Background: To study the potential role of caspase-2, we determined its cleavage site specificity and compared it with that of caspase-3 and -7.
Results: N-terminal COFRADIC analysis of native proteomes revealed that caspase-2, -3, and -7 have overlapping specificities.
Conclusion: Caspase-2, -3, and -7 share nearly indistinguishable cleavage site specificity.
Significance: This finding suggests that caspase-2 has a proapoptotic function.
Keywords: Apoptosis, Caspase, Protease, Proteolytic Enzymes, Proteomics, COFRADIC, Caspase-2, Degradomics
Abstract
Caspase-2 is considered an initiator caspase because its long prodomain contains a CARD domain that allows its recruitment and activation in several complexes by homotypic death domain-fold interactions. Because little is known about the function and specificity of caspase-2 and its physiological substrates, we compared the cleavage specificity profile of recombinant human caspase-2 with those of caspase-3 and -7 by analyzing cell lysates using N-terminal COmbined FRActional DIagonal Chromatography (COFRADIC). Substrate analysis of the 68 cleavage sites identified in 61 proteins revealed that the protease specificities of human caspases-2, -3, and -7 largely overlap, revealing the DEVD↓G consensus cleavage sequence. We confirmed that Asp563 in eukaryotic translation initiation factor 4B (eIF4B) is a cleavage site preferred by caspase-2 not only in COFRADIC setup but also upon co-expression in HEK 293T cells. These results demonstrate that activated human caspase-2 shares remarkably overlapping protease specificity with the prototype apoptotic executioner caspases-3 and -7, suggesting that caspase-2 could function as a proapoptotic caspase once released from the activating complex.
Introduction
Caspases belong to the C14 family of the CD clan of aspartate-specific cysteine proteases and are implicated in apoptosis, proliferation, differentiation, and inflammation (1, 2). Non-apoptotic family members are the inflammatory caspases-1, -4, -5, -11, -12 (3), and -14, which mediates keratinocyte differentiation (4). Apoptotic caspases are typically subdivided into an apical (initiator) group (caspases-2, -8, -9, and -10) and an effector (executioner) group (caspases-3, -6, -7) (2). However, both initiator and executioner caspases are also involved in non-apoptotic functions such as cell survival, proliferation, and differentiation (1).
All caspases exist as inactive single-chain zymogens that are activated by proteolytic processing. This activation occurs by one of two possible mechanisms depending on the presence or absence of death-fold-domain-containing prodomains (2, 5) that enable multimeric homotypic interactions (5). The large-prodomain caspases possessing a CARD6 or DED motif are autocatalytically activated in large protein complexes, such as the death-inducing signaling complex (DISC) for caspase-8 (6), the inflammasome for caspase-1 (7), the apoptosome for caspase-9 (8), and the PIDDosome for caspase-2 (9). The short-prodomain caspase-3, -6, -7, and -14 are activated by upstream proteases, mostly activated caspases (2). The activation steps of the large prodomain caspases involve recruitment to an activating complex, dimerization, cleavage in trans between the large and small catalytic subunits, and cleavage in trans after the large prodomain. The last step releases the activated caspase from the complex (11). The short-prodomain caspases do not require recruitment for activation because they exist as dimers of non-processed precursors. They are proteolytically activated by cleavage between the two subunits and removal of the short prodomain (10).
Based on the phylogenic analysis of the catalytic domain, caspase-2 clusters between the apoptotic and inflammatory caspase groups (11). The prodomain of caspase-2 contains a CARD domain that allows formation of a protein complex of high molecular mass (>670 kDa), called the PIDDosome, involved in proximity-induced activation of procaspase-2 (9). It consists of the scaffold protein PIDD (p53-induced protein with death domain) and the adaptor protein RAIDD (receptor-interacting protein (RIP)-associated ICH-1/CED-3-homologous protein with a death domain), interacting with each other through their death domains (DD). RAIDD additionally contains a CARD domain, which recruits procaspase-2 (14). Although PIDD functions as a platform, it is not clear how it senses cellular stress. PIDD is regulated on at least three levels: transcriptional up-regulation in response to DNA damage (12), alternative splicing giving rise to at least five isoforms (13), and autoproteolysis generating PIDD-C or PIDD-CC fragments of different functionalities. The former fragment functions in recruitment of RIP1 and NEMO, whereas the latter functions in binding to RAIDD and consequent activation of caspase-2 (14). However, caspase-2 activation in PIDD- or RAIDD-deficient cells points to the existence of other yet undiscovered caspase-2 activation mechanisms (15). Caspase-2 together with TRAF2 and RIP1 is also a component of another high molecular weight complex. However, assembly of this complex is independent of the catalytic activity of caspase-2 and leads to NF-κB and p38 MAPK activation upon overexpression of caspase-2 (16).
Although caspase-2 is one of the first mammalian caspases to be identified (17, 18), its physiological function remains enigmatic. Although caspase-2 was identified as a neuronal developmentally down-regulated gene (19), caspase-2-knock-out animals develop normally and do not show any overt phenotypic abnormality other than minor defects, including a slight increase in the number of oocytes and resistance of caspase-2-deficient oocytes to doxorubicin (20). Additionally, caspase-2 deficiency enhances certain aging traits in mice, such as bone loss, shortened maximum lifespan and impaired hair growth (21), and resistance of caspase-2-deficient neurons to β-amyloid-mediated death (22).
So far several functions have been attributed to caspase-2, both apoptotic and non-apoptotic (23). Caspase-2-mediated Bid cleavage has been shown to promote mitochondrial cytochrome c release in HeLa cells (24). In addition, caspase-2 has been placed downstream from mitochondria, as its processing in mouse thymocytes after γ-irradiation depends on the presence of Apaf-1 and caspase-9 (25). The response of caspase-2-deficient mouse embryonic fibroblasts to cytoskeleton-targeting agents, such as taxol or zoledronic acid, was shown to be delayed or reduced (29). In contrast, a more recent study did not show any difference in taxol-induced apoptosis between wild type and caspase-2 knock-out mouse embryonic fibroblasts (15). Furthermore, caspase-2 has been implicated in non-apoptotic processes such as proliferation, tumor suppression, and regulation of p53 activity (26, 27).
The few caspase-2 protein substrates identified so far have not clearly revealed the enzyme function. Moreover, the caspase-2 substrates HDAC4 and α-II spectrin can be cleaved by other caspases, often caspase-3 and -7, which cleave at the same site as caspase-2 (28). The only known unique caspase-2 cleavage site is in golgin-160. It is unclear whether this cleavage generates a specific biological event, but it might contribute to the disintegration of the Golgi complex during apoptosis (29). Cleavage of human Mdm2 by caspase-2 has been reported to stabilize p53 and might contribute to the increased p53-dependent drug resistance of non-small cell lung carcinoma cell lines (27). Caspase-2 does not cleave (30) or proteolytically activate executioner caspases (31), with the exception of cleaving caspase-7 at pH 2.0, which is most likely not relevant in vivo (32).
The methods originally developed to monitor caspase specificities and activities are typically based on short peptide sequences, including the use of synthetic peptide libraries (33, 34). However, the identified peptide substrates and their derived peptide-based inhibitors lack specificity when used in cells and, therefore, cannot be used to unambiguously detect particular active caspases (35). For instance, it has been reported that the assumed caspase-2-specific substrate Ac-VDVAD-AMC is cleaved almost equally well by caspase-3 and -7 (36). The development of high throughput methods based on mass spectrometry overcame some of the experimental limitations of using libraries of synthetic peptides. Global proteomic approaches now make it possible to identify and compare hundreds of cleavage events in one experimental setup (37, 38).
We used N-terminal COFRADIC (combined fractional diagonal chromatography) (39, 40) to study the degradome of human caspase-2 in native proteomes and to compare it with the degradomes of human caspase-3 and -7. We used lysates of the A549 human cell line that had been subjected to metabolic SILAC labeling (stable isotope labeling by amino acids in cell culture) (41). We directly compared peak intensities in peptide MS spectra of neo-N termini generated by caspase-2, caspase-3, or caspase-7, which allowed us to unambiguously identify caspase cleavage events and directly compare potential differences in the specificity profiles of the three enzymes. Additionally, upon overexpression in HEK 293T cells, we confirmed that one of two cleavage sites identified in eukaryotic translation initiation factor 4B (eIF4B) is preferred by caspase-2, as observed in our COFRADIC results. To the best of our knowledge, this is the first study reporting the use of a comprehensive proteomic approach to profile caspase-2 cleavage specificity.
EXPERIMENTAL PROCEDURES
Cell Culture
The A549 human non-small cell lung carcinoma cell line was maintained in Kaighn's modified version of Ham's F-12 medium (Invitrogen) supplemented with 10% fetal calf serum (HyClone), 50 IU/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Enzyme-free cell dissociation buffer (Invitrogen) was used to detach cells from the culture dish. The HEK 293T human embryonic kidney cell line was maintained in DMEM medium (Invitrogen) supplemented with 10% fetal calf serum (Sigma), 2 mm l-glutamine (Lonza), 0.4 mm sodium pyruvate (Sigma), 50 IU/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen). Cells were detached from the culture dish using a solution of 0.05% trypsin and 0.032% EDTA.
SILAC Labeling and N-terminal COFRADIC Setup
SILAC labeling was performed by growing A549 cells for at least five population doublings in customized arginine-free F-12K medium (Invitrogen) supplemented with 10% dialyzed fetal calf serum (Invitrogen), 50 IU/ml penicillin, and 50 μg/ml streptomycin. F-12K medium was supplemented with 0.3 mm l-arginine (15% of the standard concentration, at which the conversion of l-arginine to proline was not observed) using [12C6]Arg, [13C6]Arg, or l-[13C615N4]arginine.HCl (Cambridge Isotope Laboratories).
After SILAC labeling, A549 cells were harvested and lysed as described (42). After blocking the activity of endogenous caspases by iodoacetamide (42), the samples were incubated separately either with 200 nm recombinant human caspase-2 (l-[12C6]Arg), caspase-3 (l-[13C6]Arg), or caspase-7 (l-[13C615N4]Arg) or were left untreated (l-[12C6]Arg) for 1 h at 37 °C. Solid guanidinium hydrochloride was then added to a final concentration of 4 m followed by downstream analysis as described (39, 43). As part of the N-terminal COFRADIC protocol, differential acetylation was then used to distinguish further between the two light-labeled samples (N-hydroxysuccinimide trideutero-acetate was used for the caspase-2, -3, and -7 samples and N-hydroxysuccinimide acetate for the control sample) and to distinguish between in vitro and in vivo N-acetylation of the caspase-treated samples, the former indicative for proteolytic events (43). Recombinant human active caspase-2, -3, and -7 were purchased from R&D Systems (Minneapolis, MN) (43).
LC-MS/MS Analysis and Peptide Identification by Mascot
Electrospray ionization-LC-MS/MS analysis of the samples digested with human caspases was performed on a Q-TOF Premier mass spectrometer (Waters Corp., Milford, MA) operated as described (44). Peptides were identified using a locally installed version of the MASCOT database search engine searching the Swiss-Prot database with restriction to human proteins. We used the November 16, 2011 release of the UniProtKB/Swiss-Prot protein database containing 533,049 sequence entries, of which 20,326 are from human. Spectra were searched with semiArgC/P enzyme settings allowing 1 missed cleavage. Mass tolerance of the precursor ions was set to ±0.2 Da (with Mascot's C13 option set to 1) and of fragment ions to ±0.1 Da. The peptide charge was set to 1+, 2+, or 3+, and instrument settings were electrospray ionization-Q-TOF spectrometry. Separate searches were performed to allow identification of light- and heavy-labeled samples. To identify peptides from the control sample, pyroglutamate formation of N-terminal glutamine and acetylation of peptide Nα-termini were set as variable modifications, whereas methionine oxidation (sulfoxide), carbamidomethyl formation of cysteine residues, and Nϵ-acetylation of lysine residues were set as fixed modifications. The same settings were used to identify peptides from the sample treated with caspase-2, except that in this case tri-deuteroacetylation of peptide Nα termini was set as an additional variable modification, whereas Nϵ-trideutero-acetylation of lysine residues was set as fixed modification. Identical settings were used for the sample treated with caspase-3 or -7, with 13C6 Arg and 13C615N4 Arg, respectively, as additional fixed modifications. Only the peptides that were ranked one and scored above the threshold score set at 95% confidence were withheld. The false discovery rate, calculated as described before (45), was 3.37%. All peptide identifications were processed, stored, and managed using the ms_lims software (46). MS spectra and MS intensity profiles of neo-N-terminal peptides were individually inspected. Spectra of all identified neo-N termini have been submitted to PRIDE (accession number 19136) (47).
Caspase Cleavage Motif Analysis by IceLogo
For each cleavage site the surrounding amino acid sequence was retrieved (four residues C-terminal and six residues N-terminal to the scissile bond), and the sequences were aligned and centered on the Asp residue at position P1. IceLogo was used to visualize conserved sequence patterns in the multiple sequence alignments (48). The reference (negative) sets of protein sequences were generated by random sampling of amino acids in the human Swiss-Prot database. For every amino acid, IceLogo calculates the percentage difference in occurrence at every position between the two sets by probability-based methods and reports it when p ≤ 0.05. In each case the sampling size was equal to the number of peptides in each group.
In Vitro Caspase Activity Assay
The activities of human recombinant caspases-2, -3, and -7 (R&D Systems) were assayed as previously described (30) with minor modifications. Briefly, serial dilutions of caspases and solutions of substrates were prepared in cell-free system buffer (10 mm HEPES NaOH, pH 7.4, 220 mm mannitol, 68 mm sucrose, 2 mm MgCl2, 2 mm NaCl, 2.5 mm H2KPO4) freshly supplemented with 10 mm dithiothreitol. To assay the activity of caspase-2 we used Ac-VDVAD-AMC, whereas for caspase-3 and -7 we used Ac-DEVD-AMC (Peptide Institute). Immediately before the measurement, 50 μl of substrate solution was added to a 96-well plate (Nunc) containing 50 μl of caspase dilutions. The fluorescence of released AMC was measured at 37 °C every 2 min for 1 h using Fluostar Omega fluorometer (BMG Labtech) equipped with 360-nm excitation and 460-nm emission filters. Data analysis was done using MARS Data Analysis Software (BMG Labtech). Slope values are reported as the change in fluorescence (relative fluorescence units (RFU)) over time for reactions showing a linear initial phase for at least 20 min.
Expression Vectors
Expression vectors for human caspase-2, pCAGGS-E-hCasp2 (16) (LMBP 4716), and for EGFP, pEGFP-NLS (49) (LMBP 6459), were obtained from BCCM/LMBP Plasmid collection, Department of Biomedical Molecular Biology, Ghent University, Belgium. Expression vectors for human caspase-3 pcDNA3-Casp3-myc and human caspase-7 pcDNA3-Casp7-FLAG (50) were obtained from Addgene (numbers 11813 and 11815). Human EIF4B cDNA from PCD-F-EIF4B vector (LMBP 6359) was first cloned into pENTR3C vector (Invitrogen) using CloneEZ PCR cloning kit (GenScript). Expression vector pdcDNA-FLAG-EIF4B-myc was generated from pENTR3C-EIF4B and pdcDNA-FLAGMyc (LMBP 4705) backbone using Gateway LR clonase II (Invitrogen). Point mutants pdcDNA-FLAG-EIF4B D531A-myc, pdcDNA-FLAG-EIF4B D563A-myc, and pdcDNA-FLAG-EIF4B D531A D563A-myc were prepared by site-directed mutagenesis of pdcDNA-FLAG-EIF4B-myc using polymerase Pfu Turbo (Stratagene) and DpnI restriction enzyme (Promega).
Transfection and Immunoblotting
HEK 293T cells were seeded in 6-well plates at 2 × 105 cells per well and transfected by the calcium phosphate precipitation method (16) with 250 ng of DNA of caspases and EIF4B expression vectors in different combinations. Each well was additionally cotransfected with 100 ng of pEGFP-NLS vector to monitor the efficiency of transfection. After 13 h of transfection both adherent and detached cells were collected from the medium, washed with cold PBS, and lysed in Laemmli sample loading buffer. Equal volumes of lysates were separated on an 8 or 12.5% SDS-PAGE gel and electrotransferred onto nitrocellulose membranes (PerkinElmer Life Sciences). The membranes were blocked with 5% nonfat dry milk in PBS containing 0.2% Tween 20 (Sigma) and incubated with antibodies against the FLAG tag (horseradish peroxidase-conjugated, M2; Sigma), caspase-2 (11B4; Chemicon), caspase-3 (Cell Signaling Technology), caspase-7 (Santa Cruz), poly(ADP-ribose) polymerase-1 (PARP-1; C-2–10, Enzo), or GFP (JL-8, BD Biosciences). They were then incubated with secondary anti-mouse, anti-rabbit, anti-rat (GE Healthcare), or anti-goat (Santa Cruz) antibodies conjugated to horseradish peroxidase and visualized by chemiluminescence (Western Lightning ECL Plus, PerkinElmer Life Sciences).
RESULTS
Comparative Degradomics of Human Caspases on Native Proteins
To identify the cleavage sites for human caspase-2, -3, and -7 in native proteomes, we used a human non-small cell lung carcinoma cell line (A549 cells) metabolically labeled by SILAC using l-[12C6]Arg for the control and the caspase-2-treated sample, l-[13C6]Arg for the caspase-3-treated sample, and l-[13C615N4]Arg for the caspase-7-treated sample. To distinguish between control and caspase-2-treated samples, differential acetylation was introduced; N-hydroxysuccinimide trideutero-acetate was used for all caspase-treated samples, whereas N-hydroxysuccinimide acetate was used for the control sample. Lysates were further treated with 5 mm iodoacetamide to block endogenous cysteine protease activity (51) before incubation with recombinant human caspases at a concentration of 200 nm for 1 h at 37 °C (42). Proteases were then inactivated, and equal amounts of samples from the four setups were mixed and analyzed by N-terminal COFRADIC, including strong cation exchange chromatography and N-terminal peptide sorting steps (40, 41, 45) (Fig. 1A). Recorded MS/MS spectra of the sorted peptides were searched by the Mascot algorithm in the human subset of proteins in the Swiss-Prot database.
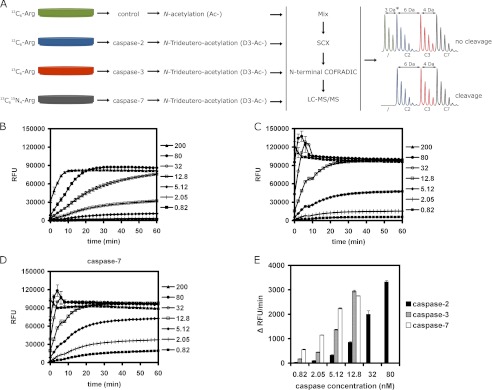
FIGURE 1.
COFRADIC setup for identifying human caspase-2 substrates and the activity of recombinant caspases used in the study. A, SILAC was used to label four differently treated A549 cell lysates. One part of the 12C6-labeled lysate served as control and another part was treated with caspase-2, whereas 13C6- and 13C615N4-labeled lysates were treated with recombinant human caspase-3 and -7, respectively. A differential acetylation step was introduced to distinguish between the control and the caspase-2-treated sample. N-Hydroxysuccinimide trideutero-acetate (D3-Ac-) was used in all caspase-treated samples and non-deuterated N-hydroxysuccinimide acetate (Ac-) in the control sample. After caspase treatment, equal amounts of samples were mixed, subjected to strong cation exchange chromatography (SCX), and N-terminal peptides were isolated by N-terminal COFRADIC and then analyzed by LC-MS/MS. Note that the actual mass difference between the control and caspase-2-treated samples (*) depends on the number of lysine residues in the peptide. Spectra are shown for a peptide without lysine residues and a Nα-(trideutero-)acetyl-group. Every additional lysine residue carried one additional (trideutero-)acetyl-group leading to an extra mass difference of 3-Da per lysine. B–D, in vitro caspase activity was measured for 60 min at 37 °C using 50 μm synthetic peptide substrates: VDVAD-AMC for caspase-2 (B) and DEVD-AMC for caspase-3 (C) and -7 (D) using the indicated caspase concentrations (nm). At 200 nm, all three caspases show maximal activation within minutes. E, results of the caspase activity assay show the initial velocity (change in fluorescence over time: Δ RFU/min) for reactions with a linear initial phase during the first 20 min. Caspase-7 was the most, and caspase-2 the least active protease.
To confirm that recombinant caspases used in the COFRADIC analysis were active, we performed an in vitro caspase activity assay. Fluorogenic substrates, namely Ac-DEVD-AMC for caspase-3 and -7 and Ac-VDVAD-AMC for caspase-2, were used at 50 μm concentration. Recombinant caspase incubations included the concentration of 200 nm used in the COFRADIC analysis and its 2.5-fold serial dilutions. Fig. 1, B–D, depict the hydrolysis activity of the three caspases as RFU values obtained in the assay. At 200 nm, the 3 caspases reached similar maximal values within minutes from the start of the assay. At this concentration the increase in fluorescence of AMC released by caspase-2 stayed linear for about 8 min (Fig. 1B), whereas for caspase-3 and -7 the linearity was not registered because the substrate cleavage reaction was very rapid (Fig. 1, C and D). To observe the linear reaction phase for caspase-3 and -7, enzyme concentrations had to be decreased to as low as 12.8 nm. To measure the initial reaction rate (the slope of the linear phase), we used caspase concentrations that displayed a linear RFU increase for at least 20 min from the start of the measurement. For caspase-2 this range was 0.82–80 nm, whereas for caspase-3 and -7 it was 0.82–12.8 nm. As seen in Fig. 1E, caspase-7 at concentrations of 0.82–5.12 nm processed the substrate more rapidly than the other two caspases. At 12.8 nm, caspase-3 and -7 showed similar activity (2944 and 2751 RFU/min, respectively), whereas caspase-2 was more than 3 times less active (850 RFU/min). Caspase-2 assayed at 80 nm had the same activity level (3323 RFU/min) as caspase-3 and -7 at 12.8 nm. The results of in vitro caspase assay collectively show that the specific enzymatic activity of caspase-2 is considerably lower than the activities of caspase-3 and -7. However, at 200 nm, which was used for the COFRADIC analysis, all three caspases reached their maximal activity within minutes.
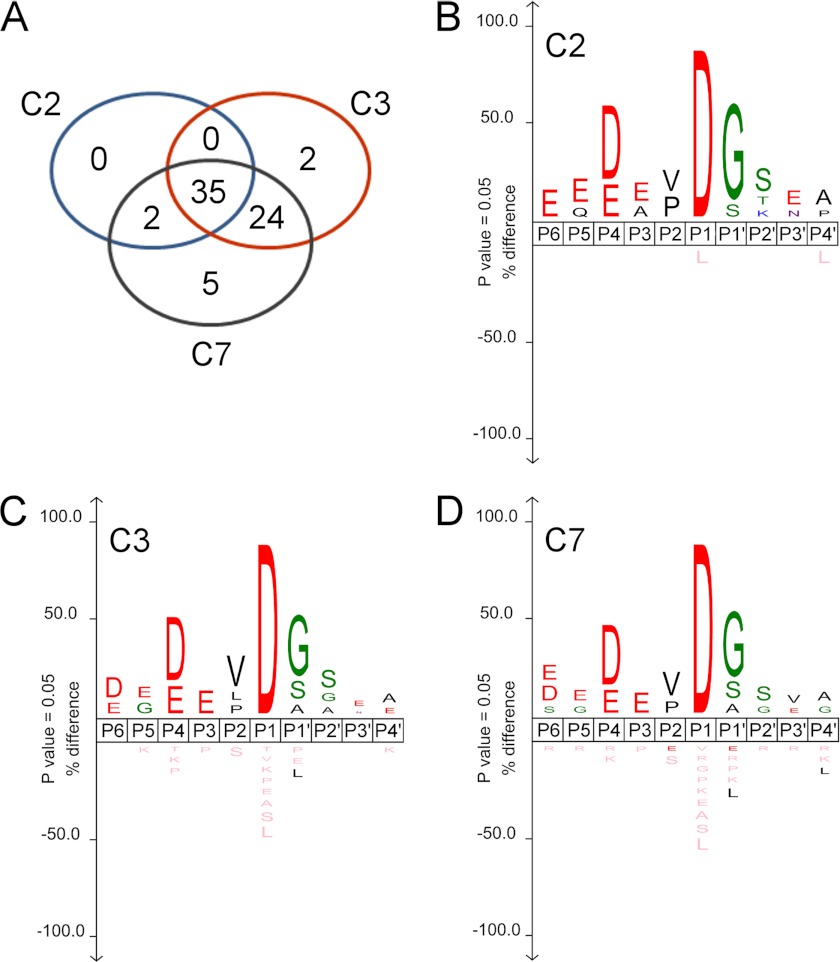
COFRADIC identification of 1768 MS/MS spectra revealed 870 unique peptides originating from 725 unique proteins. 101 MS/MS spectra were derived from 68 peptides in 61 proteins that were absent in the control sample. These peptides were trideutero-acetylated at their Nα-amine and were generated by cleavage after aspartic acid residues in the protein sequence. Therefore, these peptides were considered proteolysis reporter peptides indicative of caspase cleavage events (Table 1). Caspase-2 cleaved 37 sites, whereas caspase-3 and -7 cleaved 61 and 66 sites, respectively. No unique cleavage sites were identified for caspase-2, whereas two and five cleavage events were generated exclusively by caspase-3 and -7, respectively (Fig. 2A). Thirty-five cleavage events were shared between all three caspases, but cleavage efficiencies were different as judged from the peptide ion signals (Table 1). Two cleavage sites appeared to have been cleaved by caspase-2 and -7 but not by caspase-3. On the other hand, 24 cleavage sites were shared by caspase-3 and -7 but were not cleaved by caspase-2.
TABLE 1.
List of identified human caspase-2, caspase-3, and caspase-7 cleavage sites
Processing sites are ordered by decreasing relative cleavage efficiency of caspase-2 (C2), caspase-3 (C3), and caspase-7 (C7). Columns from the left to right contain the Swiss-Prot accession number, protein name, start and end position of the identified (neo-N-terminal) peptide in the protein sequence, sequence of the identified peptide, residues surrounding the cleavage site from P6 to P4′, relative cleavage efficiency by each caspase compared to the most efficient cleavage, cleavage ratios of caspase-2/caspase-3 (C2/C3), caspase-2/caspase-7 (C2/C7), and caspase-7/caspase-3 (C7/C3), number of identified MS/MS spectra, Mascot score and threshold of the highest scoring spectrum, and possible protein isoforms in which the same peptide sequence can be found. The last two columns indicate whether the identified site was previously reported to be cleaved by caspases, in which case the name of caspase(s) involved and reference is provided.
| Accession | Protein name | Start | End | Peptide sequence | Cleavage site | C2 | C3 | C7 | C2/C3 | C2/C7 | C7/C3 | No. spectra | Score | Threshold | Isoforms | Known caspase cleavage | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P23588 | Eukaryotic translation initiation factor 4B | 564 | 571 | GKKDQDSR | ESDRKD↓GKKD | 100 | 33.17 | 73.45 | 3.01 | 1.36 | 2.21 | 2 | 57 | 47 | |||
| P09496 | Clathrin light chain A | 77 | 102 | GVMNGEYYQESNGPTDSYAAISQVDR | GPDAVD↓GVMN | 16.74 | 100 | 94.85 | 0.17 | 0.18 | 0.95 | 4 | 77 | 41 | Caspase-3 | 59 | |
| Q13263 | Transcription intermediary factor 1-β | 686 | 702 | GADSTGVVAKLSPANQR | GSLSLD↓GADS | 0 | 100 | 92.48 | 0 | 0 | 0.92 | 3 | 110 | 46 | |||
| Q15046 | Lysyl-tRNA synthetase | 13 | 25 | GSEPKLSKNELKR | AEVKVD↓GSEP | 7.34 | 100 | 92.04 | 0.07 | 0.08 | 0.92 | 2 | 53 | 46 | Caspase-3 | 59 | |
| Q8WU90 | Zinc finger CCCH domain-containing protein 15 | 325 | 340 | DSVSVNDIDLSLYIPR | GGDEVD↓DSVS | 8.35 | 100 | 89.55 | 0.08 | 0.09 | 0.9 | 1 | 82 | 46 | |||
| Q7Z406 | Myosin-14 | 1178 | 1186 | STNAQQELR | LEDTLD↓STNA | 15.84 | 100 | 85.17 | 0.16 | 0.19 | 0.85 | 1 | 63 | 47 | |||
| Q96IZ0 | PRKC apoptosis WT1 regulator protein | 132 | 145 | GVPEKGKSSGPSAR | DEEEPD↓GVPE | 9.03 | 100 | 82.33 | 0.09 | 0.11 | 0.82 | 2 | 72 | 47 | Caspase-3 | 59 | |
| Q13148 | TAR DNA-binding protein 43 | 90 | 98 | ASSAVKVKR | KmDETD↓ASSA | 12.42 | 100 | 81.76 | 0.12 | 0.15 | 0.82 | 2 | 53 | 47 | Caspase-3 | 59 | |
| P46821 | Microtubule-associated protein 1B | 1197 | 1220 | GKDYNASASTISPPSSMEEDKFSR | KTDATD↓GKDY | 8.55 | 100 | 81.73 | 0.09 | 0.1 | 0.82 | 2 | 52 | 44 | Caspase-3, -7 | 38 | |
| P23588 | Eukaryotic translation initiation factor 4B | 532 | 545 | GMNAPKGQTGNSSR | DENKVD↓GMNA | 4.87 | 100 | 77.77 | 0.05 | 0.06 | 0.78 | 2 | 62 | 47 | Caspase-3 | 59 | |
| P35579 | Myosin-9 | 1154 | 1162 | STAAQQELR | LEDTLD↓STAA | 5.99 | 100 | 75.64 | 0.06 | 0.08 | 0.76 | 3 | 55 | 47 | Caspase-3 | 59 | |
| P05783 | Keratin, type I cytoskeletal 18 | 398 | 411 | SSNSMQTIQKTTTR | LGDALD↓SSNS | 9.46 | 100 | 74.24 | 0.09 | 0.13 | 0.74 | 4 | 137 | 46 | Caspase-3, -7 | 71 | |
| Q15056 | Eukaryotic translation initiation factor 4H | 94 | 109 | SLKEALTYDGALLGDR | EFDEVD↓SLKE | 5.04 | 100 | 73.35 | 0.05 | 0.07 | 0.73 | 3 | 105 | 46 | Caspase-3 | 59 | |
| P14625 | Endoplasmin | 29 | 39 | GTVEEDLGKSR | DEVDVD↓GTVE | 0 | 100 | 54.35 | 0 | 0 | 0.54 | 2 | 61 | 47 | |||
| P45974 | Ubiquitin carboxyl-terminal hydrolase 5 | 768 | 778 | AEAAMDISEGR | HIDDLD↓AEAA | 0 | 100 | 48.67 | 0 | 0 | 0.49 | 1 | 54 | 47 | |||
| P22102 | Trifunctional purine biosynthetic protein adenosine-3 | 952 | 965 | AGQIILQEAVPVKR | VAEDVD↓AGQI | 0 | 100 | 46.48 | 0 | 0 | 0.46 | 1 | 62 | 46 | |||
| P08670 | Vimentin | 86 | 100 | FSLADAINTEFKNTR | LQDSVD↓FSLA | 9.39 | 100 | 45.84 | 0.09 | 0.2 | 0.46 | 1 | 71 | 46 | Caspase-3, -7 | 72 | |
| Q9UIL1 | Short coiled-coil protein | 88 | 99 | AENQVELEEKTR | DMDAVD↓AENQ | 0 | 100 | 42.62 | 0 | 0 | 0.43 | 1 | 66 | 47 | |||
| P25705 | ATP synthase subunit α, mitochondrial | 160 | 169 | GKGPIGSKTR | LGNAID↓GKGP | 4.47 | 100 | 40.28 | 0.04 | 0.11 | 0.4 | 1 | 53 | 47 | Caspase-3 | 59 | |
| P60709 | Actin, cytoplasmic 1 | 12 | 28 | NGSGMCKAGFAGDDAPR | AALVVD↓NGSG | 0 | 100 | 40.23 | 0 | 0 | 0.4 | 3 | 90 | 45 | A5A3E0, AP0CG38, AP0CG39, AP63261, AQ6S8J3 | Caspase-1 | 58 |
| Q6P996 | Pyridoxal-dependent decarboxylase domain-containing protein 1 | 585 | 597 | AAELVETIAATAR | ASDNVD↓AAEL | 0 | 100 | 38.75 | 0 | 0 | 0.39 | 1 | 52 | 47 | |||
| P63313 | Thymosin β-10 | 7 | 40 | MGEIASFDKAKLKKTETQEKNTLPTKETIEQEKR | MADKPD↓MGEI | 27.95 | 100 | 37.07 | 0.28 | 0.75 | 0.37 | 1 | 64 | 42 | |||
| Q5SNT6 | Protein FAM21B | 1044 | 1059 | SGDIFSTGTGSQSVER | EADLFD↓SGDI | 0 | 100 | 35.55 | 0 | 0 | 0.36 | 1 | 87 | 46 | Q5SRD0, AQ641Q2, AQ9Y4E1 | ||
| O95801 | Tetratricopeptide repeat protein 4 | 16 | 27 | SFLEKFQSQPYR | SDDVMD↓SFLE | 0 | 100 | 34.61 | 0 | 0 | 0.35 | 1 | 61 | 47 | |||
| P06454 | Prothymosin α | 8 | 31 | TSSEITTKDLKEKKEVVEEAENGR | SDAAVD↓TSSE | 0 | 100 | 33.31 | 0 | 0 | 0.33 | 2 | 77 | 45 | Caspase-3 | 59 | |
| P14625 | Endoplasmin | 60 | 67 | GLNASQIR | EAIQLD↓GLNA | 6.86 | 100 | 32.41 | 0.07 | 0.21 | 0.32 | 1 | 64 | 48 | |||
| Q9BWU0 | Kanadaptin | 538 | 554 | AFMSEMKSGSTLDGVSR | SQDSLD↓AFMS | 0 | 100 | 28.91 | 0 | 0 | 0.29 | 2 | 56 | 46 | Caspase-3 | 59 | |
| P21333 | Filamin-A | 2319 | 2333 | SPFVVPVASPSGDAR | EEHIPD↓SPFV | 0 | 100 | 28.39 | 0 | 0 | 0.28 | 1 | 56 | 47 | |||
| P21333 | Filamin-A | 1505 | 1515 | GTQTVNYVPSR | VVDNAD↓GTQT | 13.92 | 100 | 28.39 | 0.14 | 0.49 | 0.28 | 1 | 54 | 47 | |||
| P21333 | Filamin-A | 1502 | 1515 | NADGTQTVNYVPSR | PVDVVD↓NADG | 4.74 | 100 | 23.13 | 0.05 | 0.2 | 0.23 | 1 | 101 | 47 | |||
| Q14203 | Dynactin subunit 1 | 111 | 119 | SSASKVLKR | SPETPD↓SSAS | 0 | 100 | 21.09 | 0 | 0 | 0.21 | 1 | 78 | 47 | |||
| Q12797 | Aspartyl/asparaginyl β-hydroxylase | 238 | 247 | SSEPVVEDER | EQENPD↓SSEP | 8.73 | 100 | 18.98 | 0.09 | 0.46 | 0.19 | 1 | 61 | 47 | |||
| P52888 | Thimet oligopeptidase | 14 | 25 | AASPCSVVNDLR | AGDMAD↓AASP | 12.93 | 100 | 15.5 | 0.13 | 0.83 | 0.15 | 1 | 57 | 47 | Caspase-3 | 59 | |
| P62877 | RING-box protein 1 | 9 | 21 | TPSGTNSGAGKKR | AAMDVD↓TPSG | 0 | 100 | 15.5 | 0 | 0 | 0.15 | 1 | 62 | 47 | Caspase-3 | 59 | |
| P53990 | Uncharacterized protein KIAA0174 | 198 | 211 | VGFTDDVKKGGPGR | ETDLID↓VGFT | 7.68 | 100 | 14.65 | 0.08 | 0.52 | 0.15 | 1 | 82 | 47 | |||
| Q9P265 | Disco-interacting protein 2 homolog B | 61 | 68 | SAVQKELR | QTQETD↓SAVQ | 0 | 100 | 13.34 | 0 | 0 | 0.13 | 1 | 57 | 47 | caspase-3 | 59 | |
| Q13263 | Transcription intermediary factor 1-β | 689 | 702 | STGVVAKLSPANQR | SLDGAD↓STGV | 5.47 | 100 | 9.31 | 0.05 | 0.59 | 0.09 | 2 | 77 | 47 | Caspase-3 | 59 | |
| Q5UIP0 | Telomere-associated protein RIF1 | 1524 | 1531 | KSSEKLVR | TQDIVD↓KSSE | 0 | 100 | 0 | 0 | Single C3 | 0 | 1 | 56 | 47 | |||
| O75369 | Filamin-B | 1447 | 1458 | SSKAGLAPLEVR | QSFTVD↓SSKA | 0 | 100 | 0 | 0 | Single C3 | 0 | 1 | 57 | 47 | |||
| P62269 | 40 S ribosomal protein S18 | 93 | 108 | GKYSQVLANGLDNKLR | QKDVKD↓GKYS | 7 | 0 | 100 | 0 | 0.07 | 0 | 1 | 69 | 46 | |||
| P39687 | Acidic leucine-rich nuclear phosphoprotein 32 family member A | 217 | 233 | GEVDDEEDEEELGEEER | EEGYND↓GEVD | 5.3 | 0 | 100 | 0 | 0.05 | 0 | 1 | 92 | 41 | |||
| P47712 | Cytosolic phospholipase A2 | 523 | 533 | AAVADPDEFER | DDDELD↓AAVA | 0 | 99.55 | 100 | 0 | 0 | 1 | 1 | 48 | 47 | Caspase-3, -8 | 73, 74 | |
| P63261 | Actin, cytoplasmic 2 | 245 | 254 | GQVITIGNER | SYELPD↓GQVI | 20.69 | 98.79 | 100 | 0.21 | 0.21 | 1.01 | 5 | 55 | 47 | P60709, AP62736, AP63261, AP63267, AP68032, AP68133, AQ562R1, AQ6S8J3, AQ9BYX7 | ||
| P61011 | Signal recognition particle 54-kDa protein | 392 | 402 | GAKVFSKQPGR | ELDSTD↓GAKV | 51.73 | 95.75 | 100 | 0.54 | 0.52 | 1.04 | 1 | 51 | 47 | Caspase-3 | 59 | |
| Q92917 | G patch domain and KOW motif-containing protein | 342 | 353 | GPAAKSEKAAPR | LPDRQD↓GPAA | 10.68 | 71.97 | 100 | 0.15 | 0.11 | 1.39 | 1 | 66 | 47 | |||
| Q13151 | Heterogeneous nuclear ribonucleoprotein A0 | 74 | 81 | GNTVELKR | SPHAVD↓GNTV | 7.69 | 67.1 | 100 | 0.11 | 0.08 | 1.49 | 2 | 51 | 47 | |||
| Q86V48 | Leucine zipper protein 1 | 443 | 454 | SGTQETKKTEDR | MGVSTD↓SGTQ | 0 | 55.04 | 100 | 0 | 0 | 1.82 | 1 | 56 | 47 | |||
| P63241 | Eukaryotic translation initiation factor 5A-1 | 12 | 26 | AGASATFPMQCSALR | DFETGD↓AGAS | 0 | 54.76 | 100 | 0 | 0 | 1.83 | 1 | 57 | 46 | Q6IS14 | ||
| P55209 | Nucleosome assembly protein 1-like 1 | 58 | 72 | GLVETPTGYIESLPR | LQERLD↓GLVE | 7.19 | 53.9 | 100 | 0.13 | 0.07 | 1.86 | 2 | 85 | 47 | |||
| Q7Z6Z7 | E3 ubiquitin-protein ligase HUWE1 | 2360 | 2371 | GGSGNSTIIVSR | ELLERD↓GGSG | 15.6 | 53.54 | 100 | 0.29 | 0.16 | 1.87 | 1 | 82 | 47 | |||
| P30419 | Glycylpeptide N- tetradecanoyltransferase 1 | 73 | 88 | SAQDQPVKMNSLPAER | KGSETD↓SAQD | 0 | 47.04 | 100 | 0 | 0 | 2.13 | 1 | 52 | 46 | Caspase-3 | 59 | |
| Q15233 | Non-POU domain-containing octamer-binding protein | 423 | 434 | GTLGLTPPTTER | ATMMPD↓GTLG | 5.63 | 35.09 | 100 | 0.16 | 0.06 | 2.85 | 1 | 48 | 47 | Caspase-3 | 75 | |
| O94992 | Protein HEXIM1 | 220 | 228 | LKTGLYSKR | DQEEPD↓LKTG | 5.13 | 34.66 | 100 | 0.15 | 0.05 | 2.89 | 1 | 68 | 47 | |||
| P46777 | 60 S ribosomal protein L5 | 137 | 152 | GQPGAFTCYLDAGLAR | NVESID↓GQPG | 0 | 32.48 | 100 | 0 | 0 | 3.08 | 1 | 49 | 46 | |||
| Q13435 | Splicing factor 3B subunit 2 | 777 | 791 | GSETPQLFTVLPEKR | IEEAMD↓GSET | 4.35 | 31.89 | 100 | 0.14 | 0.04 | 3.14 | 1 | 94 | 46 | |||
| Q6PKG0 | La-related protein 1 | 496 | 508 | FSQLLNCPEFVPR | DYSQTD↓FSQL | 0 | 28.54 | 100 | 0 | 0 | 3.5 | 1 | 66 | 47 | |||
| O60664 | Mannose 6-phosphate receptor-binding protein 1 | 10 | 28 | GSTQVTVEEPVQQPSVVDR | DGAEAD↓GSTQ | 0 | 26.79 | 100 | 0 | 0 | 3.73 | 1 | 70 | 46 | |||
| Q04637 | Eukaryotic translation initiation factor 4γ1 | 754 | 762 | GSKTQDLFR | GEEDAD↓GSKT | 8.73 | 22.98 | 100 | 0.38 | 0.09 | 4.35 | 1 | 53 | 47 | |||
| Q13442 | 28-kDa heat- and acid-stable phosphoprotein | 25 | 35 | AQLQAEKQKAR | SPEEID↓AQLQ | 0 | 20.67 | 100 | 0 | 0 | 4.84 | 2 | 68 | 47 | Caspase-3 | 59 | |
| Q32MZ4 | Leucine-rich repeat flightless-interacting protein 1 | 679 | 690 | FEDDTVQSSGPR | DGDTLD↓FEDD | 0 | 16.42 | 100 | 0 | 0 | 6.09 | 1 | 79 | 47 | |||
| P06454 | Prothymosin α | 77 | 90 | GDEDEEAESATGKR | DGEEED↓GDED | 18.54 | 16.31 | 100 | 1.14 | 0.19 | 6.13 | 1 | 66 | 46 | |||
| P14618 | Pyruvate kinase isozymes M1/M2 | 355 | 376 | GADCIMLSGETAKGDYPGADCIMLSGETAKGDYPLELEAVR | ANAVLD↓GADC | 4.37 | 14.11 | 100 | 0.31 | 0.04 | 7.09 | 1 | 72 | 45 | |||
| P06454 | Prothymosin α | 72 | 90 | GEEEDGDEDEEAESATGKR | EEEEGD↓GEEE | 8.51 | 10.48 | 100 | 0.81 | 0.09 | 9.54 | 1 | 115 | 42 | |||
| Q8N1G4 | Leucine-rich repeat-containing protein 47 | 526 | 554 | AVSGQLPDPTTNPSAGKDGPSLLVVEQVR | SDTEAD↓AVSG | 0 | 0 | 100 | 0 | 0 | Single C7 | 1 | 62 | 44 | |||
| P05455 | Lupus La protein | 375 | 384 | ENGATGPVKR | EHDEHD↓ENGA | 0 | 0 | 100 | 0 | 0 | Single C7 | 2 | 49 | 47 | Caspase-3 | 76 | |
| P26599 | Polypyrimidine tract-binding protein 1 | 3 | 14 | GIVPDIAVGTKR | MD↓GIVP | 0 | 0 | 100 | 0 | 0 | Single C7 | 2 | 59 | 47 | |||
| Q00839 | Heterogeneous nuclear ribonucleoprotein U | 233 | 244 | GKTEQKGGDKKR | GAPAGD↓GKTE | 0 | 0 | 100 | 0 | 0 | Single C7 | 1 | 68 | 46 | |||
| Q9NX24 | H/ACA ribonucleoprotein complex subunit 2 | 10 | 22 | GPEAQAEACSGER | IKADPD↓GPEA | 0 | 0 | 100 | 0 | 0 | Single C7 | 1 | 67 | 46 | Caspase-3 | 59 |
FIGURE 2.
Cleavage site specificities of caspase-2, -3, and -7. Schematic representation of cleavage by human caspase-2, -3, and -7 identified by COFRADIC is shown. A, a Venn diagram shows the numbers of identified peptide sequences cleaved by caspase-2 (C2), caspase-3 (C3), and caspase-7 (C7). B–D, IceLogo visualizations of alignments of the P6-P4′ amino acid sequences of cleavage sites with the human Swiss-Prot proteome as reference set show that DEVD↓G is the preferred sequence from P4 to P1 for all three caspases. The height of a stack of symbols reflects the degree of conservation, and the height of individual amino acids indicates the percentage difference in the frequency of occurrence at a given position compared with a reference set. Residues depicted in pink were never observed at a certain position.
To determine whether caspase-2, -3, and -7 differ in the preference for amino acids surrounding the scissile bond, we used the IceLogo application. This software tool was developed to statistically analyze amino acid occurrence within a given set of peptides and its deviation from random sequence occurrences as found, for instance, in databases (48). The results are plotted as sequence logos to visualize differentiating patterns in our datasets (Fig. 2, B–D). For a reference set, we used the same sample size of random sequences stored in the human Swiss-Prot database, dynamically sampled 100 times. The positive sample set used in the analysis of caspase-2 included all 38 decapeptide sequences cleaved by caspase-2 (P6-P4′) aligned at the P1 position. Caspase-3 analysis included all 63 sites identified in the caspase-3-treated sample, whereas caspase-7 analysis was based on 68 cleavage sites.
Our results show that DEVD↓G is the consensus P4-P1′ amino acid sequence preferred by all three caspases in native proteins. No caspase showed striking amino acid preference at the P5 position, although Glu was the most frequent residue in the datasets of all three caspases. Additionally, caspase-2 sites were enriched for Gln on P5, whereas both caspase-3 and -7 showed preference for the smaller Gly residue. The small preference of all three caspases for the negatively charged Glu at position P5 differs from the data of earlier reports that described a marked increase in caspase-2 catalytic activity when substrates contained the small hydrophobic Val at P5 (33, 51). Caspase-3, but not caspase-7, was reported to prefer small hydrophobic residues such as Leu or Val at the P5 position due to hydrophobicity of its S5 substrate binding pocket (52, 53). However, we did not observe any marked differences in the P5 preference between caspase-3 and -7. Both showed a small preference for Glu and Gly at this position. All three caspases prefer Asp at P4, but Glu was also found to be enriched. At P3 all caspases preferred Glu, and caspase-2 additionally preferred Ala. All three caspases preferred Val and Pro at P2, whereas caspase-3 additionally showed a small preference for Leu. For caspase-2, the most frequent residues at P1′ were Gly and, to a lesser extent, Ser. Caspases-3 and -7 cleavage sites were additionally enriched in Ala at this position. These results are in accordance with previous reports showing that caspases favor small uncharged amino acid residues, such as Gly and Ala, at P1′ (54, 55). Moreover, the newly formed protein fragments originating from cleavage sites with Gly, Ser, or Ala at P1′ might be stable in a cellular environment corresponding to the N-end rule (56).
In general, despite very subtle differences in cleavage efficiency and specificity, all three caspases clearly have a preference for the DEVD↓G substrate motif at P4-P1′ in native proteins and no marked preferences at P5. Furthermore, all identified caspase-2 cleavage sites are also cleaved by caspase-3 and -7, almost always with efficiencies higher than that of caspase-2, suggesting that caspase-2 has an overlapping specificity with executioner caspases but lower efficiency.
Caspase-2 Cleaves eIF4B at Asp563
Although our COFRADIC analysis did not reveal any substrate cleavage unique for caspase-2, we have identified its preferred cleavage site. eIF4B was cleaved at Asp563 (DRKD↓G) most efficiently by caspase-2, whereas caspase-7 cleaved it with 73% efficiency and caspase-3 with only 33% efficiency relative to caspase-2 (Table 1). When these values were expressed as cleavage ratios, caspase-2 showed a 3-fold preference over caspase-3 and a 1.36-fold preference over caspase-7 in cleaving eIF4B at Asp563 (Table 1). The second cleavage site identified in eIF4B, Asp531 (NKVD↓G), was preferred by caspase-3 and caspase-7, whereas caspase-2 showed very limited cleavage.
To verify if Asp563 truly represents a caspase-2-preferred cleavage site, we expressed eIF4B and its point mutants (eIF4B D531A, EIF4B D563A, and the double mutant) together with human procaspase-2, -3, or -7 in HEK 293T cells. The FLAG-tagged wild type eIF4B and the FLAG-tagged eIF4B mutant proteins were stably expressed in HEK 293T cells. Fig. 3 shows the result of monitoring eIF4B expression by Western blotting with anti-FLAG antibody and the caspase expression by using specific antibodies. The antibody directed against caspase-2 detected increased amounts of its proform together with processed subunits of caspase-2 upon caspase-2 overexpression as compared with the endogenous procaspase-2 in all other conditions (25). Similarly, when caspase-3 was expressed, we observed increased intensity of the proform band and an appearance of its cleaved form. The appearance of multiple cleavage bands and an intense proform band slightly larger than the endogenous band can be attributed to the presence of a myc tag in the caspase-3 expression construct. Caspase-7 expression in HEK 293T cells was confirmed by the appearance of an intense proform band, also slightly larger than the endogenous procaspase-7, possibly due to the expression of a FLAG tag. The duration of caspase expression was kept limited to 13 h to prevent extensive caspase cascade signaling and dismantling of the cell, characteristic of late apoptotic stages. We visualized poly(ADP-ribose) polymerase protein to find it is not cleaved, which confirmed that cells did not enter the late apoptotic phase. Because proteins commonly used as loading controls for the Western blot, such as tubulin, actin, and GAPDH, are reported to be caspase substrates (Table 1) (38, 57, 58), we used GFP protein as both transfection control and Western blot loading control.
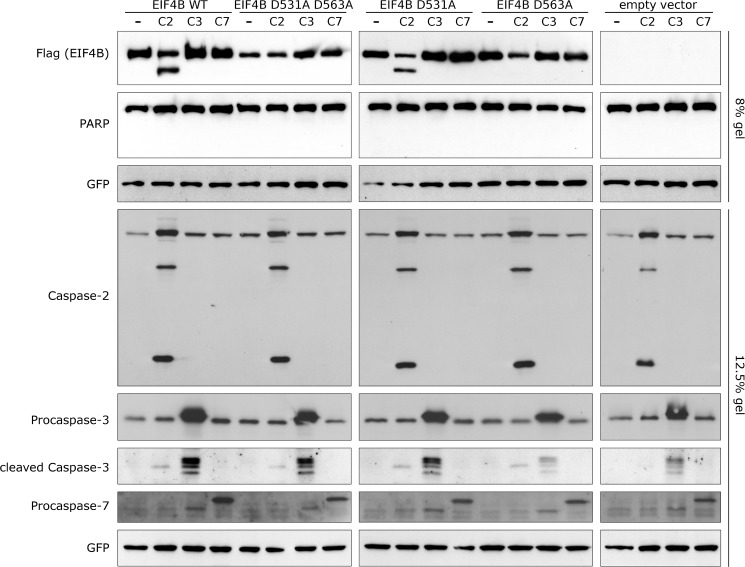
FIGURE 3.
eIF4B Asp563 is a true caspase-2 cleavage site. Processing of wild type eIF4B and its mutants upon co-expression with caspases is shown. HEK 293T cells were transiently transfected with the indicated variants of FLAG-tagged eIF4B and caspase expression vectors. Cleavage of eIF4B was assessed by Western blotting with anti-FLAG antibody. Expression of caspases was confirmed using their respective antibodies. GFP served as a loading control. Intact poly(ADP-ribose) polymerase (PARP) indicates that cells had not reached late apoptotic phase. The inability of caspase-2 to cleave the eIF4B D563A mutant (EIF4B D563A), in contrast to wild type eIF4B (EIF4B WT), indicates that Asp563 is a true caspase-2 cleavage site. Results are representative of at least three independent experiments.
Co-expression of wild type eIF4B with caspase-2, but not with caspase-3 or -7, generated an observable eIF4B cleavage fragment identified as an additional band below the full-length eIF4B protein detected with anti-FLAG antibody. As expected, mutating both Asp531 and Asp563 to Ala (Fig. 3, EIF4B D531A D563A) abolished this cleavage. The expression of single point mutants (Fig. 3) showed that although eIF4B D531A (Fig. 3, EIF4B D531A) was still cleaved, eIF4B D563A (Fig. 3, EIF4B D563A) was not. Although eIF4B processing by caspase-3 at Asp531 was previously reported (59), under these experimental conditions cleavage at Asp563 or Asp531 by caspase-3 or -7 was not detectable by Western blotting. These data confirm the results of the COFRADIC analysis; Asp563 in eIF4B protein is a caspase-2-preferred cleavage site.
DISCUSSION
We studied the specificity of human caspase-2 on native proteomes. Previously reported specificities of caspase-2 were based on combinatorial libraries of short peptide sequences (33, 34) or ad hoc substrate identification (28). The COFRADIC N-terminal peptide sorting technique (40, 41, 59) allows identification of protein cleavage events in native proteomes. Caspase-mediated substrate cleavage can be identified unambiguously by SILAC labeling of a cell culture using stable isotopes of arginine. Neo-N termini resulting from caspase treatment are identified by mass spectrometry, and matching protein sequences are retrieved from databases. We aligned the sequences of six amino acid residues before (non-prime sites) and four residues after the caspase scissile bond (prime sites) (P6-P4′) and statistically analyzed them by IceLogo to visualize residues over- or underrepresented at each of these positions in the caspase-2, -3, and -7 degradomes relative to their random and natural occurrence in Swiss-Prot. We compared the cleavage sites generated by caspase-2 to those generated by caspase-3 and caspase-7. Surprisingly, the P4-P1′ cleavage sequence of caspase-2, DEVD↓G, was nearly indistinguishable from the caspase-3 and -7 cleavage sequence (Fig. 2).
Our data are in agreement with the observation of Thornberry et al. (34) in that the caspase-2 substrate sequence resembles that of executioner caspases, namely, DExD. The only discrepancy is the residue preference at the P2 position, where we identify Val as the most common amino acid, whereas the previous analysis reported His. Additionally, according to our results, the P5 position was moderately enriched for Glu, which differs from a previous report stating that caspase-2 activity was markedly enhanced when Val was at the P5 position (33). So far, caspase-2 is the only caspase reported to require recognition of an amino acid in a peptide substrate at position P5 for optimal activity (33). Caspase-3, unlike caspase-7, was also shown to recognize hydrophobic residues at the P5 position (52). Caspase-3 cleaved pentapeptides better than tetrapeptides, but the presence of the P5 site was not a prerequisite for strong catalytic activity (53). Structural differences between caspase-3 and -7 that influence P5 binding are also found in loops 3 and 4. Although caspase-3 in loop 3 contains a Ser209 residue that binds P5 Leu of the LDESD sequence, caspase-7 contains Pro235, which does not allow such interaction. Additional amino acids constituting the S5 pocket residing in loop 4 are hydrophilic in caspase-7 (Gln-276, Ser-277, and Asp-278), whereas in caspase-3 two of the three are hydrophobic (Phe-250, Ser-251, and Leu-252) (42). Contrary to these structural data, we observed no marked difference between caspase-3 and -7 in the amino acid sequence recognized in substrates derived from whole cell proteomes. The apparent discrepancy between the results of structural studies of caspase-2 specificity and the results reported here could be related to methodological differences. Although the previous studies were based on synthetic peptides, in our COFRADIC analysis we used caspase substrates in their native state, which might impose other restraints on the accessibility of substrate cleavage sites in the catalytic pocket.
It was recently shown for caspase-1 that the specificity of caspases assayed with short synthetic peptides does not accurately represent their specificity for native proteins (12). Caspase-1 had long been considered very specific due to the scarcity of its validated substrates (IL-1β and IL-18), which was ascribed to the specificity of its catalytic pocket. However, Walsh et al. (12) observed that the promiscuity of the enzyme varies significantly with the concentration used in in vitro assays. Additionally, the specificity of caspase-1 is improved by more rapid inactivation due to the loss of quaternary structure as compared with caspases-3 and -7 (12). Moreover, enzyme regions distant from the binding site but participating or modulating substrate binding, also referred to as exosites, have been identified in other regulatory proteases, such as matrix metalloproteinases (60). Although no exosites in caspase-2 have been reported, their role in modulating the activity of other caspase family members has been postulated (61). Also of significant influence on substrate cleavage are higher-order protein structures and -folds. In general, apoptosis-specific cleavage sites are located outside structured domains (62). In fact, caspase-3 was shown to prefer extended loop regions, although cleavage within α-helices is not uncommon, whereas extended β-strands were strongly disfavored (55). This might reflect the importance of factors other than the amino acid sequence in determining caspase specificity. It is thus quite plausible that the short peptide specificities of other caspases do not accurately reflect their specificities for native proteomes.
The remarkable overlap between the specificity of caspase-2, -3, and -7 revealed in our study indicates that caspase-2 resembles executioner caspases. Its consensus cleavage sequence, DEVD↓G, matches that of caspase-3 and -7, and most of its cleavage sites are shared with the other two caspases. Caspases-3 and -7 appear to cleave shared substrates more efficiently than caspase-2. However, caspase-2 is most often placed in the initiator caspase group because it includes a long CARD-containing prodomain and is activated by a proximity-induced mechanism. But in contrast to other initiator caspases, under cellular stress and in death models caspase-2 does not process executioner caspases (31), and its known substrates are mostly structural proteins (golgin 160, αII-spectrin, and desmoplakin), and proteins that assist in dismantling the cell by transmitting a death signal to enzymes executing this process (Bid, PKCδ, and iCAD) (28). All caspase-2 cleavage sites described so far are shared with other caspases, most often with caspase-3 and -7 (e.g. αII-spectrin, desmoplakin, and iCAD) or with caspase-8 (Bid). The only exception is golgin 160, which is cleaved uniquely at ESPD59↓G by caspase-2, although it also contains cleavage sites for caspase-3 (CSTD139↓S) and caspase-7 (SEVD311↓G) (29).
Because COFRADIC analysis did not identify any cleavage sites unique to caspase-2, we further analyzed Asp563 in eukaryotic translation initiation factor eIF4B, which was the only cleavage site for which caspase-2 showed preference. Wild type eIF4B was cleaved in HEK 293T cells when co-expressed with caspase-2, but cleavage of the eIF4B D563A mutant was abolished, confirming that Asp563 is a true caspase-2 cleavage site (Fig. 3). However, the intensity of the full-length eIF4B D563A mutant band appeared to be decreased when co-expressed with caspase-2, although we did not detect any additional eIF4B cleavage band; therefore, we can fairly conclude that this cleavage site is unique for caspase-2. We have no obvious explanation for this decrease in expression of full-length eIF4B, but it is conceivable that caspase-2-mediated cleavage of factors regulating transcription or translation (Table 1) may indirectly affect eIF4B expression. Moreover, caspase-3 and -7 did not cleave the substrate in our setup, most probably because the cleavage extent of these two caspases is too little to be visualized by Western blot. A similar observation of different sensitivity between proteomic approach and Western blotting was reported earlier (63). The DRKD563↓G cleavage site in eIF4B does not fully agree with the caspase-2 consensus cleavage site identified with iceLogo (Fig. 2). This might indicate subtle differences in the substrate specificity profiles of caspase-2, -3, and -7 and explain why this site was observed to be caspase-2-preferred. In eIF4B, positions P3 and P2 are occupied by Arg and Lys, whereas iceLogo showed preferences for Glu/Ala and Val/Pro, respectively. However, iceLogo analysis is not weighted for the cleavage preference, and the caspase-2 cleavage sites were fewer (n = 37) than the cleavage sites for caspase-3 (n = 61) and caspase-7 (n = 66). These results suggest that efficient caspase-2 cleavage, in comparison to caspase-3 and -7, favors hydrophilic amino acids at P3 and P2 and that the ability of caspases to cleave certain sites might be determined not only by amino acid sequence but also by other factors regulating caspase specificity. Additionally, the physiological significance of eIF4B cleavage is unknown. Translation initiation factors are known caspase substrates and contribute to inhibition of protein synthesis during apoptosis (64). Two caspase-3 cleavage sites have already been reported in eIF4B (Asp45 and Asp531), but their physiological significance has not been established (59, 65). eIF4B is an RNA-binding protein and a co-factor of eukaryotic translation initiation complex; it interacts with and potentiates the RNA helicase activities of eIF4F and eIF4A. These functions are associated with its RRM, ARM and DRYG domains, located between amino acids 97–175, 214–327, and 367–423, respectively (66). eIF4B activity is regulated at Ser406 and Ser422 by Ras-MAPK and PI3K/mTOR signaling pathways, and its knock-down decreases proliferation and promotes apoptosis (67). The cleavage of eIF4B at Asp563 is not directly located in any of its functional domains, but it might alter protein conformation or affect its function otherwise. The in vivo role of this cleavage event requires further investigation.
The scarcity of substrates of caspase-2 and the discrepancy in the sequence preference between synthetic and natural substrates corresponds with recent findings on caspase-1 published by Walsh et al. (12). They demonstrated that the high specificity of inflammatory caspase-1 is not an intrinsic property but is achieved by regulation of the stability of the enzyme. Although caspase-1 has only two confirmed substrates (IL-1β and IL-18), these authors observed surprising promiscuity of caspase-1 at physiological concentrations toward both synthetic and natural substrates. High specificity of caspase-1 in cells is achieved by lower stability of the active enzyme, as shown by the loss of activity attributed to the loss of quaternary structure, which in THP-1 cells occurs upon activation of the inflammasome but not of the apoptosome (12). We hypothesize that caspase-2 might share features with both initiator and executioner caspases and that it operates in specific conditions, such as DNA damage (68), disruption of the spindle formation during mitosis (69), and heat shock (70) to allow apoptotic processes to dismantle the cell but with lower efficiency than executioner caspases. Our data demonstrate that the specific function of caspase-2 might not rely on the intrinsic specificity features of the enzymes, but like caspase-1, rather on the context in which caspase-2 is activated.
In conclusion, our data show that there is a large overlap between the degradome of caspase-2 and the degradomes of caspase-3 and caspase-7. Moreover, all three caspases have a preference for the P4-P1′ sequence DEVD↓G in the substrate, with the indication that caspase-2 at positions P2 and P3 prefers charged amino acids. Our analysis is the first comprehensive high-throughput screening for substrates and cleavage specificity of human caspase-2 using a native proteome. Our findings indicate that caspase-2, despite sharing structural features with initiator caspases, might functionally be considered a member of the executioner caspases group, albeit not a very efficient one. Finally our data demonstrate that the specific function of caspase-2 might not rely on its intrinsic specificity features but rather on the context in which it is activated.
Acknowledgment
We thank Amin Bredan for editing the manuscript.
This work was supported by European grants (FP6 ApopTrain, MRTN-CT-035624; FP7 EC RTD Integrated Project, Apo-Sys, FP7–200767; Euregional PACT II), Belgian grants (Interuniversity Attraction Poles, IAP 6/18), Flemish grants (Research Foundation Flanders, FWO G.0875.11 and FWO G.0973.11), Ghent University grants (MRP, GROUP-ID consortium), and grants from Flanders Institute for Biotechnology (VIB).
Spectra of all identified neo-N termini have been submitted to PRIDE (accession number 19136).
- CARD
- caspase recruitment domain
- DD
- death domain
- DED
- death effector domain
- DISC
- death-inducing signaling complex
- PIDD
- p53-induced protein with death domain
- RAIDD
- receptor-interacting protein (RIP)-associated ICH-1/CED-3-homologous protein with a death domain
- COFRADIC
- combined fractional diagonal chromatography
- SILAC
- stable isotope labeling by amino acids in cell culture
- eIF4B
- eukaryotic translation initiation factor 4B
- RFU
- relative fluorescence unit
- AMC
- amidomethylcoumarin.
REFERENCES
- 1. Lamkanfi M., Festjens N., Declercq W., Vanden Berghe T., Vandenabeele P. (2007) Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 14, 44–55 [DOI] [PubMed] [Google Scholar]
- 2. Pop C., Salvesen G. S. (2009) Human caspases. activation, specificity, and regulation. J. Biol. Chem. 284, 21777–21781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Scott A. M., Saleh M. (2007) The inflammatory caspases. Guardians against infections and sepsis. Cell Death Differ. 14, 23–31 [DOI] [PubMed] [Google Scholar]
- 4. Lippens S., Kockx M., Knaapen M., Mortier L., Polakowska R., Verheyen A., Garmyn M., Zwijsen A., Formstecher P., Huylebroeck D., Vandenabeele P., Declercq W. (2000) Epidermal differentiation does not involve the pro-apoptotic executioner caspases but is associated with caspase-14 induction and processing. Cell Death Differ 7, 1218–1224 [DOI] [PubMed] [Google Scholar]
- 5. Kersse K., Verspurten J., Vanden Berghe T., Vandenabeele P. (2011) The death-fold superfamily of homotypic interaction motifs. Trends Biochem. Sci. 36, 541–552 [DOI] [PubMed] [Google Scholar]
- 6. Muzio M., Stockwell B. R., Stennicke H. R., Salvesen G. S., Dixit V. M. (1998) An induced proximity model for caspase-8 activation. J. Biol. Chem. 273, 2926–2930 [DOI] [PubMed] [Google Scholar]
- 7. Martinon F., Burns K., Tschopp J. (2002) The inflammasome. A molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 8. Zou H., Li Y., Liu X., Wang X. (1999) An APAF-1.cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J. Biol. Chem. 274, 11549–11556 [DOI] [PubMed] [Google Scholar]
- 9. Tinel A., Tschopp J. (2004) The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 304, 843–846 [DOI] [PubMed] [Google Scholar]
- 10. Riedl S. J., Fuentes-Prior P., Renatus M., Kairies N., Krapp S., Huber R., Salvesen G. S., Bode W. (2001) Structural basis for the activation of human procaspase-7. Proc. Natl. Acad. Sci. U.S.A. 98, 14790–14795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lamkanfi M., Declercq W., Kalai M., Saelens X., Vandenabeele P. (2002) Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 9, 358–361 [DOI] [PubMed] [Google Scholar]
- 12. Walsh J. G., Logue S. E., Lüthi A. U., Martin S. J. (2011) Caspase-1 promiscuity is counterbalanced by rapid inactivation of processed enzyme. J. Biol. Chem. 286, 32513–32524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cuenin S., Tinel A., Janssens S., Tschopp J. (2008) p53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-κB and caspase-2 in response to genotoxic stress. Oncogene 27, 387–396 [DOI] [PubMed] [Google Scholar]
- 14. Tinel A., Janssens S., Lippens S., Cuenin S., Logette E., Jaccard B., Quadroni M., Tschopp J. (2007) Autoproteolysis of PIDD marks the bifurcation between pro-death caspase-2 and pro-survival NF-κB pathway. EMBO J. 26, 197–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Manzl C., Krumschnabel G., Bock F., Sohm B., Labi V., Baumgartner F., Logette E., Tschopp J., Villunger A. (2009) Caspase-2 activation in the absence of PIDDosome formation. J. Cell Biol. 185, 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamkanfi M., D'hondt K., Vande Walle L., van Gurp M., Denecker G., Demeulemeester J., Kalai M., Declercq W., Saelens X., Vandenabeele P. (2005) A novel caspase-2 complex containing TRAF2 and RIP1. J. Biol. Chem. 280, 6923–6932 [DOI] [PubMed] [Google Scholar]
- 17. Kumar S., Kinoshita M., Noda M., Copeland N. G., Jenkins N. A. (1994) Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1β-converting enzyme. Genes Dev. 8, 1613–1626 [DOI] [PubMed] [Google Scholar]
- 18. Wang L., Miura M., Bergeron L., Zhu H., Yuan J. (1994) Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 78, 739–750 [DOI] [PubMed] [Google Scholar]
- 19. Kumar S., Tomooka Y., Noda M. (1992) Identification of a set of genes with developmentally down-regulated expression in the mouse brain. Biochem. Biophys. Res. Commun. 185, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 20. Bergeron L., Perez G. I., Macdonald G., Shi L., Sun Y., Jurisicova A., Varmuza S., Latham K. E., Flaws J. A., Salter J. C., Hara H., Moskowitz M. A., Li E., Greenberg A., Tilly J. L., Yuan J. (1998) Defects in regulation of apoptosis in caspase-2-deficient mice. Genes Dev. 12, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Padalecki S. S., Chaudhuri A. R., De Waal E., Goins B. A., Grubbs B., Ikeno Y., Richardson A., Mundy G. R., Herman B. (2007) Caspase-2 deficiency enhances aging-related traits in mice. Mech. Ageing Dev. 128, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Troy C. M., Rabacchi S. A., Friedman W. J., Frappier T. F., Brown K., Shelanski M. L. (2000) Caspase-2 mediates neuronal cell death induced by β-amyloid. J. Neurosci. 20, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar S. (2009) Caspase 2 in apoptosis, the DNA damage response and tumor suppression. Enigma no more? Nat. Rev. Cancer 9, 897–903 [DOI] [PubMed] [Google Scholar]
- 24. Guo Y., Srinivasula S. M., Druilhe A., Fernandes-Alnemri T., Alnemri E. S. (2002) Caspase-2 induces apoptosis by releasing proapoptotic proteins from mitochondria. J. Biol. Chem. 277, 13430–13437 [DOI] [PubMed] [Google Scholar]
- 25. O'Reilly L. A., Ekert P., Harvey N., Marsden V., Cullen L., Vaux D. L., Hacker G., Magnusson C., Pakusch M., Cecconi F., Kuida K., Strasser A., Huang D. C., Kumar S. (2002) Caspase-2 is not required for thymocyte or neuronal apoptosis even though cleavage of caspase-2 is dependent on both Apaf-1 and caspase-9. Cell Death Differ. 9, 832–841 [DOI] [PubMed] [Google Scholar]
- 26. Ho L. H., Taylor R., Dorstyn L., Cakouros D., Bouillet P., Kumar S. (2009) A tumor suppressor function for caspase-2. Proc. Natl. Acad. Sci. U.S.A. 106, 5336–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oliver T. G., Meylan E., Chang G. P., Xue W., Burke J. R., Humpton T. J., Hubbard D., Bhutkar A., Jacks T. (2011) Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell 43, 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kitevska T., Spencer D. M., Hawkins C. J. (2009) Caspase-2. Controversial killer or checkpoint controller? Apoptosis 14, 829–848 [DOI] [PubMed] [Google Scholar]
- 29. Mancini M., Machamer C. E., Roy S., Nicholson D. W., Thornberry N. A., Casciola-Rosen L. A., Rosen A. (2000) Caspase-2 is localized at the Golgi complex and cleaves golgin-160 during apoptosis. J. Cell Biol. 149, 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van de Craen M., Declercq W., Van den brande I., Fiers W., Vandenabeele P. (1999) The proteolytic procaspase activation network. An in vitro analysis. Cell Death Differ 6, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 31. Slee E. A., Harte M. T., Kluck R. M., Wolf B. B., Casiano C. A., Newmeyer D. D., Wang H. G., Reed J. C., Nicholson D. W., Alnemri E. S., Green D. R., Martin S. J. (1999) Ordering the cytochrome c-initiated caspase cascade. Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karki P., Dahal G. R., Shin S. Y., Lee J. S., Cho B., Park I. S. (2008) Efficient cleavage of Bid and procaspase-7 by caspase-2 at lower pH. Protein Pept. Lett. 15, 1044–1049 [DOI] [PubMed] [Google Scholar]
- 33. Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., Wong W. W. (1997) Substrate specificities of caspase family proteases. J. Biol. Chem. 272, 9677–9682 [DOI] [PubMed] [Google Scholar]
- 34. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 35. McStay G. P., Salvesen G. S., Green D. R. (2008) Overlapping cleavage motif selectivity of caspases. Implications for analysis of apoptotic pathways. Cell Death Differ. 15, 322–331 [DOI] [PubMed] [Google Scholar]
- 36. Pereira N. A., Song Z. (2008) Some commonly used caspase substrates and inhibitors lack the specificity required to monitor individual caspase activity. Biochem. Biophys. Res. Commun. 377, 873–877 [DOI] [PubMed] [Google Scholar]
- 37. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schilling O., Overall C. M. (2008) Proteome-derived, database-searchable peptide libraries for identifying protease cleavage sites. Nat. Biotechnol. 26, 685–694 [DOI] [PubMed] [Google Scholar]
- 39. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 40. Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. (2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362–1370 [DOI] [PubMed] [Google Scholar]
- 41. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 42. Demon D., Van Damme P., Vanden Berghe T., Deceuninck A., Van Durme J., Verspurten J., Helsens K., Impens F., Wejda M., Schymkowitz J., Rousseau F., Madder A., Vandekerckhove J., Declercq W., Gevaert K., Vandenabeele P. (2009) Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol. Cell. Proteomics 8, 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Staes A., Impens F., Van Damme P., Ruttens B., Goethals M., Demol H., Timmerman E., Vandekerckhove J., Gevaert K. (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat. Protoc. 6, 1130–1141 [DOI] [PubMed] [Google Scholar]
- 44. Ghesquière B., Van Damme J., Martens L., Vandekerckhove J., Gevaert K. (2006) Proteome-wide characterization of N-glycosylation events by diagonal chromatography. J. Proteome Res. 5, 2438–2447 [DOI] [PubMed] [Google Scholar]
- 45. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 46. Helsens K., Colaert N., Barsnes H., Muth T., Flikka K., Staes A., Timmerman E., Wortelkamp S., Sickmann A., Vandekerckhove J., Gevaert K., Martens L. (2010) ms_lims, a simple yet powerful open source laboratory information management system for MS-driven proteomics. Proteomics 10, 1261–1264 [DOI] [PubMed] [Google Scholar]
- 47. Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., Hermjakob H., Martens L. (2010) The Proteomics Identifications database. 2010 update. Nucleic Acids Res. 38, D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 49. Saelens X., Kalai M., Vandenabeele P. (2001) Translation inhibition in apoptosis. Caspase-dependent PKR activation and eIF2α phosphorylation. J. Biol. Chem. 276, 41620–41628 [DOI] [PubMed] [Google Scholar]
- 50. Stennicke H. R., Salvesen G. S. (1997) Biochemical characteristics of caspases-3, -6, -7, and -8. J. Biol. Chem. 272, 25719–25723 [DOI] [PubMed] [Google Scholar]
- 51. Schweizer A., Briand C., Grutter M. G. (2003) Crystal structure of caspase-2, apical initiator of the intrinsic apoptotic pathway. J. Biol. Chem. 278, 42441–42447 [DOI] [PubMed] [Google Scholar]
- 52. Fang B., Boross P. I., Tozser J., Weber I. T. (2006) Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition. J. Mol. Biol. 360, 654–666 [DOI] [PubMed] [Google Scholar]
- 53. Fu G., Chumanevich A. A., Agniswamy J., Fang B., Harrison R. W., Weber I. T. (2008) Structural basis for executioner caspase recognition of P5 position in substrates. Apoptosis 13, 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stennicke H. R., Renatus M., Meldal M., Salvesen G. S. (2000) Internally quenched fluorescent peptide substrates disclose the subsite preferences of human caspases 1, 3, 6, 7 and 8. Biochem. J. 350 Pt 2, 563–568 [PMC free article] [PubMed] [Google Scholar]
- 55. Timmer J. C., Zhu W., Pop C., Regan T., Snipas S. J., Eroshkin A. M., Riedl S. J., Salvesen G. S. (2009) Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 16, 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Varshavsky A. (1997) The N-end rule pathway of protein degradation. Genes Cells 2, 13–28 [DOI] [PubMed] [Google Scholar]
- 57. Klaiman G., Petzke T. L., Hammond J., Leblanc A. C. (2008) Targets of caspase-6 activity in human neurons and Alzheimer disease. Mol. Cell. Proteomics 7, 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mashima T., Naito M., Noguchi K., Miller D. K., Nicholson D. W., Tsuruo T. (1997) Actin cleavage by CPP-32/apopain during the development of apoptosis. Oncogene 14, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 59. Impens F., Colaert N., Helsens K., Ghesquière B., Timmerman E., De Bock P. J., Chain B. M., Vandekerckhove J., Gevaert K. (2010) A quantitative proteomics design for systematic identification of protease cleavage events. Mol. Cell. Proteomics 9, 2327–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. López-Otín C., Overall C. M. (2002) Protease degradomics. A new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 3, 509–519 [DOI] [PubMed] [Google Scholar]
- 61. Fuentes-Prior P., Salvesen G. S. (2004) The protein structures that shape caspase activity, specificity, activation, and inhibition. Biochem. J. 384, 201–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 63. Enoksson M., Li J., Ivancic M. M., Timmer J. C., Wildfang E., Eroshkin A., Salvesen G. S., Tao W. A. (2007) Identification of proteolytic cleavage sites by quantitative proteomics. J. Proteome Res. 6, 2850–2858 [DOI] [PubMed] [Google Scholar]
- 64. Saelens X., Festjens N., Parthoens E., Vanoverberghe I., Kalai M., van Kuppeveld F., Vandenabeele P. (2005) Protein synthesis persists during necrotic cell death. J. Cell Biol. 168, 545–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bushell M., Wood W., Clemens M. J., Morley S. J. (2000) Changes in integrity and association of eukaryotic protein synthesis initiation factors during apoptosis. Eur. J. Biochem. 267, 1083–1091 [DOI] [PubMed] [Google Scholar]
- 66. Shahbazian D., Parsyan A., Petroulakis E., Hershey J., Sonenberg N. (2010) eIF4B controls survival and proliferation and is regulated by proto-oncogenic signaling pathways. Cell Cycle 9, 4106–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shahbazian D., Parsyan A., Petroulakis E., Topisirovic I., Martineau Y., Gibbs B. F., Svitkin Y., Sonenberg N. (2010) Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol. Cell. Biol. 30, 1478–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vakifahmetoglu H., Olsson M., Orrenius S., Zhivotovsky B. (2006) Functional connection between p53 and caspase-2 is essential for apoptosis induced by DNA damage. Oncogene 25, 5683–5692 [DOI] [PubMed] [Google Scholar]
- 69. Castedo M., Perfettini J. L., Roumier T., Valent A., Raslova H., Yakushijin K., Horne D., Feunteun J., Lenoir G., Medema R., Vainchenker W., Kroemer G. (2004) Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene 23, 4362–4370 [DOI] [PubMed] [Google Scholar]
- 70. Tu S., McStay G. P., Boucher L. M., Mak T., Beere H. M., Green D. R. (2006) In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat. Cell Biol. 8, 72–77 [DOI] [PubMed] [Google Scholar]
- 71. MacFarlane M., Merrison W., Dinsdale D., Cohen G. M. (2000) Active caspases and cleaved cytokeratins are sequestered into cytoplasmic inclusions in TRAIL-induced apoptosis. J. Cell Biol. 148, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Byun Y., Chen F., Chang R., Trivedi M., Green K. J., Cryns V. L. (2001) Caspase cleavage of vimentin disrupts intermediate filaments and promotes apoptosis. Cell Death Differ 8, 443–450 [DOI] [PubMed] [Google Scholar]
- 73. Adam-Klages S., Schwandner R., Lüschen S., Ussat S., Kreder D., Krönke M. (1998) Caspase-mediated inhibition of human cytosolic phospholipase A2 during apoptosis. J. Immunol. 161, 5687–5694 [PubMed] [Google Scholar]
- 74. Lüschen S., Ussat S., Krönke M., Adam-Klages S. (1998) Cleavage of human cytosolic phospholipase A2 by caspase-1 (ICE) and caspase-8 (FLICE). Biochem. Biophys. Res. Commun. 253, 92–98 [DOI] [PubMed] [Google Scholar]
- 75. Thiede B., Dimmler C., Siejak F., Rudel T. (2001) Predominant identification of RNA-binding proteins in Fas-induced apoptosis by proteome analysis. J. Biol. Chem. 276, 26044–26050 [DOI] [PubMed] [Google Scholar]
- 76. Ju W., Valencia C. A., Pang H., Ke Y., Gao W., Dong B., Liu R. (2007) Proteome-wide identification of family member-specific natural substrate repertoire of caspases. Proc. Natl. Acad. Sci. U.S.A. 104, 14294–14299 [DOI] [PMC free article] [PubMed] [Google Scholar]