Background: How vascular plants control phenoxy radical coupling is enigmatic.
Results: Two dirigents engendered (−)-pinoresinol formation in Arabidopsis. Coupling stereoselectivity was reversed from (+)- to (−)-pinoresinol through swapping identical regions.
Conclusion: Novel insights into stereoselective control over phenoxy radical coupling were obtained.
Significance: This is the first report of dirigent-mediated phenoxy radical coupling control leading to opposite stereoselectivities and identification of protein regions involved.
Keywords: Arabidopsis, Biosynthesis, RNA Interference (RNAi), Secondary Metabolism, Site-directed Mutagenesis, Dirigent Protein, Phenoxy Radical-Radical Coupling, Stereoselectivity
Abstract
How stereoselective monolignol-derived phenoxy radical-radical coupling reactions are differentially biochemically orchestrated in planta, whereby for example they afford (+)- and (−)-pinoresinols, respectively, is both a fascinating mechanistic and evolutionary question. In earlier work, biochemical control of (+)-pinoresinol formation had been established to be engendered by a (+)-pinoresinol-forming dirigent protein in Forsythia intermedia, whereas the presence of a (−)-pinoresinol-forming dirigent protein was indirectly deduced based on the enantiospecificity of downstream pinoresinol reductases (AtPrRs) in Arabidopsis thaliana root tissue. In this study of 16 putative dirigent protein homologs in Arabidopsis, AtDIR6, AtDIR10, and AtDIR13 were established to be root-specific using a β-glucuronidase reporter gene strategy. Of these three, in vitro analyses established that only recombinant AtDIR6 was a (−)-pinoresinol-forming dirigent protein, whose physiological role was further confirmed using overexpression and RNAi strategies in vivo. Interestingly, its closest homolog, AtDIR5, was also established to be a (−)-pinoresinol-forming dirigent protein based on in vitro biochemical analyses. Both of these were compared in terms of properties with a (+)-pinoresinol-forming dirigent protein from Schizandra chinensis. In this context, sequence analyses, site-directed mutagenesis, and region swapping resulted in identification of putative substrate binding sites/regions and candidate residues controlling distinct stereoselectivities of coupling modes.
Introduction
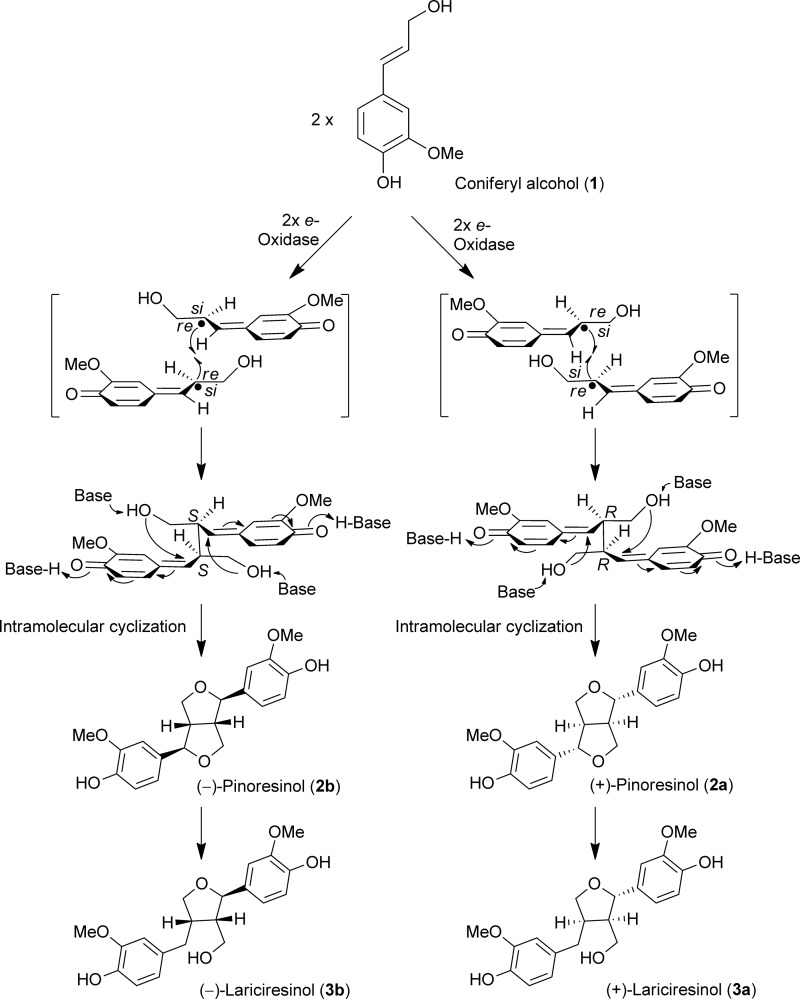
The first biochemical documentation of stereoselective control of monolignol radical-radical coupling was discovery of the (+)-pinoresinol-forming dirigent protein (DP)2 in Forsythia intermedia (1–4). These reports described the hitherto unprecedented biochemical mechanism involving a dimeric DP where each DP monomer was envisaged to capture, bind, and orientate one coniferyl alcohol (1) radical substrate in such a way that, after stereoselective coupling, only (+)-pinoresinol (2a) was formed (4) (Fig. 1). However, this gave no definitive insight as to how substrate binding and orientation occurred. Additionally, there was ample indirect evidence for formation/accumulation of the opposite antipode, (−)-pinoresinol (2b), in some other plant species including, for example, Daphne tangutica (5), and flax (Linum usitatissimum (6)), suggesting the presence of a related DP engendering formation of the opposite enantiomeric form (Fig. 1).
FIGURE 1.
Proposed mechanism for distinctive dirigent protein-mediated stereoselective coupling to either (+)- or (−)-pinoresinol (2a or 2b) with subsequent enantiospecific reduction to (+)- and (−)-lariciresinols (3a) and (3b). The generation of (+)- and (−)-pinoresinols (2a and 2b) in planta from coniferyl alcohol (1) radical intermediates via coupling at si-si and re-re faces, respectively, is also shown.
Furthermore, a plethora of other different regiospecificities and stereoselectivities are commonly encountered in various lignan skeletons throughout the plant kingdom depending upon plant species (7). Thus, this raised further important questions about how such structurally diverse coupling transformations are biochemically engendered. Moreover, there is growing recognition of regiospecific control over lignin macromolecular assembly and configuration (8–10); proteins harboring arrays of coniferyl alcohol radical binding (dirigent) sites in lignifying plant cell walls have been proposed to be involved (11, 12), e.g. to account for the near conservation of 8-O-4′ interunit linkage frequency in the specific plant lines examined (8). A DP has also been implicated in formation of the phenolic terpenoid (+)-gossypol. This was considered to occur via stereoselective control of the presumed radical-radical coupling of hemigossypol and restricted rotation around the biphenyl bond product so obtained (supplemental Fig. S1) (13). Accordingly, how vascular plant species exercise distinctive stereoselective and/or regiospecific control over radical-radical coupling reactions of monolignols and other compounds is an intriguing, timely, and important biochemical question with profound evolutionary implications.
The existence of a (−)-pinoresinol-forming DP in Arabidopsis was provisionally deduced (7) as indicated below. This was based on the observation that pinoresinol reductase homologs (AtPrR1 and AtPrR2) from Arabidopsis root tissues preferentially converted (−)-pinoresinol (2b) into (−)-lariciresinol (3b) in vitro over that of the (+)-antipode (2a) (14). When racemic pinoresinol (2a/2b) was used as substrate in vitro, the lariciresinol (3) generated was in 96% enantiomeric excess (e.e.) of the (−)-antipode (3b) using AtPrR2, whereas only 6% e.e. of the (+)-form (3a) was attained using AtPrR1. However, AtPrR1 was ∼16 times catalytically more efficient than AtPrR2 with (−)-pinoresinol (2b) as substrate.
In agreement with these presumed biochemical/physiological roles, in a loss-of-function study, the double T-DNA mutant (atprr1-1 atprr2) accumulated the AtPrR substrate (−)-pinoresinol (2b; ∼4,000 ng mg−1 of dry weight root tissue; 74% e.e.), but no lariciresinol (3) was detected. Furthermore, AtPrR mutants with a single T-DNA insertion in each gene gave phenotypes with slightly increased levels of lariciresinol (3) (∼360 versus 560–640 ng mg−1 of dry weight root tissue). Interestingly, the e.e. of (−)-lariciresinol (3b) in the wild type (WT) line was 88%, whereas in atprr1 and atprr2, it was 95 and 82%, respectively. These findings, particularly those from analysis of the double mutant, thus suggested preferential stereoselective formation of (−)-pinoresinol (2b) from coniferyl alcohol (1). Since then, discovery of a gene encoding a (−)-pinoresinol-forming DP in Arabidopsis root tissue was briefly communicated (7, 15).
In the study herein, all possible Arabidopsis DP gene expression patterns were initially examined to identify those DPs either constitutively expressed or potentially inducible in roots. From these data, specific DP genes were identified and cloned, and their recombinant proteins were determined to be (−)-pinoresinol-forming DPs; as a control, these data were compared with a S. chinensis (+)-pinoresinol-forming DP. In addition, overexpression (OE) and RNAi of the most highly expressed Arabidopsis (−)-pinoresinol-forming DP in root tissue provided definitive insight into its physiological role in planta. Moreover, how substrate binding and different stereoselectivities are presumably attained is discussed, largely based on site-directed mutagenesis and region swapping results.
EXPERIMENTAL PROCEDURES
Materials
Solvents/chemicals were either reagent or high performance liquid chromatography (HPLC) grade unless otherwise specified. The Wizard® Plus Minipreps DNA purification system (Promega, Madison, WI) was used to purify plasmid DNA. Custom oligonucleotide primers for sequencing and polymerase chain reaction (PCR) were from Invitrogen.
Instrumentation and Chromatography Materials
Chemical reactions were carried out under anhydrous conditions using dry freshly distilled solvents in an argon atmosphere. Silica gel thin-layer and column chromatography utilized Partisil® PK5F (Whatman; 20 × 20 cm, 1 mm, 150 Å), AL SIL G/UV254 (Whatman; 20 × 20 cm, 0.25 mm), and silica gel 60 (EM Science), respectively. UV light and a 10% H2SO4 spray followed by heating were used for TLC plate visualization. Gel filtration and anion exchange chromatographies were carried out on a fast protein liquid chromatography system (GE Healthcare). NMR spectra were recorded on an Inova 500 spectrometer operating at 499.85 and 125.67 MHz for 1H and 13C, respectively, with J values given in Hz. One-dimensional proton and two-dimensional 1H-1H double quantum-filtered COSY and 1H-13C heteronuclear single quantum coherence and heteronuclear multiple bond correlation spectra were acquired using BioPack pulse sequences at 293 K on an Agilent (Varian) 800-MHz VNMRS instrument equipped with an HCN (Z-gradient) cold probe. Data were processed with Felix (Felix NMR, Inc.) Samples for 800-MHz NMR were prepared by dissolving ∼250 μg of lariciresinol 4-O-β-d-glucoside (5) in 240 μl of methanol-d4 (Cambridge Isotope Laboratories) and transferring the solution to 5-mm susceptibility-matched tubes (Shigemi).
HPLC analyses were carried out on an Alliance 2690 HPLC system (Waters, Milford, MA) equipped with a photodiode array detector (Model 2990, Waters) with detection at 280 nm. Reversed-phase separations utilized a Novapak C18 column (Waters; 150 × 3.9-mm inner diameter) as described (4). The (+)- and (−)-enantiomers of pinoresinols (2) were resolved on a Chiralcel OD (Chiral Technologies, West Chester, PA) column as described (4), and antipodes of lariciresinol (3) were separated on a Chiralcel OC column eluted with hexane-EtOH (1:4; flow rate, 0.3 ml min−1).
Liquid chromatography-mass spectrometry (LC-MS) analyses of pinoresinol (2) were conducted as described (9). Gas chromatography (GC)-MS analyses of trimethylsilylated samples used an HP 6890 Series GC system equipped with a RESTEK Rtx-5Sil-MS (30 m × 0.25 mm × 0.25 μm) column and an HP 5973 MS detector (electron impact mode, 70 eV).
Time-of-flight (TOF) electrospray ionization MS analyses of lignan glucosides were conducted using a Xevo® G2 QTof/ACQUITY UPLC® system (Waters) equipped with a BEH C18 column (Waters; 2.1 × 150 mm) using the following chromatographic conditions: flow rate of 0.2 ml min−1; linear gradients of CH3CN, 3% AcOH in H2O (v/v) from 10:90 to 60:40 in 6.5 min, then to 80:20 in 2 min, and finally to 100:0 in 1 min with this composition held for an additional 1 min. Leucine-enkephalin was utilized as a lock mass standard.
PCR amplifications utilized a PTC-0220 DNA Engine Dyad Peltier thermal cycler (MJ Research, Waltham, MA), whereas real time quantitative PCR analyses of gene expression were performed on a Mx3005PTM real time PCR system (Stratagene, La Jolla, CA) with Platinum® SYBR® Green qPCR SuperMix-UDG (Invitrogen). Protein, RNA, and DNA concentrations were determined on a Response spectrophotometer (Gilford). Protein purification used an FPLC system (GE Healthcare). DNA sequencing utilized an ABI PRISM 377 automated DNA sequencer (Applied Biosystems, Foster City, CA). Biolistic transformation was carried out with a helium-driven PDS-1000/He system (Bio-Rad).
Plant Growth Conditions
Arabidopsis ecotype Columbia seeds were cold-treated at 4 °C for 48 h and grown in pots in a growth chamber maintained at 22/18 °C. Light (230 μmol m−2 s−1) was provided under a 16/8-h light/dark cycle. S. chinensis seedlings were obtained from Forest Farm, Williams, OR and maintained in Washington State University greenhouse facilities: the light intensity was 150 μmol m−2 s−1 with a 15/9-h light/dark cycle at 21/16 °C, respectively.
Chemical Syntheses
E-Coniferyl alcohol (1) was synthesized as described in Kim et al. (16), whereas (±)-pinoresinols (2a/b) and (±)-lariciresinols (3a/b) were synthesized and resolved into their enantiomeric forms as in Moinuddin et al. (17).
E-[9-2H2]Coniferyl alcohol (1) was synthesized as follows. To a solution of coniferyl aldehyde (0.5 g; 2.8 mmol) in MeOH (10 ml) was added NaB2H4 (0.235 g; 5.6 mmol) at 0 °C for 2 h. Progress of the reaction was monitored by NMR spectroscopic analysis until coniferyl aldehyde was completely reduced to deuterated coniferyl alcohol (1). After reaction completion, the resulting mixture was acidified with dilute HCl (10 ml) and extracted with EtOAc (2 × 50 ml). The EtOAc-solubles were then washed with H2O (2 × 25 ml) and brine (2 × 50 ml), dried (Na2SO4), and evaporated to dryness in vacuo. The residue so obtained was purified by silica gel column chromatography using hexane-EtOAc (6:4) as eluant to afford E-[9-2H2]coniferyl alcohol (1) (0.44 g; 2.41 mmol; 87% yield): δH (acetone-d6; 99.9 atom % D; 500 MHz) 7.6 (1H, br s, ArOH), 7.05 (1 H, d, J 2, H-2), 6.86 (1 H, dd, J 2.5 and 8.5, H-6), 6.77 (1 H, d, J 8.0, 5-H), 6.5 (1 H, d, J 16, 7-H), 6.22 (1 H, d, J 16, 8-H), 3.86 (3 H, s, 3-OMe); δC (acetone-d6; 99.9 atom % D; 125 MHz) 148.5 (C-3), 147.2 (C-4), 130.6 (C-1), 130.3 (C-7), 128.1 (C-8), 120.7 (C-6), 115.8 (C-5), 110.0 (C-2), 56.2 (C-3-OMe). C10H10D2O3, m/z = 182.
Isolation of Arabidopsis thaliana AtDIR Genes Including Their Promoters and 3′-UTR
Genomic DNA was purified from 3-week-old Arabidopsis rosette leaves using the DNeasy plant minikit (Qiagen, Valencia, CA). For each dirigent gene, the 5′-upstream region to the 3′-UTR obtained from The Arabidopsis Information Resource database was amplified using PfuTurbo® DNA polymerase (Stratagene) and DP-specific primers (supplemental Table S1). PCR amplifications were performed as follows: initial denaturation at 96 °C for 5 min, 35 cycles of denaturation at 96 °C for 30 s, annealing at 52–55 °C for 30 s, and extension at 68 °C for 3 min with an additional extension at 68 °C for 10 min. PCR products were analyzed on agarose gels, and amplified fragments of interest were purified using the QIAquick gel extraction kit (Qiagen). Next, a single deoxyadenosine (A) was added to the 3′-end of each amplified fragment using Taq polymerase (Invitrogen) at 72 °C for 15 min. Each dirigent homolog was then subcloned into a pCRII-TOPO® (Invitrogen) vector for sequencing; sequences were verified using the BioeditTM program against the NCBI database and The Arabidopsis Information Resource database. Then the 5′-region (including signal peptide sequence) and the 3′-UTR of each dirigent gene were individually amplified using the corresponding entire coding sequences as templates and region-specific primers (supplemental Tables S2 and S3).
Vector Construction
All 16 dirigent promoter (including signal peptide sequence)::uidA::3′-UTR::nopaline synthase terminator vectors were constructed as follows. First, uidA (GUSPlusTM) was excised from pCAMBIA 1305.1 and introduced into pGEM58ZNf(−) (GenBankTM accession number AF310245) using NcoI and NheI/SpeI sites to produce clone pGEM59ZNf(−). Specific restriction recognition primers (supplemental Table S2) were introduced into each amplified 5′-upstream region and confirmed by sequencing in the pCRII-TOPO vector. Using a combination of SphI/HindIII and BglII/BamHI sites, these were subcloned into shuttle vector pGEM59. Similarly, using specific restriction recognition sites (supplemental Table S2), the introduced 3′-UTRs were confirmed and subcloned into the pGEM59 vector 5′-upstream region using a combination of SalI/PstI/NotI and BamHI/SalI sites. Next, the SphI sites (2,464 nucleotides) of the pCAMBIA1305.1 right border was filled out using exonuclease I (New England Biolabs, Ipswich, MA), generating pCAMBIA 1305.1-1, which was subsequently digested with BstEII and blunted by fill-in with T4 DNA polymerase (New England Biolabs). To remove the cauliflower mosaic virus 35S promoter region, the blunted pCAMBIA 1305.1-1 vector was digested with either SphI or HindIII and purified. Each AtDIR promoter::uidA::AtDIR 3′-UTR in pGEM59 was then digested using SphI/HindIII and NruI/EcoRV, introduced into the pCAMBIA 1305.1-1 vector, and transformed into Escherichia coli XL-10 Gold (Stratagene) strains. Sequence confirmation was performed as described above.
Plant Transformation Screening and Histochemical GUS Staining
Each binary vector harboring individual GUS-DP promoter constructs was introduced into Agrobacterium tumefaciens (strain GV3101) with subsequent transformation in Arabidopsis, selection, and histochemical GUS staining as in Kim et al. (18, 19).
AtDIR6 Overexpression and RNAi Lines
Primers for AtDIR6 ORF amplification incorporated the XhoI restriction enzyme site at both the 5′- and 3′-ends (5′-GCTCGAGATGGCATTTCTAGTAGA-3′ and 5′-GCTCGAGTTAGTAACATTCATAGAG-3′; introduced restriction sites are shown in italic underlined). The amplified PCR fragment was then cloned into the pSTBlue-1 vector (Novagen) for sequencing. After sequence verification, AtDIR6 was excised from pSTBlue-1 with XhoI restriction enzyme and subcloned in pART7 vector at the XhoI site (20). The NotI fragment from the pART7 vector containing AtDIR6 was then cloned into T-DNA binary vector pART27.
The selected RNAi target region (316 bp) was amplified with forward (5′-CACCATCGTGAGCCCTCCAGGACTAG-3′) and reverse (5′-GAAATAAATCAGTCACGAAGGTAGCG-3′) primers and cloned into the pENTR/D-TOPO vector (Invitrogen). Next, the DNA fragment flanked by recombination sites (att) was transferred into the pK7GWIWG2 (II) vector (21) using Gateway® LR ClonaseTM II enzyme mixture (Invitrogen).
After sequence confirmation, each overexpression and RNAi construct was individually introduced into A. tumefaciens (strain LBA4404/pC2760) and transformed into A. thaliana as described in Kim et al. (19). Successful integration of each construct in T1 plants was confirmed by screening genomic DNA isolated from a 3-week-old rosette leaf using a REDExtract-N-AMP Plant PCR kit (Sigma-Aldrich). PCR was carried out using an initial denaturation of 94 °C for 2 min followed by 35 cycles of 94 °C (30 s), 48 °C (30 s), and 72 °C (2 min) and finally extension (72 °C for 10 min). Amplified PCR products from this screening were sequenced.
More than 40 T2 AtDIR6 OE plant lines were screened on Murashige-Skoog medium containing kanamycin (50 μg ml−1); only one line was resistant to the antibiotic. The latter was propagated, and T3 plants were used for further analyses. Using the same selection process, 48 T1 AtDIR6 RNAi lines were selected (of ∼6,000 T0 seeds). The RNAi line with the lowest level of AtDIR6 expression was subsequently used for further study (T3 plants).
Gene Expression Analysis
Total RNA was individually isolated from 2-week-old-seedlings of WT, AtDIR6 OE, and RNAi plant lines using the RNeasy plant minikit (Qiagen), and first strand cDNA was synthesized using the SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen). For real time quantitative PCR analyses, each PCR mixture contained synthesized first strand cDNA, Platinum SYBR Green qPCR SuperMix-UDG (Invitrogen) and gene-specific primers, which were designed using Primer Premier software (Biosoft International, Palo Alto, CA) (supplemental Table S4). Expression levels were normalized against the TIP41-like gene (22) with expression levels for each gene in WT set to 1 and data averaged from triplicate samples. AtDIR6 OE and RNAi expression levels were relative to WT.
Cloning of AtDIR5, AtDIR6, AtDIR10, and AtDIR13
RNA was purified from 2-week-old Arabidopsis seedlings using the RNeasy plant minikit. AtDIRs were amplified using PfuTurbo DNA polymerase (Stratagene) and gene-specific primers (supplemental Table S5) and then cloned into pCR4-TOPO vector (Invitrogen) for sequence verification.
Cloning of S. chinensis DP
Using 5 μg of mRNA isolated from leaf tissue, a cDNA library (2.3 × 106 pfu μl−1) was constructed using the Stratagene ZAP-cDNA® synthesis kit. The [32P]CTP-labeled F. intermedia DP gene (FiDIR; GenBank accession number AF210061) was used as a probe for screening. The isolated S. chinensis DP gene (ScDIR; GenBank accession number HQ428029) was reamplified using T7 and T3 primers for cloning into the pMT/V5-His-TOPO vector for recombinant protein expression in insect (Drosophila melanogaster) cells.
Heterologous Expression and Purification of AtDIR6, AtDIR10, and ScDIR in Insect Cell Cultures
The amplified AtDIR6, AtDIR10, and ScDIR genes were individually subcloned into the pMT/V5-His-TOPO vector (Invitrogen) for heterologous expression in insect cells (23). The DP genes were next stably co-transfected individually with pCoHygro vector (Invitrogen) into Drosophila Schneider2 cells using calcium phosphate, and cells were grown at 26 °C. After post-transfection for 2 days, each selection was carried out in HyQ-SFX insect cell medium (HyClone) containing 300 μg ml−1 hygromycin-B (Invitrogen), 5% fetal bovine serum (v/v; Invitrogen), and 2 mm l-glutamine (Invitrogen). Every 4–5 days, selective medium was replaced with fresh medium by centrifugation (1,000 × g for 5 min); this was carried out for 1 month. After resistant cell selection, each cell culture was gradually expanded from 5 to 500 ml with densities kept at 106–107 cells ml−1. When a volume of 500 ml and a density of ∼107 cell ml−1 were reached, cells were induced with copper sulfate (900 μm) and further incubated for 40 h at 26 °C. Before harvesting, five protease inhibitors (leupeptin, antipain, pepstatin A (in DMSO), 4-(2-aminoethyl)benzenesulfonyl fluoride (5 mm each), and trans-epoxysuccinyl-l-leucylamido-(4-guanidino)butane (1 mm)) were added to each flask. Six flasks were harvested at the same time.
Each cell culture was centrifuged twice (1,000 × g for15 min and 12,000 × g for 30 min), and supernatants (∼3 liters) were further concentrated (Amicon concentrator, Model 2000) under nitrogen pressure through a PM-10 membrane (Amicon) to less than 200 ml. Each concentrated medium was fractionated with (NH4)2SO4 with proteins precipitating between 40 and 80% (NH4)2SO4 recovered in 20 mm MES-NaOH buffer, pH 5.0 (buffer A; 20 ml). After dialysis overnight in buffer A (4 liters × 2) at 4 °C, each dialysate (∼100 mg of total protein; 20 ml) was applied to an SP-Sepharose Fast Flow® column (8 ml; GE Healthcare) pre-equilibrated in buffer A at a flow rate of ∼7 ml min−1 (via hyperbaric air pressure) at room temperature. After washing each with buffer A (25 ml) and 50 mm Na2SO4 in buffer A (50 ml), each DP was eluted with 333 mm Na2SO4 in buffer A (50 ml), and the latter eluate was concentrated and desalted using an Amicon Centricon® filtration apparatus. Each concentrate (∼25 mg of total protein) was next applied to a Mono STM 5/50 column (5 × 50 mm; GE Healthcare) pre-equilibrated in buffer A at a flow rate of 1 ml min−1. DPs were individually eluted using a linear gradient from 0 to 133 mm Na2SO4 in buffer A in 8 ml followed by Na2SO4 step gradients in buffer A as follows: 133 mm (30 ml), 167 mm (30 ml), 200 mm (30 ml), and 333 mm (20 ml). The 200 mm Na2SO4 fractions were pooled, concentrated, and desalted as above. Each desalted DP solution (∼1 mg; typically from 3 liters of culture medium) was next applied to a POROS® 20 SP column pre-equilibrated in HEPES-sodium acetate-MES buffer (33 mm each, pH 5.0; buffer B) at a flow rate of 0.75 ml min−1. After washing with buffer B (7.5 ml), the DPs were individually eluted with linear Na2SO4 gradients in buffer B as follows: 0–50 mm in 4.5 ml, 50–233 mm in 30 ml with this concentration held for an additional 33 ml, and finally from 233 to 333 mm in 10.5 ml. Recombinant DPs were eluted with 233 mm Na2SO4. Homogeneity was assessed by SDS-PAGE analysis.
Heterologous Expression and Purification of AtDIR5, AtDIR6, and AtDIR13 in Plant Cell Cultures
Cloned DP genes were reamplified with EcoRI and HindIII restriction sites containing primers (supplemental Table S5) and cloned into pCR4-TOPO. Afterward, each gene was individually excised from the vector with EcoRI and HindIII restriction enzymes and subcloned into the pART17 vector. To express the recombinant DPs in a tomato (Solanum peruvianum) (24) cell suspension culture, the pART17 vector was manipulated as follows. A kanamycin selectable marker region, nopaline synthase promoter::neomycin phosphotransferase::nopaline synthase terminator, was amplified from pART27 vector with SpeI restriction site-containing primers (5′-GGACTAGTATACATGAGAATTAAGGGAGTC-3′ and 5′-GGACTAGTATCAGCTTGCATGCCGGTCGATC-3′; introduced restriction sites are shown in italic underlined) and then introduced into the pART7 vector using the SpeI site. The generated pART17 vector was then digested with EcoRI and HindIII restriction enzymes and AtDIR5, AtDIR6, and AtDIR13 ORFs were individually cloned, and their sequences were confirmed.
AtDIR5, AtDIR6, and AtDIR13 were individually heterologously expressed in the tomato cell suspension culture maintained in a Murashige-Skoog medium (pH 5.5) containing Nitshe vitamins (110 mg liter−1), thiamine (0.2 mg liter−1), myo-inositol (20 mg liter−1), α-naphthalene acetic acid (5 mg liter−1), 6-benzylaminopurine (1 mg liter−1), and sucrose (30 g liter−1). Five-to-7-day-old cells (20 ml) were washed with fresh medium, gently swirled for 10 min, and washed once again. After decanting medium, fresh medium (10 ml) was added to make relatively packed cells, and 2 ml of cells was next applied onto a medium plate (6 cm in diameter). AtDIR5, AtDIR6, and AtDIR13 genes cloned in the pART17 vector were individually coated onto 1-μm gold microcarriers (Bio-Rad) in a calcium-spermidine precipitation. Biolistic transformation was conducted under the following conditions: 27 inches of mercury in a chamber vacuum, 1,100-p.s.i. burst helium pressure, and 6-cm target distance. Transformed cells (2 ml) were allowed to recover for 1 day on the plates, transferred to 6 ml of fresh culture medium, and incubated on an orbital shaker (100 rpm at 25 °C) for 1 week. Cells were then selected on culture plates containing kanamycin (75 μg ml−1). Initial callus growth was observed after 3–4 weeks, and individual calli were transferred and maintained on fresh selection plates for ∼8 weeks. DP gene expression levels were assessed from individual callus using RT-PCR analysis with gene-specific primers (supplemental Table S4). A callus line with the highest AtDIR5 (or AtDIR6 or AtDIR13) mRNA expression level was chosen for subsequent suspension cell culturing as follows. Each selected callus was maintained on a kanamycin plate, and cell culturing was established from the callus by inoculating 10 ml of tomato cell suspension culture medium. The suspension cell cultures were scaled up by inoculating into a new medium (up to 6 liters). Eight days after final inoculation, plant cells were harvested by vacuum filtration (Whatman filter paper Number 5), and cell wall-bound proteins were obtained by filtration after agitating the cells in potassium phosphate buffer (0.1 m, pH 5.9; buffer C) containing 75 and 150 mm KCl, respectively. Both fractions were combined and applied onto an SP-Sepharose Fast Flow column (80 ml), which was equilibrated with 75 mm KCl in buffer C. Proteins eluted with 1 m NaCl in buffer C were fractionated with (NH4)2SO4, and AtDIR5 (or AtDIR6 or AtDIR13) was purified as described above (see “Heterologous Expression and Purification of AtDIR6, AtDIR10, and ScDIR in Insect Cell Cultures”).
DP Assays
Each purified recombinant DP was assayed using DP (0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, or 6.4 μm as a monomer), Trametes versicolor laccase as oxidizing agent, and coniferyl alcohol (1; 720 μm) in buffer B in a total volume of 250 μl. The total protein concentration in the assays was kept constant (i.e. at the highest DP concentration used) with bovine serum albumin (Promega). After incubation for 4 h at 30 °C, each assay mixture was extracted with EtOAc (500 μl × 2), and the EtOAc-solubles were combined, dried in vacuo, redissolved in MeOH-H2O (1:1, v/v), and subjected to reversed-phase HPLC. Fractions containing pinoresinol (2) were collected, freeze-dried, and subjected to chiral HPLC analysis (see “Instrumentation and Chromatography Materials”).
In assays with E-[9-2H2]coniferyl alcohol (1) and 3.3 μm AtDIR5, the generated [9,9′-2H2]pinoresinol (2) was subjected to chiral HPLC as described above, and both (−)- and (+)-antipodes were individually collected and analyzed by LC-MS and GC-MS: LC-MS analyses: (−)-[9,9′-2H2]pinoresinols (2b), m/z 362 [M+] (C20H18D4O6) (tR = 3.29 min); (+)-[9,9′-2H2]pinoresinols (2a), m/z 362 [M+] (C20H18D4O6) (tR = 6.74 min); GC-MS analyses: (−)-[9,9′-2H2]pinoresinols (2b), m/z 506 [M•+] (C26H34D4O6Si2) (tR = 28.57 min); (+)-[9,9′-2H2]pinoresinols (2a), m/z 506 [M•+] (C26H34D4O6Si2) (tR = 28.59 min).
Site-directed Mutagenesis
Site-directed mutagenesis was performed on ScDIR using the QuikChange site-directed mutagenesis kit (Stratagene) and primers (supplemental Table S6) as described (17), and transformants were selected on Luria–Bertani plates containing kanamycin (50 μg ml−1). Mutations were confirmed by sequencing, individual mutated clones were transfected into insect cell cultures, and recombinant mutated DPs were purified as described above.
Region Swapping
The selected regions, Region-A and -B, were swapped using a three-step PCR procedure (25). First, Region-A was amplified from AtDIR6 using an ScDIR linker primer set (supplemental Table S7). The second PCR was conducted to amplify individual N- and C-terminal regions with a mixture of wild type ScDIR, the amplified AtDIR6 Region-A template, and specific primers to the N- and C-terminal sequences (supplemental Table S7). Both N- and C-terminal amplified products contained the ScDIR gene flanked with AtDIR6 Region-A. The final PCR was carried out using the mixture of second round PCR products of N- and C-terminal regions as template and ScDIR primers, respectively. The amplified PCR products were cloned into a PCR vector, and the sequence was verified by DNA sequencing. Region-B was swapped in the same manner. Region-A-swapped ScDIR was transfected into insect cells with recombinant proteins obtained as described above. Region-B-swapped ScDIR was reamplified and cloned into the pART17 vector for preparation of recombinant proteins in tomato cell suspension cultures as described above.
Isolation of Pinoresinols (2) and Lariciresinols (3) from Arabidopsis WT, AtDIR6 OE, and AtDIR6 RNAi Lines
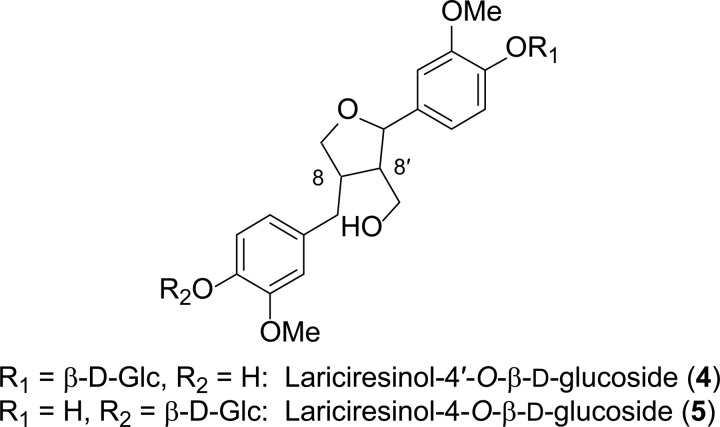
Lignans were isolated essentially as described in Nakatsubo et al. (14) and Suzuki et al. (26) from freeze-dried 4-week-old roots of WT (1.9 g), AtDIR6 OE (1.5 g), and AtDIR6 RNAi (1.7 g) plants, respectively. Following treatment with β-glucosidase (from almonds; Sigma), each EtOAc extract (∼15–20 mg) was purified by preparative silica gel thin-layer chromatography (eluant, CH2Cl2-MeOH, 9:1) and further purified by reversed-phase HPLC to afford pinoresinol (2) and lariciresinol (3), and each was subjected to chiral HPLC analyses (see “Instrumentation and Chromatography Materials”). The LC-MS and GC-MS data of 2 and 3 were in agreement with published data (27–29). Note that pinoresinol (2) was present as its aglycone, whereas lariciresinol (3) was in two different glucoside forms, 4 and 5 (Fig. 2).
FIGURE 2.
Lariciresinol glucosides (4 and 5).
Isolation of Lariciresinol 4′-O-β-d-Glucoside (4) and Lariciresinol 4-O-β-d-Glucoside (5) from Arabidopsis AtDIR6 OE Lines
The isolation of lignan glucosides (4 and 5) from freeze-dried 4-week-old AtDIR6 OE roots was carried out as described in Sugiyama and Kikuchi (30) with the following modifications. The ground roots (8 g) were extracted with hot MeOH (3 × 300 ml), centrifuged (2,000 × g for 10 min), and evaporated to dryness in vacuo. The MeOH extract (4.3 g) so obtained was suspended in a minimum amount of H2O (50 ml) and partitioned with Et2O, EtOAc, and n-BuOH, successively. The n-BuOH-soluble fraction was subjected to silica gel thin-layer chromatography (eluant, CHCl3-MeOH-H2O, 22:11:2) to obtain fractions Fr1–Fr14 (10 ml each). To ascertain which fractions contained the lignan derivatives of lignan glucosides, an aliquot (40 μl) of each fraction from Fr1 to Fr14 was treated with β-glucosidase as described above. Each hydrolysate aliquot (Fr1–Fr14) was then individually subjected to reversed-phase UPLC-MS analyses (see “Instrumentation and Chromatography Materials”) to identify the aglycone 3 by comparing its UV and MS data with an authentic lariciresinol (3) standard. After identifying which fractions contained lariciresinol (3), the lignan glucoside n-BuOH fractions (Fr7 and Fr8) were individually pooled, evaporated to dryness under N2, and freeze-dried. Each fraction (Fr7, 11.6 mg; Fr8, 17.8 mg) was individually applied to a Sephadex® LH-20® column (1.2 × 94 cm; Sigma-Aldrich) eluted with MeOH-H2O (1:1) at a flow rate of 0.3 ml min−1. The isolated fractions (Fr1–F54; 4 ml each) were next subjected to reversed-phase UPLC-MS analyses as above. Fractions 37–41 containing a mixture of lignan glucosides (4 and 5) were pooled and further purified by reversed-phase HPLC using a SymmetryShield RP18 column (Waters; 150 × 3.9-mm inner diameter) eluted as follows: flow rate of 1 ml min−1; linear gradients of CH3CN-H2O (Optima pure) from 5:95 to 15:85 in 15 min, to 40:60 in 18 min, to 80:20 in 7 min, and finally to 95:5 in 1 min with this composition held for an additional 2 min. The isolated lignan glucosides (4 and 5) were individually pooled, concentrated, and freeze-dried. The NMR, MS, and UV spectroscopic analyses of 4 and 5 were in agreement with published data (30) and also with authentic standards: TOF-electrospray ionization MS analyses: lariciresinol 4′-O-β-d-glucoside (4), m/z 545.1996 (M + Na; C26H34O11Na requires 545.1999) (tR = 4.04 min); lariciresinol 4-O-β-d-glucoside (7), m/z 545.1992 (M + Na; C26H34O11Na requires 545.1999) (tR = 4.28 min).
Homology Modeling
The “PHYRE” server was used to search for the closest structural homolog of ScDIR. AtAOC2 (Protein Data Bank code 2brj) was identified and used as the structural template for homology modeling of mature ScDIR on a bioinformatics tool server (@TOME). The modeled protein structures were visualized and analyzed using the UCSF Chimera molecular graphics program.
RESULTS AND DISCUSSION
Cellular and Organ/Tissue Specificities of AtDIR Expression
Although the putative 16 members of the DP family in Arabidopsis (31) have ∼65–29% (Table 1) sequence similarity to the F. intermedia (+)-pinoresinol-forming DP (FiDIR), no biochemical properties of the Arabidopsis homologs were reported at that time. Those with highest similarity to the (+)-pinoresinol-forming DP in F. intermedia, however, were AtDIR6 (At4g23690; ∼65%) and AtDIR5 (At1g64160; ∼61%). Nevertheless, in this study, a three-pronged approach was undertaken to systematically establish which, if any, of the DPs had (−)-pinoresinol-forming capacity in root tissue: this included use of GUS-promoter fusions, detailed analysis of various database compilations in silico, and assaying candidate recombinant Arabidopsis DP(s) in vitro and comparing the latter with an S. chinensis (+)-pinoresinol-forming DP. From the data obtained, OE and RNAi experiments were then designed.
TABLE 1.
The proposed Arabidopsis dirigent protein multigene family
AtDIR14 and AtDIR16 were originally annotated in The Arabidopsis Information Resource database as two different genes, but both are the same. AtDIR17 had very low identity to the F. intermedia DP and was not used in this study.
| Gene name | Locus number | Promoter size | Amino acid similarity/identity to F. intermedia DP |
|
|---|---|---|---|---|
| Similarity | Identity | |||
| bp | % | |||
| AtDIR1 | At5g42510 | 2113 | 42.3 | 24.7 |
| AtDIR2 | At5g42500 | 2099 | 41.4 | 24.1 |
| AtDIR3 | At5g49040 | 804 | 36.6 | 21.6 |
| AtDIR4 | At2g21110 | 1356 | 41.7 | 22.8 |
| AtDIR5 | At1g64160 | 1590 | 60.9 | 50.2 |
| AtDIR6 | At4g23690 | 1850 | 64.8 | 52.6 |
| AtDIR7 | At3g13650 | 1853 | 39.0 | 22.3 |
| AtDIR8 | At3g13662 | 1663 | 35.5 | 19.8 |
| AtDIR9 | At2g39430 | 815 | 28.8 | 17.5 |
| AtDIR10 | At2g28670 | 2015 | 30.0 | 19.4 |
| AtDIR11 | At1g22900 | 1113 | 44.8 | 28.4 |
| AtDIR12 | At4g11180 | 718 | 53.6 | 42.2 |
| AtDIR13 | At4g11190 | 2005 | 55.5 | 44.0 |
| AtDIR14 | At4g11210 | 2082 | 53.9 | 43.5 |
| AtDIR15 | At4g38700 | 1077 | 39.9 | 21.1 |
| AtDIR18 | At4g13580 | 2136 | 33.7 | 20.2 |
The GUS reporter system was first used to determine which DP genes were constitutively expressed in root tissues (18, 19). To do this, the Bacillus gusA (or uidA) which encodes a thermostable and enzymatically stable form of β-glucuronidase, was used. Promoter regions selected for each gene were also restricted to approximately 2 kb of the 5′-untranslated regions (supplemental Table S1). However, the GUS reporter gene was constructed using not only the promoter but also the signal peptide and 3′-untranslated regions. A general schematic of the vector construct is depicted in supplemental Fig. S2. Individual binary vectors were transformed into A. tumefaciens and then by the floral dip procedure into Arabidopsis (32). After harvesting, seeds from individual transformed lines were screened with hygromycin B. To confirm that transformed plant lines contained each AtDIR::GUS construct, genomic DNA was isolated from 3-week-old rosette leaf tissue from the individual Arabidopsis lines, and each of these was PCR-amplified and sequenced using promoter-specific and vector-specific primers, respectively. After verification of the presence of each vector construct, analyses were carried out using T2 plants that were grown from seed to maturity, and histochemical GUS analyses were performed at the following growth/development intervals: 3, 7, and 14 days after germination (DAG) and 3–10 weeks after germination (WAG) with the latter on a weekly basis. Of these 16 homologs, however, only AtDIR6, AtDIR10, and AtDIR13 were found to be constitutively expressed in root tissue under the growth/development conditions studied. All other putative DPs, such as AtDIR5, were not detected in root tissue.
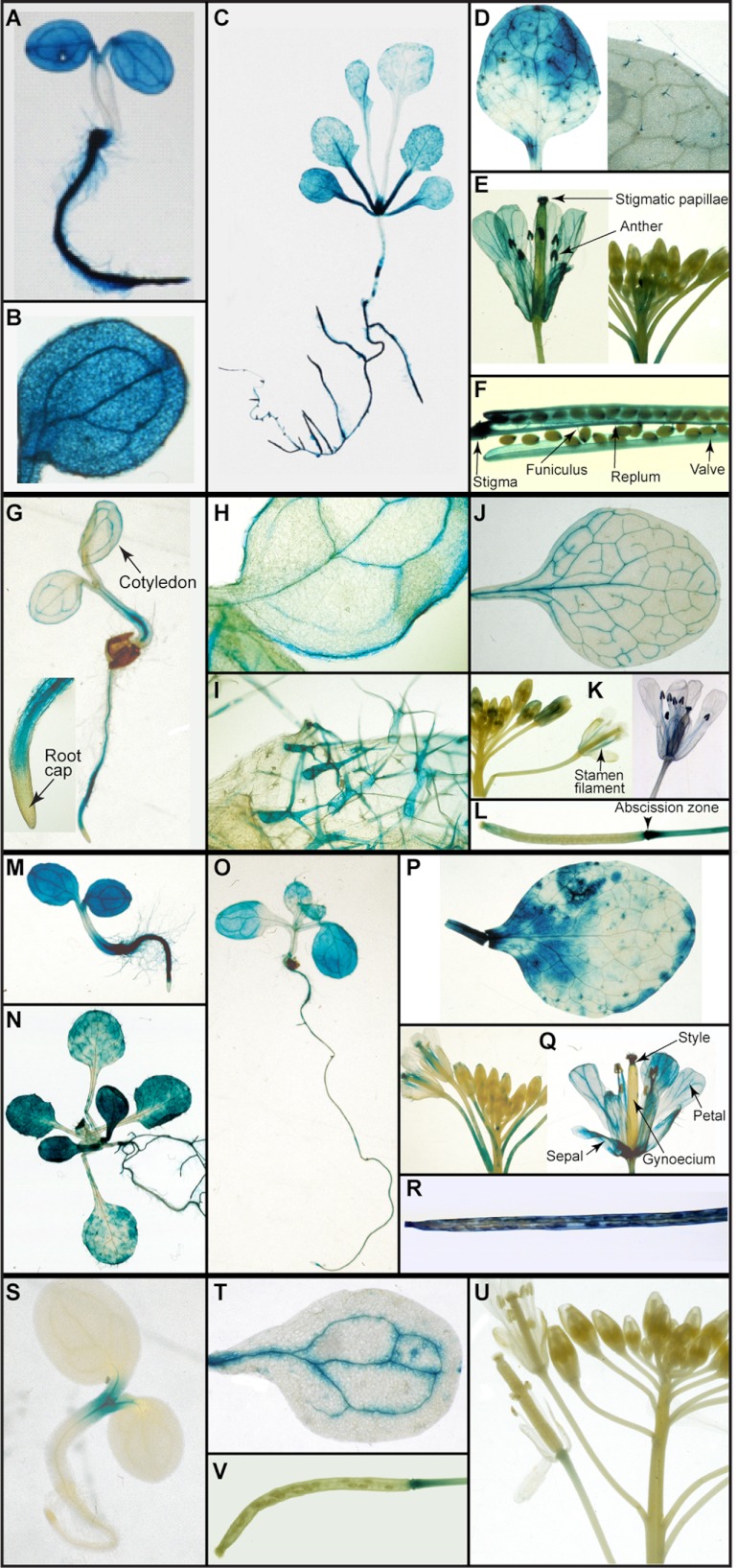
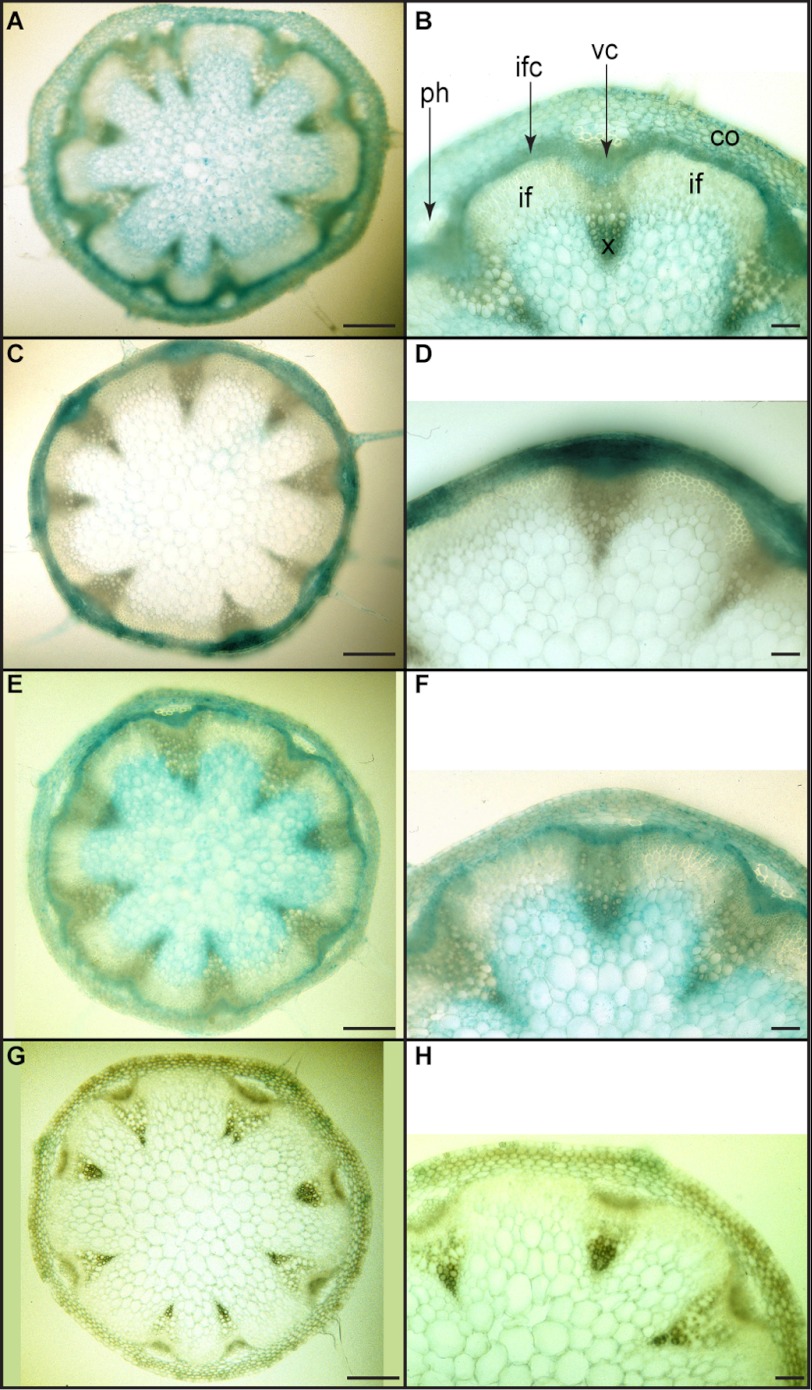
Specifically, the AtDIR6::GUS transformant displayed very strong expression 3, 7, and 14 DAG in the roots (see Fig. 3, A and C for examples). However, staining was not specific to root tissue as it was evident in cotyledon veins, leaf trichomes, and meristems at 14 DAG (Fig. 3, B and C). At 3 WAG, expression in aerial tissue was also detected in rosette (Fig. 3D) and cauline leaves (data not shown) but mainly in trichomes (Fig. 3, D and inset). By 4 WAG, there was substantial expression in the vasculature of petals, stamen filaments, and anther microsporangia as well as papillar cells of the stigma and style of mature flowers (Fig. 3E). In siliques (Fig. 3F), staining also occurred in the stigmatic region, replum, funiculus, and valve. AtDIR6::GUS expression was additionally noted in basal stem cross-sections in the interfascicular/vascular cambia and developing xylem (Fig. 4, A and B).
FIGURE 3.
Dirigent protein gene expression pattern at different stages of Arabidopsis development. A–F, AtDIR6::GUS; G–L, AtDIR10::GUS; M–R, AtDIR13::GUS; S–V, AtDIR5::GUS. A, 3 DAG, seedling. B, close-up of cotyledons in A. C, 14 DAG, seedling. D, 3 WAG, third rosette leaf and close-up of trichomes in left panel. E, 4 WAG, flower buds and mature flower. F, 5 WAG, a mature silique. G, 3 DAG, seedling and close-up of root. H, 14 DAG, seedling cotyledon. I, 14 DAG, rosette leaf. J, 3 WAG, third rosette leaf. K, 4 WAG, flower buds and mature flower. L, 5 WAG, a developing silique. M, 3 DAG, seedling. N, 14 DAG, seedling. O, 7 DAG, seedling. P, 4 WAG, third rosette leaf. Q, 4 WAG, flower buds and mature flower. R, 5 WAG, a mature silique. S, 3 DAG, seedling. T, 3 WAG, cotyledon. U, 5 WAG, flower. V, 5 WAG, a mature silique.
FIGURE 4.
Expression patterns of selected dirigent protein genes in transverse sections of Arabidopsis basal stems. A and B, 5 WAG, stem of AtDIR6::GUS. C and D, 6 WAG, stem of AtDIR10::GUS. E and F, 6 WAG, stem of AtDIR13::GUS. G and H, 4 WAG, stem of AtDIR5::GUS. Scale bars, 200 μm in A, C, E, and G and 50 μm in B, D, F, and H. co, cortex; if, interfascicular fibers; ifc, interfascicular cambium; ph, phloem; vc, vascular cambium; x, xylem.
Interestingly, AtDIR6::GUS showed GUS expression patterns comparable with bona fide cinnamyl-alcohol dehydrogenases (CADs). CADs catalyze the last enzymatic step of monolignol formation, thereby generating DP substrate as well as lignin monomers. In this regard, both AtCAD4 (At3g19450)::GUS and AtCAD5 (At4g34230)::GUS were strongly expressed in root tissues (primary/lateral) by 3–14 DAG as well as in cotyledon veins and the apical meristematic region of hypocotyls (19). Furthermore, expression patterns in stem tissues, such as in the vascular/interfascicular cambia and developing xylem, were also similar to each other.
Although not as intense as AtDIR6::GUS, the AtDIR10::GUS transformant displayed very distinct expression patterns (Fig. 3, G–L); i.e. although detected in root vascular cylinder (Fig. 3, G and inset), it was also evident in the main and peripheral cotyledon veins by 3 DAG (Fig. 3H). Staining was additionally detected in leaf margins, a rather unusual location, the significance of which is currently unknown. Interestingly, by 14 DAG, staining was observed at the basal region of rosette leaf trichomes (Fig. 3I) with expression extended throughout by 3 WAG (not shown). At 3 WAG, AtDIR10::GUS expression was evident in primary and secondary veins of rosette leaves (Fig. 3J), and it was observed in the developing xylem region (data not shown) of stems; whereas at 6 WAG, it was essentially restricted to cortex and phloem (Fig. 4, C and D). Reproductive tissues at 4 WAG displayed an interesting pattern of expression that was localized to vasculature of the stamen filament as well as in anthers and papillar cells of the stigma of mature flowers (Fig. 3, K and inset). However, in siliques, expression was associated mainly with the abscission zone (Fig. 3L). Thus, although AtDIR10::GUS expression principally occurred in the vascular strand of root tissues, it was evident in other organs as well.
Expression patterns of AtDIR13::GUS were pronounced from an early developmental stage to maturity (Fig. 3, M–R). At 3 DAG, although strong levels of expression were observed throughout root vascular regions, staining was also detected in cotyledons (Fig. 3M). However, staining was absent in root tips and hypocotyl regions. At 7 and 14 DAG (Fig. 3, O and N), a pattern fairly similar to that of AtDIR6::GUS was observed, e.g. in newly formed pairs of leaves as well as in trichomes. Interestingly, however, in the root tissue, AtDIR13::GUS activity was mainly associated with root meristematic regions with expression levels decreasing as the roots developed.
In the leaf tissue 4 WAG, expression was patchy and not restricted to vascular tissues (Fig. 3P). In floral organs, expression was noted in the vasculature of sepals, petals, and stamen filaments as well as in the gynoecium stylar region (Fig. 3Q); whereas in siliques, it was observed in the stigmatic region and epidermal layers (Fig. 3R). In the stem cross-sections, AtDIR13::GUS expression was also noted in the interfascicular/vascular cambia and developing xylem at 4–6 WAG (Fig. 4, E and F).
Additionally, GUS expression patterns of AtDIR5, which has the highest similarity/identity (∼89/78%) to AtDIR6 among all 16 AtDIRs, were examined. Interestingly, these were notably different from AtDIR6::GUS. At 3 DAG, AtDIR5::GUS was only apparently expressed in the shoot meristem (Fig. 3S), whereas expression was not detected at 7 and 14 DAG in all other tissues examined, including roots (data not shown). The AtDIR5::GUS transformant also displayed an expression pattern exclusively in the vascular region of cotyledons at 3 WAG (Fig. 3T). However, no expression of AtDIR5::GUS was noted either in flowers (Fig. 3U) or in stem cross-sections (Fig. 4, G and H). In siliques, AtDIR5::GUS expression was only observed in the abscission zone after prolonged staining (Fig. 3V).
Taken together, this phase of study indicated that AtDIR6, AtDIR10, and AtDIR13, but not AtDIR5, were constitutively expressed in root tissues, albeit not specific to only these tissues. However, this lack of tissue specificity is not unusual; e.g. the laccase AtLac15 is considered responsible for Arabidopsis seed coat pigmentation as its knock-out produces a transparent testa (33). However, AtLac15 was also expressed throughout a number of tissues and organs during growth/development (34). Thus, these three DP genes were the most likely to be potential root-associated (−)-pinoresinol-forming DPs with AtDIR6 the most probable based on levels of root tissue expression and closest similarity/identity with (+)-pinoresinol-forming DPs.
In Silico Analyses of Potential Root-specific DPs
A. thaliana trans-factor and cis-element prediction ATTED-II (35), AtProteome (36), and AtGenExpress (37) microarray databases were also examined in silico to attempt to gain additional insight into DP root expression. Using ATTED-II, it was useful to examine whether any of the DP homologs were potentially co-regulated with AtPrR1 and AtPrR2. Based on microarray data and predicted cis-elements (ATTED-II), AtDIR6 was potentially co-regulated with AtPrR1 and with various phenylpropanoid pathway-related genes, such as phenylalanine-ammonia lyase (AtPAL1), 4-coumarate-CoA ligase (At4CL1 and At4CL2), and laccases (AtLac11 and AtLac17). Regarding the latter, GUS expression of both AtLac11 (At5g03260) and AtLac17 (At5g60020) was also observed in the root vasculature (34); this is provisionally consistent with a role(s) in monolignol radical generation. By contrast, AtPrR2 was possibly co-regulated with AtDIR10 and AtDIR18.
Additionally, in the AtProteome database, AtDIR6 was detected in tissues similar to those described above with GUS staining and was the most abundantly expressed; i.e. AtDIR6 had a high number of 23 spectral hits in 10- and 23-day-old root tissues. However, AtDIR13 (15 spectral hits), AtDIR5 (three spectral hits), and AtDIR14 (three spectral hits) were also detected in 10-day-old root tissue.
Lastly, interrogation of previously reported transcriptional expression profiles from the AtGenExpress microarray database indicated that the two DP homologs, AtDIR6 and AtDIR10, had the highest degrees of intensity (above 2,000) in root tissues. AtDIR5, by comparison, showed very weak intensity in root tissue. Thus, based on the various GUS-DP promoter studies and in silico analyses, AtDIR6, AtDIR10, AtDIR13, and possibly AtDIR5 had the highest potential as (−)-pinoresinol-forming DP candidates with AtDIR6 again being the most likely.
Stereoselectivities of Recombinant AtDIR6, AtDIR5, and S. chinensis DPs
To establish potential functions of the various DPs of interest, mRNA was next obtained from 2-week-old seedlings, and amplified cDNA was individually placed into either the pMT/V5-His-TOPO expression vector containing the Drosophila metallothionein promoter (for AtDIR6 and AtDIR10) or the pART17 vector containing the cauliflower mosaic virus 35S promoter (for AtDIR5, AtDIR6, and AtDIR13), respectively. The metallothionein promoter enables inducible expression of heterologous protein production in insect cells. Production of AtDIR6 and AtDIR10 proteins was individually induced using 900 μm copper sulfate for 40 h.
By contrast, the AtDIR5, AtDIR6, and AtDIR13 genes individually cloned into the pART17 vector as indicated above were bombarded into tomato (S. peruvianum) cells (24). Afterward, suspension cell cultures were established from callus lines with the highest AtDIR5, AtDIR6, and AtDIR13 mRNA expression levels, respectively. Cell wall-bound recombinant AtDIR5, AtDIR6, and AtDIR13 were then individually obtained by filtration after agitating cells in buffer containing KCl (75 and 150 mm successively). Each DP was next purified to apparent homogeneity using strong cation exchange column chromatographic (Mono S and POROS 20 SP) steps.
For comparative purposes, an S. chinensis DP gene was also obtained from a leaf cDNA library using the FiDIR gene as a probe. As both of these genes encode (+)-pinoresinol-forming DPs, the S. chinensis recombinant DP (ScDIR) was expressed using both Drosophila and tomato cell systems and purified for control purposes.
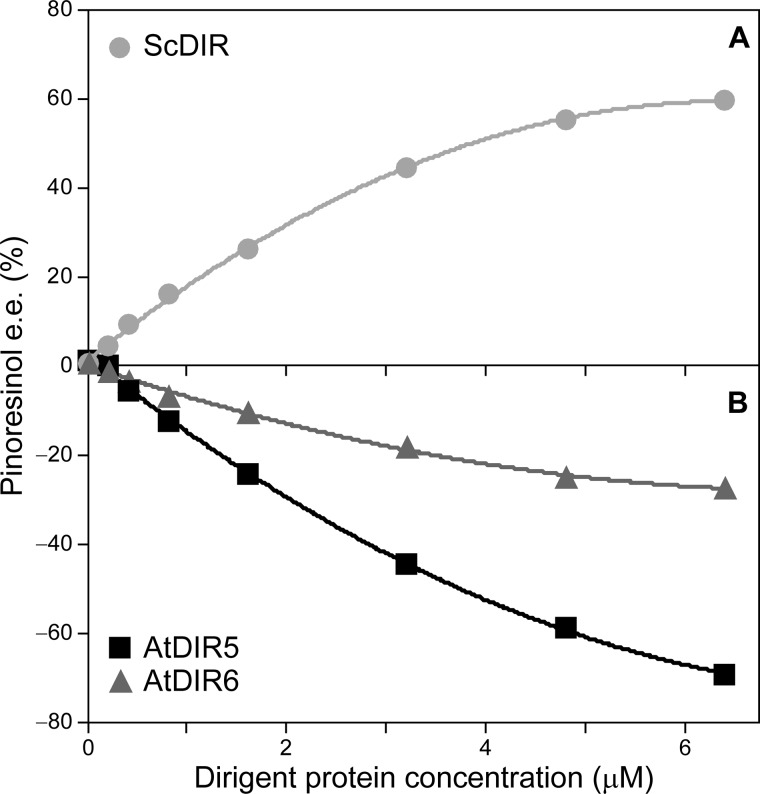
Recombinant AtDIR6, AtDIR10, AtDIR13, AtDIR5, and ScDIR were then assayed for pinoresinol (2)-forming capacity in vitro in reaction mixtures containing T. versicolor laccase (for one electron oxidation) and coniferyl alcohol (1) as substrate. Under the conditions used, neither recombinant AtDIR10 nor AtDIR13 resulted in stereoselective coupling to afford either (+)- or (−)-pinoresinols (2a or 2b) in enantiomeric excess, and they were thus no longer considered as (−)-pinoresinol-forming DPs. On the other hand, with AtDIR6, AtDIR5, and ScDIR, stereoselective coupling occurred. Of these, AtDIR6 clearly engendered preferential formation of (−)-pinoresinol (2b), reaching ∼27% e.e. with 6.4 μm DP (Fig. 5). Interestingly, although not demonstrated to be constitutively expressed in root tissue via GUS staining, AtDIR5 also engendered formation of (−)-pinoresinol (2b) in ∼70% e.e. (6.4 μm DP; Fig. 5). This corresponds to an enantiomeric ratio of 85:15 of the (−)- and (+)-antipodes, respectively. As a control, increasing recombinant ScDIR protein concentration resulted in increased (+)-pinoresinol (2a) formation, i.e. to ∼60% e.e. at 6.4 μm DP (Fig. 5).
FIGURE 5.
Dirigent protein-mediated coupling of coniferyl alcohol (1). The effect of varying ScDIR ( ) (A) and AtDIR6 (
) (A) and AtDIR6 ( ) and AtDIR5 (■) (B) concentrations on the formation of (+)-pinoresinol (2a) and (−)-pinoresinol (2b) in e.e. is shown.
) and AtDIR5 (■) (B) concentrations on the formation of (+)-pinoresinol (2a) and (−)-pinoresinol (2b) in e.e. is shown.
To further confirm and extend these observations, assays were carried out using E-[9-2H2]coniferyl alcohol (1) and AtDIR5/AtDIR6. For example, the [9,9′-2H2]pinoresinols (2) so produced from AtDIR5 was in ∼58% e.e. of the (−)-antipode (2b). Both antipodes were individually isolated by chiral HPLC, and each was subjected to LC-MS (27) and GC-MS (of the corresponding trimethylsilyl ether derivatives (28, 29)) analyses. In this regard, GC-MS analyses of (−)-[9,9′-2H2]pinoresinol (2b) (supplemental Fig. S3A) gave a molecular ion peak [M•+] at m/z 506 corresponding to the molecular formula C26H34D4O6Si2 along with fragment ions at m/z 491 [M•+ − CH3], 281 [C15H17D4O3Si+], 237 [C13H17D2O2Si+], 223 [C11H15O3Si+], 209 [C11H17O2Si+], 193 [C10H13O2Si+], 179 [C10H15OSi+], 166 [C9H14OSi+], and 73 [(CH3)3Si, base ion], respectively. In an analogous manner, GC-MS analyses of (+)-[9,9′-2H2]pinoresinols (2a) formed in a smaller amount under the assay conditions (supplemental Fig. S3B) gave molecular ion peaks and fragment ions identical to those of (−)-[9,9′-2H2]pinoresinols (2b). Furthermore, chiral liquid chromatography-mass spectrometry of (−)- and (+)-[9,9′-2H2]pinoresinols (2b and 2a) gave identical molecular ion peaks [M+] at m/z 362 corresponding to the molecular formula C20H18D4O6 (data not shown). Taken together, these assays thus unambiguously confirmed that AtDIR6 and AtDIR5 genes encoded DPs capable of stereoselective coupling leading to preferential formation of (−)-pinoresinol (2b).
Overexpression and RNAi Mutation of AtDIR6
AtDIR6 was the only DP predicted to be co-expressed with AtPrR1 and phenylpropanoid pathway genes in the trans-factor and cis-element prediction database and the most strongly expressed constitutively in Arabidopsis roots. Thus, it was selected for further study of its physiological function in vivo using both OE and RNAi strategies. In this regard, the AtDIR6 OE and RNAi constructs were individually transformed into Arabidopsis. After harvesting, seeds from the individual transgenic lines were screened with kanamycin (18, 19), and T3 plants were used for further studies.
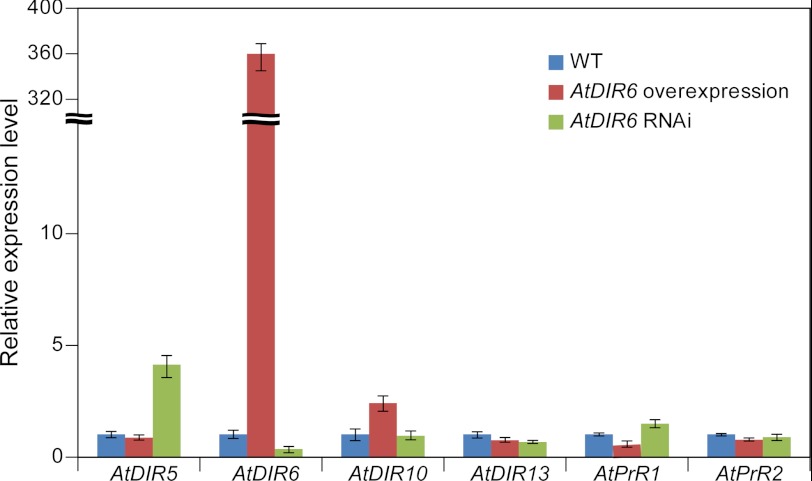
Quantitative RT-PCR analyses were next carried out to determine mRNA expression levels of DPs (i.e. AtDIR5, AtDIR6, AtDIR10, and AtDIR13) and AtPrRs in seedlings (14 DAG) of WT, OE, and RNAi lines. These analyses indicated that AtDIR6 mRNA expression was greatly up-regulated (∼375-fold higher than WT) in the AtDIR6 OE line, whereas its expression was lower (∼3-fold lower than WT) in the AtDIR6 RNAi line (Fig. 6). Expression levels of other AtDIR orthologs and AtPrR genes were not significantly affected in the OE and RNAi lines except for AtDIR5, which was apparently up-regulated ∼4-fold in the RNAi line. The difference in AtDIR5 gene expression as well as the established biochemical activities of AtDIR5 and AtDIR6 possibly suggests a limited functional redundancy of both genes. However, as indicated earlier, AtDIR5 expression was not detected in roots by promoter-GUS analysis, and AtDIR5 mRNA expression was <100 times that of AtDIR6 in the AtGenExpress microarray database. Given that AtDIR5 expression was increased ∼4-fold in the AtDIR6 RNAi line, it is perhaps not surprising then that it could not fully compensate for AtDIR6 activity in vivo because its gene expression level was still >25× lower than AtDIR6.
FIGURE 6.
Real time quantitative PCR analysis. Expression levels of AtDIR and AtPrR homologs in 2-week-old seedlings of AtDIR6 overexpression and RNAi lines compared with WT. Expression levels of each gene in WT were set to 1, and the expression of the TIP41-like (At4g34270) gene was used as an internal control. Error bars represent S.D. of three replicates.
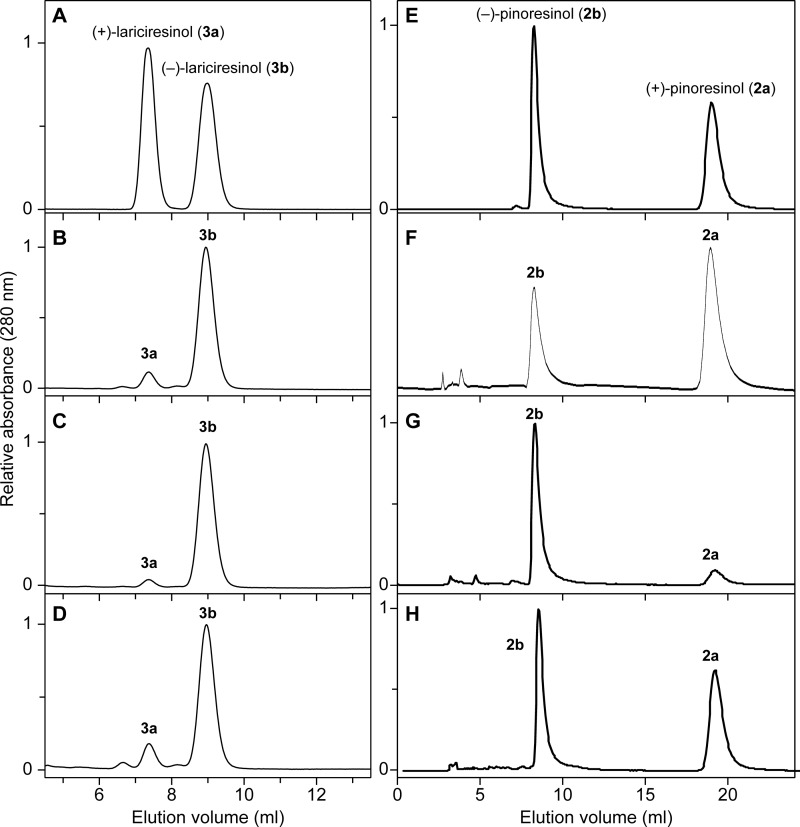
Next, stereochemical differences in pinoresinol (2)/lariciresinol (3) accumulating in WT, OE, and RNAi lines were also investigated in root tissues of 4-week-old plants (Table 2). Lariciresinol (3) (272 ng/mg of dry weight; 85% e.e. of (−)-form (3b); Fig. 7B) was obtained from WT root tissues after β-glucosidase treatment; a smaller amount of pinoresinol (2) was detected under these conditions (72 ng/mg of dry weight; 36% e.e. of (+)-form (2a); Fig. 7F). The pinoresinol (2) in all cases was present in its aglycone form. With the OE plants, however, lariciresinol (3) amounts released after β-glucosidase treatment nearly doubled (591 ng/mg of dry weight root tissue in 94% e.e. of (−)-antipode (3b); Fig. 7C), and pinoresinol (2) levels also substantially increased (585 ng/mg of dry weight root tissue in 75% e.e. (−)-form (2b); Fig. 7G). Thus, elevated AtDIR6 gene expression levels enhanced formation (or accumulation) of (−)-pinoresinol (2b) and by extension (−)-lariciresinol (3b). This was somewhat similar to the roots of the atprr1-1 atprr2 double mutant that apparently accumulated only pinoresinol (4,000 ng/mg of dry weight root tissue (released after β-glucosidase treatment) in 74% e.e. of (−)-enantiomer (2b) (14)), whereas lariciresinol (3) was not detected because the latter reductive step was “knocked out.”
TABLE 2.
Lignan contents of roots from 28-day-old WT, AtDIR6 OE, and AtDIR6 RNAi Arabidopsis
| Lines | Pinoresinola |
Lariciresinola |
||||||
|---|---|---|---|---|---|---|---|---|
| (+)-(2a) | (−)-(2b) | Total | e.e. | (+)-(3a) | (−)-(3b) | Total | e.e. | |
| ng mg−1 dry weight | ng mg−1 dry weight | ng mg−1 dry weight | % | ng mg−1 dry weight | ng mg−1 dry weight | ng mg−1 dry weight | % | |
| WT | 49 | 23 | 72 | 36 | 20 | 252 | 272 | 85 |
| AtDIR6 OE | 74 | 511 | 585 | 75 | 17 | 574 | 591 | 94 |
| AtDIR6 RNAi | 48 | 41 | 89 | 8 | 42 | 361 | 403 | 79 |
a Lariciresinol aglycone (3) (derived from glucosides 4 and 5) was released after β-glucosidase treatment. Pinoresinol (2) was present in its aglycone form.
FIGURE 7.
Chiral HPLC analyses of lariciresinol (3) and pinoresinol (2) antipodes in WT and transgenic lines. Racemic lariciresinols (3a/b; A) and racemic pinoresinols (2a/b; E) are shown. Lariciresinols (3) isolated after β-glucosidase treatment (B–D) and pinoresinols (2) (F–H) from root tissues of A. thaliana WT, AtDIR6 OE, and RNAi transgenic plant lines, respectively, are shown.
However, AtDIR6 mRNA expression was mostly repressed in RNAi transgenic plant lines. Interestingly, the pinoresinol (2) was slightly higher than in WT (89 versus 72 ng/mg of dry weight), and the level of lariciresinol (3) released after β-glucosidase treatment was slightly increased (403 ng/mg of dry weight) when also compared with WT (Table 2). However, the pinoresinol (2) that accumulated was in 8% e.e. of the (+)-antipode (2a) (Fig. 7H), whereas the lariciresinol (3) was present in a 79% e.e. of the (−)-enantiomer (3b) (Fig. 7D).
Taken together, these results clearly demonstrated that AtDIR6 was involved in preferential coupling leading to (−)-pinoresinol (2b) in Arabidopsis. In addition, our results along with those from Nakatsubo et al. (14) indicate that the combined actions of DP and PrRs are essential in defining the enantiomeric compositions of lariciresinol (3) in Arabidopsis root tissue.
It was also instructive to examine how the lariciresinol (3) was chemically bound given that it was only released following β-glucosidase treatment. To investigate this, the dried roots of Arabidopsis AtDIR6 OE plant lines were extensively extracted with hot MeOH and later partitioned with Et2O, EtOAc, and n-BuOH successively as described previously (30). The n-BuOH-solubles were further purified initially by silica gel thin-layer and Sephadex LH-20 chromatography and finally by reversed-phase HPLC as described under “Experimental Procedures.” The chemical structures of isolated lignan glucosides (4 and 5; Fig. 2) were unambiguously established using different NMR and mass spectroscopic analyses. The TOF-electrospray ionization MS of lariciresinol 4′-O-β-d-glucoside (4) in the positive ion mode gave a molecular ion peak at m/z 545.1996 [M + Na] corresponding to the molecular formula C26H34O11Na. Similarly, lariciresinol 4-O-β-d-glucoside (7) gave a molecular ion peak at m/z 545.1992 [M + Na] corresponding to the molecular formula of C26H34O11Na. β-Glucosidase treatment of lignan glucosides (4 and 5) confirmed the presence of lariciresinol (3). Furthermore, the one-dimensional and two-dimensional NMR spectra of 4 and 5 were in close agreement with the reported data in the literature (30), establishing the presence of lariciresinol conjugated with a glucosyl group (Fig. 2) (data not shown). Based upon UV spectral analyses, compound 5 predominantly accumulated (∼60–65%) compared with 4 in AtDIR6 OE roots.
DP Substrate Binding/Stereoselectivity
With both (+)- and (−)-pinoresinol-forming DPs in hand, an important question to now begin to resolve is how regiospecificity and stereoselectivity are effectuated. Based on previous kinetic data (4), it was provisionally concluded that a coniferyl alcohol (1) radical binds to each DP monomer in the DP dimer, although how substrate binding occurred and how distinct stereoselectivities were engendered were unknown. However, various attempts to obtain pinoresinol-forming DPs in crystalline form suitable for crystal structure determination have thus far been unsuccessful. Nevertheless, using site-directed mutagenesis and region swapping, some provisional insights into how this fascinating regiospecificity and stereoselectivity occur have been gained as summarized below.
Site-directed Mutagenesis
DP site-directed mutagenesis was used to begin to identify potentially important amino acid residues required for coniferyl alcohol (1) radical binding and processing. Specifically, this first entailed establishing which residues either abolished or reduced stereoselective coupling leading to (+)- or (−)-pinoresinol (2a or 2b).
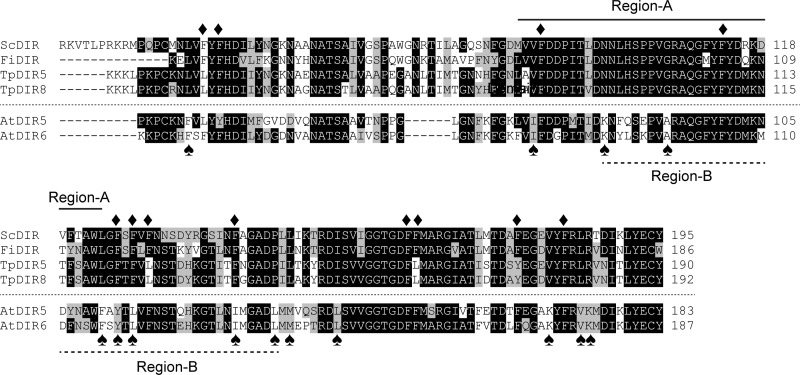
To identify potential DP amino acid residues involved in protein-substrate interactions, a bioinformatics analysis of the (+)-pinoresinol-forming DPs (FiDIR, ScDIR, TpDIR5, and TpDIR8 (31)) and (−)-pinoresinol-forming DPs (AtDIR5 and AtDIR6) was carried out. This analysis established the presence of highly conserved regions in these sequences (Fig. 8). Of these, 12 phenylalanine amino acid residues (Phe-47, Phe-49, Phe-90, Phe-113, Phe-126, Phe-128, Phe-130, Phe-141, Phe-163, Phe-164, Phe-177, and Phe-183) in S. chinensis (+)-pinoresinol-forming DP were selected as putative residues that might contribute to direct binding to or accommodating the substrate, thereby facilitating the coupling reaction. These residues were chosen as they were considered as possible candidates for π-π interactions between the bound substrate and the phenyl group of the Phe residue.
FIGURE 8.
Sequence alignments of (+)-pinoresinol and (−)-pinoresinol-forming dirigent proteins. Phenylalanine residues (Phe-47, Phe-49, Phe-90, Phe-113, Phe-126, Phe-128, Phe-130, Phe-141, Phe-163, Phe-164, Phe-177, and Phe-183) in S. chinensis used for site-directed mutagenesis are marked with ♦. Differentially conserved residues (Leu-45, Val-89, Asn-98, Gly-106, Leu-124, Phe-126, Phe-128, Phe-141, Pro-146, Leu-148, Ile-154, Val-181, Leu-185, and Arg-186) in S. chinensis are marked with ♠. Region-A and -B used for swapping in this study are also indicated. Abbreviations and locus numbers of GenBank or The Arabidopsis Information Resource database accession numbers are as follows: ScDIR (S. chinensis dirigent protein), HQ428029; FiDIR (F. intermedia dirigent protein), AF210061; TpDIR5 (Thuja plicata dirigent protein 5), AF210067; TpDIR8 (T. plicata dirigent protein 8), AF210070; AtDIR5 (A. thaliana dirigent protein 5), At1g64160; AtDIR6 (A. thaliana dirigent protein 6), At4g23690.
To investigate the role of these aromatic residues in binding, each was individually changed to an Ala residue by site-directed mutagenesis using the QuikChangeXL site-directed mutagenesis kit (Stratagene) (supplemental Table S6). The mutated cDNAs as well that of the WT ScDIR were then individually transfected into Schneider2 Drosophila expression cells (2). Next, each expressed protein was partially purified, and the ability for pinoresinol (2) formation was assayed as before.
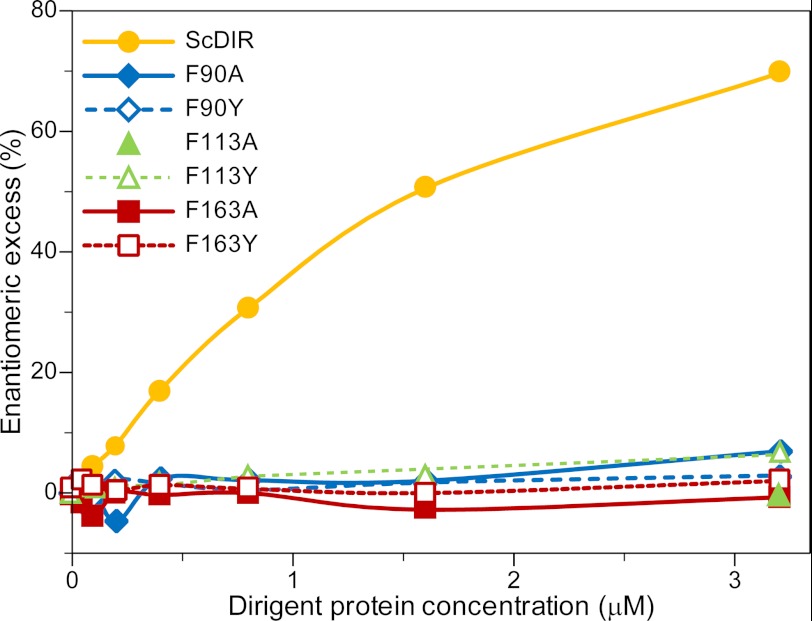
Of the mutants obtained, F47A, F49A, F126A, F128A, F130A, F141A, F164A, F177A, and F183A retained comparable with or had very minor differences in stereoselectivity from that of the WT ScDIR, indicating that these residues had no direct involvement in substrate binding/orientation. On the other hand, the F90A, F113A, and F163A ScDIR mutants produced racemic products, indicating that stereoselective coupling had been greatly reduced if not eliminated.
Following these preliminary studies, these three mutated proteins were purified to apparent homogeneity and assayed (see “Experimental Procedures”). This confirmed that mutation of these three Phe residues resulted in abolition of stereoselective coupling because the corresponding mutants F90A, F113A, and F163A only gave racemic products (Fig. 9). Interestingly, Phe-163 and Phe-164 are adjacent to each other, but their mutation had very different effects on stereoselectivity. As expected, the double mutant F163A/F164A lost the ability to engender stereoselective coupling (data not shown).
FIGURE 9.
Effects of Phe-90, Phe-113, and Phe-163 mutations on stereoselective (+)-pinoresinol (2a) formation in enantiomeric excess. (+)-Pinoresinol (2a) formation in e.e. was compared among wild-type ScDIR and F90A, F90Y, F113A, F113Y, F163A, and F163Y DPs assayed at various concentrations.
Phe to Tyr Mutations
Based on the above site-directed mutagenesis results, the Phe residues provisionally considered as important for substrate binding and/or involvement in control over stereoselectivity were individually substituted with Tyr residues (F90Y, F113Y, and F163Y) to establish whether stereoselective coupling was maintained. This was of interest because Tyr mutation at these positions might retain stereoselective properties as opposed to Ala mutants because Phe and Tyr have a similar bulkiness and side-chain. Accordingly, plasmids containing the corresponding mutated DPs were individually transformed into Schneider2 insect cells, and recombinant proteins were initially purified and assayed as before. Overall, the three Tyr mutants essentially lost the ability to engender stereoselective coupling as observed previously for the Phe to Ala mutants (Fig. 9), although in the presence of 6.4 μm F113Y protein, an excess of ∼10% of (+)-pinoresinol (2a) was detected (data not shown). However, this level of stereoselectivity conferred was negligible as compared with WT ScDIR. Tyr contains a phenolic group as compared with Phe; therefore, the phenolic group moiety might prevent proper binding of coniferyl alcohol (1) radical substrate, thereby contributing to the lack of stereoselectivity observed.
Region Swapping for Distinct DP Enantioselectivities
A bioinformatics analysis was also carried out to identify differentially conserved regions/amino acid residues that might affect stereoselective coupling preferences for the two distinct DP types. Of the 14 residues differentially conserved in the (+)- and (−)-pinoresinol-forming DPs (Fig. 8), 10 of these were distributed in the middle region of the protein, whereas only one and three were located in the N- and C-terminal regions, respectively. The middle region of the protein was then arbitrarily divided into two regions for swapping experiments.
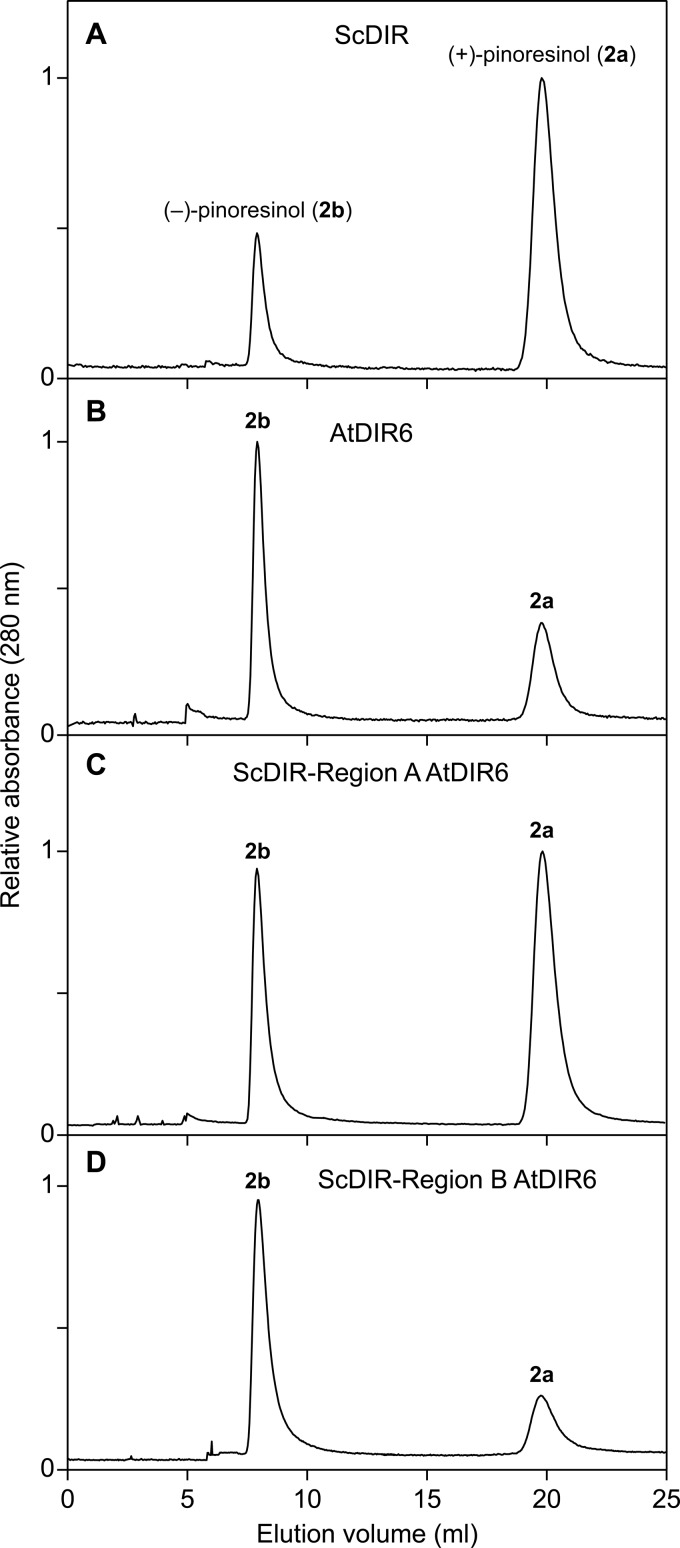
First, Region-A spanning from Met-87 to Leu-124 in ScDIR (Phe-79 to Phe-116 in AtDIR6) was selected. This region contains four residues differentially conserved for the (+)- and (−)-pinoresinol-forming DPs known thus far (Val/Ile, Asn/Lys, Gly/Ala, and Leu/Phe of (+)-DP/(−)-DP; Fig. 8, marked with spades), and this region also contains the two aforementioned residues (Phe-90 and Phe-113 in ScDIR). Using a three-step PCR procedure (25), Region-A (38 amino acids) of ScDIR was replaced by the corresponding Region-A of AtDIR6. A plasmid DNA carrying the Region-A-swapped ScDIR was then constructed and used to produce recombinant proteins. The enantiomeric excess composition of (+)-pinoresinol (2a), which was produced by the Region-A-swapped ScDIR, was only somewhat decreased (26% e.e. of the (+)-form (2a); Fig. 10C) as compared with that of WT ScDIR (60% e.e. of the (+)-form (2a); Fig. 10A). However, the overall stereoselectivity of coupling was unaffected, presumably indicating that neither Phe-90 nor Phe-113 was involved in control over the coupling mode.
FIGURE 10.
Effects of region swapping on stereoselectivity of (+)-and (−)-pinoresinol-forming dirigent proteins. Chiral HPLC analyses of pinoresinol (2) antipodes are shown. A, recombinant native ScDIR. B, recombinant native AtDIR6. C, AtDIR6 Region-A swapped into ScDIR. D, AtDIR6 Region-B swapped into ScDIR.
Next, Region-B containing 49 amino acids (Asn-98 to Pro-146 in ScDIR and Lys-90 to Leu-138 in AtDIR6) was selected. This region contains seven divergent differentially conserved residues (Asn/Lys, Gly/Ala, Leu/Phe, Phe/Tyr, Phe/Leu, Phe/Ile, and Pro/Leu of (+)-DP/(−)-DP) and Phe-113. Interestingly, the ScDIR harboring the AtDIR6 Region-B now gave a protein whose properties resulted in complete reversal of its previous stereoselectivity (50% e.e. of the (−)-form (2b); Fig. 10D). That is, the swapped ScDIR clearly resulted in an alteration of stereoselectivity from a (+)-pinoresinol (2a)-forming DP (Fig. 10A) to that producing the opposite antipode (2b) (Fig. 10D). This result suggests that pivotal residues for control of stereoselectivity are within Region-B.
Although there is currently no structural biology report for any DP with or without bound substrate, a secondary structural homology comparison with Arabidopsis allene-oxide cyclase 2 (AOC2; Protein Data Bank code 2brj) was carried out for both AtDIR6 (Protein Model Database accession number PM0078038 (38)) and ScDIR (39). This was done as a working hypothesis as it is the only protein with a possibly useful level of structural homology in the database. However, sequence alignments between AOC2 and AtDIR6/ScDIR were low with 11% identity and 12% similarity, respectively (38, 39).
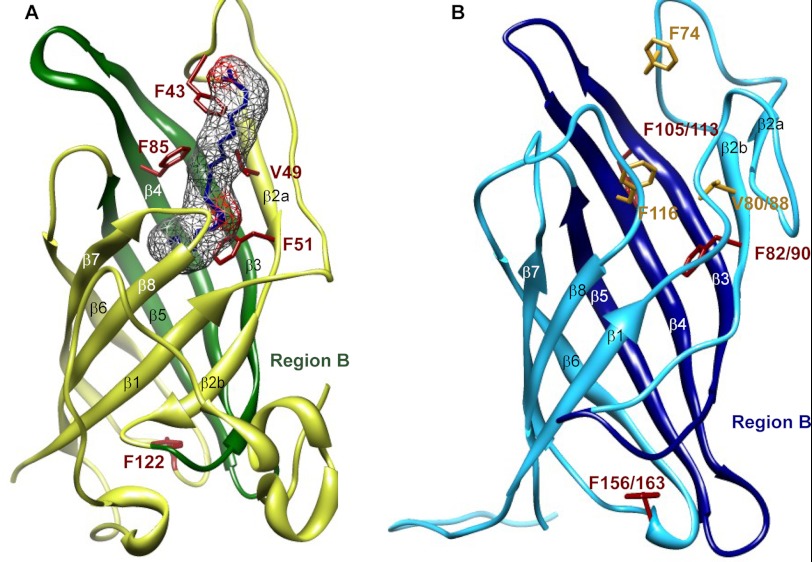
In terms of mode of action, in AOC2 (Fig. 11A), binding of the inhibitor vernolic acid is considered to be “straddled” across the three β-strands with four ligands (Phe-43, Val-49, Phe-51, and Phe-85) envisaged in substrate binding. Interestingly, this is also largely the same region in ScDIR and AtDIR6 (Asn-98 to Pro-146 in ScDIR and Lys-90 to Leu-128 in AtDIR6) where the swapping of the regions resulted in reversal of the coupling outcome (i.e. from being a (+)-pinoresinol-forming DP to a (−)-pinoresinol-forming DP) (Fig. 10, A and D). That is, swapping of the Region-B β3, β4, and β5 strands with the AtDIR6 region resulted in the opposite stereoselectivity in ScDIR, whereas swapping of the Region-A encompassing β2 and part of the β3 strand had no effect (Fig. 11B). Thus, the β2 and/or β3 strand, which provides an open entrance to the putative large β-barrel, can provisionally be considered as involved in substrate binding, and the β4 and/or β5 strands in the presumed hydrophobic pocket are more likely to be involved in controlling the stereoselectivity of coupling.
FIGURE 11.
Monomeric structure of AtAOC2 and AtDIR6. Ribbon diagrams of AtAOC2 (A; Protein Data Bank code 2dio) and AtDIR6 (B; Protein Model Database accession number PM0078038) are shown. A, the residues considered involved in interactions with the putative inhibitor, vernolic acid, are shown in red, and the electron density map is shown in gray. Region-B of AtAOC2 is colored dark green. B, the site-directed mutated residues of AtDIR6 and ScDIR are depicted in red, and the potential substrate binding residues are shown in yellow. Region-B of AtDIR6 and ScDIR is colored blue. Residue numbers are shown for both AtDIR6 and ScDIR. Images were generated using UCSF Chimera 1.5.3rc.
The overall coupling process in both cases is depicted in Fig. 1, whereby either re-re or si-si coupling can occur. Only a very small change in DP protein conformation and/or steric hindrance in this region would thus effectuate a different coupling outcome in terms of the bound substrates. Indeed, the atomic distances between re-re and si-si coupling is only approximately 1.54 Å (C8-C8′), respectively, for the quinone methide intermediates formed through radical-radical coupling of two coniferyl alcohol radicals (Fig. 1). In contrast, the (−)- and (+)-forms of pinoresinol (2a and 2b) subsequently generated via intramolecular cyclization of the coupled quinone methide intermediates have a C–C bond length of approximately 1.50 Å (C8-C8′), respectively.
Mutation of the DP residues Phe-90 in ScDIR (Phe-82 is the corresponding residue in AtDIR6) and Phe-163 in ScDIR (Phe-156 in AtDIR6) resulted in the loss of stereoselective/regiospecific coupling as did mutation of the Phe-113 residue in ScDIR. Yet of the four ligands (Phe-43, Val-49, Phe-51, and Phe-85) considered to be involved in substrate binding for AOC2, only two residues are conserved, namely Val-49/Phe-51 in AOC2, Val-88/Phe-90 in ScDIR, and Val-80/Phe-82 in AtDIR6. Therefore, it seems possible that the Phe-51/Phe-82/Phe-90 residue plays an important role in binding of the substrates as this residue is placed within the putative β2 strand of the presumed hydrophobic pocket. This residue may also help restrict the conformational freedom of coupling and/or rotation to further constrain product stereoselectivity.
In addition, although the ScDIR F113A mutant resulted in the loss of stereoselectivity, there is no corresponding Phe residue in AOC2. Moreover, in the AOC2 homology-based structure for ScDIR, the Phe-113 residue appears to project away from the face of the β-strand presumed to be involved in substrate binding/orientation. There is also no equivalent Phe residue of either AOC2 Phe-85 or Phe-43 present in ScDIR, although there are comparable Phe-116 and Phe-74 residues in AtDIR6. However, the F163A mutant in ScDIR leading to loss of stereoselectivity is conserved in AtDIR6 (Phe-156) and AOC2 (Phe-122). Yet, Phe-163 is located at the opposite end of the barrel cavity, which is masked by the N-terminal region. Therefore, mutation of this residue to Ala might confer a long range effect on the conformation of the hydrophobic pocket and thus influence stereoselectivity as well. In closing, however, it must be emphasized that the corresponding modeling above, although serving to help develop a working hypothesis, needs to be fully established with the corresponding DP structures unambiguously determined via either x-ray crystallographic and/or NMR spectroscopic analyses.
Conclusions
This study demonstrated the existence of both (+)- and (−)-pinoresinol-forming DPs in planta. In Arabidopsis, two genes (AtDIR5 and AtDIR6) encoded proteins capable of engendering formation of (−)-pinoresinol (2b) from E-coniferyl alcohol (1) in vitro when incubated in the presence of an auxiliary oxidase (laccase). Of these, however, only AtDIR6 was expressed in root tissues, which accumulated (−)-lariciresinol (3b) in the form of corresponding glucosides 4 and 5. Overexpression and RNAi approaches further confirmed the physiological role of AtDIR6 as a (−)-pinoresinol-forming DP.
Bioinformatics analyses, site-directed mutagenesis, and region swapping of various amino acid residues in the (+)-pinoresinol-forming DP when compared with the corresponding (−)-pinoresinol-forming DP and AOC2 help provide new insights into how both regiospecificity and stereoselectivity are controlled. Most importantly, region swapping clearly established the importance of Region-B (Lys-90 to Leu-138 in AtDIR6) in helping engender distinct stereoselectivities. In the future, it will be instructive to establish how these fascinating DPs fully engender these distinct stereoselectivities and thus how nature controls the outcome of distinct radical-radical coupling reactions in vivo.
Acknowledgments
We thank Greg Pearce (Institute of Biological Chemistry) for the S. peruvianum cell cultures used for heterologous expression of DPs, Julianna Gothard for growing Arabidopsis plants, and Dr. Helen Skaltsa (University of Athens, Greece) for providing authentic samples of the two lariciresinol glucosides. NMR spectra were acquired in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, Richland, WA. We gratefully acknowledge the assistance of Dr. John R. Cort in collection and processing of NMR spectra.
This work was supported by National Science Foundation Grants MCB-0417291 and MCB-1052557 and the G. Thomas and Anita Hargrove Center for Plant Genomic Research.

This article contains supplemental Figs. S1–S3 and Tables S1–S7.
- DP
- dirigent protein
- DAG
- days after germination
- e.e.
- enantiomeric excess
- GUS
- β-glucuronidase
- OE
- overexpression
- WAG
- week after germination
- PrR
- pinoresinol reductase
- tR
- retention time
- UPLC
- ultraperformance LC
- CAD
- cinnamyl-alcohol dehydrogenase
- At
- A. thaliana
- Sc
- S. chinensis
- Fi
- F. intermedia
- Tp
- T. plicata
- AOC2
- allene-oxide cyclase 2
- TOF
- time-of-flight.
REFERENCES
- 1. Davin L. B., Wang H. B., Crowell A. L., Bedgar D. L., Martin D. M., Sarkanen S., Lewis N. G. (1997) Stereoselective bimolecular phenoxy radical coupling by an auxiliary (dirigent) protein without an active center. Science 275, 362–366 [DOI] [PubMed] [Google Scholar]
- 2. Gang D. R., Costa M. A., Fujita M., Dinkova-Kostova A. T., Wang H. B., Burlat V., Martin W., Sarkanen S., Davin L. B., Lewis N. G. (1999) Regiochemical control of monolignol radical coupling: a new paradigm for lignin and lignan biosynthesis. Chem. Biol. 6, 143–151 [DOI] [PubMed] [Google Scholar]
- 3. Halls S. C., Lewis N. G. (2002) Secondary and quaternary structures of the (+)-pinoresinol forming dirigent protein. Biochemistry 41, 9455–9461 [DOI] [PubMed] [Google Scholar]
- 4. Halls S. C., Davin L. B., Kramer D. M., Lewis N. G. (2004) Kinetic study of coniferyl alcohol radical binding to the (+)-pinoresinol forming dirigent protein. Biochemistry 43, 2587–2595 [DOI] [PubMed] [Google Scholar]
- 5. Lin-gen Z., Seligmann O., Jurcic K., Wagner H. (1982) Constituents of Daphne tangutica. Planta Med. 45, 172–176 [PubMed] [Google Scholar]
- 6. Ford J. D., Huang K. S., Wang H. B., Davin L. B., Lewis N. G. (2001) Biosynthetic pathway to the cancer chemopreventive secoisolariciresinol diglucoside-hydroxymethyl glutaryl ester-linked lignan oligomers in flax (Linum usitatissimum) seed. J. Nat. Prod. 64, 1388–1397 [DOI] [PubMed] [Google Scholar]
- 7. Vassão D. G., Kim K.-W., Davin L. B., Lewis N. G. (2010) in Comprehensive Natural Products Chemistry II (Townsend C., Ebizuka Y., eds) pp. 815–928, Elsevier, Oxford, UK [Google Scholar]
- 8. Davin L. B., Jourdes M., Patten A. M., Kim K. W., Vassão D. G., Lewis N. G. (2008) Dissection of lignin macromolecular configuration and assembly: comparison to related biochemical processes in allyl/propenyl phenol and lignan biosynthesis. Nat. Prod. Rep. 25, 1015–1090 [DOI] [PubMed] [Google Scholar]
- 9. Moinuddin S. G. A., Jourdes M., Laskar D. D., Ki C., Cardenas C. L., Kim K. W., Zhang D., Davin L. B., Lewis N. G. (2010) Insights into lignin primary structure and deconstruction from Arabidopsis thaliana COMT (caffeic acid O-methyl transferase) mutant Atomt1. Org. Biomol. Chem. 8, 3928–3946 [DOI] [PubMed] [Google Scholar]
- 10. Patten A. M., Jourdes M., Cardenas C. L., Laskar D. D., Nakazawa Y., Chung B. Y., Franceschi V. R., Davin L. B., Lewis N. G. (2010) Probing native lignin macromolecular configuration in Arabidopsis thaliana in specific cell wall types: further insights into limited substrate degeneracy and assembly of the lignins of ref8, fah 1-2 and C4H::F5H lines. Mol. BioSyst. 6, 499–515 [DOI] [PubMed] [Google Scholar]
- 11. Burlat V., Kwon M., Davin L. B., Lewis N. G. (2001) Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57, 883–897 [DOI] [PubMed] [Google Scholar]
- 12. Uzal E. N., Gómez-Ros L. V., Hernández J. A., Pedreño M. A., Cuello J., Ros Barceló A. (2009) Analysis of the soluble cell wall proteome of gymnosperms. J. Plant Physiol. 166, 831–843 [DOI] [PubMed] [Google Scholar]
- 13. Liu J., Stipanovic R. D., Bell A. A., Puckhaber L. S., Magill C. W. (2008) Stereoselective coupling of hemigossypol to form (+)-gossypol in moco cotton is mediated by a dirigent protein. Phytochemistry 69, 3038–3042 [DOI] [PubMed] [Google Scholar]
- 14. Nakatsubo T., Mizutani M., Suzuki S., Hattori T., Umezawa T. (2008) Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J. Biol. Chem. 283, 15550–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pickel B., Constantin M. A., Pfannstiel J., Conrad J., Beifuss U., Schaller A. (2010) An enantiocomplementary dirigent protein for the enantioselective laccase-catalyzed oxidative coupling of phenols. Angew. Chem. Int. Ed. Engl. 49, 202–204 [DOI] [PubMed] [Google Scholar]
- 16. Kim S. J., Kim M. R., Bedgar D. L., Moinuddin S. G. A., Cardenas C. L., Davin L. B., Kang C., Lewis N. G. (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moinuddin S. G. A., Youn B., Bedgar D. L., Costa M. A., Helms G. L., Kang C., Davin L. B., Lewis N. G. (2006) Secoisolariciresinol dehydrogenase: mode of catalysis and stereospecificity of hydride transfer in Podophyllum peltatum. Org. Biomol. Chem. 4, 808–816 [DOI] [PubMed] [Google Scholar]
- 18. Kim K.-W., Franceschi V. R., Davin L. B., Lewis N. G. (2006) in Methods in Molecular Biology (Salinas J., Sanchez-Serrano J. J., eds) pp. 263–273, Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 19. Kim S. J., Kim K. W., Cho M. H., Franceschi V. R., Davin L. B., Lewis N. G. (2007) Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry 68, 1957–1974 [DOI] [PubMed] [Google Scholar]
- 20. Gleave A. P. (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207 [DOI] [PubMed] [Google Scholar]
- 21. Karimi M., Inzé D., Depicker A. (2002) GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195 [DOI] [PubMed] [Google Scholar]
- 22. Czechowski T., Stitt M., Altmann T., Udvardi M. K., Scheible W. R. (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schneider I. (1972) Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27, 353–365 [PubMed] [Google Scholar]
- 24. Cedzich A., Huttenlocher F., Kuhn B. M., Pfannstiel J., Gabler L., Stintzi A., Schaller A. (2009) The protease-associated domain and C-terminal extension are required for zymogen processing, sorting within the secretory pathway, and activity of tomato subtilase 3 (SlSBT3). J. Biol. Chem. 284, 14068–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grandori R., Struck K., Giovanielli K., Carey J. (1997) A three-step PCR protocol for construction of chimeric proteins. Protein Eng. 10, 1099–1100 [DOI] [PubMed] [Google Scholar]
- 26. Suzuki S., Sakakibara N., Umezawa T., Shimada M. (2002) Survey and enzymatic formation of lignans of Anthriscus sylvestris. J. Wood Sci. 48, 536–541 [Google Scholar]
- 27. Katayama T., Davin L. B., Lewis N. G. (1992) An extraordinary accumulation of (−)-pinoresinol in cell-free extracts of Forsythia intermedia: evidence for enantiospecific reduction of (+)-pinoresinol. Phytochemistry 31, 3875–3881 [DOI] [PubMed] [Google Scholar]
- 28. Duffield A. M. (1967) Mass spectrometric fragmentation of some lignans. J. Heterocycl. Chem. 4, 16–22 [Google Scholar]
- 29. Yamamoto S., Otto A., Simoneit B. R. T. (2004) Lignans in resin of Araucaria angustifolia by gas chromatography/mass spectrometry. J. Mass Spectrom. 39, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 30. Sugiyama M., Kikuchi M. (1993) Characterization of lariciresinol glucosides from Osmanthus asiaticus. Heterocycles 36, 117–121 [Google Scholar]
- 31. Kim M. K., Jeon J. H., Fujita M., Davin L. B., Lewis N. G. (2002) The western red cedar (Thuja plicata) 8–8′ DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 49, 199–214 [DOI] [PubMed] [Google Scholar]
- 32. Clough S. J., Bent A. F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 33. Pourcel L., Routaboul J. M., Kerhoas L., Caboche M., Lepiniec L., Debeaujon I. (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17, 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Turlapati P. V., Kim K. W., Davin L. B., Lewis N. G. (2011) The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233, 439–470 [DOI] [PubMed] [Google Scholar]
- 35. Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009) ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. (2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938–941 [DOI] [PubMed] [Google Scholar]
- 37. Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Schölkopf B., Weigel D., Lohmann J. U. (2005) A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506 [DOI] [PubMed] [Google Scholar]
- 38. Pickel B., Pfannstiel J., Steudle A., Lehmann A., Gerken U., Pleiss J., Schaller A. (2012) A model of dirigent proteins derived from structural and functional similarities with allene oxide cyclase and lipocalins. FEBS J. 279, 1980–1993 [DOI] [PubMed] [Google Scholar]
- 39. Kim K.-W., Moinuddin S. G. A., Costa M. A., Davin L. B., Lewis N. G. (2010) in Joint Annual Meeting of the American Society of Pharmacognosy and the Phytochemical Society of North America, St. Petersburg Beach, FL, July 10–14, 2010, American Society of Pharmacognosy and Phytochemical Society of North America [Google Scholar]