Background: Messenger RNA of hif-1α could be regulated by post-transcriptional mechanisms.
Results: Depletion of either JNK2 or nucleolin affected hif-1α mRNA stability.
Conclusion: JNK2 regulated nucleolin expression and might in turn stabilize hif-1α mRNA.
Significance: We provided more evidence for the oncogenic roles of JNK2 and nucleolin in regulating the cancer microenvironments by controlling HIF-1α expression.
Keywords: Hypoxia-inducible Factor (HIF), Jun N-terminal Kinase (JNK), mRNA Decay, Nickel, Signal Transduction, Nucleolin
Abstract
The mRNA of hif-1α is considered as being constitutively and ubiquitously expressed, regardless of the level of oxygen tension. However many recent reports have showed that hif-1α mRNA could be regulated by natural antisense transcripts, potential microRNAs, and low O2. In this study, it was found that a deficiency of JNK2 expression reduced HIF-1α protein induction in response to nickel treatment resulting from the impaired expression of hif-1α mRNA. Both the promoter luciferase assay and mRNA degradation assay clearly showed that depletion of JNK2 affected stability of hif-1α mRNA, rather than regulated its transcription. In addition, nucleolin, a classic histone chaperone, was demonstrated to physically bind to hif-1α mRNA and maintain its stability. Further investigation indicated that JNK2 regulated nucleolin expression and might in turn stabilize hif-1α mRNA. Collectively, we provided one more piece of evidence for the oncogenic role of JNK2 and nucleolin in regulating the cancer microenvironments by controlling HIF-1α expression.
Introduction
Nickel is a widely distributed natural metal, and humans are constantly exposed to environmental nickel because of its constitutive release from natural sources, or pollutants from nickel manufacturing industries or airborne particles from combustion of fossil fuels (1). The causality between nickel exposure and cancer development has been supported by both animal studies and epidemiologic investigations (2). It was reported that both the genetic and epigenetic factors were attributable to the carcinogenic characteristics of mode of action for nickel (3, 4). Induction of hypoxia-inducible factor-1α (HIF-1α)3 by nickel treatment was documented both in vivo and in vitro (5). The reports from our group and others have demonstrated the requirement of HIF-1α for the malignant transformation caused by nickel exposure in cultured cell system (6–8). However, the molecular mechanisms about the regulation of HIF-1α expression after nickel exposure are far from fully understood.
HIF-1 is a heterodimeric transcription factor, and plays a key role in cellular adaptations to a deficiency of oxygen supply by controlling expressions of a series of genes involved in angiogenesis, oxygen transport, and glucose metabolism (9). HIF-1 consists of an oxygen-regulated HIF-1α subunit and a constitutively expressed hydrocarbon receptor nuclear translocator (also called HIF-1β or ARNT) (7). Dysregulation of HIF-1α occurs in renal cell carcinoma, breast, lung, and ovarian cancers, with a strong correlation to tumor metastasis and a poor prognosis for patients (10). HIF-1α protein is very dynamic in normoxia, and is normally regulated by post-translational mechanism. In contrast, hif-1α mRNA is constitutively and ubiquitously expressed, regardless of the level of oxygen tension (11). However, an increasing number of recent studies have shown that hif-1α mRNA could be regulated by natural antisense transcripts (12, 13), potential microRNAs (14, 15), and low O2 pressure in the renal medulla in rats (16) and sea bass (17). The hif-1α mRNA contains 823 bp long 3′-UTR that is AU-rich and contains AUUUA pentamers that are conserved among rat, mouse, and human (15, 16). It is now known that a group of RNA-binding proteins can bind to AU-rich elements (ARE) in 3′-UTR of mRNA and regulate the mRNA turnover rate (60). In studies reported here, it was found for the first time that nucleolin could physically bind to and stabilize hif-1α mRNA in cell culture system.
Nucleolin (also known as C23) is a multifunctional phosphoprotein whose expression is abundant, i.e. it represents up to 5∼10% of nucleolar proteins in exponentially-growing cells (18). Nucleolin is also implicated in several pathologies including viral infection (19), autoimmune diseases (20), Alzheimer disease (21), Parkinson disease (22), and cancer development (40). The role of nucleolin in these pathological processes roots from its function(s) in DNA metabolism, chromatin re-modeling, ribosome biogenesis and mRNA decay (23). Nucleolin has been found to bind to the mRNA of several important genes, including p53 (24), bcl-2 (25), and bcl-xl (26), leading to regulation on mRNA turnover or translation. Moreover, nucleolin has been shown to have binding activity to a number of G-rich oligonucleotides, like the G-quadruplex structure in the promoter region of c-myc gene (27) and vegf gene (28). Gonzalea et al. showed, in an in vitro filter binding assay, that nucleolin bound to the G-quadruplex structure in 5′-UTR of hif-1α mRNA (27). However, it is still open for investigation as to whether or not nucleolin could regulate HIF-1α expression in vivo. Therefore, the goal of this study, reported here, was to obtain experimental evidence for this issue in a cell culture model.
MATERIALS AND METHODS
Cell Culture and Reagents
Mouse embryonic fibroblasts (MEFs), including WT cells and JNK2−/− cells (29) as well as their stable transfectants, mouse fibroblast NIH3T3 cells, and human embryonic kidney cells HEK293T were cultured in DMEM (Invitrogen, Grand Island, NY) supplemented with 10% FBS (Nova-Tech, Grand Island, NE). The MEFs were immortalized according to 3T3 protocol (30). Tristetraprolin (TTP) knock-out MEFs and WT MEFs (31) were kindly provided by Dr. Perry J. Blackshear (National Institute of Environmental Health Sciences). The cells were cultured in DMEM supplemented with 10% FBS, and identified by PCR, using the primers of 5′-CTG AGC TGT CAC CCT CAC CT-3′and 5′-TGG TGC TGG GGG TAG TAG AC-3′. Actinomycin D was purchased from Calbiochem. Cordycepin was purchased from Santa Cruz Biotechnology Inc. Nickel chloride was purchased from Sigma-Aldrich. Dimethyloxalylglycine (DMOG) was purchased from Frontier Scientific (Logan, UT). λ protein phosphatase was purchased from New England Biolabs (Ipswich, MA). Antibodies against HIF-1α (for Western blotting) and HIF-1β were purchased from Novus Biologicals, Inc. (Littleton, CO); Anti-JNK1 antibody was from Invitrogen (Carlsbad, CA); Antibodies against non-phosphorylated c-Jun, JNK1/2, phosphor-c-Jun at Ser-73, phosphor-JNK at Thr-183/Tyr-185 were purchased from Cell Signaling Technology (Beverly, MA); anti-β-actin, α-tubulin, HA, nucleolin, and HIF-1α (for immunoprecipitation) antibodies were purchased from Sigma; Anti-GFP antibody was purchased from Santa Cruz Biotechnology.
Constructs and Transfections
The HRE luciferase reporter was constructed by inserting the sequence of the HIF-1α binding site into the luciferase reporter vector pG12-basic, as previously described (32, 33). The vegf-luciferase reporter was constructed by inserting a 2.65-kb KpnI-BssHII fragment of the human vegf promoter sequence from −2274 to +379, relative to the transcription initiation site into the pGL2-basic vector (Promega, Madison, WI) (32, 33). Each of the above reporters (2 μg) was stably transfected into WT MEFs and JNK2−/− cells in combination with hygromycin-resistance plasmid (0.8 μg) for positive clone selection. HA-JNK2 expression construct (2 μg) (34) in combination with hygromycin-resistance plasmid (0.8 μg) was transfected into JNK2−/− cells. The stable transfectants, named as JNK2−/− (HA-JNK2), were established by hygromycin selection (400 μg/ml) and all transfectants were pooled as mass culture as described in our previous publications (35–37). The shRNA sets for Nucleolin and JNK2 were purchased from Open Biosystems (Thermo Fisher Scientific., Huntsville, AL), and were transfected stably into WT MEFs or NIH3T3 cells, respectively. The hif-1α promoter luciferase reporter (PH800) was kindly provided by Dr. Carine Michiels (Laboratory of Biochemistry and Cellular Biology, FUNDP-University of Namur, Belgium) (38). The Nucleolin promoter luciferase reporter was kindly provided by Dr. Bruno Amati (Cellular Growth Control Unit, Swiss Institute for Experimental Cancer Research, Switzerland) (39). GFP-nucleolin expression vector was kindly provided by Dr. Michael B. Kastan (Comprehensive Cancer Center, St. Jude Children's Research Hospital, Memphis, TN )(29).
RT-PCR
Total RNA was extracted from the cells using Trizol reagent (Invitrogen). Total cDNAs were synthesized by ThermoScript TM RT-PCR system (Invitrogen). The primers for mouse hif-1α were: 5′-AGC CCT AGA TGG CTT TGT GA-3′ and 5′-TAT CGA GGC TGT GTC GAC TG-3′; for mouse nucleolin were 5′-GGA GGT TGT CAT CCC TCA GA-3′ and 5′-TCC TCC TCA GCC ACA CTC TT-3′; for mouse ahif-1α were 5′-GCT GGA AGG TTT GTG GTG TT-3′and 5′-TGG AAG GTA TGG GGC ATT TA-3′; for mouse β-actin were 5′-CAT CCG TAA AGA CTC CTA TGC C-3′ and 5′-ACG CAG CTC AGT AAC AGT CC-3′ (also used in real-time PCR). The PCR products were separated over 2% agarose gels, stained with ethidium bromide. The results were imaged with Alpha Innotech SP image system (Alpha Innotech Corporation, San Leandro, CA). The densitometric analyses of the product bands were conducted using the software of ImageQuant 5.2 (GE Healthcare). The results shown were representative of three independent experiments.
Quantitative RT-PCR
The same cDNAs that were used for the above RT-PCR were also analyzed for real-time PCR using the 7900HT Fast Real-Time PCR System (Applied Biosystems). The primers for real-time PCR were: mouse hif-1α 5′-GAA GAC AAC GCG GGC ACC GA-3′ and 5′-TGC TTC GCC GAG ATC TTG CTG C-3′; mouse nucleolin 5′-GAG GAC CCC CTT CGT CGC CT-3′ and 5′-GCC TCA CCG TGG GTT TTG CCA-3′. The real-time PCR was conducted following the protocol for Fast SYBR Green Master Mix kit (Applied Biosystems). Briefly, an initial activation was performed at 95 °C for 20 s, followed by 40 cycles of denaturation at 95 °C for 1 s, and annealing and extension at 60 °C for 20 s. The relative mRNA levels were obtained by exponentially transforming ΔCT values to 2ΔΔCT, and the mean values were calculated with one standard deviation (41). The data were representative of three independent experiments.
Luciferase Reporter Assay
The cells transfected with the HRE, vegf, hif-1α, or nucleolin promoter luciferase reporters were seeded into 96-well plates (8 × 103/well) and were subjected to the specific treatments when cultures reached 70 to 80% confluence. pRL-TK vector was used as an internal control. Cellular lysates were prepared, and luciferase activities were determined using a luminometer (Wallac 1420 Victor 2 multilabel counter system), as previously described (42).
Western Blotting
MEFs and their transfectants were plated in 6-well plates and cultured in normal 10% FBS DMEM until 70–80% confluence. After various treatments, the proteins were extracted and total protein was quantified with a Dc protein assay kit (Bio-Rad). Western blotting was carried out as previously described (43). Primary antibody-bound proteins were detected by using an alkaline phosphatase-linked secondary antibody and an ECF Western blotting system (Amersham Biosciences, Piscataway, NJ).
Pulse Assays
Cells were exposed to nickel (0.5 mm) for 12 h, then incubated with methionine-cysteine-free DMEM (Invitrogen) containing 2% dialyzed fetal calf serum (Invitrogen) for 1 h. 35S-labeled methionine/cysteine (Trans 35S-Label; ICN) (250 μCi/dish) was added, and the cells were cultured for the time periods indicated. The cells were collected with lysis buffer (1% Triton X-100, 150 mm NaCl, 10 mm Tris, pH 7.4, 1 mm EDTA, 1 mm EGTA, 0.2 mm Na3VO4, 0.5% Nonidet P-40, and complete protein inhibitors mixture tablet) on ice. Total lysate of 500 μg was incubated with 2 μg of anti-HIF-1α monoclonal antibody (Sigma) or control IgG antibody for negative control for 2 h at 4 °C. Then protein-A/G plus-agarose beads (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) that were pre-cleared by 20 mg/ml BSA were added into the mixture and incubated with agitation for an additional 2 h at 4 °C. The immunoprecipitated samples were washed with the cell lysis buffer 5 times and heated at 100 °C for 5 min. Radiolabeled HIF-1α protein as well as the input whole cell lysate (WCL) were assessed using SDS-polyacrylamide gels (10%) analysis.
Dephosphorylation Assay
After exposure to the specific nickel treatment, cells were collected by centrifuge, resuspended in PBS, and homogenized by sonication. About 50 μg of whole cell lysate was used as substrate and incubated with λ phosphatase (New England Biolabs) for 40 min at 30 °C. Proteins were resolved in SDS-polyacrylamide gels (7.5%) and revealed by Western blotting.
RNA-IP
The cells were cultured in 10-cm dishes and harvested by scraping after the specific treatment. Polysome lysis buffer (PLB), containing: 10 mm HEPES pH 7; 100 mm KCl; 5 mm MgCl2; 25 mm EDTA; 0.5% IGEPAL; 2 mm DTT; 50 units/ml RNase OUT; 50 units/ml Superase IN; 0.2 mg/ml heparin; and complete proteinase inhibitor, was used to lyse the cell pellet. The cell lysate was centrifuged at 14,000 × g for 10 min at 4 °C. The anti-nucleolin antibody and agarose beads A/G were added into the supernatant and rotated overnight at 4 °C in NET2 buffer containing: 50 mm Tris-HCl; pH 7.4; 150 mm NaCl; 1 mm MgCl2; 0.05% IGEPAL; 50 units/ml RNase OUT; 50 units/ml Superase IN; 1 mm dithiothreitol; and 30 mm EDTA. The beads were washed three times, resuspended in 100 μl of NET2 and 100 μl of SDS-TE (20 mm Tris-HCl, pH 7.5, 2 mm EDTA, and 2% SDS) then incubated for 30 min at 55 °C, with occasional mixing. The RNAs in the buffer were extracted by phenol-chloroform-isoamyl alcohol. RT-PCR was performed to detect the mRNA present in the immune-complex.
RNA Pull-down Assay
The 3′-UTR of mouse hif-1α mRNA (NM_010431.2) was amplified by PCR from cDNA of WT MEFs, using the pair of primers (forward: 2994-TTG GGT TTT TGT TTC TGT TGG-3014; reverse: 4125-TTT CCT GGT CCA CAG AAG ATG-4105). The PCR product was used for in vitro transcription in the presence of biotinylated-UTP (MAXIscript® Kit, Ambion, Inc., Grand Island, NY). The biotinylated RNAs were incubated with cell lysis from WT MEFs (in PLB buffer) in a final volume of 200 μl at room temperature for 30 min and captured by streptavidin MagneSphere Paramagnetic Particle (Promega). After washing according to the manufacturer's instructions, bound proteins were eluted in Laemmli buffer and resolved by SDS-PAGE, electrically transferred to PVDF membrane, and the presence of nucleolin and HuR was detected with Western blotting assay (44).
Statistical Analysis
The significance of the difference between the treated and untreated groups, or between different cell lines, was determined with the Wilcoxon Rank Sum Test. The results are expressed as mean ± S.D. A p value of ≤ 0.05 was used to assign significance.
RESULTS
JNK2 Participated in HIF-1α Protein Expression
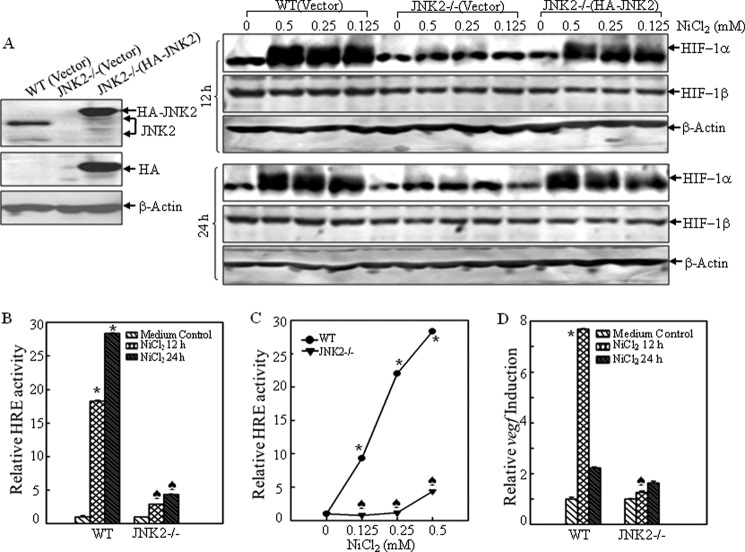
Although it is well documented that JNKs can regulate the activity of dozens of transcription factors (45), there is no report about the relationship between JNKs and HIF-1α. Our previous findings show that JNK1 is responsible for HIF-1α expression through maintaining its protein stability (7). Therefore, it is of interest to investigate whether JNK2 also plays a role in the regulation of HIF-1α expression and whether it holds the distinct underlying molecular mechanisms from JNK1. To explore this, we first compared HIF-1α protein levels among WT(vector), JNK2−/− (vector) and JNK2−/− (HA-JNK2) MEFs following nickel treatment. The results showed that depletion of JNK2 expression in MEFs attenuated HIF-1α protein accumulation upon nickel treatment at all time points and doses tested, whereas the expression level of HIF-1β (the constitutively expressed subunit of HIF-1) was comparable between JNK2−/− and WT cells (Fig. 1A). We then evaluated the HIF-1-dependent transactivation activity by transfecting a construct containing a hypoxia-responsive element (HRE) luciferase reporter into both WT and knock-out cells. The HRE luciferase activity was significantly increased in WT MEFs following nickel exposure; however, it was attenuated in JNK2−/− cells (Fig. 1, B and C), indicating that both HIF-1α protein expression and its activity were impaired in JNK2−/− cells following nickel exposure. We also determined the transcriptional induction of vascular endothelial growth factor (vegf), a well-known downstream target gene of HIF-1. As expected, the induction of vegf transcription was decreased in JNK2−/− cells as compared with that in WT cells (Fig. 1D).
FIGURE 1.
JNK2 deficiency impaired nickel-induced HIF-1α expression and transactivation. A, WT(Vector), JNK2−/− (Vector), and JNK2−/− (HA-JNK) MEFs were identified by Western blotting assay for expression of JNK2 (left panel). The induction of HIF-1α protein was compared after the indicated cells were exposed to nickel at doses and time points as indicated (right panel). B--D, WT and JNK2−/− cells that were stably transfected with HRE-luciferase reporter (B and C) or vegf-luciferase reporter (D) were exposed to 0.5 mm nickel for 12 h and 24 h (B and D), or different doses of nickel for 24 h (C). The luciferase activities were determined using a luminometer. The results are expressed as relative HRE activities or vegf inductions to medium control. Each bar indicates the mean and standard error of triplicate assay wells. The asterisk (*) indicates a significant increase as compared with medium control in WT cells (p < 0.05). The spade (♣) indicates a significant decrease as compared with that in the WT cells for the same treatment (p < 0.05).
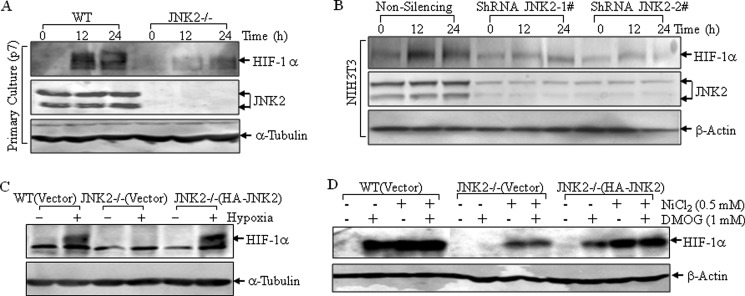
To further confirm whether the impairment of HIF-1α protein induction in JNK2−/− cells was directly due to jnk2 gene deficiency rather than changes of other genes during the establishment of immortalized cell lines, primarily cultured MEFs (at passage 7) were utilized. Consistent with the results observed in immortalized MEFs, HIF-1α protein accumulation following nickel exposure was dramatically impaired in primary JNK2−/− cells when compared with that in WT cells at the same passage (Fig. 2A). This effect was reproduced in another mouse fibroblast cell line NIH3T3, by introducing two different shRNAs of JNK2 to knockdown endogenous JNK2 expression (Fig. 2B). Also, the reconstitution of JNK2 in JNK2−/− cells restored HIF-1α protein expression upon nickel exposure (Fig. 1A).
FIGURE 2.
JNK2 regulated HIF-1α protein expression in various cells. A, primarily cultured WT and JNK2−/− MEFs at passage 7 were exposed to nickel (0.5 mm) for 12 h and 24 h. B, two separate shRNAs for JNK2 or non-silencing control plasmid were transfected into NIH3T3 cells. The stable transfectants were treated with nickel (0.5 mm) for 12 h and 24 h. C and D, WT(Vector), JNK2−/− (Vector), and JNK2−/− (HA-JNK) MEFs were exposed to hypoxia (1% O2) for 8 h (C), or DMOG in combination with or without nickel for 24 h (D). The cell extracts were subjected to Western blotting assay.
The mammalian prolyl hydroxlyase domain (PHD) enzymes are critical for VHL-dependent HIF-1α proteasomal degradation (11, 46). Deprivation of oxygen, transition metals (cobalt and nickel), and 2-oxoglutarate inhibitor (dimethyloxalylglycine, DMOG), all impair the activity of PHD enzymes. In the present studies, the hypoxia as well as several chemicals that mimick hypoxia conditions were used to investigate the accumulation of HIF-1α protein in WT(Vector), JNK2−/− (Vector) and JNK2−/− (HA-JNK2) cells. As shown in Fig. 2, C and D, the induction of HIF-1α protein was impaired upon deletion of the JNK2 gene in all the above experimental conditions, while it could be restored in JNK2−/− (HA-JNK2) cells. Therefore, it was concluded that JNK2 might play a crucial role in HIF-1α protein accumulation under hypoxic conditions.
JNK2 Expression Was Crucial for Maintaining hif-1α mRNA Stability
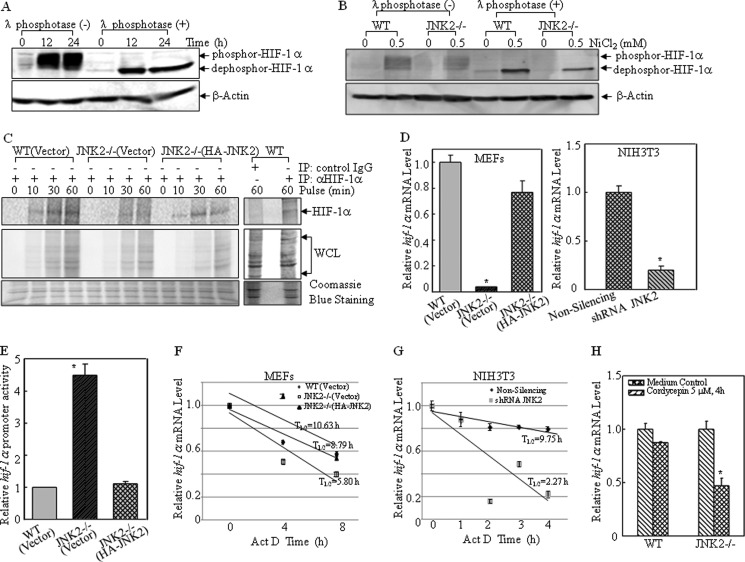
As observed in the above studies and other findings (47), induced HIF-1 α protein migrated with a diffused pattern in SDS- polyacrylamide gels, suggesting HIF-1 α protein undergoes strong post-translational modifications. It is known that protein phosphorylation can markedly modify the migration pattern in SDS-polyacrylamide gels. Therefore, we evaluated whether the diffused band of HIF-1α protein was due to phosphor-modification. As shown in Fig. 3A, incubation of whole cell lysate (WCL) with a nonspecific protein phosphatase, λ phosphatase, dramatically converted the diffused HIF-1α protein band to a clear sharp band on the same gel. This result suggested that HIF-1 α was phosphorylated in vivo and that this kind of modification was responsible for the reduced mobility on SDS-polyacrylamide gel.
FIGURE 3.
JNK2 regulated hif-1α mRNA stability. A and B, WT (A) or WT and JNK2−/− MEFs (B) (1 × 106/well) were seeded into 10-mm dish and exposed to 0.5 mm nickel for 12 h and 24 h (A), or 24 h (B). The whole cell lysate was incubated with λ phosphatase for 40 min at 30 °C. Proteins were resolved in SDS-PAGE and revealed by Western blotting assay. C, WT(Vector), JNK2−/− (Vector), and JNK2−/− (HA-JNK) MEFs were treated with nickel (0.5 mm) for 12 h in complete medium. Cells were then accommodated in methionine- and cysteine-free DMEM for 1 h. 35S-labeled methionine and cysteine was then added for the indicated times for pulse assay. Cell extracts were immunoprecipitated with anti-HIF-1α antibody or control IgG, and subjected to SDS-PAGE. Autoradiography was used to visualize 35S-labeled HIF-1α. D, hif-1α mRNA levels in the individual cells were determined by real-time PCR. The asterisk (*) indicates a significant decrease as compared with those in WT(Vector) and JNK−/− (HA-JNK2) MEFs or non-silencing control cells (p < 0.05). E, basal levels of hif-1α promoter activity were evaluated by transfecting the indicated cells with a construct containing hif-1α promoter-driven luciferase. pRL-TK vector was used as an internal control. The results are expressed as the ratios of firefly to Renilla lucifease activity, as means ± S.D. (n = 3). The asterisk (*) indicates a significant increase as compared with that of WT(Vector) cells or JNK−/− (HA-JNK2) cells (p < 0.05). F and G, mRNA degradation rate of hif-1α was detected following treatment with actinomycin D (5 μm) for the indicated time. The PCR products were separated over 2% agarose gels, stained with ethidium bromide. The densitometric analyses of the product bands were conducted using the software of ImageQuant 5.2 (GE Healthcare). The results were shown as means ± S.D. (n = 3). H, indicated cells were treated with cordycepin (5 μm) for 4 h. Real-time PCR was conducted to detect the hif-1α mRNA expression. The asterisk (*) indicates a significant decrease as compared with that in WT cells under the same treatment (p < 0.05).
To answer whether the decreased HIF-1 α protein induction in JNK2−/− cells was due to less protein synthesis or due to impaired phosphorylation, the dephosphorylated form of HIF-1 α protein was compared in WT and JNK2−/− cells in the presence of λ phosphatase. As shown in Fig. 3B, the dephosphorylated form of HIF-1 α protein was obviously reduced in JNK2−/− cells when compared with that in WT cells. This result indicated that the reduction of the modified form of HIF-1α protein observed in JNK2−/− cells was caused by impairment of nascent HIF-1α protein synthesis rather than a defect of post-translational modification. Pulse analysis was conducted in turn, using 35S-labeled methionine and cysteine to monitor HIF-1α synthesis rate. It was found that newly translated HIF-1α protein was observed at 10 min after addition of labeling medium, and gradually increased until 60 min in WT(Vector) and JNK2−/− (HA-JNK2) cells. However, in JNK2−/− (Vector) cells HIF-1α protein translation amount was dramatically reduced (Fig. 3C), suggesting that de novo HIF-1 α protein synthesis was defective in JNK2−/− (Vector) cells. It was noted that the background in JNK2−/− (HA-JNK2) lanes of Coomassie Blue staining was slightly higher than those of the JNK2−/− (Vector) lanes (Fig. 3C). Thus, we anticipated this might be due to the photographing process. The explanation was supported by our data showing that the bands of 35S-labeled WCL in JNK2−/− (HA-JNK2) cells are slightly weaker in comparison to those in JNK2−/− (Vector) cells, however the de novo HIF-1α protein bands in JNK2−/− (HA-JNK2) cells were stronger compared with those in JNK2−/− (Vector).
Next, real-time PCR was conducted to compare hif-1α mRNA content. It was found that the JNK2 deficiency caused reduction in hif-1α mRNA expression in both knock-out and knockdown systems (Fig. 3D). Therefore, hif-1α promoter-driven luciferase reporter (38, 48) was employed to compare hif-1α transcription among those three transfectants. As shown in Fig. 3E, knock-out of JNK2 increased hif-1α transcription. The higher transcription of hif-1α observed in JNK2−/− cells might result from an adaptive strategy developed by the cells to compensate for the impaired hif-1α mRNA expression. Then hif-1α mRNA stabilities were detected in WT(Vector), JNK2−/− (Vector), and JNK2−/− (HA-JNK2) cells using actinomycin D (Act D) to inhibit mRNA transcription. The half-life of hif-1α mRNA (T½) in WT(Vector) cells and JNK2−/− (HA-JNK2) cells was 8.79 h and 10.63 h, respectively. However, in JNK2−/− (Vector) cells it was reduced to 5.80 h (Fig. 3F). Also, more rapid hif-1α mRNA degradation rate was observed when JNK2 was knocked down in NIH3T3 cells (Fig. 3G). The half-life of hif-1α mRNA was about 9.75 h in non-silencing NIH3T3 cells, but it was reduced to 2.27 h in shRNA JNK2 cells (Fig. 3G). Another mRNA transcription inhibitor, cordycepin (50), was also employed at 5 μm for 4 h to confirm the findings with Act D. The results obtained from real-time PCR showed that cordycepin treatment caused a mild reduction in hif-1α mRNA (12.36%) in WT cells, while it obviously decreased hif-1α mRNA in JNK2−/− cells (52.86%; Fig. 3H). Therefore, it was concluded that JNK2 modulated hif-1α mRNA stability rather than its transcription.
Nucleolin Regulated hif-1α mRNA Stability
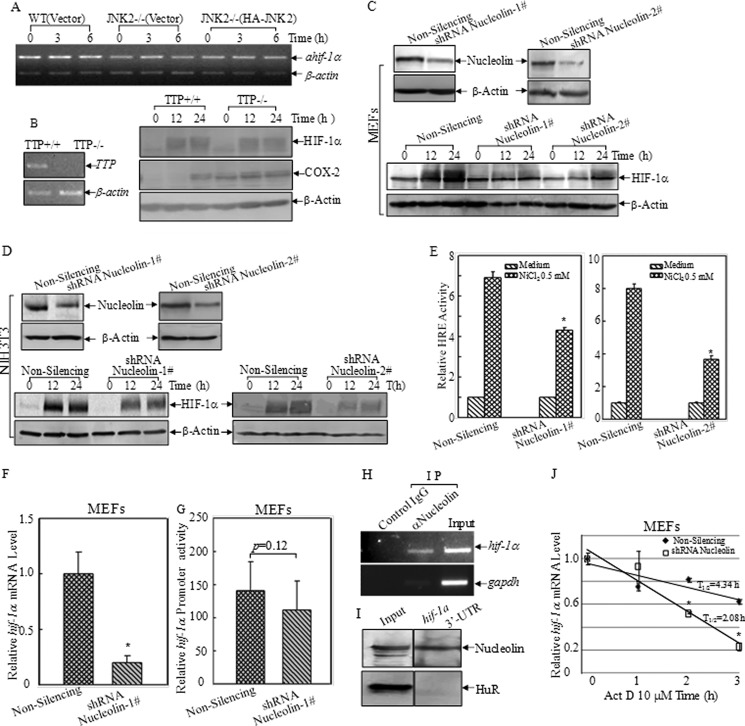
It was found that hif-1α mRNA could be regulated by a natural antisense transcript, ahif (12, 13). Part of ahif sequence is strictly complementary to the 3′-UTR of hif-1α mRNA and influences hif-1α transcript stability. However, in the current studies, ahif was not obviously elevated in JNK2−/− cells when compared with that in WT cells (Fig. 4A). Therefore ahif was not considered in our further investigation. Tristetraprolin (TTP) is an mRNA-binding protein, which was reported to bind to the 3′-UTR of several inflammation related genes such as TNF-α and TTP was subjected to the regulation by MAPKs, including JNKs (51). So the feasibility about the involvement of TTP in the regulation of hif-1α mRNA stability was determined by employing both TTP+/+ and TTP−/− MEFs (31). First the knock-out cells were identified by PCR as shown in Fig. 4B. However, knock-out of TTP gene did not affect HIF-1α protein induction by nickel, while the increased basal level of COX-2 expression was observed in TTP−/− cells as expected (52), indicating that TTP may not be a promising regulator for HIF-1α expression (Fig. 4B).
FIGURE 4.
Nucleolin bound to hif-1α mRNA and increased its stability. A, expression of a natural antisense transcript of hif-1α (ahif) was determined by RT-PCR. B, WT and TTP−/− MEFs were treated with 0.5 mm nickel for 12 and 24 h. The cell extracts were subjected to Western blotting assay. C and D, shRNA-nucleolin was stably transfected into WT MEFs (C) or NIH3T3 (D). The transfectants were identified by Western blotting. HIF-1α protein induction by nickel exposure (0.5 mm) was compared between non-silencing and shRNA-nucleolin transfectants. E, HRE-luciferase reporter and pRL-TK vector were transiently transfected into non-silencing and shRNA-nucleolin MEFs. The HRE induction was determined by luciferase assay. The results were presented as relative HRE activities by normalizing the ratios of firefly to Renilla lucifease activity, as means ± S.D. (n = 3). The asterisk (*) indicates a significant decrease as compared with that in non-silencing cells (p < 0.05). F, hif-1α mRNA expression levels were detected by real-time PCR in the indicated MEFs. The asterisk (*) indicates a significant decrease as compared with that in non-silencing cells (p < 0.05). G, hif-1α transcription was determined using hif-1α promoter-driven firefly luciferase and pRL-TK vector. The results were expressed relative hif-1α promoter activities by normalizing the ratios of firefly to Renilla lucifease activity, as means ± S.D. (n = 3). H, RNA-IP was performed to examine the binding of nucleolin to hif-1α mRNA in 293T cells. I, RNA-pull down was performed using an in vitro transcribed RNA containing mouse hif-1α 3′-UTR labeled with biotin. Magnetic streptavidin beads were used to capture the proteins bound with hif-1α 3′-UTR after incubating with cell lysate. Western blotting was carried out to detect the presence of nucleolin in pull-down complex. HuR was used as a negative control. J, hif-1α mRNA degradation rates were compared between the indicated MEFs by real-time PCR following treatment with actinomycin D (10 μm). The data were shown as means ± S.D. (n = 3). The asterisk (*) indicates a significant decrease as compared with that in non-silencing cells (p < 0.05).
Nucleolin has been reported to be involved in JNK-mediated IL-2 mRNA stabilization in activated T-cells (53). To determine whether nucleolin was required for regulation of HIF-1α expression, two sets of shRNAs targeting different sequences of mouse nucleolin mRNA were used to exclude the possibility of off-target effect. As shown in Fig. 4C (upper panel), both sets of shRNAs could efficiently reduce nucleolin expression. More importantly, the induction of HIF-1α protein by nickel treatment was greatly decreased in shRNA nucleolin transfectants at 12 and 24 h compared with that in Non-silencing transfectants (Fig. 4C, lower panel). Similar results were obtained in NIH3T3 cells using two shRNAs of nucleolin (Fig. 4D). Following these, HRE luciferase reporter was transfected into non-silencing and shRNA nucleolin MEFs. It was found that the HRE induction was attenuated in either shRNA nucloelin transfectants following nickel exposure (Fig. 4E). The result from real-time PCR further revealed a decrease of hif-1α mRNA level in shRNA nucleolin transfectants (Fig. 4F). However, the results of hif-1α promoter-driven luciferase assay showed that hif-1α transcription was at comparable levels between non-silencing cells and shRNA-nucleolin cells (Fig. 4G), indicating that nucleolin did not inhibit hif-1α gene transcription. Thus, it was anticipated that nucleolin might regulate hif-1α mRNA stability.
To test whether nucleolin could bind to hif-1α mRNA, RNA-IP assay was carried out in which anti-nucleolin antibody was used to pull down all mRNAs that physically interacted with nucleolin protein. Messenger RNA was then extracted from the precipitated complex and reverse transcript-PCR was performed to detect the presence of hif-1α mRNA. As shown in Fig. 4H, hif-1α mRNA was found in the immune-complex pulled down by anti-nucleolin antibody, but not present in the precipitation that was pulled down by control IgG, indicating that nucleolin could interact with hif-1α mRNA.
In addition, the RNA pull down assay was performed to reversely confirm the physical binding of nucleolin to hif-1α mRNA 3′-UTR. Briefly, an in vitro transcribed RNA containing 3′-UTR of hif-1α mRNA (1132 pb, 2994–4125 relative to the transcription starting site of NM_010431.2) was labeled with biotin, and incubated with WCL and captured by magnetic-streptavidin beads. The proteins bound to 3′-UTR of hif-1α mRNA were analyzed by Western blotting assay, and nucleolin was found in the magnetic particles-bound complex, indicating the interaction of nucleolin with hif-1α mRNA 3′-UTR (Fig. 4I). HuR, another RNA-binding protein, was used as a negative control to indicate the specific binding of nucleolin to hif-1α mRNA 3′-UTR (Fig. 4I). Next, hif-1α mRNA turnover rates were compared between non-silencing cells and shRNA nucleolin MEFs. As shown in Fig. 4J, knockdown of nucleolin rendered more rapid degradation of hif-1α mRNA (T½ = 2.08 h) as compared with that in non-silencing cells (T½ = 4.34 h), suggesting that nucleolin was involved in regulating hif-1α mRNA turnover. Collectively, our results demonstrated that nucleolin could bind to and stabilize hif-1α mRNA, by which nucleolin mediated HIF-1α protein induction following nickel treatment.
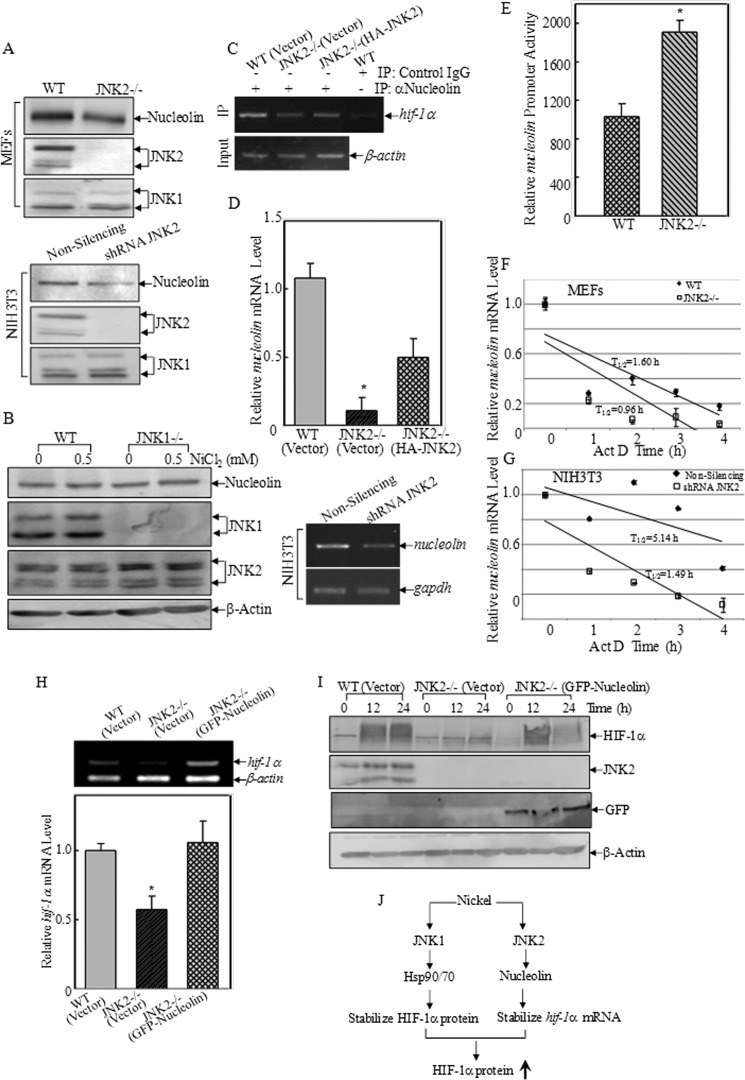
JNK2 Affected Nucleolin Expression
Nucleolin expression levels were compared between WT and JNK2−/− cells. As shown in Fig. 5A, depletion of JNK2 by either knock-out or knockdown method led to down-regulation of nucleolin expression. In contrast, JNK1 deficiency did not cause obvious change in nucleolin expression (Fig. 5B). The result from RNA-IP assay showed that binding of nucleolin to hif-1α mRNA was reduced in JNK2−/− (Vector) cells when compared with those in WT(Vector) and JNK2−/− (HA-JNK2) cells (Fig. 5C). A reduction in the mRNA level of nucleolin was detected in JNK2-deficient cells by reverse transcript-PCR (Fig. 5D), while transcription of nucleolin was increased in JNK2−/− cells, as shown in nucleolin-promoter luciferase reporter assay (Fig. 5E). The degradation of nucleolin mRNA was found more rapid in JNK2−/− cells (T½ = 0.96 h) compared with that in WT cells (T½ = 1.60 h) following Act D treatment (Fig. 5F). This result was reproducible when using shRNA JNK2 cells (Fig. 5G). The half-life of nucleolin mRNA was 3.45 times longer in non-silencing cells (T½ = 5.14 h) compared with that in shRNA JNK2 cells (T½ = 1.49 h). To confirm whether it was through nucleolin that JNK2 regulated HIF-1α expression, GFP-tagged nucleolin was transfected into JNK2−/− cells. We found that the hif-1a mRNA was elevated by overexpressing GFP-nucleolin in JNK2−/− cells (Fig. 5H), and the HIF-1α protein induction was also restored after nickel exposure (Fig. 5I). Taken together, it was concluded that JNK2 regulated nucleolin expression by stabilizing its mRNA, which further contributed to HIF-1α induction in nickel response.
FIGURE 5.
Nucleolin was regulated by JNK2, but not JNK1. A and B, expression of nucleolin protein in the indicated cell lines was analyzed by Western blotting assay. C, RNA-IP was performed to compare the binding with nucleolin to hif-1α mRNA in the indicated cell lines using anti-nucleolin antibody. D, comparison of nucleolin mRNA expression in the transfectants as indicated by real-time PCR (top panel and RT-PCR (bottom panel). The asterisk (*) indicates a significant decrease as compared with that in WT(Vector) or JNK2−/− (HA-JNK2) cells (p < 0.05). E, nucleolin transcription levels were determined using nucleolin promoter-driven firefly luciferase and pRL-TK vector. The results were expressed relative nucleolin promoter activities by normalizing the ratios of firefly to Renilla lucifease activity, as means ± S.D. (n = 3). The asterisk (*) indicates a significant increase as compared with that in WT cells (p < 0.05). F and G, mRNA degradation rate of nucleolin was determined by reverse transcript-PCR following treatment with actinomycin D (5 μm) as indicated. The densitometric analyses of the product bands were conducted using the software of ImageQuant 5.2 (GE Healthcare). The results were shown as means ± S.D. (n = 3). H, hif-1α mRNA expression levels were assessed by RT-PCR in the indicated cells. The figures shown were representative of three independent experiments. The densitometric analyses of the product bands were conducted using the software of ImageQuant 5.2 (GE Healthcare). The results were shown as means ± S.D. (n = 3). I, WT(Vector), JNK2−/− (Vector), and JNK2−/− (GFP-Nucleolin) MEFs were exposed to nickel (0.5 mm) for 12 and 24 h. The cell extracts were subjected to Western blotting as indicated. J, scheme showing the molecular mechanisms underlying JNK1 and JNK2 regulation of HIF-1α expression following nickel exposure.
DISCUSSION
JNKs are members of the superfamily of MAPKs (54). These types of enzymes are involved in the regulation of various mammalian physiological events, including cell proliferation, cell death, DNA repair, and metabolism (55). In the study reported here, JNK2 was found to be involved in the regulation of HIF-1α expression following nickel treatment via regulation of its mRNA stability. Further studies revealed that JNK2 mediated nucleolin protein expression, which could bind to hif-1α mRNA and increase its stability.
There are three genes, i.e. jnk1, jnk2, and jnk3, encoding for respective protein products; alternatively-spliced variants of the three transcripts give rise to four JNK1 isoforms, four JNK2 isoforms, and two JNK3 isoforms (56). Although JNK1 and JNK2 are presumed to operate in a redundant fashion, it is noteworthy that JNK1 and JNK2 function in opposite manners in a number of experimental systems (58, 59). For examples, JNK1, but not JNK2, was found to be responsible for cycloxygenase-2 induction following nickel exposure (49). Similarly, skin tumor formation induced by the tumor promoter 12-O-tetradecanoyl-phorbol-13-acetate (TPA) was suppressed in JNK2−/− mice (58), but the tumor incidence increased in JNK1−/− mice (59). Recently, JNK1 was found to affect the expression and the function of the Hsp90/70 chaperone proteins, thereby preventing the degradation of HIF-1α protein in a Von Hippel-Lindau (VHL)-independent manner (7). In the study reported here, it was further demonstrated that JNK2 was involved in regulating HIF-1α expression through controlling its mRNA stability, which was different from the molecular mechanism operated by JNK1 (Fig. 5J).
In the current studies, it was first noticed that the migrating pattern of HIF-1α protein on an SDS-polyacrylamide gel was different between WT and JNK2−/− cells. Specifically, a selective loss of the slower migrating band of HIF-1α species (presumed phosphorylated) was observed in JNK2−/− cells compared with that in WT cells following nickel treatment. The phenomenon was reproduced under hypoxia and other chemical mimicked hypoxia conditions, and was further confirmed using shRNA knockdown approach and restoration of the exogenous jnk2 gene back into knock-out cells. One potential cause for the loss of the slower-migrating band seen in JNK2−/− cells was due to a defect in total HIF-1α protein synthesis, which may lead to impairment in any types of modification of HIF-1α protein, including phosphorylation. An alternative explanation was based on an equal synthesis of HIF-1α protein in WT and knock-out cells, while less phosphor-groups were conjugated to HIF-1α protein in JNK2-deficient cells. To further explore this question, we removed global phosphor-groups by addition of nonspecific protein phosphatase, λ phosphatase into the whole cell lysate, and found that the amount of HIF-1α proteins synthesized in nickel-treated JNK2−/− cells was attenuated in comparison to that in WT cells, therefore the first hypothesis was supported. This supposition was further confirmed by pulse assay using 35S-labeled methionine and cysteine to monitor the de novo HIF-1α protein synthesis rate.
The result of reverse transcript-PCR showed that the expression level of hif-1α mRNA was down-regulated in JNK2−/− cells in comparison with that in WT cells. Moreover, the results from hif-1α promoter-driven luciferase assay excluded the feasibility regarding the transcriptional regulation of hif-1α expression by JNK2. Instead, a more rapid hif-1α mRNA degradation rate was observed in JNK2−/− cells following treatment with transcription inhibitors, demonstrating that JNK2 up-regulated hif-1α mRNA expression level via increasing its stability.
Nucleolin is an important nucleus protein and functions in fundamental DNA and RNA metabolism (40). Previous work revealed that nucleolin had high affinity to G-quadruplex structure (27), including the sequences in the 5′-UTR region of hif-1α mRNA. The study reported here provided in vivo evidence for the physical interaction between nucleolin and hif-1α mRNA using RNA-IP assay with nucleolin antibody. However, in contrast to the previous report from Gonzalea et al., who showed binding of nucleolin to 5′-UTR of hif-1a in in vitro filter binding assay (27), our result demonstrated that the interaction of nucleolin with hif-1α mRNA occurred at 3′-UTR region in intact cells. In addition, this kind of interaction facilitated hif-1α mRNA stability, evident from the results of mRNA decay assay, which showed that nucleolin knockdown shortened hif-1α mRNA half-life.
Nucleolin has been reported to be required for JNK-mediated IL-2 mRNA stabilization in activated T-cells (53). The details of the mechanisms responsible for JNK2 regulation of nucleolin expression are currently under investigation in our laboratory. Reverse transcript-PCR result revealed a reduction in the nucleolin mRNA amount in JNK2-deficient cells. But the nucelolin gene transcription was comparable between WT and knock-out cells, as determined by nucleolin promoter reporter. Similar with what we identified in the case of hif-1α, nucleolin mRNA degradation rate was elevated in JNK2-deficient cells. However at the present stage, we are uncertain whether JNK2 regulated global mRNA stability through a commonly shared mechanism or specifically affected mRNA stability of hif-1α and nucleolin.
Collectively, our present studies demonstrate that JNK2 was involved in regulating HIF-1α expression in nickel response through maintaining its mRNA stability. Nucleolin might be a mediator for JNK2 in controlling hif-1α mRNA decay. Therefore, it was suggested that although JNK1 and JNK2 were both involved in the regulation of HIF-1α expression (7), they performed in different mechanisms, thus providing more evidence for the distinct molecular scenarios of each isoform.
Acknowledgments
We thank Drs. Carine Michiels, Michael B. Kastan, and Bruno Amati for generous gifts of plasmids, Drs. Zheng-gang Liu and Perry J. Blackshear for providing JNK2−/− and TTP−/− MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grants CA112557 from NIH/NCI and ES000260 and ES010344 from NIH/NIEHS as well as NSFC81229002 and NSFC 30971516.
- HIF
- hypoxia-inducible factor
- MEF
- mouse embryonic fibroblasts
- UTR
- untranslated region
- TTP
- tristetraprolin
- DMOG
- dimethyloxalylglycine
- HRE
- hypoxia-responsive element
- PHD
- prolyl hydroxlyase domain
- Act
- actinomycin.
REFERENCES
- 1. Grandjean P. (1984) Human exposure to nickel. IARC. Sci. Publ. 53, 469–485 [PubMed] [Google Scholar]
- 2. Furst A. (1984) Mechanism of action of nickel as a carcinogen: needed information. IARC. Sci. Publ. 53, 245–252 [PubMed] [Google Scholar]
- 3. Salnikow K., Zhitkovich A. (2008) Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem. Res. Toxicol. 21, 28–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lu H., Shi X., Costa M., Huang C. (2005) Carcinogenic effect of nickel compounds. Mol. Cell. Biochem. 279, 45–67 [DOI] [PubMed] [Google Scholar]
- 5. Ke Q., Costa M. (2006) Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 70, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 6. Salnikow K., An W. G., Melillo G., Blagosklonny M. V., Costa M. (1999) Nickel-induced transformation shifts the balance between HIF-1 and p53 transcription factors. Carcinogenesis 20, 1819–1823 [DOI] [PubMed] [Google Scholar]
- 7. Zhang D., Li J., Costa M., Gao J., Huang C. (2010) JNK1 mediates degradation HIF-1alpha by a VHL-independent mechanism that involves the chaperones Hsp90/Hsp70. Cancer Res. 70, 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang Q., Salnikow K., Kluz T., Chen L. C., Su W. C., Costa M. (2003) Inhibition and reversal of nickel-induced transformation by the histone deacetylase inhibitor trichostatin A. Toxicol. Appl. Pharmacol. 192, 201–211 [DOI] [PubMed] [Google Scholar]
- 9. Semenza G. L. (2007) Sci. STKE 2007, cm8. [DOI] [PubMed] [Google Scholar]
- 10. Gordan J. D., Simon M. C. (2007) Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr. Opin. Genet. Dev. 17, 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bárdos J. I., Ashcroft M. (2005) Negative and positive regulation of HIF-1: a complex network. Biochim. Biophys. Acta 1755, 107–120 [DOI] [PubMed] [Google Scholar]
- 12. Rossignol F., Vaché C., Clottes E. (2002) Natural antisense transcripts of hypoxia-inducible factor 1α are detected in different normal and tumour human tissues. Gene 299, 135–140 [DOI] [PubMed] [Google Scholar]
- 13. Rossignol F., de Laplanche E., Mounier R., Bonnefont J., Cayre A., Godinot C., Simonnet H., Clottes E. (2004) Natural antisense transcripts of HIF-1α are conserved in rodents. Gene 339, 121–130 [DOI] [PubMed] [Google Scholar]
- 14. Taguchi A., Yanagisawa K., Tanaka M., Cao K., Matsuyama Y., Goto H., Takahashi T. (2008) Identification of hypoxia-inducible factor-1α as a novel target for miR-17–92 microRNA cluster. Cancer Res. 68, 5540–5545 [DOI] [PubMed] [Google Scholar]
- 15. Dolt K. S., Mishra M. K., Karar J., Baig M. A., Ahmed Z., Pasha M. A. (2007) cDNA cloning, gene organization and variant specific expression of HIF-1α in high altitude yak (Bos grunniens). Gene 386, 73–80 [DOI] [PubMed] [Google Scholar]
- 16. Zou A. P., Yang Z. Z., Li P. L., Cowley A. W., Jr. (2001) Oxygen-dependent expression of hypoxia-inducible factor-1α in renal medullary cells of rats. Physiol. Genomics 6, 159–168 [DOI] [PubMed] [Google Scholar]
- 17. Terova G., Rimoldi S., Cora S., Bernardini G., Gornati R., Saroglia M. (2008) Acute and chronic hypoxia affects HIF-1α mRNA levels in sea bass (Dicentrarchus labrax). Aquaculture 279, 150–159 [Google Scholar]
- 18. Sirri V., Roussel P., Gendron M.-C., Hernandez-Verdun D. (1997) Amount of the two major Ag-NOR proteins, nucleolin, and protein B23 is cell-cycle dependent. Cytometry 28, 147–156 [PubMed] [Google Scholar]
- 19. Greco A. (2009) Involvement of the nucleolus in replication of human viruses. Rev. Med. Virol. 19, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seko Y., Cole S., Kasprzak W., Shapiro B. A., Ragheb J. A. (2006) The role of cytokine mRNA stability in the pathogenesis of autoimmune disease. Autoimmun. Rev. 5, 299–305 [DOI] [PubMed] [Google Scholar]
- 21. Dranovsky A., Vincent I., Gregori L., Schwarzman A., Colflesh D., Enghild J., Strittmatter W., Davies P., Goldgaber D. (2001) Cdc2 phosphorylation of nucleolin demarcates mitotic stages and Alzheimer's disease pathology. Neurobiol. Aging 22, 517–528 [DOI] [PubMed] [Google Scholar]
- 22. Caudle W. M., Kitsou E., Li J., Bradner J., Zhang J. (2009) A role for a novel protein, nucleolin, in Parkinson's disease. Neurosci. Lett. 459, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ginisty H., Sicard H., Roger B., Bouvet P. (1999) Structure and functions of nucleolin. J. Cell Sci. 112, 761–772 [DOI] [PubMed] [Google Scholar]
- 24. Takagi M., Absalon M. J., McLure K. G., Kastan M. B. (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123, 49–63 [DOI] [PubMed] [Google Scholar]
- 25. Soundararajan S., Chen W., Spicer E. K., Courtenay-Luck N., Fernandes D. J. (2008) The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 68, 2358–2365 [DOI] [PubMed] [Google Scholar]
- 26. Zhang J., Tsaprailis G., Bowden G. T. (2008) Nucleolin stabilizes Bcl-X L messenger RNA in response to UVA irradiation. Cancer Res. 68, 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. González V., Guo K., Hurley L., Sun D. (2009) Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 284, 23622–23635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uribe D. J., Guo K., Shin Y. J., Sun D. (2011) Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry 50, 3796–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sabapathy K., Hu Y., Kallunki T., Schreiber M., David J. P., Jochum W., Wagner E. F., Karin M. (1999) JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 9, 116–125 [DOI] [PubMed] [Google Scholar]
- 30. Tang G., Minemoto Y., Dibling B., Purcell N. H., Li Z., Karin M., Lin A. (2001) Inhibition of JNK activation through NF-κB target genes. Nature 414, 313–317 [DOI] [PubMed] [Google Scholar]
- 31. Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F., Blackshear P. J. (1996) A pathogenetic role for TNFα in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445–454 [DOI] [PubMed] [Google Scholar]
- 32. Ding J., Li J., Chen J., Chen H., Ouyang W., Zhang R., Xue C., Zhang D., Amin S., Desai D., Huang C. (2006) Effects of polycyclic aromatic hydrocarbons (PAHs) on vascular endothelial growth factor induction through phosphatidylinositol 3-kinase/AP-1-dependent, HIF-1α-independent pathway. J. Biol. Chem. 281, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 33. Jiang B.-H., Jiang G., Zheng J. Z., Lu Z., Hunter T., Vogt P. K. (2001) Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 12, 363–369 [PubMed] [Google Scholar]
- 34. Wan J., Sun L., Mendoza J. W., Chui Y. L., Huang D. P., Chen Z. J., Suzuki N., Suzuki S., Yeh W. C., Akira S., Matsumoto K., Liu Z. G., Wu Z. (2004) Elucidation of the c-Jun N-terminal kinase pathway mediated by Estein-Barr virus-encoded latent membrane protein 1. Mol. Cell. Biol. 24, 192–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang C., Ma W. y., Bowden G. T., Dong Z. (1996) Ultraviolet B-induced activated protein-1 activation does not require epidermal growth factor receptor but is blocked by a dominant negative PKClambda/iota. J. Biol. Chem. 271, 31262–31268 [DOI] [PubMed] [Google Scholar]
- 36. Huang C., Ma W. Y., Dong Z. (1996) Requirement for phosphatidylinositol 3-kinase in epidermal growth factor-induced AP-1 transactivation and transformation in JB6 P+ cells. Mol. Cell. Biol. 16, 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huang C., Ma W., Dong Z. (1996) Inhibitory effects of ascorbic acid on AP-1 activity and transformation of JB6 cells. Int. J. Oncol. 8, 389–393 [DOI] [PubMed] [Google Scholar]
- 38. Vlaminck B., Toffoli S., Ghislain B., Demazy C., Raes M., Michiels C. (2007) Dual effect of echinomycin on hypoxia-inducible factor-1 activity under normoxic and hypoxic conditions. FEBS J. 274, 5533–5542 [DOI] [PubMed] [Google Scholar]
- 39. Greasley P. J., Bonnard C., Amati B. (2000) Myc induces the nucleolin and BN51 genes: possible implications in ribosome biogenesis. Nucleic Acids Res. 28, 446–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Storck S., Shukla M., Dimitrov S., Bouvet P. (2007) Functions of the histone chaperone nucleolin in diseases. Subcell Biochem. 41, 125–144 [DOI] [PubMed] [Google Scholar]
- 41. Guo W., Yang Z., Xia Q., Liu J., Yu Y., Li J., Zuo Z., Zhang D., Li X., Shi X., Huang C. (2011) Arsenite stabilizes HIF-1α protein through p85α-mediated up-regulation of inducible Hsp70 protein expression. Cell. Mol. Life Sci. 68, 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Song L., Li J., Ye J., Yu G., Ding J., Zhang D., Ouyang W., Dong Z., Kim S. O., Huang C. (2007) p85α acts as a novel signal transducer for mediation of cellular apoptotic response to UV radiation. Mol. Cell. Biol. 27, 2713–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang D., Song L., Li J., Wu K., Huang C. (2006) Coordination of JNK1 and JNK2 is critical for GADD45α induction and its mediated cell apoptosis in arsenite responses. J. Biol. Chem. 281, 34113–34123 [DOI] [PubMed] [Google Scholar]
- 44. Campalans A., Kondorosi A., Crespi M. (2004) Enod40, a Short Open Reading Frame-Containing mRNA, induces Cytoplasmic Localization of a Nuclear RNA Binding Protein in Medicago truncatula. The Plant Cell Online 16, 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bode A. M., Dong Z. (2007) The functional contrariety of JNK. Mol. Carcinogenesis 46, 591–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Insights into the role of the von Hippel-Lindau gene product. A key player in hypoxic regulation. Exp. Nephrol. 9, 235–240 [DOI] [PubMed] [Google Scholar]
- 47. Richard D. E., Berra E., Gothié E., Roux D., Pouysségur J. (1999) p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274, 32631–32637 [DOI] [PubMed] [Google Scholar]
- 48. Minet E., Ernest I., Michel G., Roland I., Remacle J., Raes M., Michiels C. (1999) HIF1A gene transcription is dependent on a core promoter sequence encompassing activating and inhibiting sequences located upstream from the transcription initiation site and cis elements located within the 5'UTR. Biochem. Biophys. Res. Commun. 261, 534–540 [DOI] [PubMed] [Google Scholar]
- 49. Zhang D., Li J., Wu K., Ouyang W., Ding J., Liu Z. G., Costa M., Huang C. (2007) JNK1, but not JNK2, is required for COX-2 induction by nickel compounds. Carcinogenesis 28, 883–891 [DOI] [PubMed] [Google Scholar]
- 50. Beach L. R., Ross J. (1978) Cordycepin. An inhibitor of newly synthesized globin messenger RNA. J. Biol. Chem. 253, 2628–2632 [PubMed] [Google Scholar]
- 51. Cao H., Dzineku F., Blackshear P. J. (2003) Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-α mRNA and serve as a substrate for mitogen-activated protein kinases. Arch. Biochem. Biophys. 412, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lin N. Y., Lin C. T., Chang C. J. (2008) Modulation of immediate early gene expression by tristetraprolin in the differentiation of 3T3-L1 cells. Biochem. Biophys. Res. Commun. 365, 69–74 [DOI] [PubMed] [Google Scholar]
- 53. Chen C. Y., Gherzi R., Andersen J. S., Gaietta G., Jürchott K., Royer H. D., Mann M., Karin M. (2000) Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 14, 1236–1248 [PMC free article] [PubMed] [Google Scholar]
- 54. Johnson G. L., Lapadat R. (2002) Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912 [DOI] [PubMed] [Google Scholar]
- 55. Karin M., Gallagher E. (2005) From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life 57, 283–295 [DOI] [PubMed] [Google Scholar]
- 56. Sabapathy K., Hochedlinger K., Nam S. Y., Bauer A., Karin M., Wagner E. F. (2004) Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15, 713–725 [DOI] [PubMed] [Google Scholar]
- 57.Deleted in proof
- 58. Chen N., Nomura M., She Q. B., Ma W. Y., Bode A. M., Wang L., Flavell R. A., Dong Z. (2001) Suppression of skin tumorigenesis in c-Jun NH(2)-terminal kinase-2-deficient mice. Cancer Res. 61, 3908–3912 [PubMed] [Google Scholar]
- 59. She Q. B., Chen N., Bode A. M., Flavell R. A., Dong Z. (2002) Deficiency of c-Jun-NH(2)-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res. 62, 1343–1348 [PubMed] [Google Scholar]
- 60. Dean J. L. E., Sully G., Clark A. R., Saklatvala J. (2004) The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell. Signal. 16, 1113–1121 [DOI] [PubMed] [Google Scholar]