Background: The mechanisms that cause castration-resistant prostate cancer remain unknown.
Results: Using high throughput proteomics and subsequent clinical validation, we identified Protein S as being elevated in high grade/advanced prostate cancer.
Conclusion: Protein S is elevated in aggressive prostate cancer.
Significance: Protein S expression could serve as a biomarker of aggressive prostate cancer.
Keywords: Bioinformatics, Cell Culture, Immunohistochemistry, Prostate Cancer, Proteomics, Androgen Independence, PROS1, Protein S, Castration-resistant Prostate Cancer, Androgen-independent Prostate Cancer
Abstract
Androgen deprivation constitutes the principal therapy for advanced and metastatic prostate cancers. However, this therapeutic intervention usually results in the transition to a more aggressive androgen-independent prostate cancer. The elucidation of molecular alterations during the progression to androgen independence is an integral step toward discovering more effective targeted therapies. With respect to identifying crucial mediators of this transition, we compared the proteomes of androgen-independent (PC3, DU145, PPC1, LNCaP-SF, and 22Rv1) and androgen-dependent (LNCaP and VCaP) and/or normal prostate epithelial (RWPE) cell lines using mass spectrometry. We identified more than 100 proteins that were differentially secreted in the androgen-independent cell lines. Of these, Protein S (PROS1) was elevated in the secretomes of all of the androgen-independent prostate cancer cell lines, with no detectable secretion in normal and androgen-dependent cell lines. Using quantitative PCR, we observed significantly higher (p < 0.05) tissue expression levels of PROS1 in prostate cancer samples, further indicating its importance in prostate cancer progression. Similarly, immunohistochemistry analysis revealed elevation of PROS1 in high grade prostate cancer (Gleason grade ≥8), and further elevation in castration-resistant metastatic prostate cancer lesions. We also observed its significant (p < 0.05) elevation in high grade prostate cancer seminal plasma samples. Taken together, these results show that PROS1 is elevated in high grade and castration-resistant prostate cancer and could serve as a potential biomarker of aggressive disease.
Introduction
Prostate cancer is the most commonly diagnosed solid organ tumor and the second leading cause of death due to cancer among men in North America (1). It is observed as either a slow growing indolent tumor or as a more advanced aggressive form; however, the current screening methods are not able to differentiate between these two forms. Prostate-specific antigen is the most widely used biomarker for prostate cancer. However, due to a documented lack of specificity, it may also be elevated in other non-cancerous pathologies, including benign prostate hyperplasia and prostatitis (2, 3). This inevitably leads to overdiagnosis and, ultimately, to overtreatment.
Current treatments depend on whether the tumor is localized or has metastasized. Localized prostate cancers are usually treated with either targeted radiation or radical prostatectomy. For more advanced and metastatic cancers, the mainstay treatment is androgen deprivation therapy, which is initially very effective at reducing tumor volume and growth (4, 5). However, persisting androgen deprivation often results in the selection and accumulation of cancerous cells that have acquired resistance, leading to the development of androgen-independent or castration-resistant prostate cancer (4, 5). This genotypic and phenotypic alteration is accompanied by increased mortality rates because there are no alternative targeted therapies. Thus, the understanding of molecular alterations during the progression to androgen independence becomes of utmost importance for catalyzing the development of targeted and efficient therapeutic options.
The androgen receptor (AR)3 signaling cascade has been extensively studied with respect to the progression of androgen-independent prostate cancer. Overall, AR signaling resistance has been attributed to AR gene amplifications, AR gene mutations, changes in co-regulators or steroidogenic enzymes, or alternative proteins via outlaw pathways (4–10). However, recent interest has shifted toward the identification of novel “bypass” pathways, the so-called AR-independent pathways, which can play a role in this process as well (11, 12).
High throughput proteomics is a growing field that can virtually be used to study the proteomic signature of any biological material (13). Numerous studies have been conducted using proteomics to identify biomarkers for prostate cancer using cell lines, tissues, and biological fluids, including expressed prostatic secretions (14–21).
Previously, we performed proteomic analysis of the conditioned media of three prostate cancer cell lines (LNCaP, PC3, and 22Rv1) and identified over 1000 proteins that could serve as potential biomarkers for prostate cancer diagnosis (16). In the current study, we performed proteomic analysis of the conditioned media (secretome analysis) of five androgen independent cell lines (PC3, DU145, PPC1, LNCaP-SF, and 22Rv1), two androgen-dependent cell lines (LNCaP and VCaP), and one normal prostate epithelial cell line (RWPE). In total, we identified over 3000 proteins, with over 100 proteins being differentially secreted between the AIPC and non-AIPC cell lines. Of these, Protein S (PROS1) was elevated in all five AIPC cell lines, with no observed secretion in the normal and androgen-dependent prostate cancer cell lines. Subsequently, we observed PROS1 overexpression in high grade prostate cancer tissue and seminal plasma. In addition, PROS1 expression was highly elevated in castration-resistant metastatic prostate cancer specimens. These results demonstrate that PROS1 could serve as a potential biomarker for high grade prostate cancers as well as providing new areas for the therapeutic intervention for the treatment of AIPC patients.
MATERIALS AND METHODS
Cells and Reagents
The human prostate cancer cell lines PC3, DU145, LNCaP, 22Rv1, and VCaP and the normal prostate epithelial cell line RWPE-1 were purchased from the American Type Culture Collection (ATCC, Manassas, VA). The LNCaP-SF cells were kindly provided by Dr. Atsushi Mizokami, and the PPC-1 cell line was provided by Dr. Aaron Schimmer. Cell culture media specified by the ATCC for each of the cell lines were used as follows: Dulbecco's modified Eagle's medium (DMEM) (ATCC) with 10% fetal bovine serum (Thermo Scientific) was used for PC3, DU145, and VCaP; Roswell Park Memorial Institute (RPMI) (ATCC) with 10% FBS was used for 22RV1, PPC-1, and LNCaP cells. The normal RWPE cell line was grown in keratinocyte serum-free media (ATCC) supplemented with bovine pituitary extract and recombinant epidermal growth factor. The LNCaP-SF cells were grown in DMEM supplemented with 10% charcoal-stripped fetal bovine serum (Invitrogen). All cells were maintained at 37 °C with 5% CO2 in a humidified incubator. All experiments were performed within the first five passages from the initiation of all cultures.
Cell Culture
Cells were cultured in six T-175 flasks and allowed to grow in 30 ml of their respective media until they reached 70% confluence. Generally, cells were allowed to grow for 48 h for them to achieve this confluence. The medium was then removed, and cells were washed three times with 20 ml of PBS (Invitrogen). Following the washes, 30 ml of Chinese hamster ovary serum-free medium (Invitrogen) were added to each of the flasks, which were then cultured for 2 days. After further growth, the conditioned medium was collected and centrifuged at 2000 × g for 5 min to remove cellular debris. The resulting supernatant was transferred to a fresh tube, and total protein was measured using a Coomassie Blue (Bradford) total protein assay. Each flask contained roughly 500 μg of total protein, of which two were combined to generate a total of 1 mg. Because we initially grew six T-175 flasks, we were able to combine them in three replicates containing a total protein level of roughly 1 mg. The triplicates were then further processed following the sample preparation protocol below.
Sample Preparation
Each sample containing 1 mg of total protein was initially dialyzed using a 3.5 kDa molecular mass cut-off membrane (Spectrum Laboratories, Inc., Compton, CA) in 5 liters of 1 mm ammonium bicarbonate buffer solution at 4 °C overnight. The buffer solution was changed twice, and the sample was then frozen at −80 °C and lyophilized to dryness using a Modulyo freeze dryer (Thermo Electron Corp.). The resulting dry protein product was denatured in 8 m urea and reduced with 200 mm dithiothreitol at 50 °C for 30 min. Samples were then alkylated in 500 mm iodoacetamide while shaking in the dark for 1 h. Each replicate was then further desalted using a NAP5 column (GE Healthcare), frozen, and lyophilized again to complete dryness. Finally, the samples were then digested with trypsin (Promega; sequencing-grade modified porcine trypsin) at 37 °C overnight using a 1:50 trypsin/protein concentration ratio. The samples, now containing tryptic peptides, were acidified with formic acid before strong cation exchange (SCX).
SCX on a High Pressure Liquid Chromatography (HPLC) System
To reduce sample complexity, the samples were subjected to SCX using an Agilent 1100 HPLC system. The HPLC system was connected to a PolySULPHOETHYL ATM column with a 200-Å pore size and a diameter of 5 μm (The Nest Group Inc., Southborough, MA). A total of 12 fractions per replicate containing the greatest number of peptides based on peak intensity were eluted and collected to perform mass spectrometric analysis. The HPLC fractions were C18-extracted using ZipTipC18 micropipette tips (Millipore, Billerica, MA) and eluted in 5 μl of Buffer B (90% acetonitrile, 0.1% formic acid, 10% water, and 0.02% trifluoroacetic acid). An additional 80 μl of Buffer A (95% water, 0.1% formic acid, 5% acetonitrile, and 0.02% trifluoroacetic acid) were added to each zipped fraction.
Mass Spectrometry (LC-MS/MS)
40 μl of each fraction were injected into an autosampler on the EASY-nLC system (Proxeon Biosystems, Odense, Denmark). Peptides were first collected onto a 3-cm C18 column (inner diameter of 200 μm) and were then eluted onto a resolving 5-cm analytic C18 column (inner diameter of 75 μm) with an 8-μm tip (New Objective). This HPLC system was coupled online to an LTQ-Orbitrap XL (Thermo Fisher Scientific) mass spectrometer, using a nano-electrospray ionization source (Proxeon Biosystems) in data-dependent mode. The fractions were run on a 55-min gradient, and eluted peptides were subjected to one full MS1 scan (450–1450 m/z) in the Orbitrap at 60,000 resolution, followed by six data-dependent MS2 scans in the linear ion trap. The following parameters were enabled: monoisotopic precursor selection, charge state screening, and dynamic exclusion. In addition, charge states of +1 and >4 and unassigned charge states were not subjected to MS2 fragmentation.
Protein Identification and Data Analysis
RAW files were uploaded onto Mascot Daemon (version 2.2), and extract_msn was used to generate DAT and Mascot generic files. Mascot generic files were further searched on GPM (Global Proteome Machine Manager, version 2006.06.01) against the concatenated nonredundant IPI.Human version 3.62 database with parent and fragment tolerances of 7 ppm to generate X!Tandem files. The DAT and X!Tandem files were then merged and searched with Scaffold (Proteome Software Inc., Portland, OR; version 2.0) to generate a file containing protein lists. The final Scaffold files contained normalized spectral counts for each of the cell lines, which were subsequently used as a semiquantitative measure to compare protein secretion among each cell line.
ProteinCenter (Thermo Fisher Scientific) was used to retrieve Gene Ontology annotations and pathway analyses from the Kyoto Encyclopedia of Genes and Genomes. To visualize and assess networks of overexpressed and underexpressed candidates, Ingenuity Pathway Analysis (Ingenuity Systems, Redwood City, CA) software was used. Using this software, pathway analysis was performed, obtaining information on canonical pathways and molecular networks that have been altered, which were determined by Fisher's exact test.
RNA Extraction and Quantitative PCR
Total RNA was isolated from cells using an RNeasy kit (Qiagen). cDNA was generated from 1 μg of total RNA using the Superscript II cDNA synthesis kit (Invitrogen). LuCaP 96 and LuCaP 96AI xenograft tissues were frozen in liquid nitrogen and then ground into a fine powder using a mortar and pestle. RNA was extracted from the resulting powdered tissue using an RNeasy kit (Qiagen), and the final RNA product was used for further cDNA synthesis.
Quantitative PCR was conducted using 1× SYBR reagent (Applied Biosystems), and transcript levels of PROS1 TWSG1, LTBP1, GBA, PAM, and GAS6 were measured on a 7500 ABI system. For clinical validation, the TissueScan Prostate Cancer cDNA Array II was used (Origene). Quantitative PCR was conducted on these samples using the same SYBR Green reagent as mentioned above. The following primer sequences were used: PROS1, GGCTCCTACTATCCTGGTTCTG (forward) and CAAGGCAAGCATAACACCAGTGC (reverse); GAS6, CCTTCCATGAGAAGGACCTCGT (forward) and GAAGCACTGCATCCTCGTGTTC (reverse); TBP, TGTATCCACAGTGAATCTTGGTTG (forward) and GGTTCGTGGCTCTCTTATCCTC (reverse); TWSG1, CTTTGGGACGAGTGCTGTGACT (forward) and GAGAAGGGATCGGTTCATGCAG (reverse); LTBP1, TGAATGCCAGCACCGTCATCTC (forward) and CTGGCAAACACTCTTGTCCTCC (reverse); GBA, TGCTGCTCTCAACATCCTTGCC (forward) and TAGGTGCGGATGGAGAAGTCAC (reverse); PAM, TGAAGGCACCTGGGAACCAGAA (forward) and CTCTGTGGAAAATCACCAGGTTAT (reverse).
Western Blotting
Protein expression of PROS1, TWSG1, LTBP1, GBA, and PAM was assessed using Western blot analysis. Roughly, 30 μg of total protein from LuCaP 96 and LuCaP 96AI were loaded onto an SDS-polyacrylamide gel (4–15%; Bio-Rad) and transferred onto PVDF membranes (Bio-Rad). Membranes were then incubated with 5% blocking solution (2.5 g of skim milk powder in Tris buffer solution containing 0.1% Tween (TBST)) overnight at 4 °C. Membranes were incubated with rabbit polyclonal antibody against PROS1 (1:1000), TWSG1 (1:500), LTBP1 (1:100), GBA (1:1000; Sigma), or PAM (1:1000; Protein Tech) for 1 h at room temperature. The membranes were then washed six times (three 15-min washes followed by three 5-min washes) with TBST. Membranes were then incubated with goat anti-rabbit secondary antibody conjugated to alkaline phosphatase (1:3000; Jackson Laboratories) or goat anti-mouse secondary antibody conjugated to alkaline phosphatase (1:3000; Jackson Laboratories) for 1 h at room temperature. After washing with TBST, proteins were detected using the ECL detection reagent (Siemens). The expression of GAPDH (Cell Signaling Technology) was used as an internal standard.
Immunohistochemistry
Prostate cancer tissue microarrays consisting of eight normal and 40 cancer cores were purchased from US BioMax (Rockville, MD). The metastatic prostate cancer tissue microarray was developed and provided by the GU Cancer Research laboratories at the University of Washington (Seattle, WA). Human tissue microarrays of fixed paraffin-embedded metastatic tissues from 23 rapid autopsy patients who died of prostate cancer (consisting of three tissue microarray blocks with two replicate cores per metastatic site) were used for immunohistochemical analyses. All patients had castration-resistant prostate cancer at the time of autopsy, defined by the presence of a rising serum prostate-specific antigen following medical or surgical castration.
Tissue microarrays were deparaffinized in xylene and rehydrated using ethanol. Endogenous peroxidase was inactivated using hydrogen peroxide for 10 min and washed with PBS. Antigen retrieval was then performed using citrate buffer in a microwave for 10 min. Slides were then blocked for 5 min in casein and incubated overnight with the following antibodies: PROS1 (1:500; Epitomics), TWSG1 (1:50; Abgent), LTBP1 (1:200; Abgent). Rabbit IgG was used on a duplicate slide to serve as a negative control. Following 10 min of PBS washing, slides were placed in secondary antibody for 30 min using the BGX kit (Biogenex, Fremont, CA). After a 10-min wash in PBS, slides were developed with the addition of 3,3′-diaminobenzidine for 5 min. Slides were then counterstained with hematoxylin, dehydrated, and coverslipped.
Seminal Plasma and ELISA
Semen was collected from suspected prostate cancer patients prior to biopsy, allowed to liquefy at room temperature for 1 h, and centrifuged at 13,000 rpm for 15 min to separate seminal plasma from cells and cellular debris. The seminal plasma was aliquoted into 1.5-ml Eppendorf tubes and frozen at −80 °C until further use. Clinical information for the patients can be found in supplemental Table S1.
PROS1 protein levels in seminal plasma were assessed using an enzyme-linked immunosorbent assay (American Diagnostica). Briefly, seminal plasma samples were diluted 10 times, and the ELISA was performed according to the manufacturer's instructions.
Wound Repair/Scratch Assay
To assess wound repair and cell migration, LNCaP cells were grown to full confluence in 6-well plates in RPMI with 10% FBS. Upon reaching full confluence, the cells were incubated with 10 μg/ml mitomycin-C for 2 h to inhibit cell proliferation, and a standardized scratch was made down the middle of each well using a 200-μl pipette. The medium was removed, and cells were washed three times with PBS to remove any debris caused by the initial scratch. Scratch-induced cells were then either treated with 2 μg/ml human purified PROS1 (Enzyme Research Laboratories) or left untreated. Wound closure was measured by determining the distance of cells at the ends of the wound 24 and 48 h postscratching. After 48 h, cells were fixed and stained with Crystal Violet dye, and the number of migrating cells was measured by counting the number of cells found within the middle of the wound.
Statistical Analysis
All gene expression studies on cell lines and xenografts consisting of normalized expressions were compared using a two-tailed t test (GraphPad Prism Software). Gene expression studies on human prostate cancer and normal tissue were compared using a non-parametric Mann-Whitney Test (GraphPad Prism Software). Finally, χ2 tests were used to compare different groups from the immunohistochemistry data. Differences were considered significant if p values were less than 0.05. All data are expressed as means ± S.E.
RESULTS
Proteomic Profiling of Prostate Cancer Cell Line Conditioned Media
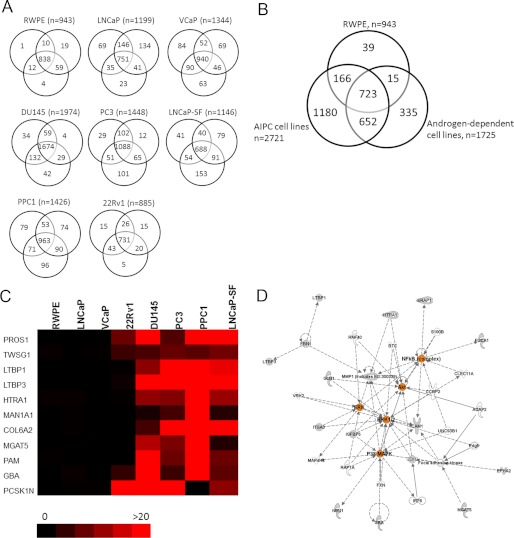
To identify modulators of androgen-independent prostate cancer that could serve as potential biomarkers of aggressive disease, we performed an in depth proteomic analysis of the conditioned media of five androgen-independent prostate cancer cell lines (DU145, PC3, LNCaP-SF, PPC-1, and 22Rv1), two androgen-dependent cell lines (LNCaP and VCaP), and one “near normal” prostate epithelial cell line (RWPE). Briefly, cells were grown in serum-free media; the conditioned media were collected, reduced, alkylated, and trypsin-digested; and peptides were subjected to two-dimensional liquid chromatography (LC), which consisted of SCX chromatography on an HPLC system, followed by reverse-phase LC, prior to tandem mass spectrometry (MS/MS). After performing the analysis, we identified, with a minimum of two peptides, 1974 proteins in DU145 cells, 1448 proteins in PC3 cells, 1146 proteins in LNCaP-SF cells, 1426 proteins in PPC-1 cells, 885 proteins in 22Rv1 cells, 1199 proteins in LNCaP cells, 1344 proteins in VCaP cells, and 943 proteins in RWPE cells, with adequate reproducibility among the triplicates (Fig. 1A). In total, we identified 3110 non-redundant proteins with at least two peptides in the conditioned media of the cell lines combined. 723 proteins (of 3110; 23.2%) were common to all cell lines. Among the androgen-independent cell lines, 1180 proteins were found to be unique, whereas 335 and 39 proteins were unique to the androgen-dependent cell lines and normal RWPE cell line, respectively (Fig. 1B). These data are summarized in Table 1, and complete lists of proteins identified within each cell line are presented in supplemental Tables S2–S9.
FIGURE 1.
Total non-redundant proteins identified and candidate selection for markers of AIPC. A, Venn diagrams displaying protein identification with a minimum of two peptides in each of the cell line conditioned media in triplicate. B, overlap of 3110 total non-redundant proteins identified in five androgen-independent cell lines (DU145, PC3, LNCaP-SF, 22Rv1, and PPC-1), two androgen-dependent cell lines (LNCaP and VCaP), and a normal prostate epithelial cell line (RWPE). C, heat map depicting the top proteins specifically found secreted in AIPC cell lines (red) but without secretion in either the androgen-dependent or normal cell lines (black). Scale represents the average spectral counts observed in triplicate experiments (full list of candidates can be found in supplemental Table S10). D, Ingenuity Pathway Analysis revealed alterations in two main networks: 1) cellular movement and 2) cellular function and development. Alterations in central nodes (depicted in orange) within ERK, AKT, NF-κB, TGFβ, and TNF signaling pathways were observed.
TABLE 1.
Total number of proteins identified in triplicate analysis of cell line conditioned medium
| RWPE | LNCaP | VCaP | 22Rv1 | PPC1 | DU145 | PC3 | LNCaP-SF | |
|---|---|---|---|---|---|---|---|---|
| Total non-redundant proteins (with ≥2 peptides) | 943 | 1199 | 1344 | 885 | 1426 | 1974 | 1448 | 1146 |
| No. of peptides identified with …a | ||||||||
| Only 2 peptides | 243 | 429 | 477 | 434 | 548 | 689 | 598 | 425 |
| Only 3 peptides | 110 | 88 | 125 | 108 | 136 | 198 | 150 | 107 |
| Only 4 peptides | 77 | 79 | 99 | 86 | 127 | 123 | 112 | 79 |
| ≥5 peptides | 513 | 603 | 643 | 227 | 615 | 964 | 588 | 535 |
a Based on the average triplicate spectral count value.
To identify gene ontology classifications, which include molecular function, biological process, and cellular components, we utilized the Protein Center database. The top cellular localization annotations of the proteins identified within each of the cell lines were cytoplasmic, membrane-bound, or nuclear. Over 67% of proteins were annotated as being either cell surface, extracellular, or membrane-bound. The majority of proteins were functionally annotated as either being protein binding or as having catalytic activity. Finally, the top biological processes of the proteins were metabolic processes, followed by regulation of biological processes and response to stimuli (supplemental Fig. 1).
Prioritization of Candidate Markers of Androgen Independence
In order to identify candidates of androgen-independent and aggressive prostate cancers, we compared the differential protein expression based on normalized spectral counts between our androgen-independent, androgen-dependent, and/or normal RWPE cell lines. Because we were interested in finding proteins that are elevated during AIPC, we decided to set as cut-offs proteins that were found with a minimum of two peptides within our androgen-independent cell line proteomes and with less than two peptides in either the androgen-dependent or normal prostate cell line proteomes. This filter resulted in the selection of 1180 proteins that were unique to at least one AIPC cell line. The top candidates included proteins that were expressed in multiple AIPC cell lines, with minimal or no spectral counts in the androgen-dependent or normal cells (Fig. 1C). To further prioritize the candidate list of proteins, a final data set of 57 proteins was derived, consisting of proteins that were secreted in at least three AIPC cell lines with a minimum of two spectral counts, and with less than one spectral count in all androgen-dependent cell lines and the normal cell line. Table 2 summarizes these candidates with respect to the observed spectral counts in each of the cell lines as well as the number of AIPC cell lines that displayed positive results. The list contains proteins that were previously studied in the context of prostate cancer progression, including MGAT5, PAM, GBA, ROBO1, CD59, MMP1, IGFBP4, CDH2, TGFB2, ICAM1, EPHA2, and IGFBP5 (22–33), thus providing further confirmation for the robustness of our quantification method.
TABLE 2.
Top candidate proteins elevated in androgen-independent cell lines based on average normalized spectral count values
Fifty-seven proteins were found with a minimum of two peptides in at least three AIPC cell lines and with less than one peptide in androgen-dependent and normal prostate epithelial cell lines. For a complete list of the 52 proteins, see supplemental Table S10.
| RWPE (normal) | LNCaP (AD)a | VCaP (AD) | 22Rv1 (AI)b | DU145 (AI) | PC3 (AI) | PPC1 (AI) | LNCaP-SF (AI) | Number of positive AIPC cell lines | Previously studied during prostate cancer progression? | |
|---|---|---|---|---|---|---|---|---|---|---|
| PROS1 | 0 | 0 | 0 | 8 | 20 | 3 | 20 | 23 | 5 | |
| TWSG1 | 1 | 0 | 0 | 5 | 6 | 8 | 7 | 9 | 5 | |
| GBA | 0 | 1 | 0 | 4 | 25 | 5 | 37 | 7 | 5 | Ref. 28 |
| PGRMC1 | 0 | 1 | 0 | 3 | 3 | 2 | 5 | 4 | 5 | |
| LTBP1 | 0 | 0 | 0 | 0 | 11 | 58 | 161 | 18 | 4 | |
| LTBP3 | 0 | 0 | 0 | 1 | 27 | 28 | 48 | 22 | 4 | |
| HTRA1 | 1 | 1 | 0 | 0 | 11 | 10 | 72 | 12 | 4 | |
| MAN1A1 | 0 | 0 | 0 | 0 | 2 | 5 | 93 | 3 | 4 | |
| COL6A2 | 0 | 0 | 0 | 0 | 5 | 38 | 23 | 30 | 4 | |
| MGAT5 | 0 | 0 | 0 | 0 | 14 | 10 | 61 | 3 | 4 | Ref. 26 |
| PAM | 0 | 0 | 0 | 0 | 32 | 8 | 34 | 8 | 4 | Ref. 27 |
| PCSK1N | 0 | 0 | 0 | 30 | 20 | 24 | 0 | 11 | 4 | |
| CHID1 | 0 | 0 | 0 | 0 | 2 | 7 | 50 | 11 | 4 | |
| ROBO1 | 0 | 1 | 0 | 2 | 0 | 9 | 25 | 10 | 4 | Ref. 29 |
| CD59 | 1 | 1 | 0 | 0 | 2 | 9 | 16 | 6 | 4 | Ref. 30 |
| B4GALT4 | 0 | 0 | 0 | 0 | 6 | 6 | 14 | 5 | 4 | |
| B3GAT3 | 0 | 0 | 0 | 3 | 7 | 1 | 10 | 3 | 4 |
a AD, androgen-dependent cell lines.
b AI, androgen-independent cell lines.
We next subjected our candidate list to preclustering pathway analysis using Ingenuity Pathway Analysis. This analysis revealed two major networks that the candidates were enriched for: 1) cellular movement and 2) cellular function and development (Fig. 1D). The candidates had central nodes in the NF-κB, AKT, ERK, p38 MAPK, TGFβ, and TNF signaling cascades, all of which have been previously documented to be associated with prostate cancer progression (34–39).
Protein S Is Elevated in Androgen-independent Prostate Cancer Cell Lines and Is Activated in the LuCaP 96AI Androgen-independent Prostate Cancer Xenograft Model
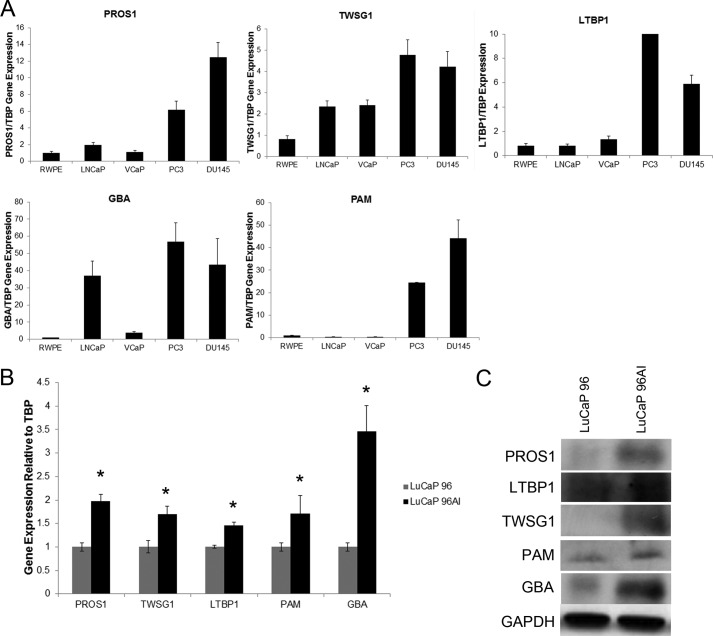
Based on our initial discovery results, the vitamin K-dependent Protein S was our most interesting candidate, because it was found to be secreted in all five androgen-independent cell lines, with no detectable secretion in any of the androgen-dependent or normal prostate epithelial cell lines (Table 2). The highest secretion was found in LNCaP-SF cells, with an average spectral count of 23, followed by 20 in DU145 cells, 20 in PPC-1 cells, 8 in 22Rv1 cells, and 3 in PC3 cells. To verify the discovery results, we performed real-time PCR of PROS1 as well as other top candidates (TWSG1, LTBP1, GBA, and PAM) on various prostate cancer cell lines to investigate if the gene expression correlated with the protein expression data. As expected, we observed significantly elevated transcript levels of each of the candidates in androgen-independent cell lines (PC3 and DU145), compared with the normal (RWPE) and androgen-dependent cells (LNCaP and VCaP) (Fig. 2A).
FIGURE 2.
Analysis of gene and protein expression levels of PROS1, TWSG1, LTBP1, GBA, and PAM on prostate cancer cell lines and LuCaP96/96AI xenografts. A, gene expression profiles of top candidates on normal (RWPE), androgen-dependent (LNCaP and VCaP), and androgen-independent (DU145 and PC3) cell lines. B and C, gene and protein expression levels of top candidates are increased in the LuCaP 96AI androgen-independent xenograft model (*, p < 0.05, Mann-Whitney test). Error bars, S.E.
To determine whether PROS1 and other top candidates may play a role during the progression of androgen-independent prostate cancer, LuCaP 96 and its androgen-independent counterpart, LuCaP 96AI, xenografts were utilized. The transcript and protein expression of PROS1, TWSG1, LTBP1, PAM, and GBA were assessed using quantitative PCR and Western blot, respectively. It was shown that all of the candidates displayed significant increases in their gene expression in the LuCaP 96AI xenograft-derived cells (p < 0.05) (Fig. 2B). Protein validation of these candidates using Western blotting supported our data, demonstrating an up-regulation of our identified enzymes in LuCaP 96AI cells (Fig. 2C). Taken together, our cell line and xenograft data clearly demonstrate that PROS1 and other top candidates are elevated at both the protein and transcript level in androgen-independent prostate cancer cells and could potentially serve as biomarkers and/or therapeutic targets for aggressive prostate cancer.
Protein S Expression Is Elevated in High Grade Prostate Cancer
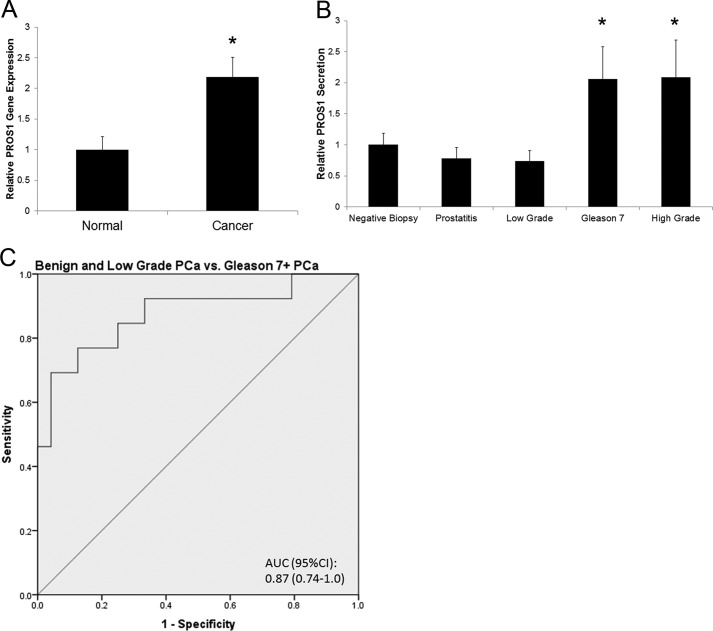
We then sought to investigate whether PROS1 was also overexpressed in prostate cancer patients. To examine this, we measured its transcript levels in normal and human tumor tissue samples. Using real-time PCR, we found that PROS1 transcript levels were significantly elevated (p = 0.017) by over 2-fold in prostate cancer compared with normal tissue (Fig. 4A).
FIGURE 4.
Expression of PROS1 in seminal plasma specimens of varying Gleason grade. A, using the TissueScan Prostate Cancer cDNA Array II consisting of eight normal and 36 prostate cancer specimens, the gene expression profiles of PROS1 showed significantly elevated mRNA expression in cancer compared with normal specimens (*, p < 0.05, Mann-Whitney test). B, ELISA analysis of PROS1 protein levels in seminal plasma from negative biopsy (n = 8) (positive prostate-specific antigen test), prostatitis (n = 8), low grade prostate cancer (Gleason ≤6) (n = 8), and intermediate and high grade prostate cancer (Gleason ≥7) (n = 13) patients (*, p < 0.05, Mann-Whitney test). C, the area under the curve (AUC) of PROS1 being able to distinguish benign (negative biopsy and prostatitis) and low grade prostate cancers from intermediate/high grade prostate cancer patients in seminal plasma was 0.875 (confidence interval (CI) = 0.744–1.0, p < 0.001).
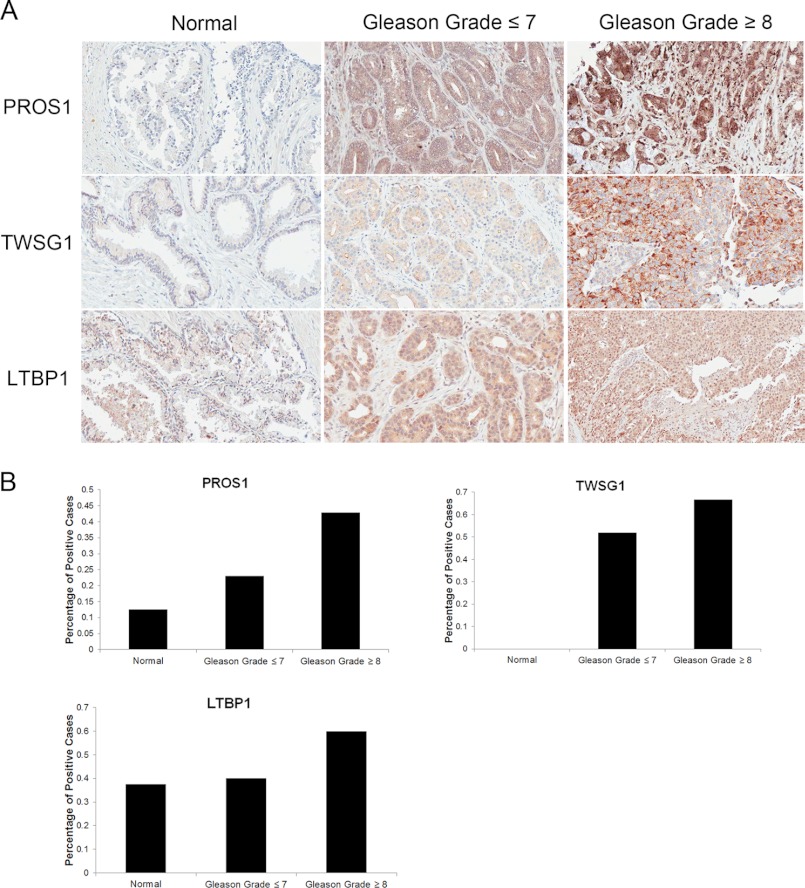
In addition, using immunohistochemistry, we assessed the protein expression level of PROS1 in normal (n = 8) and clinically localized prostate cancers of varying grade (n = 40). We devised a scoring system to assess protein expression, whereby each core was scored with a 0, 1, 2, or 3, which corresponds to no staining and low, moderate, or high staining, respectively. After analysis, we observed a stair-wise expression pattern of PROS1 staining, with minimal staining in normal cores, moderate staining in low grade prostate cancer (Gleason grade ≤7), and increased staining in high grade prostate cancer (Gleason >8) (Fig. 3A). Normal cores depicted very little positive staining, with only 13% of cores displaying a score of 2 or greater, whereas low grade prostate cancer cores had intermediate expression, staining positively in 25% of cases. Finally, high grade prostate cancer cores displayed the greatest expression levels, with 43% of cores staining positively (Fig. 3B). Similarly, we also assessed the expression of TWSG1 and LTBP1 on these samples. Both TWSG1 and LTBP1 displayed elevated expression in cases of high grade prostate cancer; however, the expression pattern was not as prominent at PROS1 (Fig. 3). These results indicate that PROS1 is overexpressed in high grade prostate cancers and may therefore play a role in the progression of prostate cancer to an advanced stage disease.
FIGURE 3.
Protein expression of PROS1, TWSG1, and LTBP1 in human prostate cancer tissues. A, representative immunohistochemistry images of cases of normal, low grade (Gleason ≤7), and high grade (Gleason ≥8) prostate cancer specimens, under light microscopy (×20). B, immunohistochemical staining was quantified using a scoring scale of 0, 1, 2, and 3 corresponding to no staining and low, moderate, and high staining, respectively, as blindly determined by a pathologist. Positive cores were determined to be ones that stained with an intensity of 2 or greater.
Protein S Is Elevated in the Seminal Plasma of Intermediate and High Grade Prostate Cancer Patients
After observing increased gene and protein expression of PROS1 in high grade prostate cancer tissue specimens, we wanted to determine if elevated PROS1 expression could also be reflected with elevated seminal plasma secretion. We chose to analyze seminal plasma because it represents the proximal fluid in which prostatic secretions should be enriched for. We assessed PROS1 levels using ELISA in a variety of seminal plasmas taken from control (n = 8), prostatitis (n = 8), low grade (Gleason ≤6, n = 8), and intermediate/high grade (Gleason ≥7, n = 13) prostate cancer patients. Clinical information of the seminal plasma samples can be found in supplemental Table S1. Based on our analysis, we observed a statistically significant (p < 0.05) elevation of PROS1 in seminal plasma from intermediate and high grade prostate cancer patients (Fig. 4B). The area under the curve of PROS1 being able to distinguish benign (negative biopsy and prostatitis) and low grade prostate cancers from intermediate/high grade prostate cancer patients in seminal plasma was 0.875 (confidence interval = 0.744–1.0, p < 0.001) (Fig. 4C). These results suggest a potential role of PROS1 as a biomarker, to assist in the differential diagnosis of high grade from low grade prostate cancer and/or benign conditions.
PROS1-stimulated Prostate Cancer Cells Have Increased Migratory Potential
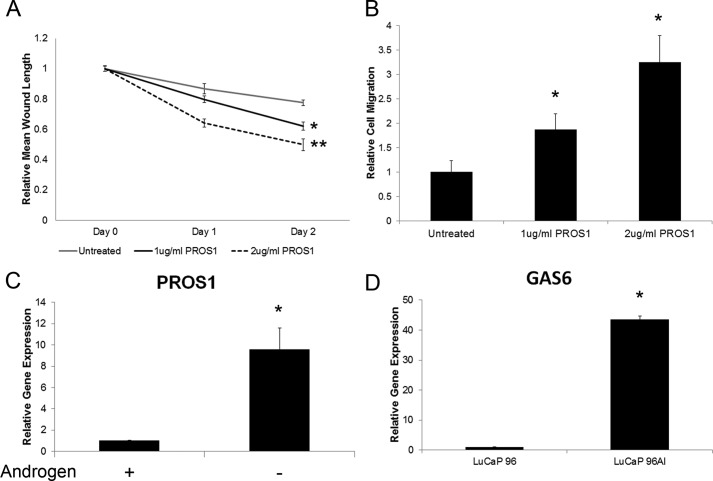
To explore whether PROS1 has a role in prostate cancer growth or migration, we performed in vitro scratch assays, in which LNCaP cells were grown to full confluence, treated with mitomycin-C for 2 h, and scratched to induce wounding. Cells were then either treated with 1–2 μg/ml of human purified PROS1 or left untreated to serve as controls, and the amount of wound closure as well as the number of migrating cells was assessed. We observed that 24 and 48 h postscratch, there was a statistically significant (p < 0.05) increase in wound closure in PROS1-treated LNCaP cells (Fig. 5A). Specifically, over 40% of the original wound was healed in PROS1-treated cells compared with less than 20% wound closure in non-treated control cells (Fig. 5B). In addition, to assess cell migration during PROS1 stimulation, we fixed and stained cells after 24 and 48 h of inducing the wound and counted cells that were found within default squares within the original wound gap. After analysis, we observed a significant increase (p < 0.05) in the number of migrating cells during PROS1 stimulation (Fig. 5B). Taken together, these data are indicative that PROS1 has a direct role on prostate cancer cellular processes, including growth and migration.
FIGURE 5.
LNCaP cells were treated with human purified PROS1, and a wound repair scratch assay was performed. PROS1-treated cells (1 or 2 μg/ml) had significantly increased wound repair (A) and an increase in migrating cells (B) compared with untreated cells 24 and 48 h postscratch (*, p < 0.05, Mann-Whitney test). C, LNCaP cells were grown in androgen-depleted conditions for a 5-day period. PROS1 gene expression levels increased the longer the cells were grown in androgen-depleted conditions (*, p < 0.05, Mann-Whitney test). A similar expression profile was observed with PC3 cells (data not shown). D, PROS1 (Fig. 2B) and its homologue GAS6 transcript levels were increased in the LuCaP 96AI androgen-independent xenograft model (*, p < 0.05, Mann-Whitney test). Error bars, S.E.
PROS1 Transcript Levels Increase after Continuous Growth in Androgen-deprived Conditions
During androgen deprivation, prostate cancer cells undergo apoptosis due to the absence of key growth stimuli, in particular androgens. To assess the effect of androgen deprivation on PROS1 expression, we grew LNCaP cells in androgen-deprived conditions for varying times, including 1, 2, and 5 days, and extracted total RNA to examine the expression levels of PROS1. Interestingly, PROS1 gene expression levels were increased in a time-dependent manner (Fig. 5C). We also observed an increase in PROS1 transcript level in PC3 androgen-deprived cells (data not shown).
Previously, we assessed PROS1 expression in the LuCaP 96/LuCaP 96AI xenograft model and observed elevated expression in the androgen-independent xenograft. We measured transcript levels of GAS6, a known homologue of PROS1, which has previously been linked to prostate cancer progression (45, 46), on these xenografts and found its elevation in LuCaP 96AI cells (p < 0.05). Taken together, these results further support the involvement of PROS1 and GAS6 in the development of AIPC.
PROS1 Is Highly Expressed in Cases of Castration-resistant Metastatic Prostate Cancers
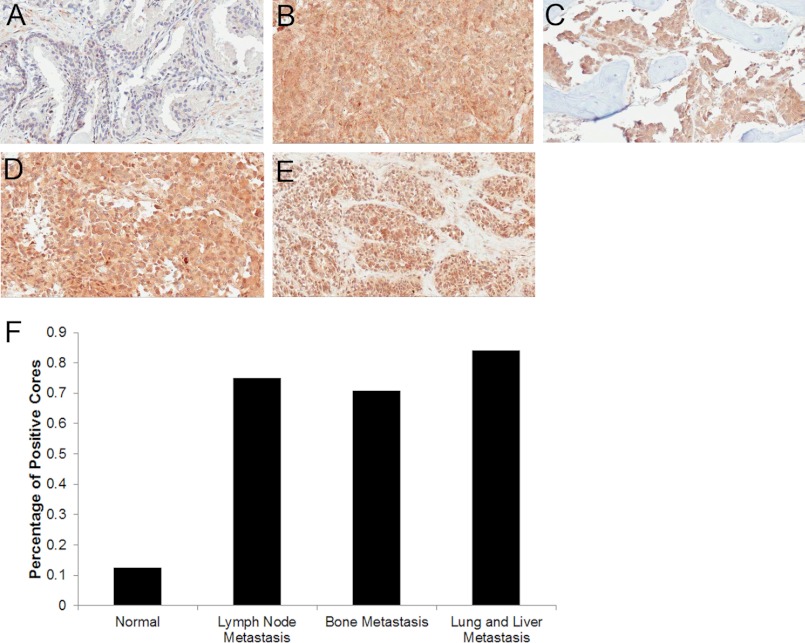
Thus far, we have demonstrated that PROS1 is elevated 1) in vitro, in androgen-independent cell lines; 2) in localized high grade disease, both at the tissue and seminal plasma level; and 3) in an androgen-independent xenograft model. Based on these findings, we aimed to examine PROS1 expression in various castration-resistant metastatic prostate cancer human samples to identify whether it is dysregulated during prostate cancer metastasis as well. Using our previous scoring system in a tissue microarray, containing castration-resistant metastatic lesions to the bone (n = 72), lymph nodes (n = 28), and lung and liver (n = 19), we demonstrated substantially increased PROS1 staining in all metastatic sites (Fig. 6). Specifically, we observed 15.8% of lung and liver metastases containing no staining or low staining and 84.2% of cores having intense staining. With respect to the lymph node metastasis cores, we found 25% with low staining and 75% with intense staining. Within bone metastatic lesions, 29% of cores displayed low staining, whereas 71% had intense staining. In contrast, in normal prostate cores, PROS1 staining was very low because the majority of the samples (87.5%) had little or no staining, and a small proportion (12.5%) had moderate PROS1 expression. Following statistical analysis, PROS1 expression was found significantly up-regulated in lung and liver (p > 0.001), lymph node (p = 0.002), and bone (p = 0.002) prostate cancer metastatic lesions compared with normal prostate samples (Table 3). In conclusion, PROS1 expression is up-regulated in castration-resistant prostate cancer metastases and, thus, could serve as a potential biomarker and therapeutic target for aggressive disease.
FIGURE 6.
Expression of PROS1 in castration-resistant metastatic prostate cancer to the bone, lymph node, liver, and lungs. Representative immunohistochemistry images of PROS1 staining in normal prostate (A), liver metastasis (B), bone metastasis (C), lymph node metastasis (D), and lung metastasis (E) of the prostate are shown. Images were taken under light microscopy (×20). F, immunohistochemical staining was quantified using a scoring scale of 0, 1, 2, and 3, corresponding to no staining and low, moderate, and high staining, respectively, as blindly determined by a pathologist. Positive cores were determined to be ones that stained with an intensity of 2 or greater.
TABLE 3.
PROS1 expression in castration-resistant metastatic prostate cancers
| Tissue type | Number of positive coresa | Staining percentage | p value compared with normalb |
|---|---|---|---|
| % | |||
| Normal prostate | 1/8 | 12.5 | NAc |
| Lung and liver metastasis | 16/19 | 84.2 | 0.0009 |
| Lymph node metastasis | 21/28 | 75.0 | 0.0026 |
| Bone metastasis | 51/72 | 70.8 | 0.0022 |
a Positive staining means a score of 2 or higher, based on the pathologist's score (see Fig. 3 legend).
b p value was calculated using a χ2 test.
c Not applicable.
DISCUSSION
In the present study, we aimed to delineate the proteomes of several prostate cancer and a near normal prostate cell line conditioned media to identify proteins that are elevated during androgen-independent prostate cancer. Specifically, by using LC-MS/MS, we performed proteomic analysis on five androgen-independent (DU145, PC3, 22Rv1, PPC1, and LNCaP-SF), two androgen-dependent (LNCaP and VCaP), and one near normal (RWPE) prostate epithelial cell line. After performing experiments in triplicates and using various protein identification search engines (X!Tandem, Mascot, and Scaffold), we were able to identify between 885 and 1974 proteins with at least two peptides in each of the cell line conditioned media. In total, we identified a 3110-non-redundant protein data set, which, to our knowledge, is the most comprehensive one to date. To internally validate our approach, we observed among our top candidates, 12 proteins (MGAT5, PAM, GBA, ROBO1, CD59, MMP1, IGFBP4, CDH2, TGFB2, ICAM1, EPHA2, and IGFBP5) that have previously been studied or implicated in prostate cancer progression (22–33). For example, MGAT5 has been shown to mediate enhanced invasion and metastatic potential for prostate cancer cells through many in vitro invasion assays and xenograft animal models.
After analyzing our candidates, we decided to further investigate the anticoagulation factor PROS1, because it was found secreted in only the androgen-independent cell lines, with no detectable secretions in androgen-dependent or normal prostate epithelial cell lines. After performing clinical validation on a variety of tissue and seminal plasma samples, we observed elevation of PROS1 specifically in high grade cases, in addition to its elevation in castration-resistant metastatic prostate cancer cases, which supported our initial goal of identifying markers of aggressive prostate cancer.
To our knowledge, this is the first study documenting a role for PROS1 with respect to prostate cancer pathogenesis. In fact, after performing multiple searches, the only other study that we could find assessing PROS1 in any form of cancer was a study evaluating various coagulation factor expressions during colorectal cancer development. In the aforementioned studies, PROS1 staining in colorectal cancer cells was performed; however, the investigators did not go into further detail aside from their immunohistochemistry analysis (40). Although PROS1 has been highly studied with respect to the coagulation cascade, recent studies have shown that PROS1 could act as a ligand for a family of receptor tyrosine kinases, consisting of Tyro3, Axl, and Mer (TAM receptors) (41–44). Interestingly, GAS6, which shares 40% amino acid identity with PROS1, is a known ligand for these TAM receptors (41). We also observed GAS6 secretion in two AIPC cell lines (DU145 and LNCaP-SF), with no detectable secretions in either the androgen-dependent or normal prostate cell lines. Previously, GAS6 has been shown to have increased affinity for the Axl protein (41). In regard to prostate cancer pathology, Sainaghi et al. (45) demonstrated that Axl could be activated by GAS6 in DU145 and PC3 cells, resulting in the phosphorylation of the MEK protein, leading to increased cell survival and proliferation. Shiozawa et al. (46) went a step further and observed that upon GAS6/Axl stimulation, prostate cancer cells had increased metastatic and invasive properties, particularly to bone. Both of these studies demonstrate that TAM receptors, specifically Axl, and their corresponding ligands (GAS6) are able to promote prostate cancer tumorogenesis. The question of whether PROS1 is able to act in a similar way has yet to be elucidated. However, from our analysis, we also observed increased cell migration upon stimulation with PROS1 as well as its elevated expression in high grade and metastatic disease, further providing evidence that it may be providing a survival advantage for prostate cancer cells. However, the mechanistic role of PROS1 and its potential downstream signaling cascade still requires further exploration.
PROS1 shares 40% amino acid identity with GAS6, and both proteins have identical structural domains, including a γ-glutamic acid domain, which is integral for vitamin K binding, four epidermal growth factor (EGF)-like modules, and two tandem laminin G domains that are structurally related to those of the sex hormone binding globulin (41). PROS1, but not GAS6, contains a unique thrombin cleavage domain, which is important for its functions within the coagulation cascade (41). Interestingly, two recent studies conducted on mouse neuronal cells showed that upon chemically induced cell injury, PROS1 was able to activate AKT and induce an anti-apoptotic cascade, resulting in reduced cell death (47, 48). The response was specific to PROS1 binding to the TYRO3 receptor and not Axl or Mer. These results marked a novel role for PROS1 outside of the coagulation cascade as a signaling molecule. Also, it was observed that the laminin G domains in particular were integral for the binding of PROS1 and subsequent activation of its downstream signaling pathway (47, 48).
The novel role of PROS1 as a signaling molecule in the neuronal mouse model provides an interesting explanation as to why it may possibly become activated in high grade and aggressive prostate cancer. During aggressive prostate cancer, androgen deprivation is usually the gold standard therapy. Many cells undergo apoptosis during this treatment; however, some are able to evade the therapy and continue growing in the absence of androgens. A possible hypothesis for our observed increase in PROS1 expression in aggressive prostate cancer could be due to its potential involvement in activating downstream anti-apoptotic pathways, which in turn provide a survival advantage for cancer cells and promote their progression to AIPC. Further experimentation needs to be conducted to determine more precisely the functional role of PROS1 and its potential downstream signaling in aggressive disease. In addition, PROS1 as a therapeutic target becomes of interest because the development of potential molecules that can possibly inhibit PROS1 signaling function without altering its coagulation properties presents an interesting avenue of therapeutic intervention.
Overall, in this study, we demonstrate that PROS1 is a novel marker of high grade and castration-resistant metastatic prostate cancer, which warrants further functional validation through the use of relevant in vitro and in vivo models. Our present findings provide sufficient evidence that PROS1 plays an important role in prostate cancer pathogenesis and suggest an interesting area for therapeutic intervention for a disease that lacks targeted treatments.
Acknowledgments
We acknowledge the support of the University Health Network Pathology Research Program for performing the immunohistochemistry experiments. We thank Apostolos Dimitromanolakis for assistance in analyzing the proteomics data and with statistical analysis. We also thank the patients and their families who were willing to participate in the Prostate Cancer Donor Program, for without them research of this nature would not be possible. We also acknowledge Lawrence True, Eva Corey, Celestia Higano, and Robert Vessella and the rapid autopsy teams in the Urology Department at the University of Washington. The donated material is the result of work supported by resources from the Pacific Northwest Prostate Cancer SPORE (P50CA97186), PO1 NIH, National Institutes of Health, Grant PO1CA085859, and the Richard M. Lucas Foundation.

This article contains supplemental Tables S1–S10 and Fig. 1.
- AR
- androgen receptor
- AIPC
- androgen-independent prostate cancer
- SCX
- strong cation exchange chromatography.
REFERENCES
- 1. Grönberg H. (2003) Prostate cancer epidemiology. Lancet 361, 859–864 [DOI] [PubMed] [Google Scholar]
- 2. Diamandis E. P. (1998) Prostate-specific antigen. Its usefulness in clinical medicine. Trends Endocrinol. Metab. 9, 310–316 [DOI] [PubMed] [Google Scholar]
- 3. Sardana G., Dowell B., Diamandis E. P. (2008) Emerging biomarkers for the diagnosis and prognosis of prostate cancer. Clin. Chem. 54, 1951–1960 [DOI] [PubMed] [Google Scholar]
- 4. Denmeade S. R., Isaacs J. T. (2002) A history of prostate cancer treatment. Nat. Rev. Cancer 2, 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Saraon P., Jarvi K., Diamandis E. P. (2011) Molecular alterations during progression of prostate cancer to androgen independence. Clin. Chem. 57, 1366–1375 [DOI] [PubMed] [Google Scholar]
- 6. Chodak G. W., Kranc D. M., Puy L. A., Takeda H., Johnson K., Chang C. (1992) Nuclear localization of androgen receptor in heterogeneous samples of normal, hyperplastic and neoplastic human prostate. J. Urol. 147, 798–803 [DOI] [PubMed] [Google Scholar]
- 7. Crawford E. D., Petrylak D. (2010) Castration-resistant prostate cancer. Descriptive yet pejorative? J. Clin. Oncol. 28, e408. [DOI] [PubMed] [Google Scholar]
- 8. Debes J. D., Tindall D. J. (2004) Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 351, 1488–1490 [DOI] [PubMed] [Google Scholar]
- 9. Ruizeveld de Winter J. A., Trapman J., Vermey M., Mulder E., Zegers N. D., van der Kwast T. H. (1991) Androgen receptor expression in human tissues. An immunohistochemical study. J. Histochem. Cytochem. 39, 927–936 [DOI] [PubMed] [Google Scholar]
- 10. Sadi M. V., Walsh P. C., Barrack E. R. (1991) Immunohistochemical study of androgen receptors in metastatic prostate cancer. Comparison of receptor content and response to hormonal therapy. Cancer 67, 3057–3064 [DOI] [PubMed] [Google Scholar]
- 11. Edwards J., Bartlett J. M. (2005) The androgen receptor and signal transduction pathways in hormone-refractory prostate cancer. Part 1. Modifications to the androgen receptor. BJU Int. 95, 1320–1326 [DOI] [PubMed] [Google Scholar]
- 12. Edwards J., Bartlett J. M. (2005) The androgen receptor and signal transduction pathways in hormone-refractory prostate cancer. Part 2. Androgen receptor cofactors and bypass pathways. BJU Int. 95, 1327–1335 [DOI] [PubMed] [Google Scholar]
- 13. Karagiannis G. S., Pavlou M. P., Diamandis E. P. (2010) Cancer secretomics reveal pathophysiological pathways in cancer molecular oncology. Mol Oncol 4, 496–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alaiya A. A., Al-Mohanna M., Aslam M., Shinwari Z., Al-Mansouri L., Al-Rodayan M., Al-Eid M., Ahmad I., Hanash K., Tulbah A., Bin Mahfooz A., Adra C. (2011) Proteomics-based signature for human benign prostate hyperplasia and prostate adenocarcinoma. Int. J. Oncol. 38, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 15. Khan A. P., Poisson L. M., Bhat V. B., Fermin D., Zhao R., Kalyana-Sundaram S., Michailidis G., Nesvizhskii A. I., Omenn G. S., Chinnaiyan A. M., Sreekumar A. (2010) Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics 9, 298–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sardana G., Jung K., Stephan C., Diamandis E. P. (2008) Proteomic analysis of conditioned media from the PC3, LNCaP, and 22Rv1 prostate cancer cell lines. Discovery and validation of candidate prostate cancer biomarkers. J. Proteome Res. 7, 3329–3338 [DOI] [PubMed] [Google Scholar]
- 17. Skvortsov S., Schäfer G., Stasyk T., Fuchsberger C., Bonn G. K., Bartsch G., Klocker H., Huber L. A. (2011) Proteomics profiling of microdissected low and high grade prostate tumors identifies Lamin A as a discriminatory biomarker. J. Proteome Res. 10, 259–268 [DOI] [PubMed] [Google Scholar]
- 18. Tyson D. R., Ornstein D. K. (2008) Proteomics of cancer of hormone-dependent tissues. Adv. Exp. Med. Biol. 630, 133–147 [DOI] [PubMed] [Google Scholar]
- 19. Vellaichamy A., Dezso Z., JeBailey L., Chinnaiyan A. M., Sreekumar A., Nesvizhskii A. I., Omenn G. S., Bugrim A. (2010) “Topological significance” analysis of gene expression and proteomic profiles from prostate cancer cells reveals key mechanisms of androgen response. PLoS One 5, e10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zheng Y., Xu Y., Ye B., Lei J., Weinstein M. H., O'Leary M. P., Richie J. P., Mok S. C., Liu B. C. (2003) Prostate carcinoma tissue proteomics for biomarker discovery. Cancer 98, 2576–2582 [DOI] [PubMed] [Google Scholar]
- 21. Drake R. R., Elschenbroich S., Lopez-Perez O., Kim Y., Ignatchenko V., Ignatchenko A., Nyalwidhe J. O., Basu G., Wilkins C. E., Gjurich B., Lance R. S., Semmes O. J., Medin J. A., Kislinger T. (2010) In depth proteomic analyses of direct expressed prostatic secretions. J. Proteome Res. 9, 2109–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tsui K. H., Chang P. L., Feng T. H., Chung L. C., Sung H. C., Juang H. H. (2008) Evaluating the function of matriptase and N-acetylglucosaminyltransferase V in prostate cancer metastasis. Anticancer Res. 28, 1993–1999 [PubMed] [Google Scholar]
- 23. Trendel J. A., Ellis N., Sarver J. G., Klis W. A., Dhananjeyan M., Bykowski C. A., Reese M. D., Erhardt P. W. (2008) Catalytically active peptidylglycine α-amidating monooxygenase in the media of androgen-independent prostate cancer cell lines. J. Biomol. Screen. 13, 804–809 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Kim K. H., Zhang W., Guo Y., Kim S. H., Lü J. (2012) Galbanic acid decreases androgen receptor abundance and signaling and induces G1 arrest in prostate cancer cells. Int. J. Cancer 130, 200–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Latil A., Chêne L., Cochant-Priollet B., Mangin P., Fournier G., Berthon P., Cussenot O. (2003) Quantification of expression of netrins, slits and their receptors in human prostate tumors. Int J Cancer 103, 306–315 [DOI] [PubMed] [Google Scholar]
- 26. Jarvis G. A., Li J., Hakulinen J., Brady K. A., Nordling S., Dahiya R., Meri S. (1997) Expression and function of the complement membrane attack complex inhibitor protectin (CD59) in human prostate cancer. Int. J. Cancer 71, 1049–1055 [DOI] [PubMed] [Google Scholar]
- 27. Cao J., Chiarelli C., Richman O., Zarrabi K., Kozarekar P., Zucker S. (2008) Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J. Biol. Chem. 283, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 28. Damon S. E., Maddison L., Ware J. L., Plymate S. R. (1998) Overexpression of an inhibitory insulin-like growth factor-binding protein (IGFBP), IGFBP-4, delays onset of prostate tumor formation. Endocrinology 139, 3456–3464 [DOI] [PubMed] [Google Scholar]
- 29. Jennbacken K., Tesan T., Wang W., Gustavsson H., Damber J. E., Welén K. (2010) N-cadherin increases after androgen deprivation and is associated with metastasis in prostate cancer. Endocr. Relat. Cancer 17, 469–479 [DOI] [PubMed] [Google Scholar]
- 30. Blanchère M., Mestayer C., Saunier E., Broshuis M., Mowszowicz I. (2001) Transforming growth factor β in the human prostate. Its role in stromal-epithelial interactions in non-cancerous cell culture. Prostate 46, 311–318 [DOI] [PubMed] [Google Scholar]
- 31. Wolff J. M., Stephenson R. N., Chisholm G. D., Habib F. K. (1995) Levels of circulating intercellular adhesion molecule-1 in patients with metastatic cancer of the prostate and benign prostatic hyperplasia. Eur. J. Cancer 31A, 339–341 [DOI] [PubMed] [Google Scholar]
- 32. Taddei M. L., Parri M., Angelucci A., Onnis B., Bianchini F., Giannoni E., Raugei G., Calorini L., Rucci N., Teti A., Bologna M., Chiarugi P. (2009) Kinase-dependent and -independent roles of EphA2 in the regulation of prostate cancer invasion and metastasis. Am. J. Pathol. 174, 1492–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miyake H., Pollak M., Gleave M. E. (2000) Castration-induced up-regulation of insulin-like growth factor-binding protein-5 potentiates insulin-like growth factor-I activity and accelerates progression to androgen independence in prostate cancer models. Cancer Res. 60, 3058–3064 [PubMed] [Google Scholar]
- 34. Suh J., Rabson A. B. (2004) NF-κB activation in human prostate cancer. Important mediator or epiphenomenon? J. Cell. Biochem. 91, 100–117 [DOI] [PubMed] [Google Scholar]
- 35. Blando J. M., Carbajal S., Abel E., Beltran L., Conti C., Fischer S., DiGiovanni J. (2011) Cooperation between Stat3 and Akt signaling leads to prostate tumor development in transgenic mice. Neoplasia 13, 254–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kinkade C. W., Castillo-Martin M., Puzio-Kuter A., Yan J., Foster T. H., Gao H., Sun Y., Ouyang X., Gerald W. L., Cordon-Cardo C., Abate-Shen C. (2008) Targeting AKT/mTOR and ERK MAPK signaling inhibits hormone-refractory prostate cancer in a preclinical mouse model. J. Clin. Invest. 118, 3051–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Recchia A. G., Musti A. M., Lanzino M., Panno M. L., Turano E., Zumpano R., Belfiore A., Andò S., Maggiolini M. (2009) A cross-talk between the androgen receptor and the epidermal growth factor receptor leads to p38MAPK-dependent activation of mTOR and cyclin D1 expression in prostate and lung cancer cells. Int. J. Biochem. Cell Biol. 41, 603–614 [DOI] [PubMed] [Google Scholar]
- 38. Zhang F., Lee J., Lu S., Pettaway C. A., Dong Z. (2005) Blockade of transforming growth factor-β signaling suppresses progression of androgen-independent human prostate cancer in nude mice. Clin. Cancer Res. 11, 4512–4520 [DOI] [PubMed] [Google Scholar]
- 39. Davis J. S., Nastiuk K. L., Krolewski J. J. (2011) TNF is necessary for castration-induced prostate regression, whereas TRAIL and FasL are dispensable. Mol. Endocrinol. 25, 611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sierko E., Wojtukiewicz M. Z., Zawadzki R., Zimnoch L., Kisiel W. (2010) Expression of protein C (PC), protein S (PS) and thrombomodulin (TM) in human colorectal cancer. Thromb. Res. 125, e71–e5 [DOI] [PubMed] [Google Scholar]
- 41. Hafizi S., Dahlbäck B. (2006) Gas6 and protein S. Vitamin K-dependent ligands for the Axl receptor tyrosine kinase subfamily. FEBS J. 273, 5231–5244 [DOI] [PubMed] [Google Scholar]
- 42. Hafizi S., Dahlbäck B. (2006) Signaling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev. 17, 295–304 [DOI] [PubMed] [Google Scholar]
- 43. Hall M. O., Obin M. S., Heeb M. J., Burgess B. L., Abrams T. A. (2005) Both protein S and Gas6 stimulate outer segment phagocytosis by cultured rat retinal pigment epithelial cells. Exp. Eye Res. 81, 581–591 [DOI] [PubMed] [Google Scholar]
- 44. Lemke G., Rothlin C. V. (2008) Immunobiology of the TAM receptors. Nat. Rev. Immunol. 8, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sainaghi P. P., Castello L., Bergamasco L., Galletti M., Bellosta P., Avanzi G. C. (2005) Gas6 induces proliferation in prostate carcinoma cell lines expressing the Axl receptor. J. Cell. Physiol. 204, 36–44 [DOI] [PubMed] [Google Scholar]
- 46. Shiozawa Y., Pedersen E. A., Patel L. R., Ziegler A. M., Havens A. M., Jung Y., Wang J., Zalucha S., Loberg R. D., Pienta K. J., Taichman R. S. (2010) GAS6/AXL axis regulates prostate cancer invasion, proliferation, and survival in the bone marrow niche. Neoplasia 12, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guo H., Barrett T. M., Zhong Z., Fernández J. A., Griffin J. H., Freeman R. S., Zlokovic B. V. (2011) Protein S blocks the extrinsic apoptotic cascade in tissue plasminogen activator/N-methyl d-aspartate-treated neurons via Tyro3-Akt-FKHRL1 signaling pathway. Mol. Neurodegener. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhong Z., Wang Y., Guo H., Sagare A., Fernández J. A., Bell R. D., Barrett T. M., Griffin J. H., Freeman R. S., Zlokovic B. V. (2010) Protein S protects neurons from excitotoxic injury by activating the TAM receptor Tyro3-phosphatidylinositol 3-kinase-Akt pathway through its sex hormone-binding globulin-like region. J. Neurosci. 30, 15521–15534 [DOI] [PMC free article] [PubMed] [Google Scholar]