Background: Current strategies to alleviate protein misfolding include manipulation of chaperones, proteasomes, or autophagy.
Results: Mild translation inhibition disproportionally blocked production of misfolded proteins and improved mutant CFTR function.
Conclusion: Slowing down translation improves folding of newly synthesized proteins in mammalian cells and recovers mutant protein function.
Significance: Attenuation of translation could be a novel approach to treatment of protein-misfolding disorders.
Keywords: CFTR, Chaperone Chaperonin, Protein Degradation, Protein Misfolding, Protein Synthesis
Abstract
Protein homeostasis depends on a balance of translation, folding, and degradation. Here, we demonstrate that mild inhibition of translation results in a dramatic and disproportional reduction in production of misfolded polypeptides in mammalian cells, suggesting an improved folding of newly synthesized proteins. Indeed, inhibition of translation elongation, which slightly attenuated levels of a copepod GFP mutant protein, significantly enhanced its function. In contrast, inhibition of translation initiation had minimal effects on copepod GFP folding. On the other hand, mild suppression of either translation elongation or initiation corrected folding defects of the disease-associated cystic fibrosis transmembrane conductance regulator mutant F508del. We propose that modulation of translation can be used as a novel approach to improve overall proteostasis in mammalian cells, as well as functions of disease-associated mutant proteins with folding deficiencies.
Introduction
Maintenance of the functional cellular proteome relies on the intricate balance of protein synthesis, folding, and degradation. In the absence of stresses, the newly synthesized misfolded proteins pose the main challenge to cellular protein homeostasis. Molecular chaperones, the ubiquitin-proteasome system, and autophagy play an important role in handling these species (1).
Because abnormal proteins cause many life-threatening diseases, there has been an ongoing interest in approaches that reduce the accumulation of misfolded species. In fact, a number of these diseases could be partially alleviated by induction of autophagy (2, 3), and some inducers of autophagy have shown efficacy in animal models (4, 5). Additionally, certain protein-misfolding pathologies can be alleviated by overexpression of heat shock proteins (6, 7), and numerous attempts have been made to develop inducers of molecular chaperones (8, 9).
This approach can be used for another group of disorders associated not with gain of toxicity of misfolded polypeptides but with insufficient function of mutant proteins. For example, the adverse effects of mutant lysosomal glucocerebrosidase in Gaucher disease (10) can be alleviated by an increase in the chaperone capacity of cells (10). Misfolded molecules of the mutant glucocerebrosidase are rapidly degraded via the endoplasmic reticulum (ER)2-associated protein degradation pathway. However, folded species of glucocerebrosidase escape ER-associated protein degradation, proceed to lysosomes, and function normally. Accordingly, induction of ER chaperones improves folding of glucocerebrosidase and increases its levels (10). Analogous effects were seen with mutant β-hexosaminidase A, the causative agent of Tay-Sachs disease (10). This approach may be useful in the rescue of other mutant proteins transported via the ER, such as mutant cystic fibrosis transmembrane conductance regulator (CFTR), which causes cystic fibrosis. The most prevalent disease-causing mutation in CFTR is F508del, which results in poor folding and rapid degradation of the polypeptide via the ER-associated protein degradation pathway (11). However, a fraction of molecules that have acquired the correct fold escape ER degradation and are functional (12, 13). Correcting the folding defect of the F508del-CFTR mutant by chemical chaperones seems to be a successful strategy in therapeutics discovery.
Here, we propose that, in addition to induction of chaperones, slowing down translation may have beneficial effects on the production of functional mutant proteins. It might decrease the pool of chaperone substrates and thereby increase the capacity of the folding system. Moreover, it may improve co-translational folding (14). This possibility is supported by the following circumstantial lines of evidence: (a) slow-translating stretches of mRNA provide for proper domain folding in vitro and in Escherichia coli (15); (b) a mutation that reduces elongation rate of E. coli ribosomes improves folding of recombinant eukaryotic proteins, which have evolved to be translated by comparatively slower eukaryotic ribosomes (16); and (c) it is a common practice to improve production of correctly folded recombinant proteins in E. coli by reducing growth temperature. However, there is no direct evidence that slowing down translation can improve folding in eukaryotic cells.
Here, we demonstrate that mild inhibition of translation significantly improves overall protein folding in mammalian cells. Furthermore, slowing down translation improves folding of mutant proteins, suggesting a novel approach to a cure for protein-misfolding disorders.
MATERIALS AND METHODS
Reagents and Antibodies
MG132 was purchased from BIOMOL (Farmingdale, NY). Emetine, forskolin, cycloheximide, 3-isobutyl-1-methylxanthine (IBMX), and genistein were from Sigma. Hippuristanol was a kind gift of Dr. J. Pelletier. Shield-1 from was obtained from Cheminpharma LLC (Farmington, CT). Anti-p21 and anti-p53 antibodies were purchased from Pharmingen and Santa Cruz Biotechnology (Santa Cruz, CA). Anti-multiubiquitin antibody (FK2) from MBL International (Woburn, MA). Anti-HA and anti-β-actin antibodies were from Cell Signaling (Danvers, MA). Mouse anti-GAPDH antibody was purchased from Millipore. Anti-CFTR antibody (clone 59) was a kind gift from Dr. Jack Riordan (University of North Carolina, Chapel Hill, North Carolina, via the Cystic Fibrosis Foundation).
Constructs
The retroviral expression construct with C-terminally tagged synphilin-1 (synphilin-1-GFP) subcloned into the pCXbsr vector was described previously (17).
Copepod GFP (copGFP) was amplified from the vector pMaxGFP (Lonza, Allendale, NJ) and fused with FBP12 (FK506-binding protein 12; from the vector ppTuner, Clontech, Mountain View, CA) by overlapping PCR. The product was then cloned into the lentiviral vector pTRIPz (Open Biosystems, Huntsville, AL). A C-terminal hemagglutinin tag was also inserted downstream of copGFP by PCR. copGFP expression was controlled by the upstream tetracycline-responsive element.
Cell Cultures and Growth
HeLa (cervix carcinoma) cells were grown in DMEM supplemented with 10% fetal bovine serum, and MCF-10A (human breast epithelial) cells were grown in 50:50 DMEM/F-12 medium supplemented with 5% horse serum, 20 ng/ml epidermal growth factor, 0.5 μg/ml hydrocortisone, 10 μg/ml human insulin, and 100 ng/ml cholera toxin. All cultures were supplemented with l-glutamine as well as penicillin and streptomycin and grown at 37 °C in an atmosphere of 5% CO2. copGFP was induced by the addition of 1 μg/ml doxycycline and further stabilized by the addition of 5 μm Shield-1 (18).
The Fischer rat thyroid (FRT) cell line stably expressing F508del-CFTR was a generous gift from Prof. Luis Galietta (University of Genoa, Genoa, Italy). Cells were grown as described previously (19).
For production of retroviruses, HEK293T cells were co-transfected with a lentiviral plasmid and helper plasmids expressing lentiviral proteins (psPAX2 and pMD2.G). Supernatants containing the virus were collected 48 h after transfection.
Cell Lysis and Analysis
For analysis of ubiquitination levels, cells were lysed with lysis buffer (40 mm HEPES (pH 7.5), 50 mm KCl, 1% Triton X-100, 2 mm DTT, 1 mm Na3VO4, 50 mm β-glycerophosphate, 50 mm NaF, 5 mm EDTA, 5 mm EGTA, 1 mm PMSF, 1 mm benzamidine, 5 μg/ml leupeptin, 5 μg/ml pepstatin A, and 5 μg/ml aprotinin). Samples were adjusted to have an equal concentration of total protein and subjected to SDS PAGE, followed by immunoblotting. The immunoblots represent a typical experiment repeated three times.
Aggresome Counting Microscopy
For analysis with a fluorescence microscope, cells were grown on Lab-Tek chambered cover glasses (Nunc) pretreated with poly-l-lysine (Sigma). Fluorescence microscopy was performed at room temperature with a Zeiss Axiovert 200 microscope using a 40×/0.75 or 100×/1.30 oil Objectives and AxioVision 4 software. GFP-tagged proteins were observed with an Axio FITC filter set. Images were obtained using a high resolution AxioCam MRm microscope camera. To assess the fraction of cells with a detectable aggresome, the fluorescent cells were blindly counted in 10 randomly chosen fields to obtain a total of >200 cells. Each counting experiment was repeated three times to ensure reproducibility of the results.
copGFP Fluorescence and Levels
HeLa cell clones were plated at a density of 2 × 104 cells/well and allowed to adhere. copGFP expression was induced by the addition of 1 μg/ml doxycycline and 5 μm Shield-1. Simultaneously, emetine or hippuristanol was titrated by 2-fold serial dilutions as indicated. Cells were incubated for 6 h at 37 °C and then prepared for analysis by flow cytometry (FACSCalibur, BD Biosciences). For immunoblotting, cells were plated on 12-well plates and lysed in buffer containing 1% Nonidet P-40. Protein was quantified by BCA (Pierce), and equivalent total protein was loaded per well. copGFP expression was monitored using anti-HA tag antibody (Clontech), whereas protein loading was monitored by β-actin levels.
FRT Cell Conductance Assay
FRT cells were seeded at a density of ∼125,000 cells/cm2 on HTS Transwell 24-well filter inserts (catalog number 3378, Corning) and grown into epithelial cell monolayers as described previously (19). Prior to the assay, monolayers were treated on both sides with compound or vehicle (negative control) for 24 h in a cell/tissue incubator (37 °C, 5% CO2).
After 24 h of treatment, the cell incubation medium was replaced with HEPES-buffered physiological saline as the serosal (bottom well) and mucosal (insert top) solutions with a composition of 137 mm NaCl, 4.0 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, and 10 mm glucose (with pH adjusted to 7.4 with NaOH). The Transwell 24-well plate was then mounted onto a heated plate, and transepithelial resistance was measured using a four-channel transepithelial current clamp (EP Design, Bertem, Belgium). The assay was carried out at a well temperature of ∼35–36 °C. Resistance values were collected at ∼10–15-min intervals. Four data points were measured to determine base-line resistance, and three data points were measured to determine transepithelial resistance after each addition of agonist (final concentrations of 10 μm forskolin, 100 μm IBMX, and 20 μm genistein) and antagonist (final concentration of 20 μm CFTR(inh)-172). IBMX and genistein were applied together. The agonists and antagonist were added to both the serosal and mucosal sides and prediluted to 10-fold concentrations in HEPES-buffered physiological saline.
Transepithelial conductance (Gt) was calculated from series resistance-subtracted transepithelial current clamp measurements. Because CFTR mediates chloride ion passage across the epithelial monolayer, activation/inhibition of functional CFTR transport proteins results in a change in transepithelial conductance (ΔGt). The magnitude of ΔGt therefore is a measure of functional CFTR surface expression.
To study the effect of test compounds on CFTR surface expression, the dose-response characteristic of the compound was determined and compared with negative controls. ΔGt mean values were calculated for each treatment condition, and the ΔGt ratio (compound/vehicle) for the CFTR-specific inhibitor response was plotted for each test concentration.
FRT Cell Ussing Chamber Assay
Cells were grown into electrically tight monolayers on Snapwell filter supports (catalog number 3801, Corning) as described previously (19). Both serosal and mucosal membranes were exposed to compound or vehicle (negative control) in a cell/tissue incubator (37 °C and 5% CO2) for 24 h prior to the assay. The inserts were then transferred to a Ussing chamber (catalog number P2302, Physiologic Instruments Inc., San Diego, CA) and superfused with 5 ml of HEPES-buffered physiological saline as the serosal solution with a composition of 137 mm NaCl, 4.0 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, and 10 mm glucose (with pH adjusted to 7.4 with NaOH). The mucosal solution was 5 ml of low Cl− physiological saline with a composition of 137 mm sodium gluconate, 4 mm KCl, 1.8 mm CaCl2, 1 mm MgCl2, 10 mm HEPES, and 10 mm glucose (with pH adjusted to 7.4 with N-methyl-d-glucamine to create a transepithelial chloride ion gradient). The transepithelial voltage was clamped to 0 mV, and the short circuit current, reflecting the net ion (Cl−) transport across the epithelial cell monolayer, was measured using a VCC MC8 epithelial voltage clamp (Physiologic Instruments Inc.). The assay was carried out at 37 °C. Base-line activity was recorded for 10 min before the agonists (final concentrations of 10 μm forskolin, 100 μm IBMX, and 20 μm genistein) and antagonist (final concentration of 20 μm CFTR(inh)-172) were applied sequentially and cumulatively at ∼10-min intervals to both serosal and mucosal epithelial surfaces. The agonists and antagonist were added as 200–1000-fold stock solutions to both the serosal and mucosal sides.
RESULTS
Translation Rate Influences Generation of Abnormal Polypeptides
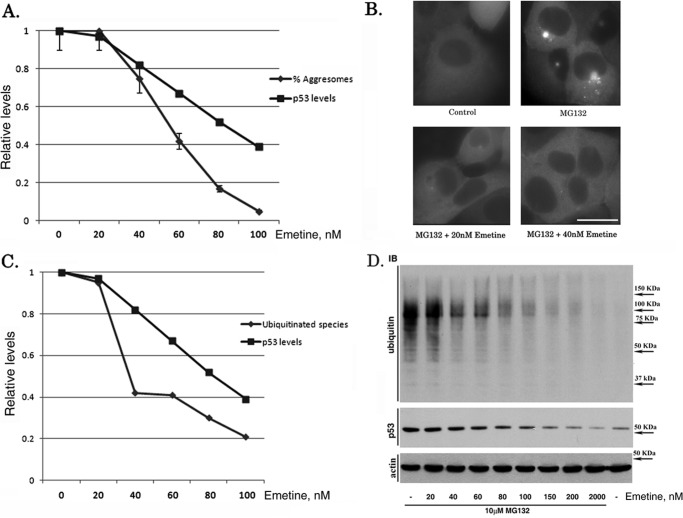
In a previous study, we investigated formation of the aggresome, an organelle that recruits small protein aggregates to the centrosome location (20, 21), using synphilin-1-GFP as a model (17). We found that this process is triggered by the buildup of newly synthesized aberrant proteins (22), which allowed assessing the levels of these species by monitoring aggresome formation. Moreover, with HeLa cells, we observed that aggresome formation is extremely sensitive to inhibition of translation. To account for this unexpected effect, we hypothesize that partial inhibition of translation disproportionally reduces the generation of aberrant polypeptides. We explored this possibility with another cell line, MCF-10A. Cells expressing synphilin-1-GFP were incubated with the proteasome inhibitor MG132, which led to a rapid formation of an aggresome (Fig. 1B, upper right panel). The translation inhibitor emetine suppressed the aggresome formation in a dose-dependent manner (Fig. 1, A and B), indicating inhibition of the production of misfolded polypeptides. To test whether emetine reduces generation of misfolded species preferentially, we quantitatively compared the effects of emetine on aggresome formation and on protein synthesis. In the presence of the proteasome inhibitor, accumulation of a de novo synthesized short-lived protein, e.g. p53, directly reflects the rate of translation. Accordingly, to assess the effects of emetine on general translation, we measured inhibition of the accumulation of p53 (note that accumulation of a distinct short-lived protein p21 (data not shown), as well as of inducible copGFP (see below), displayed similar to p53 dose dependence). As shown in Fig. 1A, inhibition of p53 synthesis was markedly less sensitive to emetine than inhibition of aggresome formation. For example, 80 nm emetine inhibited accumulation of p53 by <2-fold but decreased the fraction of cells forming detectable aggresomes by almost 10-fold. Moreover, at low concentrations, emetine markedly reduced the sizes of the remaining aggresomes (Fig. 1B). For example, the diameters of aggresomes formed in the presence of 20 nm emetine were, on average, three times smaller than those seen without translation inhibition, demonstrating that even very mild inhibition of translation suppresses growth of an aggresome. Because aggresome formation depends on accumulation of abnormal newly synthesized polypeptides (22), these data suggest that mild inhibition of translation disproportionally reduces generation of abnormal polypeptides.
FIGURE 1.
Emetine disproportionally inhibits accumulation of ubiquitinated species and aggresome formation. A and B, effect of emetine on translation and aggresome formation. MCF-10A cells stably expressing synphilin-1-GFP were incubated with 5 μm MG132 and the indicated concentrations of emetine for 2 h, and the fraction of cells with an aggresome were counted under a fluorescence microscope. In parallel, cell lysates were prepared after 4 h of incubation, and the levels of p53 were measured by immunoblotting. A, quantification of the results of aggresome counting and immunoblotting. B, effect of low doses of emetine on aggresome appearance. Scale bar = 20 μm. C and D, effect of emetine on accumulation of ubiquitinated species. MCF-10A cells were incubated with 5 μm MG132 and the indicated concentrations of emetine, and the amounts of ubiquitinated species and p53 were assessed by immunoblotting. C, quantification of immunoblotting. D, immunoblots. All experiments were reproduced three times, and data are representative. Error bars represent S.E.
To further evaluate the effects of translation inhibitors on generation of abnormal polypeptides, we took advantage of the observation that the majority of proteins that are ubiquitinated and degraded by the ubiquitin-proteasome pathway are newly synthesized. Indeed, the levels of ubiquitinated species notably increased upon proteasome inhibition, but blocking translation with emetine prevented their buildup by 90–95% (Fig. 1, C and D). A large fraction of newly synthesized species that are degraded by proteasome and therefore accumulate upon its inhibition are polypeptides that cannot fold normally. According to our hypothesis, improved protein folding due to the presence of low doses of emetine should disproportionally inhibit generation of ubiquitinated species compared with inhibition of translation. To test this possibility, MCF-10A cells were incubated for 3.5 h with MG132, and the effects of various concentrations of emetine on the cellular levels of ubiquitin conjugates and on the buildup of p53 were evaluated. Fig. 1 (C and D) demonstrates that the buildup of ubiquitinated species was significantly more sensitive to emetine. This result supports our hypothesis that low concentrations of emetine specifically reduce a fraction of abnormal species among newly synthesized polypeptides. Taken together, the data on aggresome formation and on the levels of ubiquitinated polypeptides indicate that inhibition of translation by ∼50% almost entirely blocks generation of broad-spectrum misfolded polypeptides.
The described effect on cellular proteostasis was not specific to emetine but rather reflected mild translation inhibition. Indeed, another inhibitor (cycloheximide) similar to emetine preferentially inhibited both aggresome formation and accumulation of ubiquitinated species (Fig. 2A) (22). Like most of the available ribosome inhibitors, these two reagents inhibited elongation of translation and thus slow downed the growth of each polypeptide chain in ribosomes.
FIGURE 2.

Different effects of cycloheximide and hippuristanol on translation, accumulation of ubiquitinated species and aggresome formation. A, effects of cycloheximide on translation, accumulation of ubiquitinated species, and aggresome formation. Experiments were performed as described in the legend to Fig. 1. B, effects of hippuristanol on translation, accumulation of ubiquitinated species and aggresome formation. Experiments were performed as described in the legend to Fig. 1. All data were reproduced several times and are representative. Error bars represent S.E.
We next tested whether comparable reduction in the output of translation achieved without slowing down polypeptide growth has a similar effect on generation of abnormal species. Accordingly, we employed hippuristanol, which reduces protein synthesis by inhibiting translation initiation and thus reducing the number of active ribosomes (24). Hippuristanol elicited the same dose responses for generation of ubiquitinated species and for translation output (Fig. 2B). Unexpectedly, at low concentrations, this inhibitor appeared to slightly increase protein synthesis, resulting in the minor accumulation of both p53 and ubiquitinated species (Fig. 2B). Nonetheless, low concentrations of hippuristanol did not cause an increase in aggresome formation, which was already close to maximum. Therefore, the dose-dependence curves for production of p53 and ubiquitinated species are seen shifted above the curve for aggresome formation. However, starting from 150 nm hippuristanol (at which translation output peaked), all three curves are parallel (Fig. 2B). These data suggest that the rate of translation affects production of abnormal proteins, and in this model, the rate of polypeptide growth is more important for generation of defective ribosome products than the number of translating ribosomes.
Rate of Translation Affects Folding of a Model Protein
To investigate the effects of the rate of translation on protein folding, we utilized a recombinant GFP from the copepod Pontellina plumata (copGFP) as a reporter. copGFP was fused to FBP12, which facilitates its rapid degradation; however, the fusion protein can be stabilized by the addition to the cells of the small molecule Shield-1. (This system, which controls protein stability, has been described previously (18).) Both the fluorescence (which reflects folding) and protein levels of this polypeptide could be easily quantified, making it a useful folding reporter.
Mutant copGFP was expressed in HeLa cells under the control of a tetracycline-regulated promoter using the retroviral expression system. Incubation with doxycycline for 6 h in the presence of Shield-1 led to >10-fold induction of copGFP (Fig. 3B, compare the first two lanes). Various concentrations of emetine were added to the cells at the start of induction, and 6 h later, copGFP levels and fluorescence were measured. As expected, the buildup of copGFP, which also reflects the overall rate of translation, was reduced by emetine in a dose-dependent manner (Fig. 3, A and B), similar to the inhibition of p53 accumulation (see Fig. 1). However, paradoxically, the fluorescence was affected in a very different manner, showing a steady increase that peaked at ∼40 nm emetine, reaching 150% of the control. Importantly, at this concentration, emetine already reduced the levels of copGFP by ∼20% (Fig. 3, A and B). Even stronger divergence between the copGFP protein levels and copGFP fluorescence was seen with 80 nm emetine (Fig. 3A). The effect of emetine on the fluorescence of copGFP normalized to its levels demonstrates that slowing down translation improved folding of this protein by >4-fold (Fig. 3C).
FIGURE 3.
Inhibition of translation elongation enhances folding of mutant copGFP in HeLa cells. A, effects of emetine on the fluorescence and levels of mutant copGFP. copGFP was induced for 6 h in the presence of the indicated concentrations of emetine, and its fluorescence and levels normalized to total protein were measured as described under “Materials and Methods.” B, levels of copGFP in the experiment presented in A assessed by immunoblotting. The levels of this inducible protein also reflect the degree of translation inhibition. C, effects of emetine on the ratio of copGFP fluorescence to copGFP level. Calculation was based on data in A. D, effects of hippuristanol on the fluorescence and levels of mutant copGFP. copGFP was induced for 6 h in the presence of the indicated concentrations of hippuristanol, and its fluorescence and levels were measured as described under “Materials and Methods.” E, effects of hippuristanol on the ratio of copGFP fluorescence to copGFP level. Calculation was based on data in D. All experiments were reproduced three times, and data are representative. Error bars represent S.E.
We next investigated whether inhibition of translation initiation also improves folding of the copGFP reporter (Fig. 3D). As shown in Fig. 3 (compare C and E for the dose dependence of specific activity), the difference between the inhibitory effects of hippuristanol on copGFP levels and on copGFP fluorescence was insignificant. Overall, these experiments demonstrate that partial inhibition of translation elongation, while reducing expression, promotes folding of certain polypeptides, thus resulting in an overall increase in their activity.
Partial Inhibition of Translation Can Improve the Activity of Mutant CFTR
The finding that mild inhibition of translation improves the function of certain proteins suggested that this treatment may be employed to correct folding defects of disease-related mutant proteins. To test this possibility, we chose the CFTR mutant F508del. Normally, CFTR functions as a chloride channel in the plasma membrane of epithelial cells, but the F508del mutation and certain other mutations jeopardize CFTR folding and trafficking, ultimately causing cystic fibrosis. To study the effect of translation inhibition on the functional surface expression of F508del-CFTR, we used FRT cells stably expressing recombinant F508del-CFTR. CFTR activity was measured by two independent methods, an Ussing chamber assay (25) and a conductance assay in a 24-well format (26), both of which measure CFTR-dependent transepithelial Cl− transport. The Ussing chamber assay was designed to measure this transport in the presence of a chloride ion gradient. Cells were exposed to various concentrations of emetine or left untreated, and CFTR activity was measured 24 h after the beginning of the treatment. During the measurement, CFTR was sequentially activated by compounds that increase cAMP levels and stimulate CFTR transport activity: 10 μm forskolin, 100 μm IBMX, and then 20 μm genistein (Fig. 4A). To confirm that the activated Cl− transport was CFTR-dependent, at the end of the measurement, we added CFTR(inh)-172, which blocked the CFTR-dependent component of the conductance.
FIGURE 4.
Inhibition of translation elongation corrects folding defects of mutant CFTR. A, left panel, effects of emetine on CFTR-mediated short-circuit currents in the Ussing chamber assay. Cells were plated on filter supports and incubated with the indicated concentrations of emetine for 24 h. Inserts were transferred to Ussing chambers. After acquisition of the base-line current, the agonists (10 μm forskolin (Fsk), 100 μm IBMX, and 20 μm genistein (Gen)) and antagonist (20 μm CFTR(inh)-172) were added sequentially to both epithelial surfaces, and short-circuit currents (Isc) were recorded. To better illustrate the emetine concentration effect on the CFTR inhibitor response, the post-inhibitor base-line current was subtracted from the raw current trace. Right panel, emetine dose response. Shown are the means (n = 2) ± S.D. of the CFTR-specific Cl− current normalized to CFTR(inh)-172. B, left panel, conductance assay of transepithelial conductance changes in response to CFTR agonists (10 μm forskolin, 100 μm IBMX, and 20 μm genistein) and antagonist (20 μm CFTR(inh)-172) additions. Traces represent averaged records (n = 3) from FRT cell monolayers treated for 24 h with the indicated concentrations of emetine. Right panel, emetine dose-dependence curve reflective of F508del-CFTR transport activity normalized to activity in the presence of CFTR inhibitor. The means ± S.D. from three independent experiments are shown. mS, millisiemens. C, effect of emetine on expression of F508del-CFTR. The levels of F508del-CFTR in cells treated as described in the legend to Fig. 3C were measured by immunoblotting. Band C, mature form; Band B, core-glycosylated form. Error bars represent S.E.
In line with our conjecture, we observed a dose-dependent increase in the activity of mutant CFTR in cells exposed to low concentrations of emetine (Fig. 4A). An almost 2-fold increase over vehicle in the CFTR-specific inhibitor response was reached at 0.5 μm emetine (Fig. 4A, right panel). The higher effective emetine concentrations used in this experiment compared with those used with HeLa or MCF-10A cells indicates a lower susceptibility of FRT cells to emetine (data not shown).
The effect of 24 h of emetine incubation on CFTR activity at the cell surface was independently confirmed in the FRT cell conductance assay, in which CFTR-dependent transepithelial (Cl−) conductance was measured in the absence of a chloride gradient (Fig. 4B). In this experiment, cells were also pretreated with various doses of emetine for 24 h. During the measurements, CFTR was first activated with 10 μm forskolin and then further stimulated with a combination of 100 μm IBMX and 20 μm genistein. CFTR(inh)-172 was added at the end of the experiment. Again, the dose dependence of the CFTR-specific inhibitor response peaked at 0.5 μm emetine as illustrated in Fig. 4B (right panel). At the peak concentration, the CFTR-specific inhibitor response was increased by >2-fold compared with the negative control.
We further investigated the effects of low doses of emetine on CFTR levels. Misfolded F508del-CFTR molecules are rapidly degraded by the ubiquitin-proteasome machinery. Therefore, we predicted that a mild inhibition of translation could paradoxically lead to elevated cellular levels of F508del-CFTR because folded molecules would escape degradation. Indeed, as shown in Fig. 4C, at low doses of emetine, we observed increased expression levels of CFTR represented by both the mature form (Band C) and the core-glycosylated form (Band B). The buildup of the core-glycosylated form of F508del-CFTR indicates that emetine enhances folding of this mutant protein in the ER, where it passes quality control and is sorted for either degradation (misfolded form) or for the membrane (folded form). Therefore, the rate of translation affects folding of newly synthesized proteins not only in cytoplasm, as seen with copGFP, but in the ER as well.
To assess the effects of the translation initiation inhibitor hippuristanol on CFTR folding, dose-response experiments were performed in FRT cell Ussing chambers. CFTR activity peaked at 0.6 μm inhibitor, culminating in an almost 3-fold increase in the CFTR(inh)-172 response compared with vehicle (Fig. 5A). As with emetine, the increase in activity was parallel to the increase in the CFTR protein levels, which also peaked at 0.6 μm (Fig. 5B). Therefore, with F508del-CFTR, inhibition of both translation elongation and initiation can improve folding. Together, the data described above indicate that modulation of translation presents a novel approach to treatment of disorders associated with protein misfolding.
FIGURE 5.

Inhibition of initiation of translation alleviates folding defects of mutant CFTR. A, Ussing chamber assay: hippuristanol concentration effect on CFTR-mediated short-circuit currents recorded in duplicates from FRT cell monolayers incubated for 24 h with the inhibitor. The experiment was done as described in the legend to Fig. 4A. B, effect of hippuristanol on expression of F508del-CFTR. The levels of F508del-CFTR in cells were measured as described in the legend to Fig. 4C. Band C, mature form; Band B, core-glycosylated form. Error bars represent S.E.
DISCUSSION
A critical finding here is that mild inhibition of translation can correct folding defects of mutant disease-associated proteins. There is a wide range of diseases beside cystic fibrosis that may be beneficially affected by partial inhibition of translation, e.g. Gaucher, Tay-Sachs, various conditions associated with collagen mutations, and others. These diseases result from insufficient folding and function of important enzymes and structural proteins. Another class of conditions that might be improved by this approach includes disorders associated with the buildup of toxic abnormal proteins, e.g. amyotrophic lateral sclerosis, inclusion body myositis, and others. Because all of these conditions are chronic, it will be necessary to develop nontoxic compounds that inhibit translation only mildly.
Another finding of this work is that the rate of translation has a significant impact on the quality of newly synthesized proteins and the overall proteostasis. Recently, it was demonstrated that newly synthesized polypeptides are most vulnerable to various stresses, e.g. heat shock or oxidative stress (27). It was argued that there are normal proteins, usually multisubunit proteins of high molecular weight, that fold slowly and often with low efficiency, and these partially unfolded species are especially sensitive to stresses. Because low doses of translation inhibitors reduce the overall production of abnormal polypeptides probably via enhancing folding, we hypothesize that partial inhibition of translation may significantly protect against proteotoxic stresses.
Several mutations that reduce translation initiation and elongation in Caenorhabditis elegans and Drosophila have been shown to have strong anti-aging effects (28, 29). Our data suggest that these anti-aging effects may be relevant to improved protein folding due to slower translation. Indeed, some of these mutations have been shown to improve overall proteostasis (30). Therefore, there is a possibility that minor suppression of translation may provide an anti-aging effect and be beneficial for a variety of age-related disorders.
In this work, we assessed translation rates by measuring de novo synthesis of short-lived proteins upon inhibition of their degradation. Independently, we measured accumulation of a stable reporter protein upon induction of its synthesis. The results obtained with these two distinct methods were similar. We found that slowing down elongation of translation improves general protein folding. These effects suggest either that the rate of growth of the polypeptide chain influences the kinetics of co-translational folding (e.g. allows association with ribosome-bound chaperones prior to co-translational misfolding or reduces the probability of improper interactions between the emerging domains) or that it reduces the overall translation output, thus increasing the number of available cytoplasmic chaperones (for both co-translational and post-translational folding). On the other hand, inhibitors of translation initiation do not affect growth of the polypeptides chain, but only the number of translating ribosomes. Therefore, these inhibitors cannot influence the kinetics of co-translational folding, but only the translation output.
The fact that only inhibitors of translation elongation reduced the overall production of misfolded species, whereas hippuristanol was not effective, indicates that, for the bulk of polypeptides, translation influences the kinetics of co-translational folding. Indeed, in vitro studies suggested that the slowing down of translation can improve co-translational folding (14, 15).
On the other hand, with F508del-CFTR, we observed folding improvement with both emetine and hippuristanol, indicating that the overall translation output is critical for CFTR folding and suggesting that the availability of chaperones is limiting. These findings may reflect the compartmental difference, where chaperones are not limiting for cytoplasmic proteins (which represent the bulk of the translation products in HeLa cells), and translation affects mainly the kinetics of co-translational folding. In contrast, for CFTR, which transits through the ER, ER chaperones may be limiting. Overall, the findings presented here can be considered a proof of principle that partial inhibition of translation could be developed as a novel therapeutic modality for treatment of diseases associated with inefficient folding of mutant proteins.
Acknowledgment
We thank Dr. J. Pelletier for reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM086890 (to M. Y. S.), R01 AI20248 (to K. L. R.), and DK32520 (to the Diabetes Endocrinology Research Center) and Training Grant AI007272-25 (to J. D. C.).
- ER
- endoplasmic reticulum
- CFTR
- cystic fibrosis transmembrane conductance regulator
- IBMX
- 3-isobutyl-1-methylxanthine
- copGFP
- copepod GFP
- FRT
- Fischer rat thyroid.
REFERENCES
- 1. Sherman M. Y., Goldberg A. L. (2001) Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron 29, 15–32 [DOI] [PubMed] [Google Scholar]
- 2. Ravikumar B., Rubinsztein D. C. (2006) Role of autophagy in the clearance of mutant huntingtin: a step towards therapy? Mol. Aspects Med. 27, 520–527 [DOI] [PubMed] [Google Scholar]
- 3. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 4. Sarkar S., Rubinsztein D. C. (2008) Small molecule enhancers of autophagy for neurodegenerative diseases. Mol. Biosyst. 4, 895–901 [DOI] [PubMed] [Google Scholar]
- 5. Sarkar S., Ravikumar B., Floto R. A., Rubinsztein D. C. (2009) Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 16, 46–56 [DOI] [PubMed] [Google Scholar]
- 6. Koyama S., Arawaka S., Chang-Hong R., Wada M., Kawanami T., Kurita K., Kato M., Nagai M., Aoki M., Itoyama Y., Sobue G., Chan P. H., Kato T. (2006) Alteration of familial ALS-linked mutant SOD1 solubility with disease progression: its modulation by the proteasome and Hsp70. Biochem. Biophys. Res. Commun. 343, 719–730 [DOI] [PubMed] [Google Scholar]
- 7. Brown I. R. (2007) Heat shock proteins and protection of the nervous system. Ann. N.Y. Acad. Sci. 1113, 147–158 [DOI] [PubMed] [Google Scholar]
- 8. Putcha P., Danzer K. M., Kranich L. R., Scott A., Silinski M., Mabbett S., Hicks C. D., Veal J. M., Steed P. M., Hyman B. T., McLean P. J. (2010) Brain-permeable small-molecule inhibitors of Hsp90 prevent α-synuclein oligomer formation and rescue α-synuclein-induced toxicity. J. Pharmacol. Exp. Ther. 332, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paris D., Ganey N. J., Laporte V., Patel N. S., Beaulieu-Abdelahad D., Bachmeier C., March A., Ait-Ghezala G., Mullan M. J. (2010) Reduction of β-amyloid pathology by celastrol in a transgenic mouse model of Alzheimer disease. J. Neuroinflammation 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mu T. W., Ong D. S., Wang Y. J., Balch W. E., Yates J. R., 3rd, Segatori L., Kelly J. W. (2008) Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell 134, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farinha C. M., Amaral M. D. (2005) Most F508del-CFTR is targeted to degradation at an early folding checkpoint and independently of calnexin. Mol. Cell. Biol. 25, 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turnbull E. L., Rosser M. F., Cyr D. M. (2007) The role of the UPS in cystic fibrosis. BMC Biochem. 8, S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amaral M. D. (2004) CFTR and chaperones: processing and degradation. J. Mol. Neurosci. 23, 41–48 [DOI] [PubMed] [Google Scholar]
- 14. Craig E. A., Eisenman H. C., Hundley H. A. (2003) Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr. Opin. Microbiol. 6, 157–162 [DOI] [PubMed] [Google Scholar]
- 15. Zhang G., Ignatova Z. (2011) Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 21, 25–31 [DOI] [PubMed] [Google Scholar]
- 16. Siller E., DeZwaan D. C., Anderson J. F., Freeman B. C., Barral J. M. (2010) Slowing bacterial translation speed enhances eukaryotic protein folding efficiency. J. Mol. Biol. 396, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 17. Zaarur N., Meriin A. B., Gabai V. L., Sherman M. Y. (2008) Triggering aggresome formation. Dissecting aggresome-targeting and aggregation signals in synphilin-1. J. Biol. Chem. 283, 27575–27584 [DOI] [PubMed] [Google Scholar]
- 18. Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., Wandless T. J. (2006) A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zegarra-Moran O., Romio L., Folli C., Caci E., Becq F., Vierfond J. M., Mettey Y., Cabrini G., Fanen P., Galietta L. J. (2002) Correction of G551D-CFTR transport defect in epithelial monolayers by genistein but not by CPX or MPB-07. Br. J. Pharmacol. 137, 504–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnston J. A., Ward C. L., Kopito R. R. (1998) Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143, 1883–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wigley W. C., Fabunmi R. P., Lee M. G., Marino C. R., Muallem S., DeMartino G. N., Thomas P. J. (1999) Dynamic association of proteasomal machinery with the centrosome. J. Cell Biol. 145, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meriin A. B., Zaarur N., Sherman M. Y. (2012) Association of translation factor eEF1A with defective ribosomal products generates a signal for aggresome formation. J. Cell Sci. 125, 2665–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deleted in proof
- 24. Lindqvist L., Oberer M., Reibarkh M., Cencic R., Bordeleau M. E., Vogt E., Marintchev A., Tanaka J., Fagotto F., Altmann M., Wagner G., Pelletier J. (2008) Selective pharmacological targeting of a DEAD box RNA helicase. PLoS ONE 3, e1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kalid O., Mense M., Fischman S., Shitrit A., Bihler H., Ben-Zeev E., Schutz N., Pedemonte N., Thomas P. J., Bridges R. J., Wetmore D. R., Marantz Y., Senderowitz H. (2010) Small-molecule correctors of F508del-CFTR discovered by structure-based virtual screening. J. Comput. Aided Mol. Des. 24, 971–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thakerar A., Van Driessche W., Bridges R. J. (2009) The conductance assay: A simple assay to measure ΔF508-CFTR channel activity. Pediatr. Pulmonol. 44, 305 [Google Scholar]
- 27. Medicherla B., Goldberg A. L. (2008) Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 182, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mehta R., Chandler-Brown D., Ramos F. J., Shamieh L. S., Kaeberlein M. (2010) Regulation of mRNA translation as a conserved mechanism of longevity control. Adv. Exp. Med. Biol. 694, 14–29 [DOI] [PubMed] [Google Scholar]
- 29. Syntichaki P., Troulinaki K., Tavernarakis N. (2007) Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann. N.Y. Acad. Sci. 1119, 289–295 [DOI] [PubMed] [Google Scholar]
- 30. Demontis F., Perrimon N. (2010) FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143, 813–825 [DOI] [PMC free article] [PubMed] [Google Scholar]