Background: The memory-associated protein KIBRA regulates cell polarity and migration in non-neuronal cells and a cellular function of KIBRA in mitosis is not defined.
Results: KIBRA activates Aurora kinases and is required for precise chromosome alignment and proper spindle organization in mitosis.
Conclusion: KIBRA plays a role in mitosis.
Significance: The findings mark the importance of KIBRA in association with the mitotic machinery.
Keywords: Cellular Regulation, Mitosis, Protein Kinases, Protein Phosphorylation, Signal Transduction, Aurora Kinase, Hippo Pathway, KIBRA
Abstract
The Hippo pathway controls organ size and tumorigenesis by inhibiting cell proliferation and promoting apoptosis. KIBRA was recently identified as a novel regulator of the Hippo pathway. Several of the components of the Hippo pathway are important regulators of mitosis-related cell cycle events. We recently reported that KIBRA is phosphorylated by the mitotic kinases Aurora-A and -B. However, the role KIBRA plays in mitosis has not been established. Here, we show that KIBRA activates the Aurora kinases and is required for full activation of Aurora kinases during mitosis. KIBRA also promotes the phosphorylation of large tumor suppressor 2 (Lats2) on Ser83 by activating Aurora-A, which controls Lats2 centrosome localization. However, Aurora-A is not required for KIBRA to associate with Lats2. We also found that Lats2 inhibits the Aurora-mediated phosphorylation of KIBRA on Ser539, probably via regulating protein phosphatase 1. Consistent with playing a role in mitosis, siRNA-mediated knockdown of KIBRA causes mitotic abnormalities, including defects of spindle and centrosome formation and chromosome misalignment. We propose that the KIBRA-Aurora-Lats2 protein complexes form a novel axis that regulates precise mitosis.
Introduction
Mitosis is tightly controlled to achieve proper separation of chromosomes during cell division. Aberration in mitosis often causes genome instability or aneuploidy, a phenotype that many human malignant tumors exhibit (1). Various cellular surveillance mechanisms ensure the fidelity of cell cycle progression (1, 2). The spindle assembly checkpoint ensures that mitosis proceeds accurately by arresting the cells in mitosis until all chromosomes are properly aligned at the metaphase plate (3). Defects in mitosis such as chromosome misalignment or abnormal spindle formation will, therefore, result in activation of the spindle assembly checkpoint and subsequent cell cycle arrest in metaphase. Thus, several anti-mitotic drugs have been developed, and they induce abnormal or prolonged cell cycle arrest in mitosis by perturbing the microtubule dynamics, leading to mitotic catastrophe or cell death (4–6).
Large tumor suppressor 2 (Lats2)4 is a serine/threonine kinase and was originally identified as a tumor suppressor in Drosophila (the kinase is known as Warts in Drosophila) (7, 8). Overexpression of Lats2 arrests HeLa cells in G2/M phase through inhibiting the activity of the mitotic kinase cyclin-dependent kinase 1 (9) and induces apoptosis via down-regulation of anti-apoptotic proteins such as Bcl2 and Bcl-xL (10). Mouse embryonic fibroblasts from Lats2-deficient mice show strong mitotic defects including centrosome fragmentation, multinucleation, chromosome misalignment, cytokinesis failure, and accelerated mitotic exit (11). Interestingly, at the centrosome Lats2 is phosphorylated on Ser83 by Aurora-A (a mitotic kinase that plays critical roles in spindle assembly, centrosome function, and mitotic progression (2, 12)) and the phosphorylation of Lats2 on Ser83 is required for its centrosome localization during mitosis (13). Therefore, Lats2 functions as a tumor suppressor, at least partially, through regulating mitotic progression.
Lats2 and its homolog Lats1 are core kinases of the Hippo signaling pathway, which plays critical roles in controlling organ size, tumorigenesis, stem cell self-renewal, and cell contact inhibition by regulating both cell proliferation and apoptosis (14–16). Interestingly, recent studies demonstrated that some other members of the Hippo pathway such as Mst1, Mst2, Mob1/Mats, and WW45 are also involved in mitotic regulation (17–22).
The WW domain-containing protein KIBRA (enriched in kidney and brain (23)) was recently identified as a novel regulator of the Hippo pathway in both Drosophila and mammalian cells (24–27). KIBRA was originally identified as a memory performance-associated protein in humans (28–32), and this function was recently confirmed in mice (33). The physiological function of KIBRA in non-neuronal cells is much less defined, although KIBRA has been shown to be involved in cell migration in podocytes (34) and NRK cells (35) and in epithelial cell polarity (36). KIBRA also interacts with the motor protein dynein light chain 1 to positively regulate cell growth in breast cancer cells (37). Interestingly, KIBRA expression is frequently down-regulated by promoter methylation in B-cell acute lymphocytic leukemia (38) and chronic lymphocytic leukemia (39) but not in epithelial cancers, including breast, colorectal, kidney, lung, and prostate, suggesting a potential cell type-specific tumor suppressive function of KIBRA. However, a role of KIBRA in cancer (including leukemia) development has not been established.
We recently reported that KIBRA associates with Aurora-A (40) and Lats2 (27). Furthermore, we showed that KIBRA is phosphorylated by Aurora-A and -B kinases during mitosis (40). Functions of Aurora kinases and Lats2 in mitosis are well defined, but whether KIBRA has a mitotic role is currently unknown. It is largely unclear how KIBRA, Aurora, and Lats2 proteins regulate each other within the KIBRA-Aurora-Lats2 axis. In this report, we show that KIBRA activates Aurora kinases and stimulates the phosphorylation of Lats2 on Ser83 through activating Aurora-A kinase. Lats2, in turn, inhibits Aurora-mediated phosphorylation of KIBRA. Importantly, KIBRA knockdown causes mitotic defects. We propose that KIBRA, in conjunction with Aurora-A and Lats2 proteins, is a novel mitotic component that regulates proper mitosis.
EXPERIMENTAL PROCEDURES
Plasmids
The human KIBRA, Mst1, Lats1, Lats2, Aurora-A, and Aurora-B constructs and their corresponding derivatives have been described previously (27, 40). Truncated constructs were made by PCR and verified by sequencing and restriction enzyme digestion. Point mutations were generated by the QuikChange site-directed PCR mutagenesis kit (Stratagene, La Jolla, CA) and verified by sequencing.
Cell Culture and Transfection
HEK293T, HeLa, and MCF-7 cell lines (purchased from American Type Culture Collection (ATCC), Manassas, VA) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics (Clontech Laboratories, Mountain View, CA). Transfection, immunoprecipitation, and Western blotting were done as described previously (40). Aurora-A siRNA (40) (SMARTpool) and siRNA against Lats2 (SMARTpool) were purchased from Dharmacon, Inc. (Lafayette, CO). PP1c siRNA (40) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). siRNA-1 and -2 against KIBRA have been described previously (27). All other chemicals were either from Sigma or Thermo Fisher (Waltham, MA).
Establishment of Tet-On-inducible Cell Lines
The parental HeLa-rtTA cell line was purchased from Clontech Laboratories. The cell lines expressing wild-type KIBRA or KIBRA S539A (both are siRNA-resistant constructs) were established as described previously (40). Cells were maintained in medium containing Tet system-approved fetal bovine serum (Clontech Laboratories).
Cell Cycle Synchronization
A double thymidine block was used as described previously with slight modification (41). Briefly, thymidine was added to subconfluent HeLa cells (2.5 mm final), and the culture was incubated for 17 h. Cells were washed three times with PBS and allowed to recover with fresh medium for 10 h. The cells were then incubated with 2.5 mm thymidine for another 18 h. The culture medium was replaced with fresh medium without the drug to release the cells from the block.
Antibodies
Rabbit polyclonal and mouse monoclonal antibodies against human KIBRA have been described (40). The rabbit polyclonal phospho-specific antibody against KIBRA Ser539 has been described (40). Anti-FLAG, anti-HA, and anti-Myc antibodies were from Sigma. Anti-HA and anti-Myc antibodies from Santa Cruz Biotechnology were also used. Anti-β-actin, anti-cyclin B, anti-PP1c (pan), and anti-GFP antibodies were also from Santa Cruz Biotechnology. Mouse monoclonal anti-Aurora-A antibody was from Sigma. Anti-Lats2 was purchased from Bethyl Laboratories (Montgomery, TX). Rabbit polyclonal anti-α-tubulin and mouse monoclonal anti-γ-tubulin were from Abcam (Cambridge, MA). Anti-phospho-Thr288 Aurora-A/Thr232 Aurora-B was from Cell Signaling Technology (Danvers, MA). Mouse monoclonal anti-phospho-Ser83 Lats2 (13) was obtained from Abnova (Taipei, Taiwan).
Immunofluorescence Staining and Confocal Microscopy
Cells were fixed for 10 min with 100% methanol at −20 °C, and then permeabilized with 1% Triton X-100 in PBS for 15 min at room temperature. Nonspecific epitopes were blocked with 4% BSA in PBS for 1 h. After three washes with PBS (each for 10 min), cells were incubated with the primary antibodies diluted in 4% BSA in PBS for 2 h at room temperature or overnight at 4 °C. Texas Red (GE Healthcare) and/or Alexa Fluor 594-conjugated (Molecular Probes, Eugene, OR) anti-rabbit/mouse IgG were incubated with the cells for 40 min with 4% BSA in PBS at room temperature. After washing the cells three times (each wash for 10 min, with DAPI added in the final wash) with PBS, the stained cells were mounted with Fluoromount (Vector Laboratories, Burlingame, CA) and visualized with an upright, inverted, Axiovert 200 m Zeiss fluorescence microscope (Carl Zeiss, New York, NY). The Slidebook software (version 4.2, Intelligent Imaging Innovations, Denver, CO) was used for analyzing and processing all immunofluorescence images. For phenotypic analysis, we independently analyzed and scored the mitotic defects in each experiment.
Statistical Analysis
Statistical significance was performed using a two-tailed, unpaired Student's t test.
RESULTS
KIBRA Activates Aurora Kinases
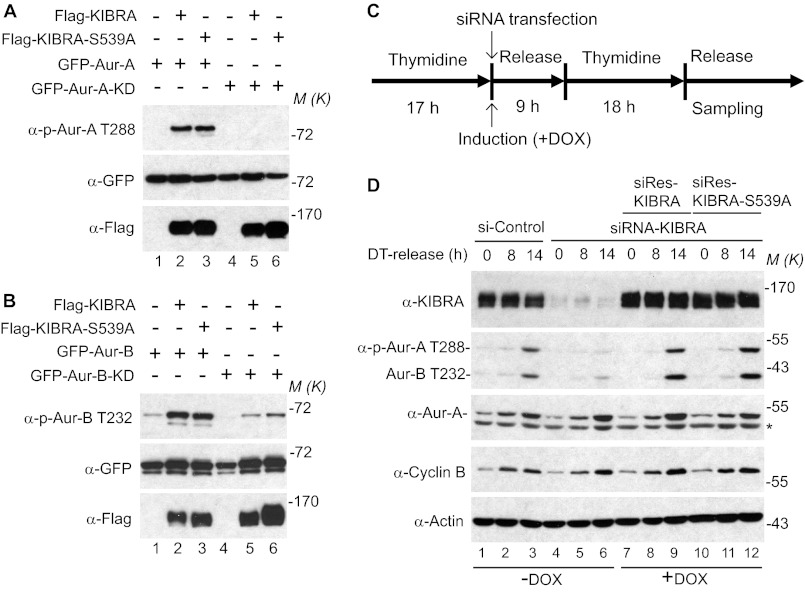
We previously identified Ser539 of KIBRA as a major phosphorylation site for Aurora kinases in mitosis (40). As many Aurora substrates also function as activators of the kinase, we tested whether this is also the case for KIBRA. To this end, we examined Aurora kinase activity by using the phospho-specific antibody against the autophosphorylation sites (Thr288 for Aurora-A, Thr232 for Aurora-B, and Thr198 for Aurora-C). As shown in Fig. 1A, overexpression of KIBRA strongly stimulated Aurora-A kinase activation, as indicated by an increase of phosphorylation of Aurora-A on Thr288. As expected, the phosphorylation of Thr288 of Aurora-A-KD (kinase dead/inactive) form was not increased by KIBRA. Interestingly, KIBRA S539A, a mutant that is not phosphorylated by Aurora-A, also promoted the phosphorylation of Aurora-A on Thr288 and did so as well as wild-type KIBRA, suggesting that Aurora-mediated phosphorylation is not required for KIBRA to activate Aurora-A. Similarly, overexpression of KIBRA enhanced the phosphorylation of Aurora-B on Thr232 (Fig. 1B). We noticed that there was still some phosphorylation of Thr232 when Aurora-B KD was used (Fig. 1B, lanes 4–6), suggesting the existence of another kinase that phosphorylated Aurora-B on Thr232.
FIGURE 1.
KIBRA activates Aurora kinases. A, various plasmids were transfected into HEK293T cells as indicated. At 48 h after transfection, cells were lysed, and proteins were separated by SDS-PAGE and transferred onto PVDF membranes, followed by Western blot analysis with the indicated antibodies. In all of the figures, M(K) indicates positions where the relevant molecular markers migrated. B, transfection and Western blotting were done as described in A. C, schematic diagram for D. Double thymidine block was employed as described under “Experimental Procedures.” siRNA transfection and doxycycline (DOX) addition were done after the first thymidine block. D, the doxycycline-inducible HeLa cell lines expressing siRNA-resistant (siRes) wild-type KIBRA or KIBRA S539A were established as described previously (39), and the cells were treated as described in C. The samples were probed with the indicated antibodies. DT, double thymidine block. The asterisk marks the incompletely stripped actin. Doxycycline (Sigma) was used at 50–100 ng/ml to induce exogenous siRNA-resistant KIBRA. Aur-B, Aurora-B.
KIBRA Is Required for Aurora Activation During Mitosis
The expression of Aurora-A is diminished in interphase cells, whereas Aurora-A is stabilized and activated by phosphorylation during mitosis (41). To further explore the involvement of KIBRA in the activation of Aurora kinase, we established doxycycline-inducible HeLa cells expressing siRNA-resistant KIBRA or KIBRA S539A and employed a double thymidine block to synchronize these cells in mitosis (Fig. 1C). As shown in Fig. 1D, Aurora kinases were clearly activated in control cells (revealed by an increase of phosphorylation of Aurora-A Thr288 and Aurora-B Thr232) 14 h after being released from the double thymidine block (compare lane 3 with lane 1). However, activation of Aurora kinases is largely diminished in KIBRA knockdown cells at the same time point, indicating that KIBRA is required for full activation of Aurora kinases when cells enter mitosis (Fig. 1D, compare lane 6 with lane 3). Aurora-A and cyclin B levels are increased similarly in both control and KIBRA knockdown cells when the cells are released into mitosis, suggesting that KIBRA knockdown did not affect the overall entry into mitosis at the time points examined. Importantly, the defect caused by KIBRA knockdown was completely rescued by re-expression of either siRNA-resistant wild-type KIBRA or KIBRA S539A, further confirming that Aurora-mediated phosphorylation is not required for KIBRA to promote activation of Aurora (Fig. 1D, compare lanes 9 and 12 with lane 6). Taken together, the data show KIBRA activates Aurora-A and -B kinases by stimulating their autophosphorylation.
KIBRA Promotes Phosphorylation of Lats2 on Ser83 through Aurora-A
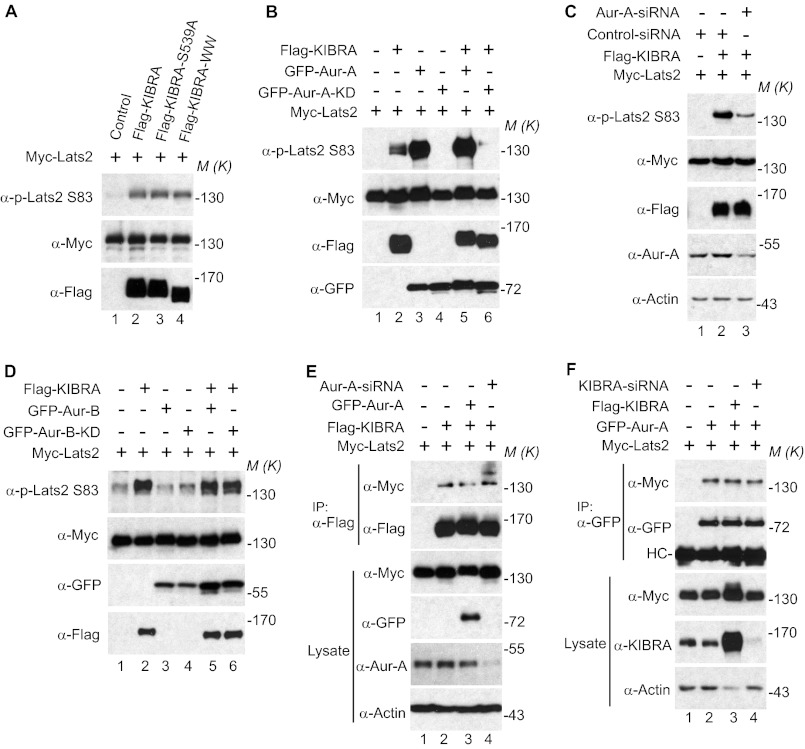
Ser83 of Lats2 was shown to be phosphorylated during mitosis by Aurora-A (13). We recently reported that KIBRA associates with both Lats2 (27) and Aurora-A (40). These findings, along with the data from Fig. 1, led us to determine whether KIBRA is involved in controlling phosphorylation of Ser83 on Lats2. Fig. 2A shows that enhanced expression of either KIBRA or KIBRA S539A similarly promoted the phosphorylation of Lats2 on Ser83. However, deletion of the WW domains (which abolishes the interaction between KIBRA and Lats2 (27)) did not affect the ability of KIBRA to stimulate the phosphorylation on Ser83 of Lats2. At this point, we reasoned that KIBRA promotes the phosphorylation of Lats2 on Ser83 by activating Aurora-A kinase. To test this hypothesis, we introduced Aurora-A-KD (kinase dead/inactive) or Aurora-A siRNA to determine the role of Aurora-A in mediating the KIBRA-dependent phosphorylation of Lats2 on Ser83. As shown in Fig. 2, B and C, overexpression of Aurora-A-KD or knocking down Aurora-A greatly impaired the phosphorylation of Ser83 on Lats2 induced by KIBRA, suggesting that KIBRA promotes Ser83 phosphorylation of Lats2 by activating Aurora-A kinase and that the Aurora-A-KD form has a dominant-negative function. Interestingly, although Aurora-A robustly phosphorylated Lats2 on Ser83 (Fig. 2B, lane 3), overexpression of Aurora-B did not increase the phosphorylation of Lats2 on Ser83 and expression of Aurora-B-KD had no effect on the ability of KIBRA to promote Ser83 phosphorylation of Lats2 (Fig. 2D).
FIGURE 2.
KIBRA promotes phosphorylation of Lats2 on Ser83 through Aurora-A. A, Myc-tagged Lats2 was transfected into HEK293T cells with FLAG-tagged KIBRA or KIBRA mutants as indicated. At 48 h after transfection, cells were lysed, and proteins were immunoprecipitated with Myc antibody followed by Western blot analysis with the indicated antibodies. Lysates were also probed with FLAG antibody to check the expression of transfected KIBRA. B, HEK293T cells were transiently transfected with plasmids as indicated. At 48 h after transfection, Myc-Lats2 was immunoprecipitated and probed with p-Lats2 Ser83 and Myc antibodies. Lysates without immunoprecipitation were also probed with FLAG and GFP antibodies to check the expression of KIBRA and Aurora-A (Aur-A). C, HEK293T cells were transiently transfected with control siRNA (lanes 1 and 2) or siRNA against Aurora-A (lane 3) and plasmids as indicated. Immunoprecipitation and Western blotting were done as described in B. D, transfection, immunoprecipitation, and Western blot analysis were performed as described in B except GFP-Aurora-B and GFP-Aurora-B-KD were used. E, HEK293T cells were transiently transfected with control siRNA (lanes 1–3) or siRNA against Aurora-A (lane 4) and plasmids as indicated. At 48 h post-transfection, cells were lysed and immunoprecipitated with FLAG antibody. The immunoprecipitates were probed with the indicated antibodies. Total cell lysates were used to check the expression of Aurora-A and Myc-Lats2. F, HEK293T cells were transiently transfected with control siRNA (lanes 1–3) or siRNA targeting KIBRA and various plasmids as indicated. Cells were harvested at 48 h post-transfection. The immunoprecipitates and total cell lysates without immunoprecipitation were probed with the indicated antibodies. HC, IgG heavy chain. M(K) indicates positions where the relevant molecular markers migrated.
Because KIBRA, Aurora-A, and Lats2 associate with each other and Aurora-A is required for KIBRA to promote Lats2 phosphorylation, we further explored whether the interaction between KIBRA and Lats2 is Aurora-A-dependent. Surprisingly, neither overexpression of Aurora-A nor Aurora-A knockdown affected the association between KIBRA and Lats2 (Fig. 2E). In addition, neither knockdown nor enhanced expression of KIBRA affected the interaction between Lats2 and Aurora-A (Fig. 2F). These data suggest that KIBRA, Aurora-A, and Lats2 interact with each other in an independent or mutually exclusive manner.
Lats2 Overexpression Enhances KIBRA Mobility
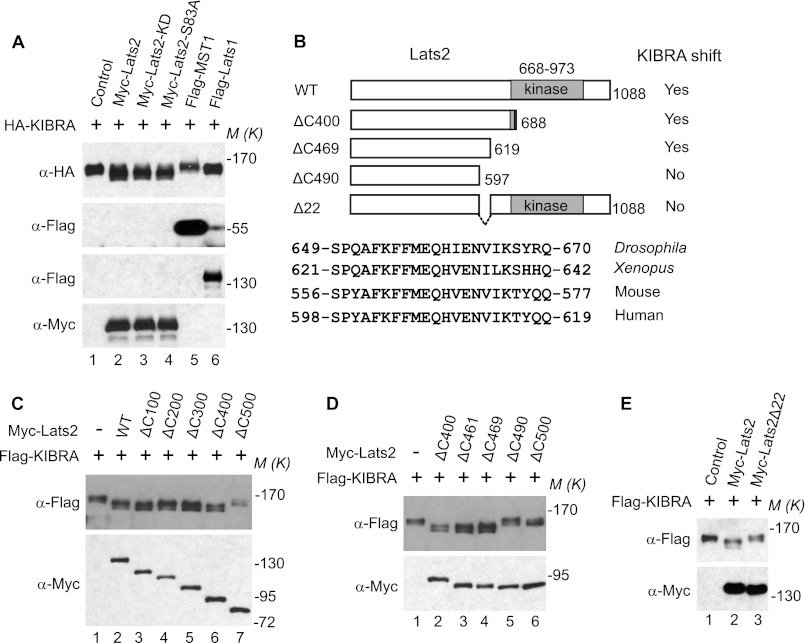
During our experiments, we noticed the migration of KIBRA increased on SDS gels when Lats2 was overexpressed ((27); Fig. 3A, compare lane 2 with lane 1). The kinase activity and Aurora-A-mediated phosphorylation on Ser83 were not required for this function of Lats2 (Fig. 3A, compare lanes 2–4 with lane 1). Interestingly, Lats2, but not Mst1 or its close homolog Lats1, possessed this function (Fig. 3A, compare lanes 5 and 6 with lanes 2–4), confirming the specificity. To further explore which domain/region is required for Lats2 to enhance the mobility of KIBRA, we generated a series of truncated Lats2 constructs (Fig. 3B). Deletion of the C-terminal 400 amino acids did not significantly alter the ability of Lats2 to enhance the mobility of KIBRA (Fig. 3C). However, deletion of an additional 100 amino acids abolished the ability of Lats2 to increase the mobility of KIBRA, suggesting that the region encompassing amino acids 588–689 is required for Lats2 to perform this function. Additional truncated constructs were made with deletions within this region, and our data suggest that the highly conserved region (amino acids 598–619 of human Lats2, Fig. 3B) is required for Lats2 to enhance the mobility of KIBRA (Fig. 3D). Internal deletion of these 22 amino acids in Lats2 (Lats2Δ22) largely abolished its function to increase KIBRA mobility (Fig. 3E).
FIGURE 3.
Overexpression of Lats2 enhances mobility shift of KIBRA. A, HA-tagged KIBRA was transfected into HEK293T cells with empty vector or various DNAs as indicated. At 48 h post-transfection, total cell lysates were probed with the indicated antibodies. B, schematic diagram of various Lats2 constructs used for C--E. C–E, FLAG-tagged KIBRA was transfected into HEK293T cells with empty vector or plasmids as indicated. At 48 h post-transfection, total cell lysates were probed with the indicated antibodies. M(K) indicates positions where the relevant molecular markers migrated.
Lats2 Inhibits Phosphorylation of KIBRA on Ser539
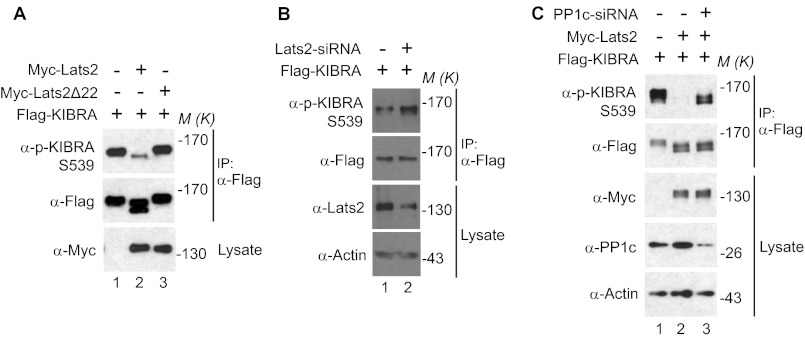
We previously reported that during mitosis Ser539 of KIBRA is phosphorylated by Aurora kinases and that KIBRA migrates differently on SDS-polyacrylamide gels depending on its phosphorylation status (40). Thus, we tested whether expression of Lats2 might inhibit the phosphorylation of KIBRA using phospho-specific antibodies. Overexpression of Lats2, but not Lats2Δ22, strongly decreased the phosphorylation of KIBRA on Ser539 (Fig. 4A). In addition, knockdown of Lats2 increased the phosphorylation of transfected KIBRA on Ser539 (Fig. 4B). Taken together, these data suggest that during mitosis Lats2 antagonizes Aurora-mediated phosphorylation of KIBRA on Ser539.
FIGURE 4.
Lats2 inhibits the phosphorylation of KIBRA on Ser539 via PP1. A, HEK293T cells were transfected with FLAG-KIBRA and plasmids as indicated. At 48 h post-transfection, the cells were lysed and immunoprecipitated with FLAG antibody. The immunoprecipitates were probed with anti-phospho-Ser539 KIBRA and subsequent anti-FLAG antibodies. B, HeLa cells were transfected with FLAG-KIBRA and control siRNA (lane 1) or siRNA against Lats2 (lane 2) for 48 h. FLAG-KIBRA was immunoprecipitated and probed with the indicated antibodies. Total cell lysates without immunoprecipitation were also analyzed. C, HeLa cells were transfected with control siRNA (lanes 1 and 2) or siRNA targeting PP1 (lane 3) and plasmids as indicated. FLAG-KIBRA was immunoprecipitated and probed with phospho-KIBRA Ser539 and subsequent anti-FLAG antibodies. Total cell lysates without immunoprecipitation were also analyzed. M(K) indicates positions where the relevant molecular markers migrated.
We recently reported that PP1 can dephosphorylate Ser539 of KIBRA (40). Thus, we explored whether PP1 is required for the Lats2-dependent reduction of phosphorylation of KIBRA Ser539. As shown in Fig. 4C, in the presence of siRNA against PP1c (catalytic subunit), Lats2 inhibited the phosphorylation of KIBRA on Ser539 less efficiently (compare lane 3 with lane 2), indicating that Lats2 may inhibit KIBRA phosphorylation on Ser539, at least partially through regulating PP1c.
KIBRA Knockdown Causes Mitotic Defects
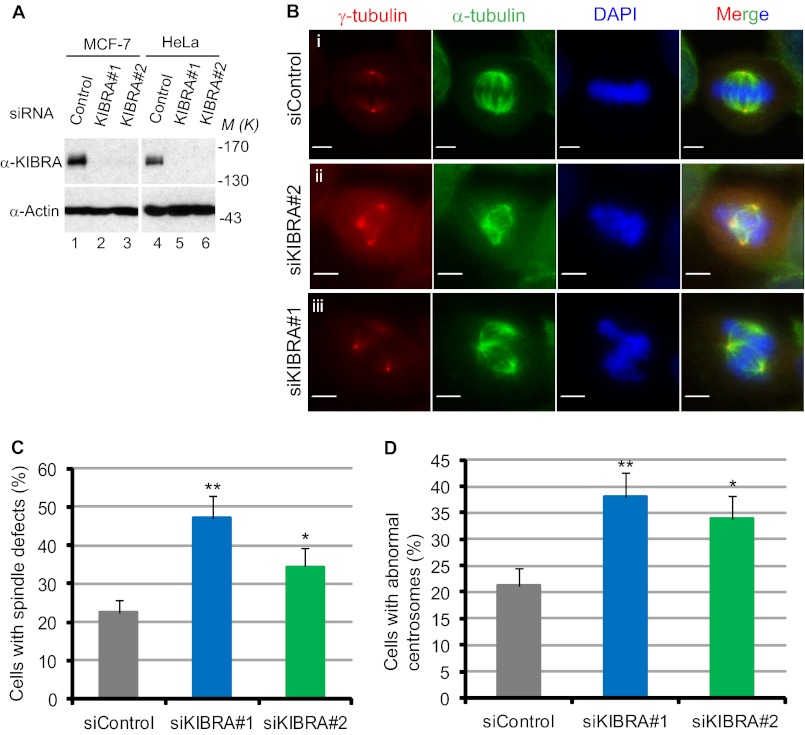
We found that KIBRA activates the important mitotic kinase, Aurora-A (Fig. 1). Moreover, KIBRA is a verified substrate of both Aurora- A and -B (40). Therefore, we expected KIBRA to play an important role in the process of mitosis. To test the function of KIBRA in mitosis, we knocked down KIBRA in both MCF-7 and HeLa cells using two different siRNA oligonucleotides. As seen in Fig. 5A, 48 h after transfection, both oligonucleotides efficiently depleted KIBRA in HeLa as well as MCF-7 cells. We first depleted KIBRA in MCF-7 cells and used immunofluorescence to identify any mitotic defects. The depletion of KIBRA in MCF-7 cells caused striking defects in spindle assembly (Fig. 5, B and C) as well as the centrosome number (Fig. 5, B and D). KIBRA activates Aurora-A and Aurora-A activity is known to be required for proper spindle assembly and centrosome function. Hence, it is likely for these reasons that depleting KIBRA caused defects in spindle assembly and centrosome number. We observed that the knockdown of KIBRA strongly affected the spindle structure (Fig. 5B). The spindle microtubules were abnormally organized in KIBRA siRNA cells (Fig. 5B, middle panels). Furthermore, the centrosomes appeared fragmented (Fig. 5B, lowest panels). About 48% of the cells that were transfected with KIBRA siRNA-1 and 35% of the cells that were transfected with KIBRA siRNA-2 displayed abnormally assembled metaphase spindles (Fig. 5C). Furthermore, 38% of the cells that were transfected with KIBRA siRNA-1 and >33% of the cells that were transfected with KIBRA siRNA-2 exhibited defects in centrosome numbers (Fig. 5D). These data show that KIBRA plays a crucial role in mitosis by regulating centrosome function and spindle assembly, possibly via regulating Aurora-A activity.
FIGURE 5.
KIBRA knockdown causes mitotic defects in MCF-7 cells. A, MCF-7 and HeLa cells were treated with control siRNA (20 nm, lanes 1 and 4) and siRNA targeting KIBRA (20 nm, lanes 2, 3, 5, and 6) for 48 h, and knockdown efficiency was analyzed by Western blotting. B, MCF-7 cells were transfected with KIBRA siRNA. At 24–48 h post-transfection, cells were fixed, permeabilized, and stained (see “Experimental Procedures”) with antibodies as indicated. Representative confocal images are shown. Scale bar, 10 μm. C, quantification of spindle defects in KIBRA knockdown cells. The graph represents the percentage of cells from three independent experiments, and at least 150 mitotic cells were counted in each group. Error bars represent S.E. **, p < 0.01; *, p < 0.05(t test). D, quantification of centrosome defects in KIBRA knockdown cells. The graph represents the percentage of cells from three independent experiments, and at least 150 mitotic cells were counted in each group. Error bars represent S.E. **, p < 0.01; *, p < 0.05(t test). M(K) indicates positions where the relevant molecular markers migrated.
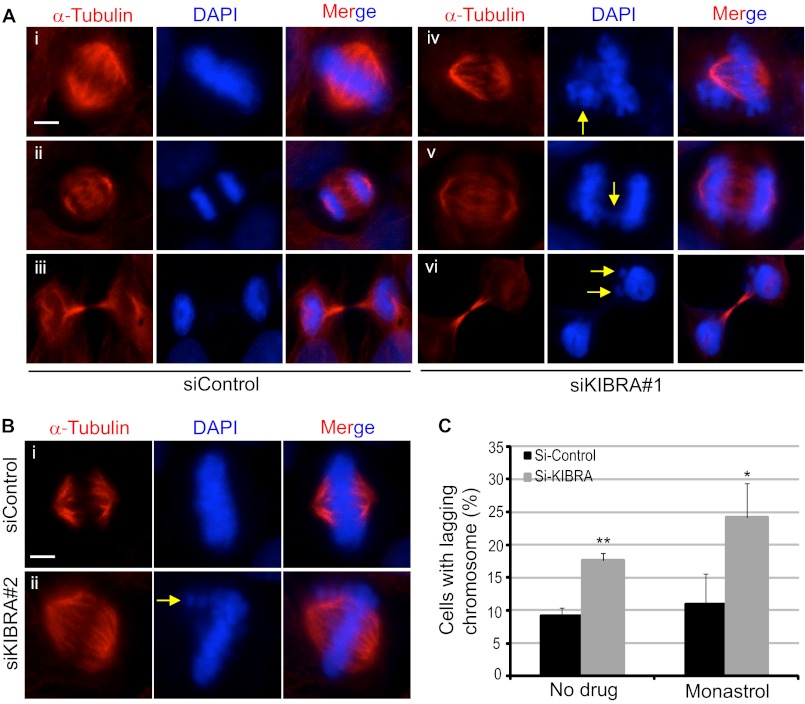
Because we detected abnormal spindles in KIBRA RNAi MCF-7 cells, we expected that the knockdown of KIBRA would also impair chromosome alignment during mitosis. To test this hypothesis, HeLa cells were transfected with either a scrambled, non-targeting siRNA or with siRNA against KIBRA. Furthermore, these cells were either treated with dimethyl sulfoxide (control) or with monastrol (an Eg5 inhibitor that arrests cells in mitosis). The monastrol was then washed out, and the cells were allowed to proceed normally through mitosis (42, 43). All cells were then subjected to immunofluorescence analysis to visualize abnormalities in chromosome alignment. Remarkably, the depletion of KIBRA from HeLa cells caused the appearance of lagging chromosomes (Fig. 6A, panel iv), chromosome bridges (Fig. 6A, panel v), and micronuclei (Fig. 6A, panel vi) during different stages of mitosis. Additionally, we observed that the knockdown of KIBRA by another siRNA in HeLa cells also yielded abnormal metaphase chromosome alignment (Fig. 6B). In addition, we observed that the enrichment of mitotic cells by monastrol treatment further increased the percentage of cells with lagging chromosomes that were obtained upon knockdown of KIBRA (Fig. 6C). All these data establish a very important role for KIBRA for the proper progression of mitosis.
FIGURE 6.
KIBRA knockdown causes chromosome misalignment in HeLa cells. A and B, HeLa cells were transfected with the siRNA oligonucleotides as indicated (20 nm). At 48 h post-transfection, cells were treated with monastrol (an Eg5 inhibitor that arrests cells in mitosis) for 2 h. The monastrol was then washed out, and the cells were allowed to proceed normally through mitosis (42, 43). These cells were then fixed and stained with α-tubulin antibody. DAPI was used to visualize the DNA. Cells at various mitotic phases are shown. Yellow arrows mark the abnormalities in KIBRA knockdown cells. Scale bar, 10 μm. C, quantification of chromosome misalignment (lagging chromosome) in KIBRA knockdown cells. The graph represents the percentage of cells from three independent experiments, and at least 150 mitotic cells were counted in each group. Error bars represent S.E. **, p < 0.01; *, p < 0.05(t test).
Taken together, our results revealed a novel axis in which KIBRA controls Lats2 phosphorylation by regulating Aurora kinase activity, and Lats2, in turn, inhibits phosphorylation of KIBRA, probably through PP1 (Fig. 7). In response to drugs (such as nocodazole, a microtubule destabilizing agent that arrests cells in mitosis)-induced mitosis or when cells enter mitosis under physiological conditions, the KIBRA-Aurora-Lats2 axis regulates the phosphorylation and/or activities of each other to control proper mitotic events during progression.
FIGURE 7.
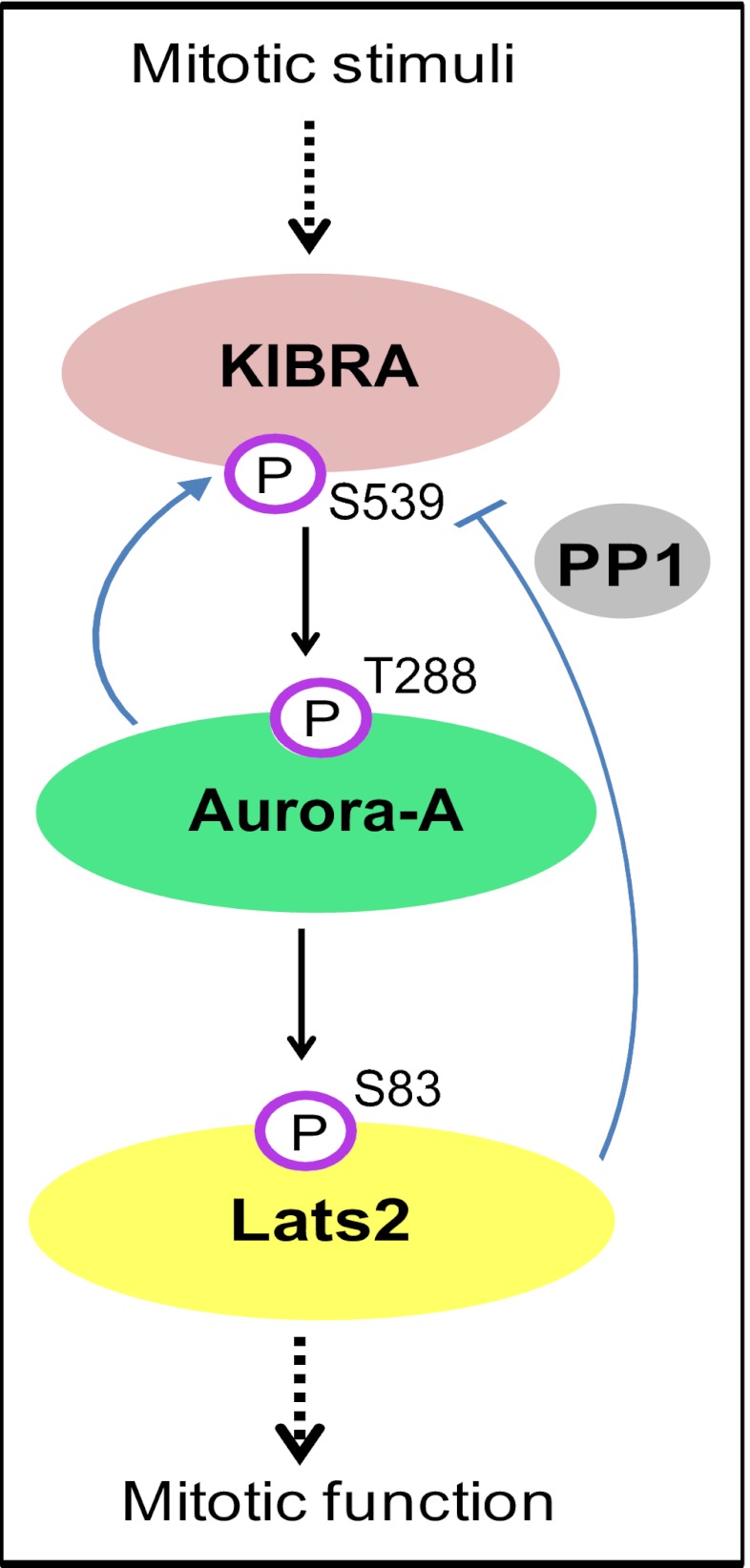
A model for the KIBRA-Aurora-Lats2 axis in mitosis. In mitosis, KIBRA, as an Aurora-A interacting partner, regulates Aurora-A kinase activity. Meanwhile, activated Aurora-A phosphorylates KIBRA Ser539, which in turn is dephosphorylated by PP1 during mitotic exit. KIBRA contributes to the phosphorylation of Lats2 on Ser83 by Aurora-A kinase. Lats2 reduces the phosphorylation of KIBRA Ser539, probably through regulating PP1 activity. Arrows and bars indicate positive and negative regulations, respectively.
DISCUSSION
Aurora kinases are important regulators of cell cycle progression and are potential oncogenes (2, 44, 45). Thus, identification of modulators and/or substrates of Aurora kinases is important for understanding the function and mechanisms of action of Aurora kinase family proteins and the basic principles of cell cycle regulation. In fact, many regulators or substrates of Aurora kinase have been implicated in controlling mitotic entry, chromosome alignment/segregation, and cytokinesis (2, 46). We previously showed that KIBRA is phosphorylated by Aurora kinases in mitosis (40). In the present study, we have further demonstrated that KIBRA is required for full activation of Aurora kinases during mitosis (Fig. 1). Future studies are needed to examine whether Aurora-mediated phosphorylation of KIBRA is involved in the mitotic defects induced by knocking down KIBRA.
Both Aurora-A and Lats2 are localized to the centrosome during mitosis (13), raising the possibility that KIBRA or phosphorylated KIBRA is also localized to this mitotic structure, but this has not been investigated and demonstrated. Interestingly, a previous report showed that KIBRA associates with the microtubule motor protein dynein light chain 1 (37). These findings, along with the demonstration of mitotic defects induced by KIBRA knockdown, strongly suggest that KIBRA may also be required for proper construction of the mitotic apparatus. We are currently investigating the spatial and temporal localization of KIBRA and phosphorylated KIBRA, and such studies are anticipated to further strengthen the importance of KIBRA in cell cycle progression, especially in mitosis.
The mechanism through which Lats2 regulates KIBRA phosphorylation is currently unknown. Phosphorylation of KIBRA on Ser539 is regulated by Aurora kinase and PP1 (40). Thus, it is possible that overexpression of Lats2 stimulates dephosphorylation of KIBRA by inhibiting Aurora kinase activity and/or activating PP1. However, although we showed that PP1 is required for Lats2 to inhibit phosphorylation of KIBRA on Ser539 (Fig. 4), a solid connection between Lats2 and PP1 has not been established. We previously demonstrated that KIBRA also associates with PP1 (40). Therefore, it will be interesting to explore whether Lats2 or Lats2Δ22 affects PP1 activity or the interaction between KIBRA and PP1. Moreover, it is of particular interest to determine the difference between Lats2 and Lats1 with regards to their activity toward inhibiting the phosphorylation of KIBRA.
We noticed that cells with Lats2 knockdown or knock-out also exhibit defects similar to those caused by knocking down KIBRA, including failure of centrosome maturation, spindle disorganization, and chromosome misalignment (11), which further supports the notion that KIBRA-Aurora-Lats2 may form a novel signaling axis that regulates mitosis. It will be interesting to explore to what extent these proteins regulate mitosis in a mutually dependent way. Interestingly, recent reports have also connected other members of the Hippo pathway with mitosis. For example, the tumor suppressors Mst1 and Mob1 are involved in centrosome duplication (17), and Mob1 also localizes to the centrosome during mitosis (21). WW45 and Mst2 control centrosome disjunction and the localization of Nek2 to centrosomes (20). In addition, Mats (Drosophila ortholog of Mob1) is required for proper chromosomal segregation in developing embryos (22). Thus, it may be a common feature that Hippo pathway components control mitotic-related events and that deregulation of their function may result in mitotic defects, contributing to genome instability/aneuploidy and subsequent tumorigenesis. One would expect that YAP (yes-associated protein) and TAZ (transcriptional co-activator with PDZ binding domain), downstream effectors in the Hippo pathway, may also have a mitotic role. Therefore, it is worth investigating whether Hippo pathway activity is cell cycle-regulated. Indeed, Lats1 kinase activity peaks during mitosis (47) and increases upon treatment with nocodazole (48), supporting the hypothesis.
Acknowledgments
We thank the imaging core facility at Nebraska Center for Cellular Signaling for providing assistance with confocal microscopy. We also thank Drs. Joyce Solheim, Robert Lewis and Keith Johnson for critical reading and comments on the manuscript.
This work was supported by Grant 5P20GM103489 from the National Center for Research Resources, National Institutes of Health and a grant from the Nebraska Cancer and Smoking Disease Research Program (to J. D.).
- Lats2
- large tumor suppressor 2
- KD
- kinase dead
- PP1
- protein phosphatase 1.
REFERENCES
- 1. Storchova Z., Pellman D. (2004) From polyploidy to aneuploidy, genome instability, and cancer. Nat. Rev. Mol. Cell. Biol. 5, 45–54 [DOI] [PubMed] [Google Scholar]
- 2. Nigg E.A. (2001) Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell. Biol. 2, 21–32 [DOI] [PubMed] [Google Scholar]
- 3. Musacchio A., Salmon E.D. (2007) The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell. Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 4. Jackson J.R., Patrick D.R., Dar M.M., Huang P.S. (2007) Targeted anti-mitotic therapies: Can we improve on tubulin agents?. Nat. Rev. Cancer. 7, 107–117 [DOI] [PubMed] [Google Scholar]
- 5. Janssen A., Medema R.H. (2011) Mitosis as an anti-cancer target. Oncogene 30, 2799–2809 [DOI] [PubMed] [Google Scholar]
- 6. Jordan M.A., Wilson L. (2004) Microtubules as a target for anticancer drugs. Nat. Rev. Cancer. 4, 253–265 [DOI] [PubMed] [Google Scholar]
- 7. Justice R.W., Zilian O., Woods D.F., Noll M., Bryant P.J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 [DOI] [PubMed] [Google Scholar]
- 8. Xu T., Wang W., Zhang S., Stewart R.A., Yu W. (1995) Identifying tumor suppressors in genetic mosaics: The Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 9. Kamikubo Y., Takaori-Kondo A., Uchiyama T., Hori T. (2003) Inhibition of cell growth by conditional expression of kpm, a human homologue of Drosophila warts/lats tumor suppressor. J. Biol. Chem. 278, 17609–17614 [DOI] [PubMed] [Google Scholar]
- 10. Ke H., Pei J., Ni Z., Xia H., Qi H., Woods T., Kelekar A., Tao W. (2004) Putative tumor suppressor Lats2 induces apoptosis through downregulation of Bcl-2 and Bcl-x(L). Exp. Cell Res. 298, 329–338 [DOI] [PubMed] [Google Scholar]
- 11. Yabuta N., Okada N., Ito A., Hosomi T., Nishihara S., Sasayama Y., Fujimori A., Okuzaki D., Zhao H., Ikawa M., Okabe M., Nojima H. (2007) Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 282, 19259–19271 [DOI] [PubMed] [Google Scholar]
- 12. Bischoff J.R., Plowman G.D. (1999) The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 9, 454–459 [DOI] [PubMed] [Google Scholar]
- 13. Toji S., Yabuta N., Hosomi T., Nishihara S., Kobayashi T., Suzuki S., Tamai K., Nojima H. (2004) The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells 9, 383–397 [DOI] [PubMed] [Google Scholar]
- 14. Halder G., Johnson R.L. (2011) Hippo signaling: Growth control and beyond. Development 138, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan D. (2010) The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao B., Li L., Lei Q., Guan K.L. (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hergovich A., Kohler R.S., Schmitz D., Vichalkovski A., Cornils H., Hemmings B.A. (2009) The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 19, 1692–1702 [DOI] [PubMed] [Google Scholar]
- 18. Chiba S., Ikeda M., Katsunuma K., Ohashi K., Mizuno K. (2009) MST2- and Furry-mediated activation of NDR1 kinase is critical for precise alignment of mitotic chromosomes. Curr. Biol. 19, 675–681 [DOI] [PubMed] [Google Scholar]
- 19. Oh H.J., Kim M.J., Song S.J., Kim T., Lee D., Kwon S.H., Choi E.J., Lim D.S. (2010) MST1 limits the kinase activity of aurora B to promote stable kinetochore-microtubule attachment. Curr. Biol. 20, 416–422 [DOI] [PubMed] [Google Scholar]
- 20. Mardin B.R., Lange C., Baxter J.E., Hardy T., Scholz S.R., Fry A.M., Schiebel E. (2010) Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 12, 1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Florindo C., Perdigão J., Fesquet D., Schiebel E., Pines J., Tavares A.A. (2012) Human Mob1 proteins are required for cytokinesis by controlling microtubule stability. J. Cell Sci. 125, 3085–3090 [DOI] [PubMed] [Google Scholar]
- 22. Shimizu T., Ho L.L., Lai Z.C. (2008) The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics 178, 957–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kremerskothen J., Plaas C., Büther K., Finger I., Veltel S., Matanis T., Liedtke T., Barnekow A. (2003) Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 300, 862–867 [DOI] [PubMed] [Google Scholar]
- 24. Genevet A., Wehr M.C., Brain R., Thompson B.J., Tapon N. (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baumgartner R., Poernbacher I., Buser N., Hafen E., Stocker H. (2010) The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev. Cell 18, 309–316 [DOI] [PubMed] [Google Scholar]
- 26. Yu J., Zheng Y., Dong J., Klusza S., Deng W.M., Pan D. (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev. Cell 18, 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xiao L., Chen Y., Ji M., Dong J. (2011) KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. J. Biol. Chem. 286, 7788–7796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Papassotiropoulos A., Stephan D.A., Huentelman M.J., Hoerndli F.J., Craig D.W., Pearson J.V., Huynh K.D., Brunner F., Corneveaux J., Osborne D., Wollmer M.A., Aerni A., Coluccia D., Hänggi J., Mondadori C.R., Buchmann A., Reiman E.M., Caselli R.J., Henke K., de Quervain D.J. (2006) Common Kibra alleles are associated with human memory performance. Science 314, 475–478 [DOI] [PubMed] [Google Scholar]
- 29. Almeida O.P., Schwab S.G., Lautenschlager N.T., Morar B., Greenop K.R., Flicker L., Wildenauer D. (2008) KIBRA genetic polymorphism influences episodic memory in later life but does not increase the risk of mild cognitive impairment. J. Cell. Mol. Med. 12, 1672–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bates T.C., Price J.F., Harris S.E., Marioni R.E., Fowkes F.G., Stewart M.C., Murray G.D., Whalley L.J., Starr J.M., Deary I.J. (2009) Association of KIBRA and memory. Neurosci. Lett. 458, 140–143 [DOI] [PubMed] [Google Scholar]
- 31. Schaper K., Kolsch H., Popp J., Wagner M., Jessen F. (2008) KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol. Aging. 29, 1123–1125 [DOI] [PubMed] [Google Scholar]
- 32. Schneider A., Huentelman M.J., Kremerskothen J., Duning K., Spoelgen R., Nikolich K. (2010) KIBRA: A new gateway to learning and memory?. Front. Aging Neurosci. 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Makuch L., Volk L., Anggono V., Johnson R.C., Yu Y., Duning K., Kremerskothen J., Xia J., Takamiya K., Huganir R.L. (2011) Regulation of AMPA receptor function by the human memory-associated gene KIBRA. Neuron 71, 1022–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Duning K., Schurek E.M., Schlüter M., Bayer M., Reinhardt H.C., Schwab A., Schaefer L., Benzing T., Schermer B., Saleem M.A., Huber T.B., Bachmann S., Kremerskothen J., Weide T., Pavenstädt H. (2008) KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 19, 1891–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rosse C., Formstecher E., Boeckeler K., Zhao Y., Kremerskothen J., White M.D., Camonis J.H., Parker P.J. (2009) An aPKC-exocyst complex controls paxillin phosphorylation and migration through localized JNK1 activation. PLoS Biol. 7, e1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshihama Y., Sasaki K., Horikoshi Y., Suzuki A., Ohtsuka T., Hakuno F., Takahashi S., Ohno S., Chida K. (2011) KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr. Biol. 21, 705–711 [DOI] [PubMed] [Google Scholar]
- 37. Rayala S.K., den Hollander P., Manavathi B., Talukder A.H., Song C., Peng S., Barnekow A., Kremerskothen J., Kumar R. (2006) Essential role of KIBRA in co-activator function of dynein light chain 1 in mammalian cells. J. Biol. Chem. 281, 19092–19099 [DOI] [PubMed] [Google Scholar]
- 38. Hill V.K., Dunwell T.L., Catchpoole D., Krex D., Brini A.T., Griffiths M., Craddock C., Maher E.R., Latif F. (2011) Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics 6, 326–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shinawi T., Hill V., Dagklis A., Baliakas P., Stamatopoulos K., Agathanggelou A., Stankovic T., Maher E.R., Ghia P., Latif F. (2012) KIBRA gene methylation is associated with unfavorable biological prognostic parameters in chronic lymphocytic leukemia. Epigenetics 7, 211–215 [DOI] [PubMed] [Google Scholar]
- 40. Xiao L., Chen Y., Ji M., Volle D.J., Lewis R.E., Tsai M.Y., Dong J. (2011) KIBRA protein phosphorylation is regulated by mitotic kinase aurora and protein phosphatase 1. J. Biol. Chem. 286, 36304–36315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. (2003) Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114, 585–598 [DOI] [PubMed] [Google Scholar]
- 42. Kim Y., Holland A.J., Lan W., Cleveland D.W. (2010) Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell 142, 444–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kapoor T.M., Lampson M.A., Hergert P., Cameron L., Cimini D., Salmon E.D., McEwen B.F., Khodjakov A. (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311, 388–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bischoff J.R., Anderson L., Zhu Y., Mossie K., Ng L., Souza B., Schryver B., Flanagan P., Clairvoyant F., Ginther C., Chan C.S., Novotny M., Slamon D.J., Plowman G.D. (1998) A homologue of Drosophila Aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 17, 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhou H., Kuang J., Zhong L., Kuo W.L., Gray J.W., Sahin A., Brinkley B.R., Sen S. (1998) Tumor-amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy, and transformation. Nat. Genet. 20, 189–193 [DOI] [PubMed] [Google Scholar]
- 46. Marumoto T., Zhang D., Saya H. (2005) Aurora-A: A guardian of poles. Nat. Rev. Cancer. 5, 42–50 [DOI] [PubMed] [Google Scholar]
- 47. Iida S., Hirota T., Morisaki T., Marumoto T., Hara T., Kuninaka S., Honda S., Kosai K., Kawasuji M., Pallas D.C., Saya H. (2004) Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene 23, 5266–5274 [DOI] [PubMed] [Google Scholar]
- 48. Praskova M., Xia F., Avruch J. (2008) MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr. Biol. 18, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]