Background: INF2 is a formin protein that accelerates both actin polymerization and depolymerization.
Results: Mutations to highly conserved FH2 residues have surprising effects on INF2 biochemical activity.
Conclusion: It is dangerous to assume that mutation of conserved FH2 domain residues will have equivalent effects on all formins.
Significance: These studies provide mechanistic insight into the role of INF2's FH2 domain in polymerization and depolymerization.
Keywords: Actin, Cell Biology, Confocal Microscopy, Cytoskeleton, Formin, Formin FH2 Domain
Abstract
INF2 (inverted formin 2) is a formin protein with unusual biochemical characteristics. As with other formins, the formin homology 2 (FH2) domain of INF2 accelerates actin filament assembly and remains at the barbed end, modulating elongation. The unique feature of INF2 is its ability to sever filaments and enhance depolymerization, which requires the C-terminal region. Physiologically, INF2 acts in the secretory pathway and is mutated in two human diseases, focal and segmental glomerulosclerosis and Charcot-Marie-Tooth disease. In this study, we investigate the effects of mutating two FH2 residues found to be key in other formins: Ile-643 and Lys-792. Surprisingly, neither mutation abolishes barbed end binding, as judged by pyrene-actin and total internal reflection (TIRF) microscopy elongation assays. The I643A mutation causes tight capping of a subset of filaments, whereas K792A causes slow elongation of all filaments. The I643A mutation has a minor inhibitory effect on polymerization activity but causes almost complete abolition of severing and depolymerization activity. The K792A mutation has relatively small effects on polymerization, severing, and depolymerization. In cells, the K792A mutant causes actin accumulation around the endoplasmic reticulum to a similar extent as wild type, whereas the I643A mutant causes no measurable polymerization. The inability of I643A to induce actin polymerization in cells is explained by its inability to promote robust actin polymerization in the presence of capping protein. These results highlight an important point: it is dangerous to assume that mutation of conserved FH2 residues will have equivalent effects in all formins. The work also suggests that both mutations have effects on the mechanism of processive elongation.
Introduction
Formin proteins are actin assembly factors and are expressed in almost all eukaryotes examined (1–3). Budding and fission yeasts have two and three formins, respectively, with well defined cellular functions (4, 5). Mammals have 15 formins, and the cellular roles of these proteins are less well defined (6).
The common feature uniting formins is the formin homology 2 (FH2)2 domain. This ∼400-residue sequence forms an anti-parallel “donut-shaped” dimer in all formins examined and is central to formin-mediated actin assembly (7–9). The FH2 domain can accelerate actin nucleation, perhaps by stabilizing dimeric or trimeric intermediates, and then can modulate subsequent filament growth by remaining processively attached to the elongating barbed end (10). The actin monomer-binding protein profilin accelerates barbed end elongation from FH2-bound barbed ends when bound to the proline-rich FH1 domain, situated N-terminal to the FH2 (Fig. 1A and Ref. 11).
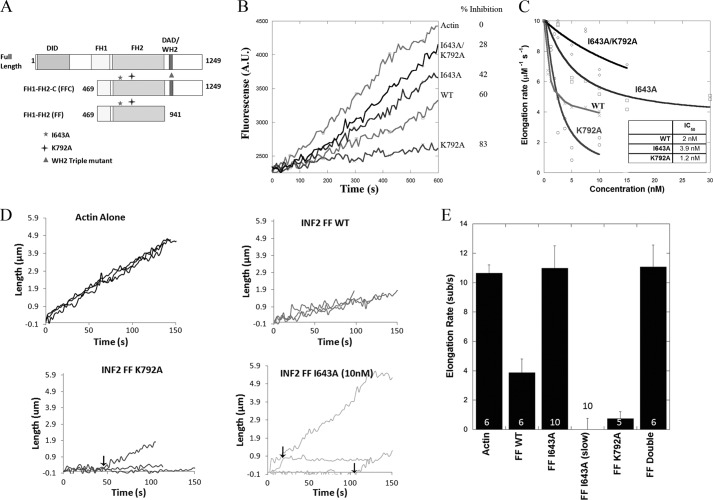
FIGURE 1.
FH2 mutations do not abolish INF2-mediated elongation inhibition. A, domain architecture of full-length human INF2 (CAAX) and the FFC and FF constructs used in this study. The approximate locations of the I643A and K792A mutations are shown. B, elongation of 0.5 μm actin monomers (25% pyrene) from phalloidin-stabilized seeds in the presence of 10 nm of the indicated FF construct. The percentage of inhibition is shown on the side of the graph. C, concentration dependence of filament elongation inhibition by FH2 mutants. The slopes were taken at 10% completion of elongation reactions and were converted to elongation rates. D, traces of individual filaments from TIRF assays with various mutants. 1 μm actin monomers (20% Alexa 488-labeled) and 1 nm of the formin were used for WT and the K792A mutant. 10 nm was used for the I643A mutant. Arrows indicate points where the elongation rate changes, suggesting that the formin is binding to or releasing from the barbed end. E, quantification of elongation rates for the various INF2-FF constructs shown in D. The error bars represent standard deviation (numbers on the bars represent n).
FH2 domains, however, vary widely in their biochemical activities. Although some FH2 domains are potent nucleation accelerators (mDia1 and mDia2), others are much less potent (FMNL1, FMNL3, and DAAM1) (12–14). The speed of barbed end elongation also varies widely depending on the formin bound to it, from an elongation rate equivalent to actin alone (mDia1) to almost complete suppression of barbed end elongation (Cdc12) in the absence of profilin (15). Some FH2 domains bundle actin filaments (FMNL1–3, mDia2, and Bnr1p), whereas others do not (mDia1 and Bni1p) (12,13,16–18).
Formin effects on actin are further influenced by sequences C-terminal to the FH2. The C termini of several formins (mDia1, DAAM1, Bni1p, Bnr1p, and FMNL3) accelerate FH2-mediated actin polymerization (13, 19). In the case of FMNL3, the C terminus contains a WH2-like motif that can bind both actin monomers and barbed ends and can modulate barbed end elongation independently of the FH2 (13). Perhaps the most dramatic change affected by the C terminus is in the mammalian formin INF2, in which the C terminus allows potent filament severing and depolymerization when present with the FH2 domain (20). The combination of activities causes the INF2 FH1-FH2-C construct to have a biphasic effect on actin polymerization, first causing rapid polymerization of actin monomers then causing their disassembly (20). The switch between assembly and disassembly is due to ATP hydrolysis and phosphate release by actin, with phosphate release being necessary for severing and depolymerization (20). The C terminus of INF2 contains an actin monomer binding WH2 motif, which is crucial for these activities (20).
Recent studies have shown INF2 to play interesting cellular roles and to have important physiological implications. In cells, INF2 can exist in two C-terminal splice variants that dramatically change cellular localization and function. The CAAX variant binds tightly to endoplasmic reticulum (ER) and overexpression causes ER collapse (21). The non-CAAX variant localizes in a cytoplasmic, actin-dependent meshwork pattern, and its suppression causes Golgi fragmentation (22). Suppression of INF2 in hepatocytic culture cells results in defects in transcytosis of apical proteins (23), whereas suppression in lymphocytic cells inhibits delivery of Lck to the plasma membrane (24). Physiologically, mutations in INF2 have been strongly linked to two human diseases: focal and segmental glomerulosclerosis (25–28), a kidney disease, and Charcot-Marie Tooth disease, a peripheral neuropathy (29).
One question regarding INF2 is the relative importance of its polymerization and severing activities to cellular function. In cells, does INF2 act to create filaments, to depolymerize filaments, or perhaps to create highly transient filaments, which it also depolymerizes? An approach to answering this question has been to use “polymerization” and “depolymerization” mutants to rescue effects of INF2 suppression in cells (23, 24). The depolymerization mutant targeted the WH2 motif and has previously been shown biochemically to block depolymerization but not polymerization (20). The mutation also blocks the autoinhibitory interaction of INF2 (21). The polymerization mutant targeted a lysine (Lys-792) found to be crucial for polymerization in Bni1p (30) but not tested in INF2. It was found that neither the K792A nor the WH2 mutant of INF2 rescued endogenous INF2 suppression, suggesting that both polymerization and depolymerization activities were necessary (23, 24).
To understand the biochemical mechanism behind the remarkable polymerization/depolymerization activity of INF2, we have made point mutations to the FH2 domain similar to those used for inactivation of other formins: K792A and I643A. Surprisingly, we find that the K792A mutation has minimal effects on biochemical activity, with small effects on both polymerization and severing/depolymerization activity. The I643A mutation has larger effects on polymerization activity and causes an almost complete abolition of severing/depolymerization. These results demonstrate that the importance of individual FH2 residues differs between formins. Also, the fact that the K792A mutation actually slows elongation rate compared with wild type suggests that this mutation affects the rate of processive elongation.
EXPERIMENTAL PROCEDURES
DNA Constructs
Human INF2 ORF clone (catalog number SC313010) was obtained from OriGene Technologies, Inc. (Rockville, MD). INF2 FH1-FH2-C (amino acids 469–1249) CAAX and INF2 FH1-FH2 (469–941) were amplified using a GC-rich PCR system (Roche Applied Science) and subcloned into pGEX-KT vector (16) with a modified tobacco etch virus protease site for bacterial expression as a glutathione S-transferase fusion protein. The QuikChange kit (Stratagene) was used for all mutagenesis. The ER-Green construct, containing the ER-targeting sequence (amino acids 233–250) of budding yeast UBC6 (31), was a kind gift from Victoria Allan (University of Manchester, Manchester, UK). The mCherry-Sec61β was a kind gift from Jennifer Lippincott-Schwartz (National Institutes Health, Bethesda, MD). For cellular studies, INF2 FH1-FH2-C CAAX (amino acids 429–1249) was subcloned into eGFP-C1 (Clontech).
Protein Preparation
All formin constructs were expressed in bacteria as GST fusion proteins, following procedures used previously (20). The constructs used were INF2-FH1-FH2-C (human CAAX variant, amino acids 469–1249) and INF2-FH1-FH2 (human, 469–941). After expression, extracts were passed over glutathione-Sepharose, cleaved with tobacco etch virus protease to elute the formin construct from GST, and further purified by ion exchange chromatography on SourceQ. All proteins were stored at −80 °C in K50MEID (50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm imidazole, pH 7.0, 1 mm DTT). Rabbit skeletal muscle actin was purified from acetone powder (32) and labeled with pyrenyliodoacetamide (33). Both unlabeled and labeled actin were gel-filtered on Superdex 200 (34) and stored in G buffer (2 mm Tris-HCl, pH 8.0, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2, 0.01% sodium azide) at 4 °C. All chromatographic resins were from GE Biosciences. Human profilin I was expressed in bacteria and purified as described (35). Mouse capping protein (α1β2) was expressed from pRSFDuet1 vector (Novagen) and purified on a HisTrap HP column (GE Healthcare).
Buffers Used for Biochemical Studies
The following buffers were used frequently: G buffer (2 mm Tris, pH 8, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2, and 0.01% NaN3), G-Mg buffer (same as G buffer but with 0.1 mm MgCl2 instead of CaCl2), 10× KMEI (500 mm KCl, 10 mm MgCl2, 10 mm EGTA, and 100 mm imidazole, pH 7.0), and polymerization buffer (G-Mg buffer plus 1× KMEI). For depolymerization assays, polymerization buffer without ATP was used. 10E/1M was 10 mm EGTA, 1 mm MgCl2, pH 7–8. Fluorescence buffer was 25 mm imidazole, pH 7, 25 mm KCl, 250 mm NaCl, 4 mm MgCl2,1 mm EGTA, 100 mm DTT, 0.5% methylcellulose, 18 μg/ml catalase, 3 μg/ml glucose, and 100 μg/ml glucose oxidase.
Actin Polymerization by Fluorescence Spectroscopy
Unlabeled and pyrene-labeled actin were mixed in G buffer to produce a 5% pyrene-actin stock. This stock was converted to Mg2+ salt by 2 min of incubation at 23 °C in 1 mm EGTA, 0.1 mm MgCl2 immediately prior to polymerization. Polymerization was induced by the addition of 10× KMEI to a concentration of 1×, with the remaining volume made up by G-Mg. Pyrene fluorescence (excitation, 365 nm; emission, 410 nm) was monitored in a 96-well fluorescence plate reader (Tecan Infinite M1000, Mannedorf, Switzerland). The time between mixing of final components and the start of fluorimeter data collection was measured for each assay and ranged between 15 and 20 s. INF2 does not affect the fluorescence of pyrene actin filaments (20).
Barbed End Elongation Assays
Unlabeled actin (10 μm) was polymerized for 1 h at 23 °C, followed by the addition of 20 μm phalloidin and then centrifuged at 100,000 rpm for 20 min in a TLA-120 rotor. The pellet was resuspended to 6 μm in 3× polymerization buffer (G-Mg with 3× KMEI) and then sheared by five passes through a 30-gauge needle. The resuspended polymerized actin was allowed to reanneal overnight at 23 °C. 30 μl of 1× polymerization buffer with or without formin protein was added to 30-μl filaments in a 96-well plate, shaken for 10 s, and then centrifuged for 2 min at 1200 rpm. After 2 min at 23 °C, 60 μl of 1.05 μm monomers (5% pyrene, Mg2+-converted) were mixed with the filaments with a cut pipette tip. Fluorescence (365/410 nm) was recorded for 2700 s. Elongation velocity was obtained by linear fitting the initial 10% of elongation. Final concentrations in the assay were 1.5 μm phalloidin-stabilized polymerized actin and 0.525 μm monomer. Slopes of pyrene fluorescence from elongation time courses were taken at 10% completion using Kaleidagraph (Synergy Software, Reading, PA) and were converted to elongation rates (assuming 10 μm−1 s−1 for actin alone).
Calculating Filament Concentration
Slopes were measured at 50% polymerization and converted to filament concentration, either under the assumption of unrestricted ATP-actin monomer addition to barbed ends with a rate constant of 10 m−1 s−1 (5, 36, 37) or with the observed elongation rates in the presence of the specific INF2 construct as measured above, according to the following equation: [F] = S′/(K+ × M0.5), where [F] is filament concentration (μm), S′ is slope converted to polymerization rate (μm/s), and M0.5 is monomer concentration (μm) at 50% polymerization. S′ is calculated by the equation S′ = S × (Mt/(fmax − fmin), where S is the raw slope in arbitrary units/s, Mt is concentration of total polymerizable monomer (μm), and fmax and fmin are fluorescence (arbitrary units) of fully polymerized and unpolymerized actin, respectively.
Filament Depolymerization Kinetics
Actin (1.05 μm, 5% pyrene) was polymerized for 16 h at 23 °C in polymerization buffer in the dark. Actin stock (95 μl) was gently mixed using a cut tip with 5 μl of protein (at various concentrations) solution in polymerization buffer. Pyrene fluorescence was monitored within 15 s of dilution in a PC1 spectrofluorometer (ISS Inc., Champaign, IL). The initial slopes were measured and used to calculate the depolymerization rates.
Severing Assay
Actin (4 μm) was polymerized for 1 h at 23 °C in polymerization buffer. Polymerized actin (2.5 μl) was incubated with 2.5 μl of polymerization buffer containing INF2 for varying times at 23 °C. 10 μl of dilution buffer (25 mm imidazole, pH 7.0, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 0.5% methylcellulose) containing TRITC-phalloidin (Sigma-Aldrich; final concentration, 2 μm) was added, and samples were immediately diluted 50-fold with fluorescence buffer. Samples (2 μl) were adsorbed on 12-mm round glass coverslips coated with 0.01% poly-l-lysine. Cut pipette tips were used to minimize filament shearing. Filaments were visualized through a TRITC filter using a Nikon-Eclipse TE-2000 microscope with a 60 × 1.4 numerical aperture objective. The images were acquired using a Roper Cool-Snap camera. Filament length was measured using NIS-Elements software (Nikon), and 200–300 filaments were measured from at least five different fields.
Cell Culture and Plasmid Transfection
Human osteosarcoma cells (U2OS) were maintained in DMEM with 4.5 g/liter glucose, 584.0 mg/liter l-glutamine, 110.0 mg/liter sodium pyruvate, and 10% calf serum (Atlanta Biologicals, Lawrenceville, GA) and were a kind gift from Duane Compton (Dartmouth Medical School). The cells were maintained at 37 °C and 5% CO2. Lipofectamine 2000 (Invitrogen) was used for all plasmid transfections as per the manufacturer's protocol. A total of 100 ng of each plasmid DNA was used for all transfections, and the cells were analyzed 24 h post-transfection.
Fluorescence Microscopy
The cells were fixed with 4% formaldehyde (Electron Microscopy Sciences, Hatfield, PA) in PBS for 1 h at room temperature. After washing with PBS, the cells were permeabilized on ice with 0.25% Triton X-100 in PBS for 15 min. The cells were then washed with PBS prior to blocking with 2.5% calf serum in PBS for 1 h at room temperature. Actin was stained using 100 nm TRITC-phalloidin for 15 min prior to mounting on polyvinyl alcohol-DABCO. Images were captured using a Wave FX spinning disk confocal system (Quorum Technologies, Guelph, Canada) on a Nikon (Melville, NY) Eclipse microscope, using a 491-nm laser and a 525/20 filter for GFP, a 403-nm laser and a 460/20 filter for DAPI, and a 561-nm laser and a 593/40 filter for Texas Red. The images were acquired using MetaMorph and were processed using Nikon Elements and Photoshop CS (Adobe, San Jose, CA).
TIRF Microscopy
Glass flow chambers were prepared from VWR microcover glass (24 × 60 mm No 1.5) and Gold Seal Rite-On micro slides (3 × 1 inches) to hold 10 μl of volume. The chambers were treated with 5K mPEG-Silane (Laysan Bio) and washed with BSA containing buffer (1% BSA, 50 mm Tris, pH 7.5, 150 mm NaCl) and then equilibrated in TIRF buffer (10 mm imidazole, pH 7.0, 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 100 mm DTT, 0.2 mm ATP, 15 mm glucose, 0.5% methyl cellulose, 0.01 mg/ml catalase (Sigma), 0.05 mg/ml glucose oxidase ((Sigma), 0.1% BSA).
Unlabeled actin monomers were mixed with 20% Alexa 488-labeled (Molecular Probes) actin monomers (38). Alexa 488 monomers were diluted with 2× TIRF buffer, mixed with formin, and then flowed under glass flow chambers. The filaments were visualized immediately on a Nikon Eclipse TE-2000 microscope. The images were acquired every 2 s with TIRF objective (60× 1.49 N.A.) and a Roper Cool-Snap camera.
The images were processed with NIS-Elements software (Nikon) and Adobe Photoshop. Filament elongation rates were quantified using ImageJ (National Institutes of Health, Bethesda, MD).
RESULTS
FH2 Mutations Have Varying Effects on INF2-mediated Inhibition of Barbed End Elongation
In an attempt to abolish the polymerization activity of INF2, we made mutations to highly conserved FH2 residues (Ile-643 and Lys-792 to Ala) in INF2 FH1-FH2 (FF) and INF2 FH1-FH2-C (FFC) constructs (Fig. 1A). Homologous FH2 mutations have previously been shown to be important for barbed end binding of other formins (5, 12–14, 39). In addition, the INF2 K792A mutant has been used as a putative polymerization-blocking mutant in cellular studies (23, 24).
We first examined the abilities of these FH2 mutants to inhibit barbed end elongation, using the INF2-FF constructs to minimize severing-mediated effects (20). We measured elongation using pyrene-actin spectrofluorimetry, which allows facile analysis of changes in formin concentration. As observed previously for the mouse protein, the WT-FF construct slows the elongation rate by 60%. Surprisingly, both I643A and K792A mutants maintain effects on INF2-mediated inhibition of elongation, but to different degrees, with the I643A mutant slowing elongation by 42%, whereas the K792A mutant slows elongation by 83% (Fig. 1B). The apparent barbed end affinities of all these constructs are similar (The IC50 values for WT and the I643A and K792A mutants are 2, 3.9, and 1.2 nm, respectively (Fig. 1C)). The I643A/K792A double mutant is severely affected both in terms of extent of elongation inhibition and apparent barbed end affinity (Fig. 1, B and C).
We next assessed barbed end elongation using TIRF microscopy, which allows direct measurement of elongation rates from single filaments. The elongation rates of WT and K792A constructs are similar to those determined by pyrene-actin assay, with WT slowing elongation by 63%, whereas K792A slows elongation by 91% (Fig. 1, D and E). Surprisingly, the I643A mutant results in two filament populations: one that elongates at a rate of actin alone and one that has an unmeasurably low elongation rate. There are examples of periodic transitions between the slow and fast elongation states (Fig. 1D, arrows).
The existence of a pool of filaments that elongates at actin-alone rate in the presence of I643A raises two possibilities: 1) I643A is not bound to the barbed end of these filaments, or 2) I643A is bound but does not change the elongation rate. We tested these possibilities by conducting TIRF assays in the presence of capping protein. Past experiments have shown that formins block barbed end binding by capping protein, allowing continued elongation. WT FF construct displays this ability, whereas the I643A construct does not (data not shown), suggesting that the I643A mutant is not tightly bound to the elongating population of filaments.
FH2 Mutations Have Varying Effects on Actin Polymerization
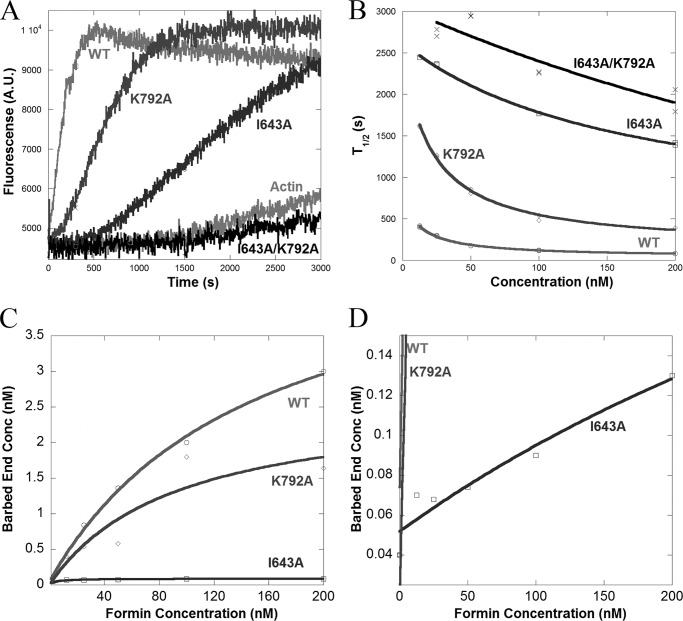
We next performed pyrene-actin polymerization assays with the INF2-FF constructs, which lack the C-terminal WH2 domain and thus lack severing and depolymerization activity. In these assays, both the I643A and K792A mutants display slower polymerization, suggesting that both residues are important for nucleation through the FH2 domain. However, the effect of the K792A mutation is relatively minor compared with the I643A mutation, which itself still maintains some polymerization activity (Fig. 2A). The double mutant (I643A/K792A) is completely impaired for actin polymerization (Fig. 2A). To analyze the polymerization efficiencies of these mutants in greater detail, we plotted the time to reach half-maximal polymerization activity (T½) as a function of the formin concentration (Fig. 2B). Despite having lower polymerization activity, the K792A mutant displays an EC50 for polymerization (20.5 nm) that is similar to that of WT (13.5 nm). In contrast the I643A mutant and the double I643A/K792A mutant do not display saturating polymerization activities. We also calculated the concentration of barbed ends created by the FH2 mutants in these assays, using the elongation rates measured upon full INF2 barbed end binding determined from Fig. 1. WT creates <2-fold more barbed ends than K792A (Fig. 2C). The I643A mutant is severely defective in barbed end production, although it is still significantly better than actin alone (Fig. 2D).
FIGURE 2.
FH2 mutations have varying effects on the polymerization activity of INF2-FF. A, pyrene-actin polymerization assays containing 1 μm actin monomers (5% pyrene) with 200 nm of the indicated FF construct of INF2. B, plot showing time to 50% completion (T½) versus formin concentration for polymerization assays conducted with various concentrations of INF2 (data not shown). The EC50 values for WT and K792A and I643A mutants are 13.9, 20.5, and >165 nm, respectively. C, plot showing barbed end concentration versus formin concentration. D, expanded view of C, showing barbed end concentration versus formin concentration in greater detail for the I643A mutant.
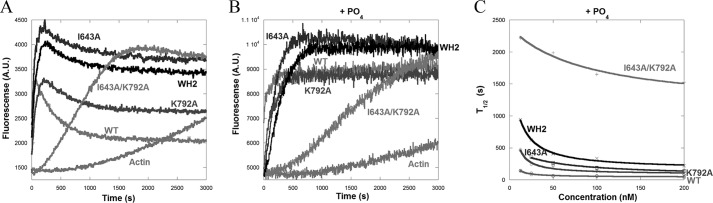
We next examined the effects of these mutations in the INF2-FFC constructs using similar pyrene actin polymerization assays. The FFC construct contains the C-terminal WH2 motif, which is required for INF2-mediated severing and causes rapid biphasic polymerization/depolymerization in pyrene-actin assays (Ref. 20 and Fig. 3A). To our surprise, the I643A and K792A mutants still potently accelerate actin polymerization. The I643A mutant lacks apparent depolymerization activity, whereas the K792A mutant has reduced depolymerization activity (Fig. 3A). As observed previously (20), the WH2 mutant is a potent polymerizer but is defective in depolymerization. The double mutant (I643A/K792A) has a pronounced polymerization defect (Fig. 3A).
FIGURE 3.
FH2 mutations have varying effects on the polymerization ability of INF2-FFC. A, pyrene-actin polymerization assays containing 1 μm actin monomers (5% pyrene) with 200 nm of the indicated FFC construct of INF2. B, pyrene-actin polymerization assays containing 1 μm actin monomers (5% pyrene) with 200 nm of the indicated FFC construct of INF2 in the presence of 10 mm sodium phosphate. C, plot showing time to 50% completion (T½) versus formin concentration for polymerization assays conducted various concentrations of INF2 (data not shown). The EC50 values for WT and I643A, K792A, double (I643A/K792A), and WH2 mutants are 2, 43, 1.2, 87, and 7.7 nm, respectively.
Because the competing activities of polymerization and depolymerization were problematic for analysis of these curves, we performed polymerization assays in the presence of phosphate, which abolishes INF2 depolymerization activity (Ref. 20 and Fig. 3B). We once again plotted the time to reach half-maximal polymerization (T½) as a function of formin concentration. By this analysis, the K792A and WH2 mutants have similar concentration dependence to the WT (EC50 values of <10 nm). In contrast, I643A has a significantly higher EC50 (43 nm; Fig. 3C), although its potency (maximal rate of polymerization) is in the same range as WT and the other mutants. The double mutant (I643A/K792A) is dramatically deficient both in EC50 and in potency.
FH2 Mutations Have Varying Effects on INF2-mediated Severing and Depolymerization
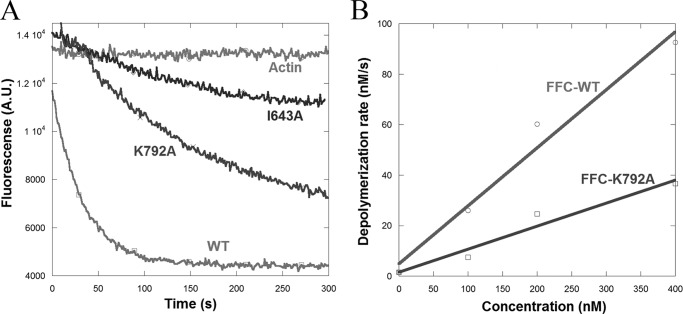
To analyze INF2-mediated depolymerization efficiency directly, we performed assays in which we mixed prepolymerized pyrene-actin filaments with INF2-FFC and measured the kinetics of pyrene fluorescence decrease, similar to those conducted previously for the mouse protein (20). At 400 nm, the I643A and K792A mutants are less efficient than WT at depolymerization, the I643A mutant being more so (Fig. 4A). To analyze the K792A mutant in detail, we plotted the depolymerization rates as a function of INF2 concentration (Fig. 4B). Both the WT and K792A mutant display concentration-dependent increases in depolymerization rate, but the WT is more efficient than the K792A mutant at all concentrations.
FIGURE 4.
Depolymerization assays in the presence of INF2-FFC constructs. A, actin monomers (1.05 μm, 5% pyrene-labeled) were polymerized 16 h at 23 °C in polymerization buffer and then diluted to 1 μm in the same buffer containing 400 nm of the indicated FFC construct; the filament depolymerization rate was measured by the decrease in pyrene fluorescence intensity with time. A.U., arbitrary units. B, depolymerization rates (derived from initial slopes of depolymerization curves) for WT and K792A mutant plotted as a function of FFC concentration.
To test whether the decreased depolymerization rates in the I643A and K792A mutants were due to severing defects, we performed severing assays. WT INF2-FFC severs actin filaments potently, whereas the WH2 triple mutant and FF construct do not (Fig. 5). The I643A mutant also displays a severe defect in severing activity, similar to the WH2 mutant and the FF construct. In contrast, the K792A mutant severs with similar efficiency to the WT under the conditions used (2 μm actin, 200 nm INF2, 2 min; Fig. 5).
FIGURE 5.
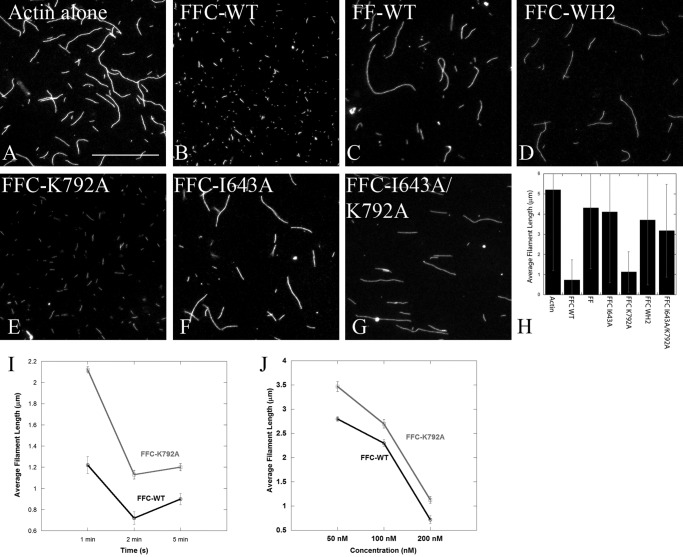
Severing assays in the presence of FFC constructs. A–G, severing assays were performed using 2 μm polymerized actin and 200 nm of various INF2 constructs for 2 min. Scale bar, 5 μm. H, bar graph shows average actin filament lengths (μm) in the presence 200 nm of various INF2 constructs. At least 200 filaments were measured for each construct in two independent experiments. The error bars represent standard deviation. I, graph showing average actin filament length measured from severing assays using 200 nm of INF2-FFC-WT or K792A. The protein was incubated with filaments for 1, 2, or 5 min prior to mounting and imaging. The error bars represent standard error. J, graph showing average actin filament length measured from severing assays using various concentrations of INF2-FFC-WT or K792A. Average length of actin filaments without any added protein was 5.2 ± 0.17 μm. The protein was incubated with filaments for 2 min prior to mounting and imaging. The error bars represent standard error.
The apparent similarity between WT and K792A in severing could be due to the limitations of our assay system, such that the assay cannot distinguish between minor severing defects. To compare the K792A mutant and WT in greater detail, we conducted severing assays with varying time and INF2 concentration (Fig. 5, I and J). WT INF2-FFC severing is both time- and concentration-dependent in these assays, showing the assay is appropriate for comparing efficiency. The K792A mutant displays longer filaments at all times and concentrations, suggesting that the severing efficiency is decreased (Fig. 5, I and J).
FH2 Mutants Display Variable Effects on Actin Polymerization in Cells
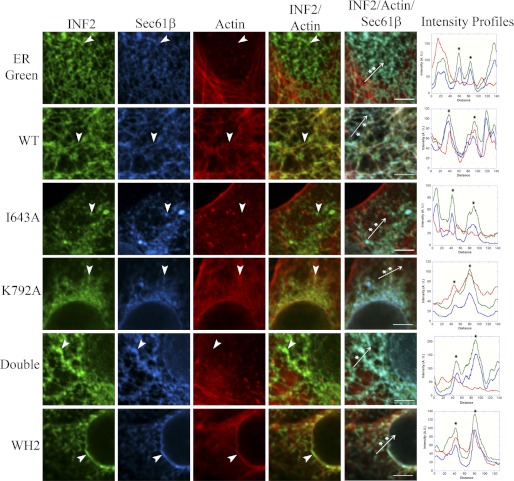
To examine the effects of these mutations in cells, we made GFP-tagged versions of the FFC constructs as the CAAX splice variant, which localizes to the ER (21). We co-transfected U2OS cells with these GFP-tagged constructs, along with CFP-Sec61β as an ER marker, and stained with rhodamine phalloidin to detect actin filaments. As a control protein that localizes to the ER but should not cause actin polymerization, we transfected cells with ER-Green.
As expected, ER-Green does not cause actin polymerization around the ER (Fig. 6, showing the close ups of a single Z-section of a confocal Z-stack. The entire cell is shown in supplemental Fig. S1). WT INF2 causes robust actin polymerization around the ER. Similar ER-associated actin polymerization occurs with the WH2 mutant, which is expected because this construct is compromised for depolymerization activity in vitro. The K792A mutant also causes ER-associated actin polymerization (Fig. 6). In contrast, the I643A mutant and the I643A/K792A double mutant do not cause measurable actin polymerization around ER (Fig. 6).
FIGURE 6.
The INF2-FFC I643A mutant does not polymerize actin in cells. U2OS cells were co-transfected with CFP-Sec61β (blue) and various GFP-tagged INF2 mutants (green). 18–24 h post-transfection, the cells were fixed and stained with rhodamine-phalloidin (red) to visualize actin filaments. ER-Green was used as a negative control. The images show small sections of single Z-slices of confocal images. The entire cell is shown in supplemental Fig. S1. The arrowheads indicate regions where the GFP and Sec61β signals co-localize. Intensity line scans along the length of the arrow are shown to the right. Asterisks indicate a peak on the Sec61β intensity profile that corresponds to an ER tubule. Scale bar, 5 μm.
To quantify these results, we measured the overall actin intensity around the nucleus (supplemental Fig. S1), because a high density of the ER accumulates in this region. Although the average actin intensities in cells transfected with ER-Green, INF2-FFC I643A, and INF2-FFC I643A/K792A are relatively low (5200, 4700, and 5100, respectively; n > 12), the actin intensities for cells with WT INF2-FFC, INF2-FFC-K792A, and INF2-FFC-WH2 are significantly higher (25,000, 37,000, and 12,000, respectively; n > 12; supplemental Fig. S1).
The I643A Mutant Is Deficient in Polymerization in the Presence of Capping Protein
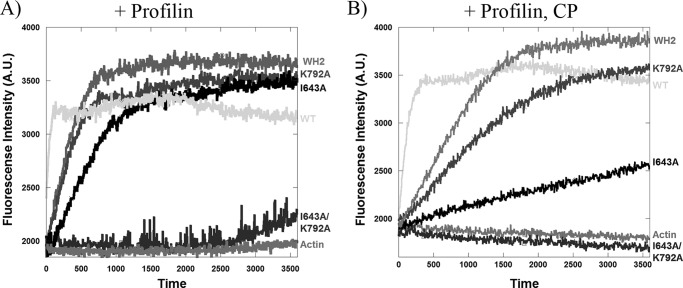
The cellular results contradicted our biochemical results showing that FFC-I643A maintained potent polymerization activity. We would have expected cellular actin filament accumulation with this mutant, especially because its depolymerization activity was severely compromised. One possible explanation is that the I643A mutant is polymerization-deficient in the presence of other actin binding proteins. We first tested profilin, because of its high abundance and its known association with formins. In pyrene-actin polymerization assays, the I643A mutant maintains potent polymerization activity in the presence of profilin (Fig. 7A). Another important actin binding protein is heterodimeric capping protein, present at mm concentration and binding barbed ends with nm affinity (40). Formins and capping protein compete for barbed end binding (8, 41, 42). In the presence of capping protein and profilin, both WT and the K792A mutant display potent polymerization activity. In contrast, the I643A mutant is severely deficient (Fig. 7B).
FIGURE 7.
Pyrene-actin polymerization assays with various INF2 FFC constructs in the presence of profilin and capping protein. 3 μm actin (5% pyrene labeled), 9 μm profilin, 20 nm capping protein, and 25 nm of each formin construct was used was used in these assays. A, with profilin. B, with profilin and capping protein (CP).
DISCUSSION
In this study, we show that mutation of highly conserved FH2 residues (Ile-643 and Lys-792) in INF2 have surprising effects on the biochemical activity of this particular FH2 domain. These residues have previously been shown to be crucial for barbed end binding and/or actin polymerization acceleration in several other formins (5, 12, 13, 30). Neither mutation abolishes barbed end binding, as judged by elongation assays, even though the extent of elongation inhibition differs. The I643A mutation causes significant changes to polymerization activity in the FF construct but has less significant polymerization effects on the FFC construct. However, the I643A mutation does severely inhibit the severing and depolymerization activities of FFC. The K792A mutation has much more minor effects on polymerization, severing, and depolymerization.
These results show that, in keeping with a number of studies by our lab and others (Refs. 5, 12, 13, 18, and 43 and Table 1), formin proteins are highly individualistic in terms of their specific biochemical activities. An important conclusion from this work is that FH2 mutations abolishing/inhibiting the activity of some formins cannot be relied upon to do the same in others. In the remainder of the discussion, we will compare the effects of these FH2 mutations in a range of formins and address how the FH2 domain might contribute to severing by INF2.
TABLE 1.
Summary of the effects of conserved FH2 mutations in various formins
+++ indicates strong effect; ++ indicates medium effect; + indicates mild effect; − indicates no effect. ND indicates that the value was not determined. The type of assay used in the study is also indicated.
| Formin | Construct | Isoleucine mutation | Lysine mutation | Reference | Type of assay |
|---|---|---|---|---|---|
| Bni1 | FH2 | +++ | +++ | 30 | Polymerization |
| Cdc12 | FH2 | +++ | − | 5 | Polymerization, elongation |
| Fus1 | FH2 | +++ | +++ | 5 | Polymerization |
| For3 | FH2 | +++ | +++ | 5 | Polymerization |
| FMNL1 | FH1-FH2-C | −a | ND | 12 | Polymerization, elongation |
| FMNL3 | FH1-FH2 | +++ | − | 13 and unpublished data | Polymerization, elongation |
| mDia1 | FH2 | +++ | ND | 12 | Polymerization, elongation |
| mDia2 | FH2 | +++ | ND | 12 | Polymerization, elongation |
| DAAM1 | FH2-C | +++ | ND | 14 | Polymerization |
| INF2 | FH1-FH2 | ++ | + | This study | Polymerization, elongation |
| INF2 | FH1-FH2-C | − | − | This study | Polymerization (isoleucine mutation affects depolymerization) |
a This value was not accurately determined due to the presence of the C terminus.
The Ile-643 residue of INF2 corresponds to an almost universally conserved isoleucine in the knob region of the FH2. This isoleucine is present in all formins examined in yeast and metazoans (Ref. 1 and further unpublished sequence analysis), and the side chain protrudes toward the center of the FH2 donut from an α helix in the knob, according to crystal structures of Bni1p, DAAM1, mDia1, and FMNL3 (14, 30, 39, 44).3 In the Bni1p/actin co-crystal (44), the isoleucine side chain engages in interactions with residues at the “barbed end cleft” of actin between subdomains 1 and 3, a similar position to that occupied by WH2 domains (45).
Mutation of this isoleucine to an alanine severely reduces or abolishes the barbed end binding affinity of most FH2 domains, including fission yeast Cdc12 (5) and mammalian mDia1, mDia2, and FMNL3 (12, 13). In contrast, the Ile → Ala mutation had much less effect on barbed end binding by FMNL1-FFC (12), but subsequent work might provide a reason for this result. In FMNL3, which is highly homologous to FMNL1, the Ile → Ala mutation abolishes barbed end binding in the FF construct, whereas significant barbed end affinity is maintained in the FFC construct (13). The C terminus of FMNL3 contains a barbed end-binding WH2-like motif, which substitutes for the FH2 on the barbed end in the Ile → Ala mutant. Because FMNL1 contains a similar WH2-like motif, the situation is probably similar for this protein.
INF2 is unusual in that the I643A mutation does not abolish its ability to slow barbed end elongation or to accelerate polymerization. Comparing these results to the complete absence of detectable barbed end binding for the Ile → Ala mutants of mDia2 (12) and FMNL3 (13) or the strongly reduced binding of Cdc12 and mDia1 Ile → Ala mutants (5, 12), the difference is dramatic. Similar mutants of Fus1, For3, DAAM1, and Bni1p have been tested in pyrene-actin assembly assays and are strongly deficient for polymerization acceleration (Refs. 5, 14, and 30 and Table 1). What might be the reason for the relative insensitivity of INF2 to this Ile → Ala mutation compared with other formins? Possibly, other residues within this knob helix might serve redundant actin binding functions in INF2. Another question is why the I643A mutant of the INF2-FFC construct maintains significantly more polymerization activity than the I643A-FF construct. The WH2 domain of INF2 might aid in actin nucleation, in a manner perhaps similar to FMNL3 (13) and other formins (19). We note here that, although the INF2 WH2 domain binds actin monomers (20), it does not have appreciable affinity for barbed ends, unlike the WH2-like motif in the C terminus of FMNL3 (13).
The Lys-792 residue of INF2 corresponds to a highly conserved basic residue in a loop within the post region of the FH2, which is mostly lysine (but sometimes arginine) in every yeast and metazoan formin we have examined (Ref. 1 and subsequent unpublished analysis). Biochemical effects of mutation to this residue have been more variable than for the Ile → Ala mutation. For Bni1p, Fus1, and For3, mutation of this residue severely reduces polymerization activity (5, 30). In contrast, this mutation neither affects barbed end binding nor polymerization activity for Cdc12 (5). We have found a similar absence of effect for FMNL3.4 Here, we find that the K792A mutant has similar barbed end affinity as WT, as judged by elongation inhibition assays, and actually inhibits elongation to a greater extent than does WT. In actin polymerization assays, the activity of the K792A mutant is only mildly reduced. As with the Ile → Ala mutation, one explanation for the variable sensitivity of formins to mutation of this basic residue might be the presence functionally redundant residues elsewhere in this region of the protein.
The elongation effects of these mutants suggest effects on the processive elongation mechanism. Current models suggest that the FH2 domain transitions between two or possibly three barbed end-bound states (11, 44). The position of Lys-792 near the lasso/post dimerization region might suggest an effect in the speed of transition between these two states. The effect of I643A is more intriguing. Does this mutation block barbed end binding in one FH2 conformation but allow it in the other, resulting in filaments that are either completely capped or that are unbound by FH2? Does this mutation slow or abolish the transition between these two states?
An interesting difference between the I643A and K792A mutations is the differential effect they have on the severing and depolymerization activities of INF2-FFC. Our previous studies showed that the presence of the FH2 in cis with the C terminus was required for severing/depolymerization (20). The relevant features of the FH2 domain for severing are not clear. Because both mutations maintain some barbed end affinity, the barbed end binding ability of the FH2 domain does not seem to be the difference here. One intriguing possibility is that FH2 binding to the filament is able to change the filament conformation during the severing process, and the I643A mutation affects this ability significantly.
One result that contrasts with our earlier study is that the WH2 mutant strongly inhibits severing in our current results, whereas we previously found much less effect on severing (20). This result is puzzling, but one possibility is the difference in species used in this study (human) as opposed to our previous work (mouse). The C terminus of INF2 is the least conserved region between mouse and human. Another possibility is that we were incorrect in our past interpretation when concluding that the slightly longer filaments observed for the mouse WH2 mutant were due to deficient depolymerization activity (20) subsequent to severing. The human protein might be much more dependent on the WH2 for severing, which might explain the difference in degree of effect in the severing assays between human and mouse.
Another interesting finding is that the I643A mutation maintains significant polymerization activity for the FFC construct in vitro but does not cause measurable actin polymerization on ER in cells when the FFC construct is overexpressed. This difference appears to be due to the absence of capping protein in our in vitro polymerization assays. When capping protein is present, the I643A is severely deficient in polymerization activity. This result agrees with the decreased ability of the I643A mutant to bind barbed ends stably, shown in the TIRF assays.
An important implication of our findings is that neither the isoleucine nor the lysine mutant can be used as “loss of function” mutations in cellular studies without first testing the biochemical effects of that mutation in the formin being studied. For INF2, the K792A mutation has been used specifically as a polymerization-blocking mutation in cellular studies (23, 24), to discriminate between the polymerization and severing/depolymerization activities of INF2. Interestingly, this mutation affects neither polymerization nor severing/depolymerization strongly. In the cellular studies, however, K792A mutant INF2 could not rescue defects caused by knockdown of the endogenous protein (23, 24). This result implies that the relatively minor effects that we observe in vitro might be significant in cells, although it is difficult to determine which defects (to polymerization or severing/depolymerization) are most relevant.
Finally, one might be tempted to use the I643A mutation of INF2 as a polymerization-blocking construct in cells. However, we show here that it has a major effect on depolymerization activity. Yet again this result shows us that, in the world of actin assembly factors, INF2 remains militantly non-conFORMINist.
Acknowledgments
We thank Duane Compton (The Geisel School of Medicine at Dartmouth), Victoria Allan (University of Manchester, Manchester, UK), and Jennifer Lippincott-Schwartz (National Institutes of Health) for cell lines and plasmids. We thank Christopher Bahl (The Geisel School of Medicine at Dartmouth) for help with threading and crystal structure analysis. Our special thanks to Ernest Heimsath for help with data analysis and critical reading of the manuscript and to Evin Serg for being a cut above.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM069818 and R01 DK088826. This work was also supported by the Hitchcock Foundation.

This article contains supplemental Fig. S1.
M. E. Thompson, E. G. Heimsath, T. J. Gauvin, H. N. Higgs, and F. J. Kull, manuscript in preparation.
E. G. Heimseth and H. N. Higgs, unpublished results.
- FH
- formin homology
- ER
- endoplasmic reticulum
- TRITC
- tetramethylrhodamine isothiocyanate
- FF
- FH1-FH2
- FFC
- FH1-FH2-C.
REFERENCES
- 1. Higgs H. N., Peterson K. J. (2005) Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chalkia D., Nikolaidis N., Makalowski W., Klein J., Nei M. (2008) Origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol. 25, 2717–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grunt M., Zárský V., Cvrcková F. (2008) Roots of angiosperm formins. The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 8, 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moseley J. B., Goode B. L. (2006) The yeast actin cytoskeleton. From cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70, 605–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scott B. J., Neidt E. M., Kovar D. R. (2011) The functionally distinct fission yeast formins have specific actin-assembly properties. Mol. Biol. Cell 22, 3826–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Campellone K. G., Welch M. D. (2010) A nucleator arms race. Cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Higgs H. N. (2005) Formin proteins. A domain-based approach. Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 8. Goode B. L., Eck M. J. (2007) Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76, 593–627 [DOI] [PubMed] [Google Scholar]
- 9. Chesarone M. A., Goode B. L. (2009) Actin nucleation and elongation factors. Mechanisms and interplay. Curr. Opin Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pring M., Evangelista M., Boone C., Yang C., Zigmond S. H. (2003) Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42, 486–496 [DOI] [PubMed] [Google Scholar]
- 11. Paul A. S., Pollard T. D. (2009) Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 13. Heimsath E. G., Jr., Higgs H. N. (2012) The C terminus of formin FMNL3 accelerates actin polymerization and contains a WH2 domain-like sequence that binds both monomers and filament barbed ends. J. Biol. Chem. 287, 3087–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu J., Meng W., Poy F., Maiti S., Goode B. L., Eck M. J. (2007) Structure of the FH2 domain of Daam1. Implications for formin regulation of actin assembly. J. Mol. Biol. 369, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435 [DOI] [PubMed] [Google Scholar]
- 16. Harris E. S., Gauvin T. J., Heimsath E. G., Higgs H. N. (2010) Assembly of filopodia by the formin FRL2 (FMNL3). Cytoskeleton 67, 755–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaillant D. C., Copeland S. J., Davis C., Thurston S. F., Abdennur N., Copeland J. W. (2008) Interaction of the N- and C-terminal autoregulatory domains of FRL2 does not inhibit FRL2 activity. J. Biol. Chem. 283, 33750–33762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moseley J. B., Goode B. L. (2005) Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J. Biol. Chem. 280, 28023–28033 [DOI] [PubMed] [Google Scholar]
- 19. Gould C. J., Maiti S., Michelot A., Graziano B. R., Blanchoin L., Goode B. L. (2011) The formin DAD domain plays dual roles in autoinhibition and actin nucleation. Curr. Biol. 21, 384–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chhabra E. S., Higgs H. N. (2006) INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J. Biol. Chem. 281, 26754–26767 [DOI] [PubMed] [Google Scholar]
- 21. Chhabra E. S., Ramabhadran V., Gerber S. A., Higgs H. N. (2009) INF2 is an endoplasmic reticulum-associated formin protein. J. Cell Sci. 122, 1430–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramabhadran V., Korobova F., Rahme G. J., Higgs H. N. (2011) Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol. Biol. Cell 22, 4822–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Madrid R., Aranda J. F., Rodríguez-Fraticelli A. E., Ventimiglia L., Andrés-Delgado L., Shehata M., Fanayan S., Shahheydari H., Gómez S., Jiménez A., Martín-Belmonte F., Byrne J. A., Alonso M. A. (2010) The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev. Cell 18, 814–827 [DOI] [PubMed] [Google Scholar]
- 24. Andrés-Delgado L., Antón O. M., Madrid R., Byrne J. A., Alonso M. A. (2010) Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116, 5919–5929 [DOI] [PubMed] [Google Scholar]
- 25. Brown E. J., Schlöndorff J. S., Becker D. J., Tsukaguchi H., Tonna S. J., Uscinski A. L., Higgs H. N., Henderson J. M., Pollak M. R. (2010) Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat. Genet. 42, 72–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boyer O., Benoit G., Gribouval O., Nevo F., Tête M. J., Dantal J., Gilbert-Dussardier B., Touchard G., Karras A., Presne C., Grunfeld J. P., Legendre C., Joly D., Rieu P., Mohsin N., Hannedouche T., Moal V., Gubler M. C., Broutin I., Mollet G., Antignac C. (2011) Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J. Am. Soc. Nephrol. 22, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee H. K., Han K. H., Jung Y. H., Kang H. G., Moon K. C., Ha I. S., Choi Y., Cheong H. I. (2011) Variable renal phenotype in a family with an INF2 mutation. Pediatr. Nephrol. 26, 73–76 [DOI] [PubMed] [Google Scholar]
- 28. Gbadegesin R. A., Lavin P. J., Hall G., Bartkowiak B., Homstad A., Jiang R., Wu G., Byrd A., Lynn K., Wolfish N., Ottati C., Stevens P., Howell D., Conlon P., Winn M. P. (2012) Inverted formin 2 mutations with variable expression in patients with sporadic and hereditary focal and segmental glomerulosclerosis. Kidney Int. 81, 94–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G., Cong E. H., Arrondel C., Tête M. J., Montjean R., Richard L., Karras A., Pouteil-Noble C., Balafrej L., Bonnardeaux A., Canaud G., Charasse C., Dantal J., Deschenes G., Deteix P., Dubourg O., Petiot P., Pouthier D., Leguern E., Guiochon-Mantel A., Broutin I., Gubler M. C., Saunier S., Ronco P., Vallat J. M., Alonso M. A., Antignac C., Mollet G. (2011) INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N. Engl. J. Med. 365, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 30. Xu Y., Moseley J. B., Sagot I., Poy F., Pellman D., Goode B. L., Eck M. J. (2004) Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell 116, 711–723 [DOI] [PubMed] [Google Scholar]
- 31. Woźniak M. J., Bola B., Brownhill K., Yang Y. C., Levakova V., Allan V. J. (2009) Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J. Cell Sci. 122, 1979–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spudich J. A., Watt S. (1971) The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 33. Cooper J. A., Walker S. B., Pollard T. D. (1983) Pyrene actin. Documentation of the validity of a sensitive assay for actin polymerization. J. Muscle Res. Cell Motil 4, 253–262 [DOI] [PubMed] [Google Scholar]
- 34. MacLean-Fletcher S., Pollard T. D. (1980) Mechanism of action of cytochalasin B on actin. Cell 20, 329–341 [DOI] [PubMed] [Google Scholar]
- 35. Harris E. S., Li F., Higgs H. N. (2004) The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 36. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pollard T. D. (1986) Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103, 2747–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Isambert H., Venier P., Maggs A. C., Fattoum A., Kassab R., Pantaloni D., Carlier M. F. (1995) Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270, 11437–11444 [DOI] [PubMed] [Google Scholar]
- 39. Yamashita M., Higashi T., Suetsugu S., Sato Y., Ikeda T., Shirakawa R., Kita T., Takenawa T., Horiuchi H., Fukai S., Nureki O. (2007) Crystal structure of human DAAM1 formin homology 2 domain. Genes Cells 12, 1255–1265 [DOI] [PubMed] [Google Scholar]
- 40. Pollard T. D., Cooper J. A. (1986) Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55, 987–1035 [DOI] [PubMed] [Google Scholar]
- 41. Zigmond S. H., Evangelista M., Boone C., Yang C., Dar A. C., Sicheri F., Forkey J., Pring M. (2003) Formin leaky cap allows elongation in the presence of tight capping proteins. Curr. Biol. 13, 1820–1823 [DOI] [PubMed] [Google Scholar]
- 42. Kovar D. R., Wu J. Q., Pollard T. D. (2005) Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell 16, 2313–2324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaillard J., Ramabhadran V., Neumanne E., Gurel P., Blanchoin L., Vantard M., Higgs H. N. (2011) Differential interactions of the formins INF2, mDia1, and mDia2 with microtubules. Mol. Biol. Cell 22, 4575–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Otomo T., Tomchick D. R., Otomo C., Panchal S. C., Machius M., Rosen M. K. (2005) Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433, 488–494 [DOI] [PubMed] [Google Scholar]
- 45. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]