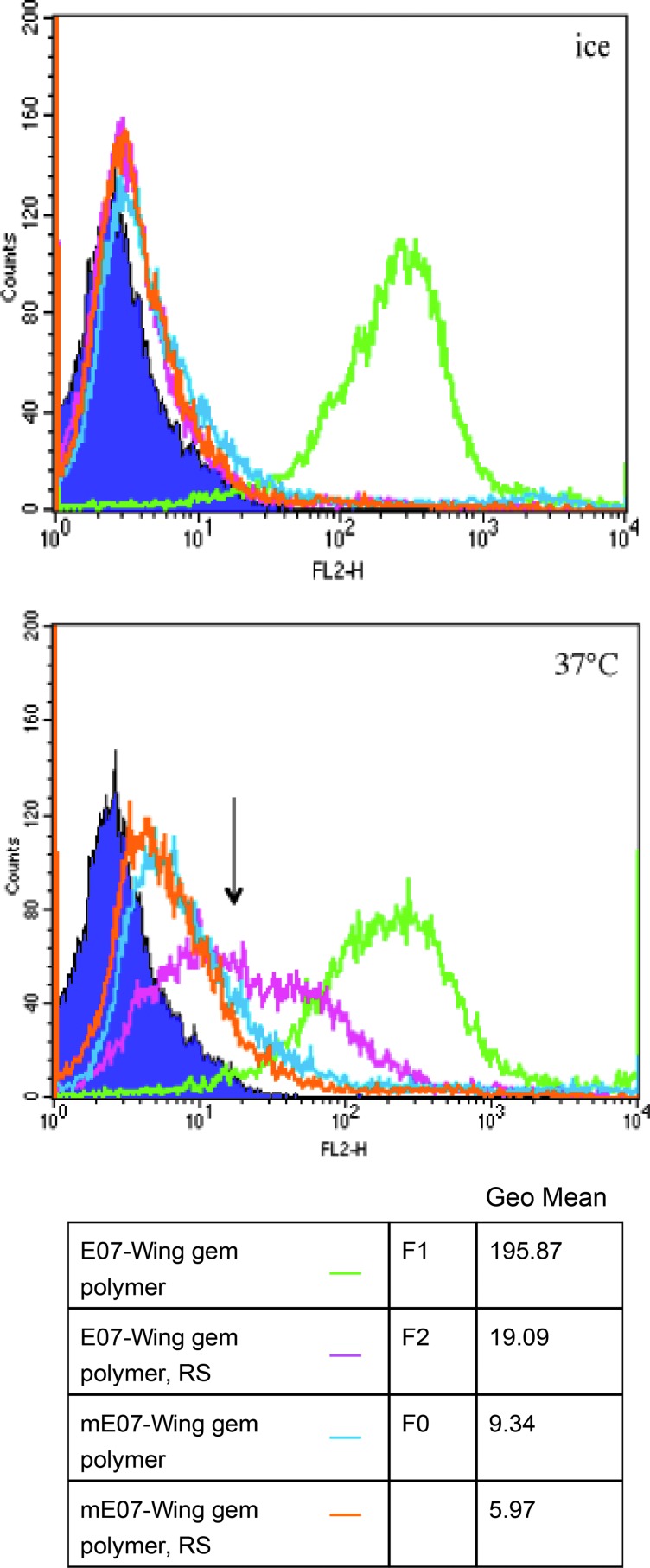
In order to use the E07 aptamer as a tool to deliver cargo to EGFR-expressing pancreatic cancer cells, E07–Gem polymer must be internalized. We performed an assay previously employed by Li et al. to demonstrate the internalization of the aptamer–Gem complex into pancreatic cancer cells (Li et al., 2010; Li et al., 2011). Briefly, the assay relies on two facts: (1) the process of endocytosis functions at the physiological temperature of 37°C and is inhibited at 4°C, and (2) 2′-fluoro-pyrimidine–modified RNA-aptamers, while relatively resistant to nuclease degradation under physiological conditions (Knudsen et al., 2002), can be cleaved by treatment with RiboShredder™—a harsh cocktail of RNases (Li et al., 2010)—in vitro. Thus, E07–Gem polymer complex that is internalized by MiaPaCa-2 cells at 37°C should be protected from the nuclease treatment. However, when the internalization is inhibited at 4°C, the E07–Gem polymer complex would be exposed at the cell surface and susceptible to the nuclease. Indeed, when the internalization experiment was carried at 4°C, the E07–Gem polymer complex was completely degraded upon the Riboshredder treatment. When MiaPaCa-2 cells were incubated with 400nM of E07–Gem polymer for 30 minutes at 37°C, 6% of the complex was internalized (Fig. 3). The absolute amount of E07 aptamer internalized and protected (F2-F0) from the Riboshredder at 37°C was similar at 100nM, although the percentage of total aptamer internalized was higher (17%) than at the 400nM concentration (Supplementary Fig. S2). The mutant aptamer (mE07)–Gem polymer complex was completely degraded at 37°C and 4°C (Fig. 3), evidence that the process of complex internalization is target cell–specific and not a random uptake of oligonucleotides by the cells.
FIG. 3.
E07–Gem polymer is internalized by the MiaPaCa-2 cells. E07–Gem polymer (green and pink) and the mE07–Gem polymer (blue and orange) labeled with the fluorophore PE were incubated with the MiaPaCa-2 cells either on ice (upper panel) or at 37°C (lower panel) for 30 minutes. After the binding reactions, the cells were treated with Riboshredder (RS) for 10 minutes at room temperature (pink and orange). The cells were analyzed by using FACSCalibur. The arrow indicates the E07-gem polymer bound to the cells after RS treatment at 37°C (pink), presumably due to aptamer internalization. Quantitation of the histograms is presented in tabular form as Geo Mean for the corresponding peaks.
EGFR aptamer (E07) delivers gemcitabine to MiaPaCa-2 cells and induces cell death
Gem (dFdC) is a cytotoxic nucleoside analog, and we reasoned that it could be amenable to polymerization and hybridization to the E07 aptamer for targeted delivery into the pancreatic cancer cells. Gem is commercially available as Gemzar® (Eli Lilly), a 1:1 wt/wt mixture of dFdC and the sugar alcohol D-mannitol. We first developed a derivative-based synthetic strategy to separate dFdC from D-mannitol and then synthesized and purified the triphosphate analog of dFdC, that is, dFdCTP [ESI-MS (infusion) M− m/z calculated for C9H14F2N3O13P3 503.0, found 501.8)] using a modified one-pot phosphorochloridite procedure originally developed by Ludwig and Eckstein and modified by our group (Ludwig and Eckstein, 1989; He et al., 1998).
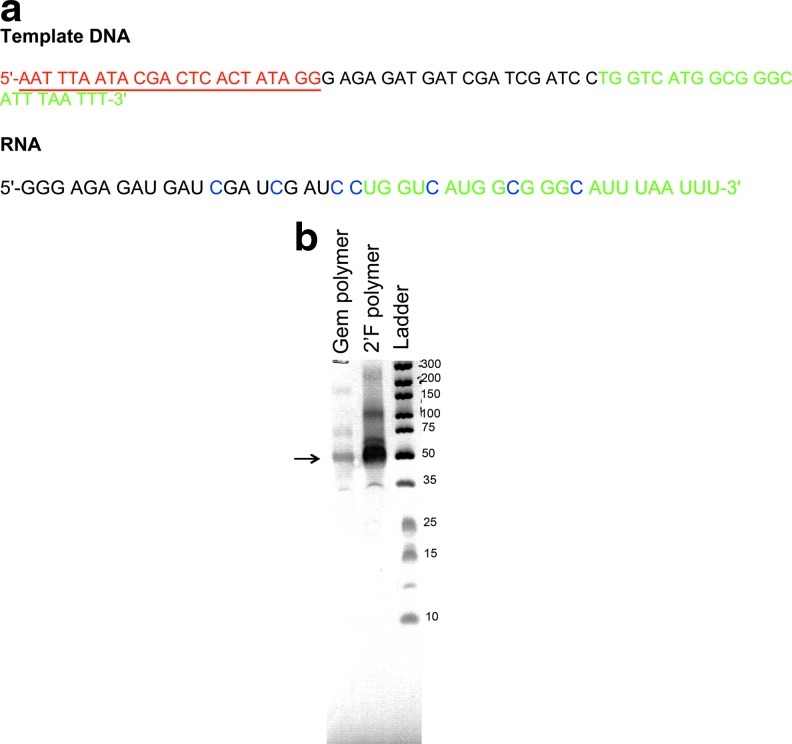
The purified dFdCTP was then used to synthesize Gem polymers in a template-dependent manner by using a mutant RNA polymerase. Wild type and mutant prokaryotic RNA polymerases (T7) are routinely used for the in vitro synthesis of RNA containing synthetic 2′-modified pyrimidine nucleotides (Milligan et al., 1987; Gaur and Krupp, 1993; Conrad et al., 1995; Padilla and Sousa, 1999). This strategy requires a DNA template and can therefore be used to produce very specific sequences and chain lengths. As a cytidine analog, we assumed that dFdCTP would base pair with guanine nucleotides (GTPs), and—since its ribose sugar conformation is similar to a ribonucleotide (3′ endo) when bound to the active site of a polymerase—that it should be incorporated with catalytic efficiencies similar to standard 2′-fluoro-modified pyrimidine (Ruiz van Haperen et al., 1993; Fowler et al., 2008). We therefore designed a DNA construct to encode a consensus T7 promoter followed by a sequence free of dFdCTP (to facilitate initiation of transcription) followed by a sequence that included seven dFdCTPs separated by 3 to 4 natural NTP “spacers” followed by a short 24-nucleotide “wing” sequence that could be used to anneal the Gem polymer to the E07 aptamer. The 45-nt Gem polymer could be visualized as a distinct band of appropriate size when run on a 10% urea-polyacrylamide gel (Fig. 4).
FIG. 4.
Sequence of the Gem-containing polymer. (a) The template DNA used for in vitro transcription to synthesize the Gem-containing polymer. The T7-promoter sequence (red) is underlined. The 3′-“wing” sequence used for annealing the gem-containing polymer to the E07 aptamer is in green font color. The Gem molecules are denoted as C (blue) in the RNA sequence. (b) The Gem polymer and the corresponding 2′F polymer (control) were run on a 10% denaturing gel to visualize the products. The arrow denotes the Gem polymer and the 2′F polymer of approximate 45 nt length.
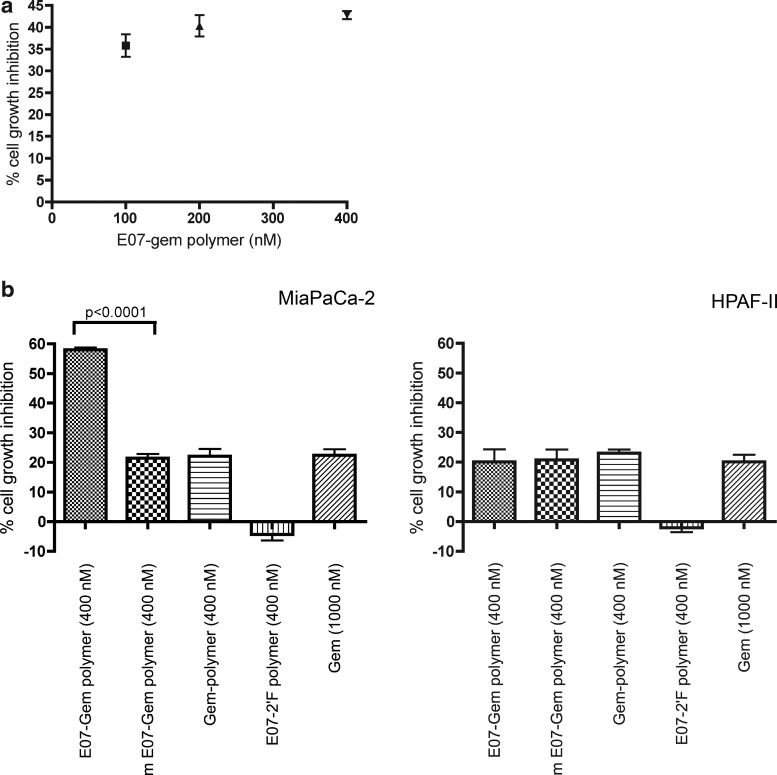
First, to confirm efficacy and establish an appropriate dose for larger scale experiments, MiaPaCa-2 cells were seeded the day before the experiment and treated with increasing concentrations of E07–Gem polymer. CellTiter-Glo assay was performed to assess cell viability after 48 hours. Maximal cell growth inhibition was observed at 400 nM E07–Gem polymer. Thus, 400 nM concentration was chosen for the subsequent assay (Fig. 5a).
FIG. 5.
E07-gem polymer inhibits MiaPaCa-2 cell growth. (a) MiaPaCa-2 cells were treated with the indicated concentrations of E07-Gem polymer to establish a dose-response curve. The percent inhibition {[(untreated – treated)/untreated]×100} is ploted in the y-axis. (b) MiaPaCa-2 and HPAF-2 cells were treated with E07-Gem polymer (400 nM), mE07–Gem polymer (400 nM), Gem-polymer (400 nM), E07–2′F polymer (400 nM), and unpolymerized Gem (Gemzar, 1000 nM) and analyzed for the percentage of cell growth inhibition.
Next, in order to test whether E07–Gem polymer is capable of targeted cell growth-inhibition we performed a similar assay to the one described above with MiaPaCa-2 (EGFR positive) and HPAF-2 (EGFR negative) cells. Cells were treated with 400nM of E07–Gem polymer. Mutant aptamer annealed to the Gem polymer (mE07–Gem polymer), Gem polymer alone (without the escort E07 aptamer), E07 annealed to transcript with 2′F CTP instead of Gem polymer (E07–2′F polymer), and unpolymerized Gem were used in control experiments. A minimal increase in cell growth was observed with the E07 aptamer annealed to control polymer. A small degree of inhibition was observed with mE07–Gem polymer, and a similar degree of inhibition was observed with the unescorted Gem polymer alone. None of these differences were statistically significant. The inhibition seen with the unescorted polymer is likely due to nuclease-mediated degradation of the Gem polymer at the sites of unmodified nucleotides and nonspecific uptake of the free nucleotides by the cells (Fig. 5b). However, in the MiaPaCa-2 cell line, there was significantly greater inhibition with the E07–Gem polymer than with the mE07–Gem polymer or with the unescorted Gem polymer, a difference that is not seen with non-EGFR-expressing pancreatic cancer cell line HPAF-2 (Fig. 5b). The HPAF-2 and MiaPaCa-2 cell lines demonstrated similar responses to unpolymerized Gem, suggesting that these findings are not due to intrinsic differences in sensitivity to Gem. These experiments demonstrate that the E07-aptamer is mediating Gem delivery to EGFR-expressing pancreatic cancer cells in vitro.
Discussion
Gemcitabine (Gemzar) monotherapy has been approved by the U.S. Food and Drug Administration for treating locally advanced and metastatic pancreatic adenocarcinoma and for the adjuvant treatment of resected pancreatic adenocarcinoma. However, the modest benefits associated with standard chemotherapy leave much room for improvement. Gemcitabine (Gem) nucleoside (dFdC) is a pro-drug that is imported into the cell by human equilibrative nucleoside transporter-1 and is initially converted to Gem monophosphate (dFdCMP). This rate-limiting step is catalyzed by the enzyme deoxycytidine kinase. Subsequently, dFdCMP is converted to diphosphate (dFdCDP) and ultimately to triphosphate (dFdCTP). dFdCDP and dFdCTP are the active metabolites that inhibit ribonucleotide reductase and DNA chain elongation, respectively (Huang et al., 1991; Ruiz van Haperen et al., 1993). Gem resistance has been associated with decreased tumor perfusion (Olive et al., 2009), cellular uptake (Mackey et al., 1998; Giovannetti et al., 2006), and metabolism (Kroep et al., 2002; Nakano et al., 2007). The proposed gem delivery strategy could help to overcome some of these chemoresistance mechanisms.
An ideal target for aptamer-mediated delivery would be one that is expressed and efficiently internalized only by pancreatic cancer cells but not by normal cells. Such an ideal target has not yet been identified. However, EGFR is one of several potential targets that are more highly expressed on cancer cells than on normal cells (Rocha-Lima et al., 2007). We utilized a nuclease resistant RNA aptamer that had previously been selected to bind EGFR (Li et al., 2011).
A phase-3 clinical trial of Gem with or without the small molecule EGFR inhibitor erlotinib (Tarceva®, Genentech) demonstrated a statistically but arguably not clinically significant survival benefit of 2 weeks in patients with advanced pancreatic cancer (Moore et al., 2007). Another phase-3 trial of Gem with or without the chimeric anti-EGFR antibody cetuximab (Erbitux®, ImClone Systems) demonstrated a similar survival difference that did not achieve statistical significance (Philip et al., 2007). The disappointing performance of EGFR inhibitors in pancreatic cancer has been attributed to the fact that EGFR protein expression is not synonymous with EGFR pathway activation.
While EGFR antagonists may have a direct effect on pancreatic cancer cell growth, the goal of EGFR-targeted delivery is to exploit the fact that EGFR is considered the “model” for receptor-mediated endocytosis (Ullrich and Schlessinger, 1990). Patra et al. have previously demonstrated that cetuximab, an EGFR-specific antibody, can target gold nanoparticles loaded with Gem to pancreatic cancer cells (Patra et al., 2008). Tumor growth in an orthotopic xenograft model was inhibited by the targeted Gem nanoparticles but not by free Gem at the same low dose. Our approach differs both in the targeting agent and in the method for delivery of Gem. Cetuximab and many other EGFR antibodies block dimerization (Gan et al., 2007). Aptamers may therefore interact differently with EGFR and be internalized more efficiently than EGFR antibodies. Furthermore, like other monoclonal antibody therapeutics, there are toxicities associated with the administration of cetuximab, including severe infusion reactions, which have been reported in 3% of patients during clinical trials (Hansel et al., 2010).
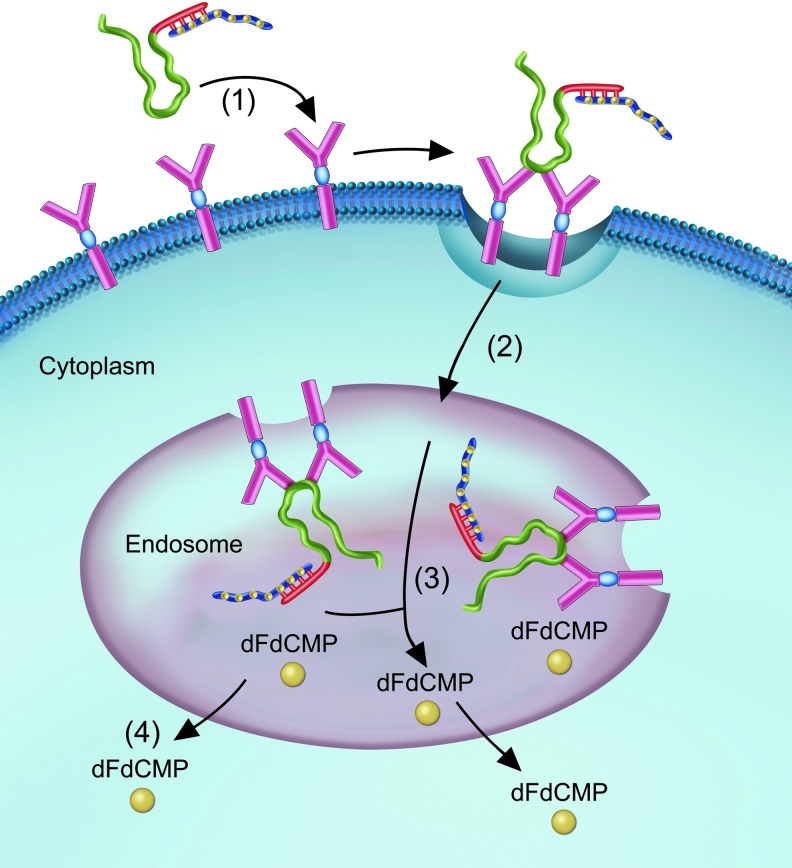
Since Gem is a nucleoside analog, we hypothesized that its monophosphate form would be incorporated by the same mutant RNA polymerases that are frequently used for in vitro transcription with other modified nucleotides. We generated a Gem-containing oligonucleotide that could be annealed to the EFGR aptamer, thus creating an “all nucleic acid” reagent with 2 functional domains (Fig. 6). The aptamer portion allows for specific binding of EGFR on the cell surface and drives the second portion, the Gem-containing polymer, into the targeted pancreatic cancer cells via clathrin-mediated endocytosis (Vieira et al., 1996). The aptamer and its cargo are trafficked via the endosome to the lysosome, where the acidic environment should promote degradation of the oligonucleotide (WICKSTROM, 1986). The current approach has the potential to improve efficacy by delivering multiple Gem monophosphate molecules per aptamer, bypassing requirements for cellular uptake by human equilibrative nucleoside transporter-1 and phosphorylation by deoxycytidine kinase. This approach also has the potential to decrease toxic side effects by minimizing the total dose of Gem required and by reducing uptake into normal cells.
FIG. 6.
Model for the E07 mediated delivery of the GEM-containing polymer. E07 aptamers are represented in green with the 3′ wing extension in red. The GEM-containing polymers (blue) are annealed by Watson-Crick base paring to the E07 aptamers via the wing extension, represented by short vertical lines. Gemcitabine monophosphates (dFdCMP) are represented by the yellow circles distributed along the blue strand. (1) E07–Gem containing polymer binds to the EGFR expressing cells on the plasma membrane and is (2) internalized to the endosome, where the Gem-containing polymer is (3–4) degraded to release dFdCMP into the cytoplasm.
Using flow cytometry and microscopy, we first established that the EGFR aptamer (E07) binds to the high EGFR-expressing pancreatic cancer cell line MiaPaCa-2 to a much greater extent than to the low EGFR-expressing pancreatic cancer cell line HPAF-2 (Figs. 1, 2). Furthermore, the annealing of Gem polymer to the E07 aptamer did not interfere with cell binding. We next established that E07 annealed to the Gem polymer is internalized by MiaPaCa-2 cells (Fig. 3). Then, using cell viability assays, we demonstrated “proof of concept” that the E07 aptamer inhibits proliferation in MiaPaCa-2 cells but not in HPAF-2 cells (Fig. 5). However, this construct is a prototype that needs to be optimized with respect to both specificity and efficacy in order to be useful clinically. With the mE07–Gem polymer and unescorted Gem polymer, we observed approximately 20% inhibition in both the high- and low-EGFR-expressing cell lines in vitro. This is presumably due to nuclease-mediated partial degradation of the Gem polymer and nonspecific uptake of the nucleotides. RNA aptamers that incorporate 2′-modified pyrimidines have been shown to be stable in serum for hours (Knudsen et al., 2002). The introduction of additional modified nucleotides into the polymer—which has only one modified nucleotide (dFdCTP) in its current form—would therefore be expected to improve nuclease stability and specificity. If this nonspecific inhibition is subtracted from total inhibition, we are left with a relatively modest 30% specific inhibition with 1 application of 400 nM of the E07–Gem polymer in MiaPaCa2 cells. Li et al. did demonstrate inhibition of proliferation in A431 epidermoid cells by the E07 aptamer alone at a dose of 1 μM (Li et al., 2011), but we did not observe any inhibition with E07 annealed to control polymer that could be attributed to the aptamer itself.
Although our internalization experiment may underestimate the degree of internalization that would occur in vivo, our data demonstrate that only a small proportion of the E07–Gem polymer complex is internalized by MiaPaCa-2 cells in vitro. The absolute amounts of aptamer–Gem polymer internalized at 100 nM and 400 nM were similar (Fig. 3 and Supplementary Fig. S2) as were the degrees of proliferation inhibition at 100 nM and 400 nM (Fig. 5a). This might be the result of saturation of the receptor-mediated uptake process and further indicates that the internalization of E07 in EGFR expressing MiaPaCa-2 cells is an active receptor-mediated transport and not a random process of oligonucleotide uptake by cells.
This low rate of internalization might be overcome by increasing the toxicity of the cargo (i.e., more Gem or more potent drugs). However, the optimal length and composition of the polymer will need to be determined empirically, as it is possible that increased length and/or higher Gem content may be offset by decreased internalization. Ultimately, in order to maximize efficacy, we also need to identify aptamers that are internalized more efficiently and/or ways to improve the internalization of existing aptamers.
Although not a new concept, aptamers and other nucleic acid-based therapeutics have only become realistic clinical agents as methods for their efficient synthesis have improved, similar to monoclonal antibodies 30 years ago. The first aptamer to enter clinical trials for cancer therapy is a DNA aptamer (AS1411) that binds nucleolin, a protein that is expressed in the nuclei of all cells but is over-expressed in the cytoplasm and on the plasma membrane of cancer cells relative to normal cells (Soundararajan et al., 2009). Phase 2 trials of AS1411 in acute myeloid leukemia (NCT00512083) and renal cell carcinoma (NCT00740441) have recently completed enrollment after phase-1 studies demonstrated clinical activity and no serious adverse events (Bates et al., 2009). Relatively high (40 mg/kg/day) doses of AS1411 were administered, demonstrating that synthesis of clinically relevant doses of oligonucleotides is feasible. Once the optimal aptamer–Gem polymer construct has been determined, it should also be amenable to large-scale chemical synthesis via standard phosphoramidite technology.
There is a dire need for more effective therapies for pancreatic cancer. Although identification of new, more effective drugs is certainly an important goal of pancreatic cancer research, the development of methods to target existing drugs to cancer cells and to overcome mechanisms of drug resistance should be parallel goals. The use of aptamers for cell-type-specific delivery is a novel approach to pancreatic cancer therapy and one that can be more globally applied to other cell surface receptors and different types of therapeutic cargo, including other drugs, toxins, siRNAs, radioisotopes, and photosensitizing agents.
Supplementary Material
Acknowledgments
We thank Yasheng Gao, PhD of Light microscopy Core facility, Duke University and Duke University Medical Center for his guidance in imaging. This work was supported by the National Institutes of Health (K08 CA142903-01A1 to RRW). The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Author Disclosure Statement
No competing financial interests exist.
References
- ACHIWA H. OGURI T. SATO S. MAEDA H. NIIMI T. UEDA R. Determinants of sensitivity and resistance to gemcitabine: the roles of human equilibrative nucleoside transporter 1 and deoxycytidine kinase in non-small cell lung cancer. Cancer Sci. 2004;95:753–757. doi: 10.1111/j.1349-7006.2004.tb03257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BAGALKOT V. FAROKHZAD O.C. LANGER R. JON S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- BATES P.J. LABER D.A. MILLER D.M. THOMAS S.D. TRENT J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUNKA D.H. PLATONOVA O. STOCKLEY P.G. Development of aptamer therapeutics. Curr. Opin. Pharmacol. 2010;10:557–562. doi: 10.1016/j.coph.2010.06.009. [DOI] [PubMed] [Google Scholar]
- BURRIS H.A., 3rd MOORE M.J. ANDERSEN J. GREEN M.R. ROTHENBERG M.L. MODIANO M.R. CRIPPS M.C. PORTENOY R.K. STORNIOLO A.M. TARASSOFF P., et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- CERCHIA L. GIANGRANDE P.H. MCNAMARA J.O. DE FRANCISCIS V. Cell-specific aptamers for targeted therapies. Methods Mol. Biol. 2009;535:59–78. doi: 10.1007/978-1-59745-557-2_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHU T.C. MARKS J.W., 3RD LAVERY L.A. FAULKNER S. ROSENBLUM M.G. ELLINGTON A.D. LEVY M. Aptamer:toxin conjugates that specifically target prostate tumor cells. Cancer Res. 2006;66:5989–5992. doi: 10.1158/0008-5472.CAN-05-4583. [DOI] [PubMed] [Google Scholar]
- CONRAD F. HANNE A. GAUR R.K. KRUPP G. Enzymatic synthesis of 2′-modified nucleic acids: identification of important phosphate and ribose moieties in RNase P substrates. Nucleic Acids Res. 1995;23:1845–1853. doi: 10.1093/nar/23.11.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONROY T. DESSEIGNE F. YCHOU M. BOUCHE O. GUIMBAUD R. BECOUARN Y. ADENIS A. RAOUL J.L. GOURGOU-BOURGADE S. DE LA FOUCHARDIERE C., et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011;364:1817–1825. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- DASSIE J.P. LIU X.Y. THOMAS G.S. WHITAKER R.M. THIEL K.W. STOCKDALE K.R. MEYERHOLZ D.K. MCCAFFREY A.P. MCNAMARA J.O., 2nd GIANGRANDE P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ELLINGTON A.D. SZOSTAK J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- FAROKHZAD O.C. CHENG J. TEPLY B.A. SHERIFI I. JON S. KANTOFF P.W. RICHIE J.P. LANGER R. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FOWLER J.D. BROWN J.A. JOHNSON K.A. SUO Z. Kinetic investigation of the inhibitory effect of gemcitabine on DNA polymerization catalyzed by human mitochondrial DNA polymerase. J. Biol. Chem. 2008;283:15339–15348. doi: 10.1074/jbc.M800310200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAN H.K. WALKER F. BURGESS A.W. RIGOPOULOS A. SCOTT A.M. JOHNS T.G. The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor AG1478 increases the formation of inactive untethered EGFR dimers. Implications for combination therapy with monoclonal antibody 806. J. Biol. Chem. 2007;282:2840–2850. doi: 10.1074/jbc.M605136200. [DOI] [PubMed] [Google Scholar]
- GAUR R.K. KRUPP G. Modification interference approach to detect ribose moieties important for the optimal activity of a ribozyme. Nucleic Acids Res. 1993;21:21–26. doi: 10.1093/nar/21.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GIOVANNETTI E. DEL TACCA M. MEY V. FUNEL N. NANNIZZI S. RICCI S. ORLANDINI C. BOGGI U. CAMPANI D. DEL CHIARO M., et al. Transcription analysis of human equilibrative nucleoside transporter-1 predicts survival in pancreas cancer patients treated with gemcitabine. Cancer Res. 2006;66:3928–3935. doi: 10.1158/0008-5472.CAN-05-4203. [DOI] [PubMed] [Google Scholar]
- HANSEL T.T. KROPSHOFER H. SINGER T. MITCHELL J.A. GEORGE A.J. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Disc. 2010;9:325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- HE K. HASAN A. KRZYZANOWSKA B. SHAW B.R. Synthesis and separation of diastereomers of ribonucleoside 5′-(alpha-P-borano) triphosphates. J. Org. Chem. 1998;63:5769–5773. doi: 10.1021/jo972002g. [DOI] [PubMed] [Google Scholar]
- HUANG P. CHUBB S. HERTEL L.W. GRINDEY G.B. PLUNKETT W. Action of 2′,2′-difluorodeoxycytidine on DNA synthesis. Cancer Res. 1991;51:6110–6117. [PubMed] [Google Scholar]
- KINDLER H.L. FRIBERG G. SINGH D.A. LOCKER G. NATTAM S. KOZLOFF M. TABER D.A. KARRISON T. DACHMAN A. STADLER W.M., et al. Phase II trial of bevacizumab plus gemcitabine in patients with advanced pancreatic cancer. J. Clin. Oncol. 2005;23:8033–8040. doi: 10.1200/JCO.2005.01.9661. [DOI] [PubMed] [Google Scholar]
- KNUDSEN S.M. ROBERTSON M.P. ELLINGTON A.D. In vitro selection using modified or unnatural nucleotides. Curr. Protoc. Nucleic Acid Chem. 2002;9 doi: 10.1002/0471142700.nc0906s56. Chapter. Unit 9.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KROEP J.R. LOVES W.J. VAN DER WILT C.L. ALVAREZ E. TALIANIDIS I. BOVEN E. BRAAKHUIS B.J. VAN GROENINGEN C.J. PINEDO H.M. PETERS G.J. Pretreatment deoxycytidine kinase levels predict in vivo gemcitabine sensitivity. Mol. Cancer Ther. 2002;1:371–376. [PubMed] [Google Scholar]
- LEVY-NISSENBAUM E. RADOVIC-MORENO A.F. WANG A.Z. LANGER R. FAROKHZAD O.C. Nanotechnology and aptamers: applications in drug delivery. Trends Biotechnol. 2008;26:442–449. doi: 10.1016/j.tibtech.2008.04.006. [DOI] [PubMed] [Google Scholar]
- LI N. LARSON T. NGUYEN H.H. SOKOLOV K.V. ELLINGTON A.D. Directed evolution of gold nanoparticle delivery to cells. Chemical communications. 2010;46:392–394. doi: 10.1039/b920865h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI N. NGUYEN H.H. BYROM M. ELLINGTON A.D. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUDWIG J. ECKSTEIN F. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphate)s, 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J. Org. Chem. 1989;54:631–635. [Google Scholar]
- MACKEY J.R. MANI R.S. SELNER M. MOWLES D. YOUNG J.D. BELT J.A. CRAWFORD C.R. CASS C.E. Functional nucleoside transporters are required for gemcitabine influx and manifestation of toxicity in cancer cell lines. Cancer Res. 1998;58:4349–4357. [PubMed] [Google Scholar]
- MCNAMARA J.O., 2ND ANDRECHEK E.R. WANG Y. VILES K.D. REMPEL R.E. GILBOA E. SULLENGER B.A. GIANGRANDE P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- MILLIGAN J.F. GROEBE D.R. WITHERELL G.W. UHLENBECK O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOORE M.J. GOLDSTEIN D. HAMM J. FIGER A. HECHT J.R. GALLINGER S. AU H.J. MURAWA P. WALDE D. WOLFF R.A., et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 2007;25:1960–1966. doi: 10.1200/JCO.2006.07.9525. [DOI] [PubMed] [Google Scholar]
- NAKANO Y. TANNO S. KOIZUMI K. NISHIKAWA T. NAKAMURA K. MINOGUCHI M. IZAWA T. MIZUKAMI Y. OKUMURA T. KOHGO Y. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br. J. Cancer. 2007;96:457–463. doi: 10.1038/sj.bjc.6603559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OETTLE H. POST S. NEUHAUS P. GELLERT K. LANGREHR J. RIDWELSKI K. SCHRAMM H. FAHLKE J. ZUELKE C. BURKART C., et al. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. Jama. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- OLIVE K.P. JACOBETZ M.A. DAVIDSON C.J. GOPINATHAN A. MCINTYRE D. HONESS D. MADHU B. GOLDGRABEN M.A. CALDWELL M.E. ALLARD D., et al. Inhibition of hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PADILLA R. SOUSA R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutantT7 RNA polymerase (RNAP) Nucleic Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PATRA C.R. BHATTACHARYA R. WANG E. KATARYA A. LAU J.S. DUTTA S. MUDERS M. WANG S. BUHROW S.A. SAFGREN S.L., et al. Targeted delivery of gemcitabine to pancreatic adenocarcinoma using cetuximab as a targeting agent. Cancer Res. 2008;68:1970–1978. doi: 10.1158/0008-5472.CAN-07-6102. [DOI] [PubMed] [Google Scholar]
- PHILIP P. BENEDETTI J. FENOGIO-PREISER C. ZALUPSKI M. LENZ H. O'REILLY E. WONG R. ATKINS J. ABBRUZZESE J. BLANKE C. Phase III study of gemcitabine plus cetuximab versus gemcitabine in patients with locally advanced or metastatic pancreatic adenocarcinoma: SWOG S0205 study. J. Clin. Oncol. 2007;25:LBA4509. [Google Scholar]
- ROCHA-LIMA C.M. SOARES H.P. RAEZ L.E. SINGAL R. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- RUIZ VAN HAPEREN V.W. VEERMAN G. VERMORKEN J.B. PETERS G.J. 2′,2′-Difluoro-deoxycytidine (gemcitabine) incorporation into RNA and DNA of tumour cell lines. Biochem. Pharmacol. 1993;46:762–766. doi: 10.1016/0006-2952(93)90566-f. [DOI] [PubMed] [Google Scholar]
- SIEGEL R. WARD E. BRAWLEY O. JEMAL A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- SOUNDARARAJAN S. WANG L. SRIDHARAN V. CHEN W. COURTENAY-LUCK N. JONES D. SPICER E.K. FERNANDES D.J. Plasma membrane nucleolin is a receptor for the anticancer aptamer AS1411 in MV4-11 leukemia cells. Mol. Pharmacol. 2009;76:984–991. doi: 10.1124/mol.109.055947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SOUSA R. PADILLA R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TUERK C. GOLD L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- ULLRICH A. SCHLESSINGER J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- VIEIRA A.V. LAMAZE C. SCHMID S.L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science. 1996;274:2086–2089. doi: 10.1126/science.274.5295.2086. [DOI] [PubMed] [Google Scholar]
- WICKSTROM E. Oligodeoxynucleotide stability in subcellular extracts and culture media. J. Biochem. Biophys. Methods. 1986;13:97–102. doi: 10.1016/0165-022x(86)90021-7. [DOI] [PubMed] [Google Scholar]
- XIONG H.Q. ROSENBERG A. LOBUGLIO A. SCHMIDT W. WOLFF R.A. DEUTSCH J. NEEDLE M. ABBRUZZESE J.L. Cetuximab, a monoclonal antibody targeting the epidermal growth factor receptor, in combination with gemcitabine for advanced pancreatic cancer: a multicenter phase II trial. J. Clin. Oncol. 2004;22:2610–2616. doi: 10.1200/JCO.2004.12.040. [DOI] [PubMed] [Google Scholar]
- ZHANG Y. CHEN Y. HAN D. OCSOY I. TAN W. Aptamers selected by cell-SELEX for application in cancer studies. Bioanalysis. 2010;2:907–918. doi: 10.4155/bio.10.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHOU J. ROSSI J.J. Cell-specific aptamer-mediated targeted drug delivery. Oligonucleotides. 2011;21:1–10. doi: 10.1089/oli.2010.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.