Background: The multidomain protein GIT1 is involved in cytoskeletal rearrangements and cell motility.
Results: GIT1 phosphorylation on serine 46 by PKD3 regulates localization of GIT1-paxillin complexes.
Conclusion: Paxillin trafficking and cellular protrusive activity are controlled by GIT1 serine 46 phosphorylation.
Significance: Selective phosphorylation of GIT1 by PKD3 implicates a specific function for this PKD isoform in the regulation of cell shape and motility.
Keywords: Cell Migration, Cell Signaling, Mass Spectrometry (MS), Phosphorylation, Protein Kinase D (PKD), G-protein-coupled Receptor Kinase-interacting Protein (GIT1), Focal Adhesion
Abstract
The continuous assembly and disassembly of focal adhesions is required for efficient cell spreading and migration. The G-protein-coupled receptor kinase-interacting protein 1 (GIT1) is a multidomain protein whose dynamic localization to sites of cytoskeletal remodeling is critically involved in the regulation of these processes. Here we provide evidence that the subcellular localization of GIT1 is regulated by protein kinase D3 (PKD3) through direct phosphorylation on serine 46. GIT1 phosphorylation on serine 46 was abrograted by PKD3 depletion, thereby identifying GIT1 as the first specific substrate for this kinase. A GIT1 S46D phosphomimetic mutant localized to motile, paxillin-positive cytoplasmic complexes, whereas the phosphorylation-deficient GIT1 S46A was enriched in focal adhesions. We propose that phosphorylation of GIT1 on serine 46 by PKD3 represents a molecular switch by which GIT1 localization, paxillin trafficking, and cellular protrusive activity are regulated.
Introduction
G-protein coupled receptor kinase-interacting protein 1 (GIT1)3 is a multidomain protein that plays an important role in actin cytoskeletal remodeling and membrane transport processes (1, 2). GIT1 comprises an N-terminal Arf GTPase-activating protein (GAP) domain, three ankyrin repeats, a Spa2 homology domain that mediates binding to the Rac/Cdc42-specific exchange factor PIX, a coiled-coil domain, and a C-terminal paxillin binding site (PBS). In addition to its ArfGAP domain with specificity mainly for the small GTPase Arf6, various protein binding partners have been identified to interact with the different domains of GIT1, including paxillin, focal adhesion kinase (FAK), phospholipase Cγ, and extracellular signal-regulated kinase (3). GIT1 thus acts as a scaffolding protein that coordinates the spatial activation of a range of signaling pathways. Furthermore, via the coiled-coil domain, GIT1 homodimerizes, and it can also heterodimerize with the structurally conserved GIT2 protein, forming large oligomeric signaling protein complexes together with PIX proteins within the cell (3).
GIT1 has been observed at different subcellular locations, including focal adhesions (FAs), membrane protrusions, adherens junctions, and the centrosome (1, 2). Binding of paxillin to the PBS domain of GIT1 is primarily responsible for the localization of GIT1 to FAs. Paxillin is a multidomain scaffolding protein that functions in the recruitment of both signaling and structural molecules to focal adhesions (4). Paxillin contains five leucine- and aspartate-rich LD motifs, of which LD4 directly binds GIT1-PIX signaling complexes. Fibroblasts that express a paxillin mutant deficient in GIT1 binding exhibit abnormal membrane protrusion dynamics that are caused by sustained global Rac activity (5). GIT1 has been suggested to restrict local Rac activation by inhibiting Arf6 which in turn down-regulates Rac activity (6). These cells are also defective in polarized cell migration (5, 7) and focal adhesion disassembly (8). GIT1 expression has also been found to promote membrane protrusions and motility by PIX-mediated recruitment and activation of p21-activated kinase (PAK) and Rac independently of GIT GAP activity (9–11), suggesting that the precise effects of GIT1 may depend on the cell type and experimental setup.
PAK is a serine/threonine kinase that is activated by the small GTPases Rac and Cdc42 and promotes the recruitment of GIT-PIX oligomers to focal complexes (1, 3). Since PAK lacks a focal adhesion targeting motif, its recruitment to FAs requires binding to PIX, which, besides its nucleotide exchange activity for Rac and Cdc42, functions as an adaptor protein. A PAK mutant that is unable to bind PIX blocks localization of GIT-PIX to focal complexes (12). The down-regulation of Rac by FA-localized GIT1 could then act as a negative feedback preventing prolonged Rac activation. It thus appears that the dynamically controlled localization of GIT1 is crucial for cellular activity. GIT1 was found to cycle between the leading edge and FAs in motile punctate cytoplasmic structures (10). These structures were described as supramolecular protein complexes containing paxillin, PIX, and PAK that were not associated with any membrane markers (10). However, in other studies GIT1 was observed in vesicular structures that stained positively for the endosomal markers Rab11 and transferrin (13–15). Regardless of the precise nature of these structures, a major function of GIT1 is the delivery of signaling complexes to cell adhesions and membrane protrusions to regulate cell shape and motility.
Mass spectrometry analysis identified multiple serine, threonine, and tyrosine residues to be phosphorylated in GIT1 (16). It was recently reported that GIT1 is phosphorylated on tyrosine 321 downstream of FAK and Src kinases in platelet-derived growth factor-stimulated osteoblasts, leading to increased association with and activation of FAK and cell motility (17). Another report describes that phosphorylation of GIT1 on serine 709, which is located within the PBS, increases paxillin binding and cellular protrusive activity (18) but in most cases the functional significance of GIT1 phosphorylations and the upstream kinases are unknown. PKD is a family of serine/threonine kinases comprising three family members, PKD1/PKCμ, PKD2, and PKD3/PKCν (19, 20). The PKD isoforms are known to localize to the Golgi complex and the plasma membrane, and they have also been reported to shuttle to the nucleus. At the Golgi complex, PKD regulates vesicular traffic to the plasma membrane, whereas at the plasma membrane PKD is involved in the regulation of cell shape, movement, and invasion. In line with such functions, substrates associated with actin cytoskeletal remodeling including cortactin, DLC1, RIN1, SSH1L, and Pak4 were recently identified to be phosphorylated by PKD (21–26). Most of these studies focused on PKD1 and/or PKD2 isoforms, whereas relatively little is known about PKD3. The current notion is that the PKD isoforms possess overlapping substrate specificity and function redundantly. Here we provide evidence that the cellular activity of GIT1 via serine 46 phosphorylation is exclusively regulated by PKD3, thus identifying GIT1 as the first specific substrate of this particular PKD isoform.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies used in this study were: monoclonal mouse anti-GFP mAb (Roche Biosciences), monoclonal mouse anti-Flag and anti-tubulin mAbs (Sigma-Aldrich), polyclonal rabbit anti-PKD1 (C20, Santa Cruz Biotechnology), anti-PKD2 (Calbiochem), and anti-PKD3 pAbs (Cell Signaling), polyclonal goat anti-GST pAb (GE Healthcare), monoclonal mouse anti-GIT1 mAb (BD), polyclonal rabbit anti-paxillin pAb (Santa Cruz), monoclonal mouse anti-vinculin mAb (Sigma), polyclonal rabbit anti-EEA1 pAb (Cell Signaling), monoclonal mouse anti-transferrin receptor mAb (Invitrogen), and monoclonal mouse anti-c-Myc mAb (9E10) (Sigma). The phospho-(Ser/Thr) PKD substrates antibody was from Cell Signaling. The phosphospecific antiserum that crossreacted with phosphorylated GIT1 was raised by immunizing rabbits with NH2-CRRHGpSMVSL-CONH2 peptide (Pineda-Abservice), corresponding to amino acids 128–136 in CERT (Q9Y5P4). HRP-labeled secondary anti-mouse and anti-rabbit IgG antibodies were from GE Healthcare, HRP-labeled secondary anti-goat was from Santa Cruz Biotechnology. Alexa Fluor 546-labeled secondary anti-mouse and anti-rabbit IgG antibodies were from Invitrogen. Gö6976 and Gö6983 were from Calbiochem, phorbol-12,13-dibutyrate (PDBu) from Enzo Life Sciences and staurosporine from Alexis Biochemicals. GST-tagged recombinant human PKD3 was from Invitrogen. Anti-Flag M2 Affinity Gel was from Sigma.
Plasmids and Cloning of GIT1 and PKD3 Expression Constructs
pEGFPN1-GIT1-Flag and pEGFPC1-GIT1 were obtained from Addgene (10). The GIT1 S46A and S46D mutations were introduced by site-directed PCR mutagenesis using the following forward primers: GIT1-S46A-for (5′-CTGGGACGCCACATCGCCATTGTCAAGCACC-3′) and GIT1-S46D-for (5′-GCCTGGGACGCCACATCGACATTGTCAAGCACC-3′). The full-length PKD3 cDNA was PCR-amplified using cDNA from HEK293T cells as a template and the following forward and reverse primers: 5′-ggcctcgagatgtctgcaaataattcc-3′ and 5′-ggctctagattaaggatcttcttccat-3′. The PCR product was cloned into the pCR3.V62-Met-Flag vector by XhoI/XbaI restriction. pEGFPN1-PKD3 was generated by PCR amplification of the PKD3 cDNA using pCR3.V62-Met-Flag-PKD3 as a template and the forward and reverse primers 5′-gatctcgagctcatgtctgcaaataattcc-3′ and 5′-tggatcccgggaaggatcttcttccatatc-3′, and cloned into the pEGFPN1 vector by SacI/XmaI restriction. To obtain the PKD3-ca cDNA, sequential site-directed PCR mutagenesis was carried out using PKD3-S731E-for (5′-atcattggtgaaaaggaattcaggagatctgtg-3′), followed by PKD3-S735E-for (5′-aaggaattcaggagagaagtggtaggaactcca-3′) as forward primers, and the PKD3-kd cDNA was obtained using PKD3-K605W-for (5′-gatgtggctatttgggtaattgataag-3′) as a forward primer. All amplified cDNAs were verified by sequencing. Oligonucleotides were purchased from MWG Biotech. The Flag-betaPIXa expression plasmid was obtained from Addgene (no. 15234) (27) and pCMV6M-Myc-PAK1 was kindly provided by Dr. Ora Bernard.
Cell Culture, Transfection, and Generation of Stable Cell Lines
MCF7 and HEK293T cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FCS (PAA) and incubated in a humidified atmosphere of 5% CO2 at 37 °C. HEK293T cells were transiently transfected with TransIT (Mirus Bio) and MCF-7 cells with Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Stable MCF7 sublines cells were generated by nucleofection (program P020, Kit V; Amaxa) with pEGFPC1 GIT1 expression vectors followed by selection with 1 mg/ml G418 (Calbiochem) and FACS sorting of GFP-positive cells (medium expression intensity). Stable expression of the GFP-tagged GIT1 proteins in the MCF7 sublines was verified by FACS analysis and immunoblotting. For RNAi, cells were transfected with siRNA using Oligofectamine (Invitrogen) according to manufacturer's instructions. The siRNAs used were: 5′-GCA GAG UGA AAG UUC CAC ACA CAU U-3′ (siPKD3–1), 5′-GCU GCU UCU CCG UGU UCA AGU CCU A-3′ (siPKD3–2), 5′-GUC GAG AGA AGA GGU CAA ATT-3′ (siPKD1) and 5′-GCA AAG ACU GCA AGU UUA ATT-3′ (siPKD2), and 5′-GCGGCUGCCGGAAUUUACCTT-3′ (siLacZ). PKD3-specific siRNAs were from Invitrogen and those for LacZ, PKD1 and PKD2 were synthesized by MWG Biotech. PKD1 and PKD2 On-Target Plus and control Smartpools were from Dharmacon.
Cell Lysis, Immunoprecipitation, SDS-PAGE, and Western Blotting
Cells were lysed in RIPA buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton-X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm sodium orthovanadate, 10 mm sodium fluoride, and 20 mm β-glycerophosphate plus Complete protease inhibitors (Roche)) or TEB buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton-X-100, 1 mm sodium orthovanadate, 10 mm sodium fluoride, and 20 mm β-glycerophosphate plus Complete protease inhibitors (Roche)), and lysates were clarified by centrifugation at 16,000 × g for 10 min. For immunoprecipitation, equal amounts of protein were incubated with specific antibodies for 2–4 h at 4 °C. Immune complexes were collected with protein G-Sepharose (KPL) and washed 3× with TEB buffer. Precipitated proteins were released by boiling in sample buffer, separated by SDS-PAGE and transferred to PVDF membrane (Roth). The membrane was blocked with 0.5% blocking reagent (Roche) in PBS containing 0.1% Tween-20 and then incubated with primary antibodies, followed by HRP-conjugated secondary antibodies. Visualization was with the ECL detection system (Pierce).
Mass Spectrometry Analysis
HEK293T cells transiently expressing Flag-tagged GIT1 (5 × 10 cm dishes) were stimulated with PDBu, lysed in 1% TEB, and GIT1 was immunoprecipitated with anti-Flag M2-agarose (Sigma Aldrich). The beads were washed with TEB and PBS, followed by elution with 0.1 m glycine, pH 2.5 and neutralization with 1/10 volume of 1 m Tris, pH 8. Eluted protein was subjected to SDS-PAGE and the gel stained with Colloidal Coomassie Blue (Roth). GIT1 expression and phosphorylation was verified by parallel Western blotting. The Coomassie-stained Flag-tagged GIT1 band was in-gel digested with trypsin as described (28). Acetonitrile was added to the peptide mixture to a final concentration of 30% and the pH was adjusted to 2–3. Enrichment of phosphopeptides by titanium dioxide chromatography was done as described previously (29) with the following modifications: phosphopeptide elution from the beads was performed three times with 100 μl of 40% ammonia hydroxide solution in 60% acetonitrile at a pH of >10.5. Analysis of the phosphopeptides was done on a Proxeon Easy-LC system (Proxeon Biosystems) coupled to a LTQ-Orbitrap-XL (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Proxeon Biosystems) as described previously (30). The five most intense precursor ions were fragmented by activation of neutral loss ions at −98, −49, and −32.6 relative to the precursor ion (multistage activation). Additionally an inclusion list containing the doubly charged ions of tryptic GIT1 peptides that include serine 46 in the phosphorylated form was applied. Mass spectra were analyzed using the software suite MaxQuant, version 1.0.14.3 (31). The data were searched against a target-decoy human database (ipi.HUMAN.v3.77) containing 182695 forward protein sequences and 262 frequently observed contaminants. Trypsin was set as protease in which two missed cleavage sites were allowed. Beside acetylation at the N terminus and oxidation of methionine, phosphorylation of serine, threonine, and tyrosine were set as variable modifications. Carbamidomethylation of cysteine was set as fixed modification. Initial precursor mass tolerance was set to 7 parts per million (ppm) at the precursor ion and 0.5 Da at the fragment ion level. Phosphorylation events with a localization probability of at least 0.75 were considered to be assigned to a specific residue. Spectra of modified peptides were manually validated.
Phosphorylation Assay
HEK293T cells were transiently transfected with plasmids encoding Flag-GIT1 wt and S46A. Proteins were immunoprecipitated from whole cell lysates with Flag-specific mouse mAb and protein G-agarose and washed with kinase buffer (50 mm Tris, pH 7.5, 10 mm MgCl2, 2 mm DTT). Immunoprecipitated proteins were incubated in the presence (+) or absence (−) of purified GST-tagged PKD3 with 20 μm ATP in kinase buffer for 30 min at 37 °C. The reaction was stopped by addition of sample buffer and proteins were separated by SDS-PAGE and analyzed by immunoblotting.
Immunofluorescence Microscopy and Live Cell Imaging
MCF7 cells transiently transfected with expression vectors and grown on glass coverslips coated with 25 μg/ml collagen (Serva) were fixed with 4% PFA for 10 min, permeabilized with PBS containing 0.1% Triton X-100 for 5 min, and blocked with 5% goat serum (Invitrogen) in PBS containing 0.1% Tween-20 for 30 min. Cells were then incubated for 2 h with primary antibodies in blocking buffer, followed by washing steps with PBS containing 0.1% Tween-20 and incubation with secondary antibody in blocking buffer for 1 h. Coverslips were mounted in Fluoromount G (Southern Biotechnology) and analyzed on a confocal laser-scanning microscope (TCS SL, Leica) using 488 nm, 543 nm and 633 nm excitation and a 63× HCX Plan Apo CS oil objective lens. Images were processed with Adobe Photoshop. Alternatively, a Zeiss LSM 700 confocal laser-scanning microscope using 488, 561, and 633 nm excitation with the oil objective lens Plan-Apochromat 63x/1.40 DIC M27 was used and images were processed with the ZEN software. For live cell imaging, MCF7 cells stably expressing GFP-GIT1 S46D were plated at low density on collagen-coated glass bottom cell culture dishes (35, 0/10 mm, Greiner BIO-ONE). After overnight cell adhesion, recording was started by acquiring fluorescence (488 nm) images every 40 s for a duration of 20 min with a Zeiss Axiovision system equipped with a heated incubation chamber (5% CO2, 37 °C) using a Plan-Apochromat 63×/1.4 oil DIC M27 objective lens and the Axiovision software.
Spreading Assay
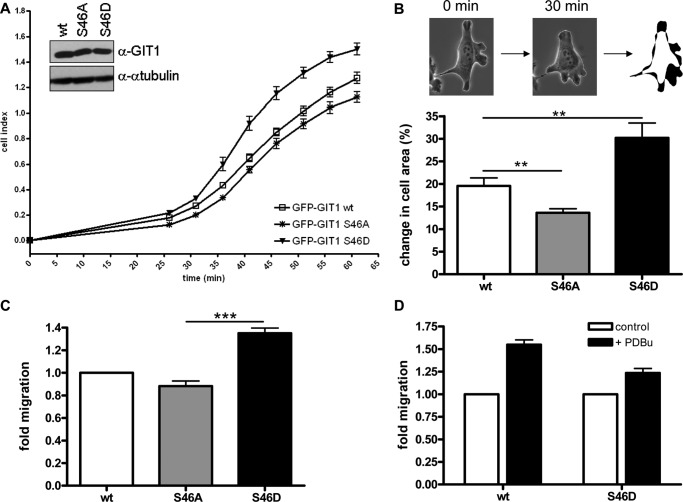
MCF7 cells stably expressing GIT1 wt, S46A, and S46D were seeded at low density (10,000 cells) into collagen-coated (10 μg/ml) 96-well E-plates (Roche). For quantitative initial spreading analysis, the impedance of cells was measured using an xCELLigence device (Roche), and the mean cell index was plotted.
Protrusion Assay
MCF7 cells stably expressing GIT1 wt, S46A, and S46D were plated at low density on collagen-coated 6-well glass bottom cell culture dishes (MatTek corporation). After overnight cell adhesion, recording of single cells was started by acquiring fluorescence (488 nm) and bright field (phase contrast) images every 30 min for several hours with a Zeiss Observer system equipped with a heated incubation chamber (5% CO2, 37 °C) using a Plan-Apochromat 40x/0.8 NA M27 objective and Zeiss Axiovision software. Then, GFP-positive cells were outlined using a paintboard (Waco) at the beginning and the end of a 30 min time interval as described previously (10). The outlined images were overlaid and the change in total cell area normalized to the initial cell size was calculated using Image J software.
Migration Assay
MCF7 cells in medium containing 0.5% FCS were seeded into Transwells (0.8 μm pore size; Costar) coated with 10 μg/ml collagen on the underside and allowed to migrate overnight. The bottom chamber contained medium supplemented with 10% FCS. Cells on the underside of the membranes were fixed, stained with crystal violet, and counted in five independent microscopic fields at a 20-fold magnification.
RESULTS
GIT1 Is Phosphorylated Downstream of PKD3
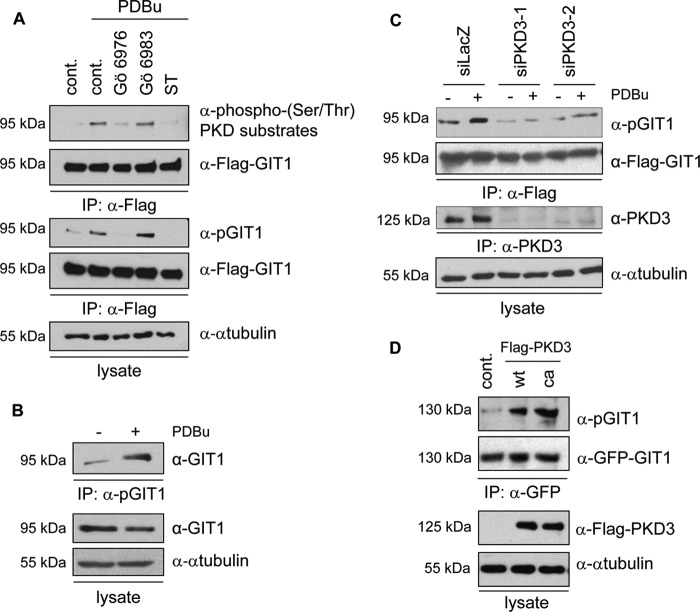
PKD substrates have successfully been identified in the past using a phosphospecific antibody that recognizes the PKD consensus motif (32). To investigate whether GIT1 is a substrate for the PKD family members, we used phorbol-12,13-dibutyrate (PDBu), an analog of diacylglycerol, to stimulate cellular PKD activity. PDBu activates novel PKCs, which in turn phosphorylate PKD family members on two specific serines within the activation loop. In addition, PDBu activates PKD directly by high-affinity binding to cysteine-rich motifs within the protein. HEK293T cells transiently expressing Flag-tagged GIT1 were stimulated with PDBu, followed by immunoblotting of GIT1 immunoprecipitates with a commercially available phospho-(Ser/Thr) PKD substrate antibody. A basal signal corresponding to GIT1 was obtained that increased upon PDBu stimulation (Fig. 1A, top panel). Preincubation of cells with specific pharmacological inhibitors of the PKC/PKD pathway abrogated GIT1 detection with the phosphospecific antibody (Fig. 1A, top panel), indicating that GIT1 was recognized in a phosphorylation-dependent manner. Gö6976 inhibits PKD family members directly, whereas Gö6983 can inhibit PKD indirectly by inhibiting novel PKCs. Staurosporine, which blocks a wide range of kinases including PKCs and PKD, was used as a control. Since preincubation of cells with Gö6983 did not affect GIT1 recognition by the phosphospecific antibody, this indicates that GIT1 phosphorylation downstream of PKD occurs independently of novel PKCs. Identical results were obtained when GIT1 immunoprecipitates were immunoblotted with a phosphospecific antiserum that was originally raised against the PKD consensus site in the lipid transfer protein CERT (33) (Fig. 1A, middle panels). This antibody recognizes CERT only when phosphorylated on serine 132 (supplemental Fig. S1). Because of its crossreactivity, in all further experiments, this antiserum was used for the detection of GIT1.
FIGURE 1.
GIT1 is downstream of PKD3. A, HEK293T cells were transiently transfected with an expression vector encoding Flag-GIT1. The next day, cells were treated with 5 μm Gö6976 and Gö6983, and 1 μm staurosporine (ST) for 3 h. DMSO was used as a control. Cells were then stimulated with 1 μm PDBu for 15 min. Cells were lysed, and GIT1 was immunoprecipitated with Flag-specific antibody and analyzed by immunoblotting with a phospho(Ser/Thr) PKD substrates antibody (Cell Signaling; top panel) and a phosphospecific antiserum originally raised against the PKD consensus sequence in CERT (pGIT1; middle panel), followed by detection with Flag-specific antibody to ensure equal GIT1 expression and precipitation. Equal loading was confirmed by immunoblotting of whole cell lysates with anti-α-tubulin antibody (bottom panel). B, MCF7 cells were stimulated with 1 μm PDBu for 15 min and endogenous GIT1 was immunoprecipitated with the pGIT1 antibody and analyzed by immunoblotting with a GIT1-specific antibody (top panel). Equal GIT1 expression and loading were confirmed by immunoblotting of whole cell lysates with anti-GIT1 and α-tubulin antibodies, respectively (middle and bottom panels). C, HEK293T cells were transiently transfected with two independent PKD3-specific and LacZ-specific siRNA. One day later, cells were transiently transfected with a Flag-GIT1 expression vector. After 24 h, cells were stimulated with PDBu, lysed, and GIT1 was immunoprecipitated with Flag-specific antibody and analyzed by immunoblotting with the pGIT1 antibody, followed by detection with Flag-specific antibody. To confirm the knockdown of PKD3, PKD3 was immunoprecipitated with a PKD3-specific antibody, followed by immunoblotting using a PKD3-specific antibody. Equal loading was verified by immunoblotting of whole cell lysates with α-tubulin-specific antibody. D, GFP-GIT1 was co-expressed with empty vector as a control, Flag-tagged PKD3 wild type (wt) and catalytically active (ca, S731/735E) in HEK293T cells. GIT1 was immunoprecipitated with GFP-specific antibody and analyzed by immunoblotting with the pGIT1 antibody, followed by detection with GFP-specific antibody. Expression of the PKD3 variants was confirmed by immunoblotting of whole cell lysates with Flag-specific antibody. Equal loading was verified by immunoblotting of whole cell lysates with α-tubulin-specific antibody.
To investigate whether GIT1 phosphorylation could also be detected at the endogenous level, MCF7 cells were stimulated with PDBu, followed by incubation of cell lysates with the phosphospecific antiserum. Immunoblotting of precipitated proteins with a GIT1-specific antibody revealed increased GIT1 protein levels upon PDBu stimulation (Fig. 1B), providing evidence that endogenous GIT1 is phosphorylated in a PDBu-dependent manner.
To identify the PKD isoform mediating GIT1 phosphorylation, we used specific siRNAs to knockdown the different PKD isoforms. Cells transfected with a β-galactosidase-specific siRNA (siLacZ) were used as a negative control. Knockdown of PKD1 and PKD2, either singly or in combination, did not affect the detection of basal and PDBu-stimulated GIT1 with the phosphospecific antibody (supplemental Fig. S2). Interestingly, knockdown of PKD3 with two independent siRNAs reduced the detection of GIT1 with the phosphospecific antibody in response to PDBu stimulation (Fig. 1C, top). Efficient silencing of PKD3 was confirmed by immunoblotting of immunoprecipitated PKD3 with a PKD3-specific antibody (Fig. 1C, bottom). Next, we co-expressed wild type (wt) and a catalytically active (ca) PKD3 variant (S731/735E) together with GIT1 in HEK293T cells (Fig. 1D). Co-expression of both PKD3-wt and PKD3-ca strongly enhanced the recognition of immunoprecipitated GIT1 by the phosphospecific antibody, indicating that ectopic PKD3 expression is sufficient to trigger GIT1 phosphorylation. Taken together, these data provide strong evidence that GIT1 is specifically phosphorylated downstream of PKD3.
PKD3 Directly Phosphorylates GIT1 on Serine 46
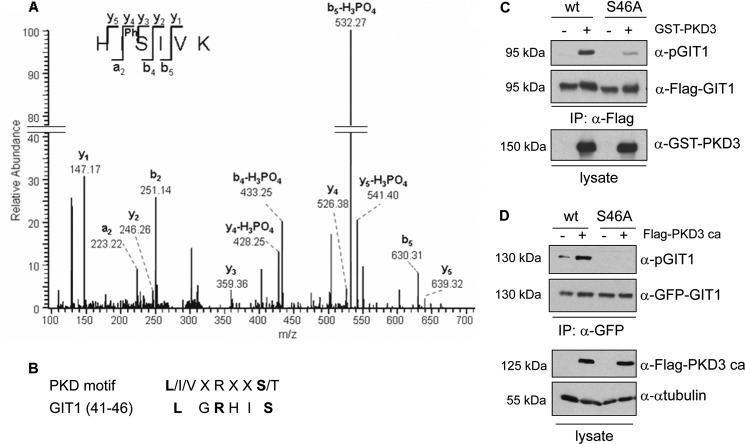
To identify the PKD3-specific phosphorylation site within GIT1, we subjected GIT1 to mass spectrometry analysis with a specific focus on identifying phosphopeptides that match the PKD consensus motif. To this end, Flag-tagged GIT1 was transiently expressed in HEK293T cells and cells were stimulated with PDBu prior to lysis and isolation of Flag-GIT1 with anti-Flag immunoaffinity resin (M2 agarose). Phosphorylated peptides were enriched by titanium dioxide chromatography and then subjected to mass spectrometry. A singly phosphorylated peptide with a parent mass of 388.71 ([M+H]2+), corresponding to amino acids 44–49 (HIpSIVK) in GIT1, was detected and fragmented. The resulting fragmentation spectrum showed good sequence coverage by y- and b-ions and the a2-ion, unambiguously identifying serine 46 to be phosphorylated (Fig. 2A). This serine residue is contained within a perfect PKD consensus motif, namely L/I/VXRXXS/T, with X denoting any amino acid (Fig. 2B; note that the PKD motif is fully conserved in mouse, rat, bovine, and avian GIT1 (data not shown)). Additional phosphosites identified are listed in supplemental Table S1.
FIGURE 2.
GIT1 is directly phosphorylated by PKD3 on serine 46. A, Flag-tagged GIT1 was purified from HEK293T cell lysates transiently expressing the protein with M2-agarose and subjected to SDS-PAGE. The colloidal Coomassie-stained band corresponding to GIT1 was in-gel digested with trypsin. Phosphopeptides were enriched by titanium dioxide chromatography and then subjected to mass spectrometry. The spectrum shows the fragmentation pattern of the phosphopeptide HIpSIVK corresponding to amino acids 44–49. B, consensus motif of the PKD family of proteins and GIT1 alignment (amino acids 41–46). C, GIT1 was immunoprecipitated with anti-Flag antibody from HEK293T cell lysates transiently expressing Flag-tagged GIT1 wt or S46A and incubated with ATP in the absence (−) or presence (+) of purified GST-tagged PKD3. Samples were subjected to SDS-PAGE, proteins were transferred to membrane and analyzed by Western blotting with pGIT1-, Flag-, and GST-specific antibodies. In D, GFP-GIT1 wt and the mutant S46A were co-expressed with Flag-tagged PKD3 ca in HEK293T cells. GIT1 was immunoprecipitated with GFP-specific antibody and analyzed by immunoblotting with the pGIT1 antibody (top panel), followed by detection with GFP-specific antibody to ensure equal GIT1 expression and precipitation. Expression of PKD3 was confirmed by immunoblotting of whole cell lysates with Flag-specific antibody. Equal loading was confirmed by immunoblotting of whole cell lysates with anti-α-tubulin antibody.
To prove that PKD3 directly phosphorylates GIT1 on serine 46, an in vitro kinase assay was performed. Flag-tagged wild type GIT1 and a serine-to-alanine point mutant (S46A) were expressed in HEK293T cells, immunoprecipitated from cell lysates, and incubated with recombinant purified PKD3 in the presence of ATP (Fig. 2C). Compared with the wild type protein, the detection of GIT1 S46A with the phosphospecific antibody was strongly reduced (Fig. 2C, top panel), indicating that the antibody binds GIT1 when phosphorylated on serine 46. To finally test if PKD3 also phosphorylates GIT1 on serine 46 in intact cells, GIT1 wild type and S46A were co-expressed with or without PKD3-ca in HEK293T cells, followed by analysis of GIT1 phosphorylation with the phosphospecific antibody (Fig. 2D). As shown in Fig. 2D, substitution of serine 46 with alanine abrogated the PKD3-induced phospho-signal (top panel). Together these data suggest that PKD3 directly phosphorylates GIT1 on serine 46 both in vitro and in intact cells.
GIT1 Phosphorylated on Serine 46 Locates to Motile Cytoplasmic Paxillin-positive Complexes
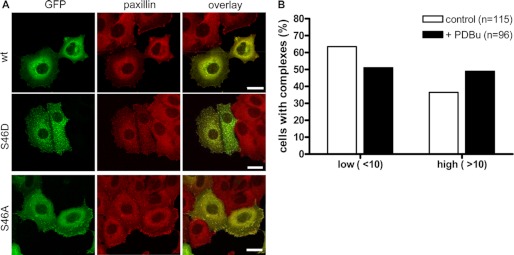
Depending on the cell type and condition, different subcellular localizations have been reported for GIT1. We therefore analyzed whether GIT1 phosphorylation at serine 46 affects its localization within the cell. GFP-tagged variants of GIT1 (wt, S46A, S46D) were expressed in MCF7 breast cancer cells and fixed cells were analyzed by confocal laser scanning microscopy (Fig. 3A). The wild type GIT1 protein was found to localize to paxillin-positive structures at the cell periphery, which showed a typical focal adhesion appearance at the bottom of the cell. GIT1 also localized to a few distinct paxillin-positive cytoplasmic structures, which have been described previously (10, 14) (Fig. 3A, top). Interestingly, cells expressing GIT1 S46D, which contains a negatively charged aspartic acid introduced to mimic the phosphorylated state, were filled with these cytoplasmic structures that further appeared to recruit cytosolic paxillin (Fig. 3A, middle). By contrast, the focal adhesion adaptor protein vinculin did not co-localize with GIT1 S46D in cytoplasmic complexes, indicating a specific effect on the distribution of paxillin and ruling out the disruption of focal adhesions in general (supplemental Fig. S3). The GFP-GIT1 S46A mutant localized only to focal adhesions and no cytoplasmic complexes were observed (Fig. 3A, bottom), indicating that dephosphorylation of serine 46 favors association with cytoskeletal structures. PKD3 was observed to localize to VAMP2-positive vesicles in HEK 293 and HCC1806 cells (34), but PKD3 did not colocalize with GIT1 S46D in MCF7 cells (data not shown). PDBu stimulation of cells expressing GFP-GIT1 wild type induced the formation of cytoplasmic complexes, as determined by scoring cells with a low (<10) and a higher number (>10) of complexes (Fig. 3B), supporting the notion that GIT1 subcellular localization is regulated by serine 46 phosphorylation.
FIGURE 3.
GIT1 S46D localizes to paxillin-positive cytoplasmic complexes. A, MCF7 cells were plated onto collagen-coated coverslips and transiently transfected with expression vectors encoding GFP-GIT1 wt, S46A, and S46D as indicated. The next day, cells were fixed and stained with paxillin-specific primary antibody, followed by Alexa Fluor 546-conjugated secondary antibody (red). The images shown are projections of several confocal sections. Scale bar, 20 μm. B, MCF7 cells were plated onto collagen-coated coverslips and transiently transfected with an expression vector encoding GFP-GIT1 wt. Prior fixation, cells were left unstimulated or stimulated with 1 μm PDBu for 15 min. Cells were analyzed by confocal microscopy and scored according to their number of GIT1 cytoplasmic complexes.
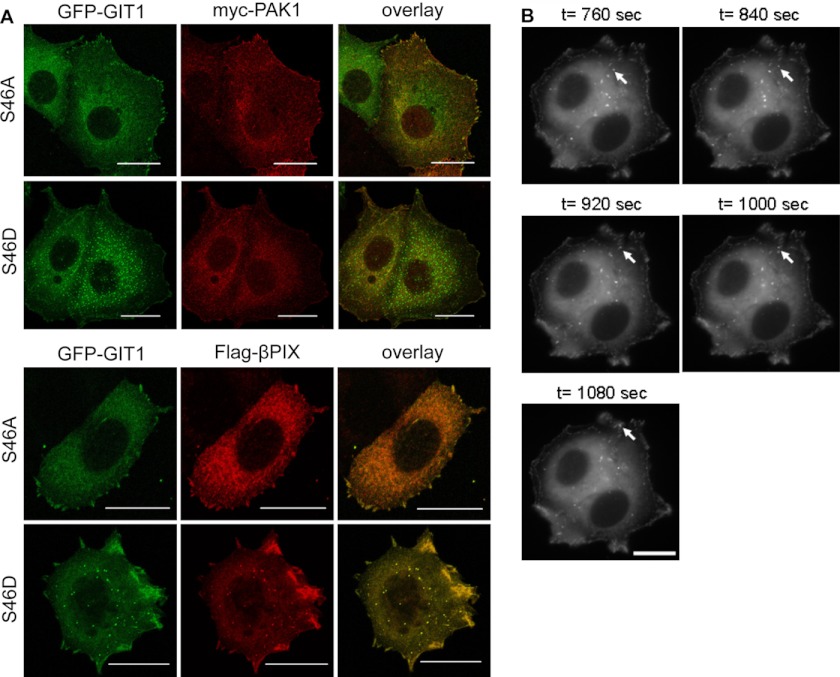
The precise nature and function of the cytoplasmic GIT1 structures, as to whether they are recycling endosomes or motile supramolecular protein complexes, has been discussed controversially. Because we could not detect any colocalization with endosomal markers such as EEA1 and the transferrin receptor (supplemental Fig. S3), the GIT S46D-containing structures observed in MCF7 cells most likely resemble the supramolecular complexes described by Manabe et al. (10). However, costainings revealed that PAK was absent from GIT1 S46D cytoplasmic complexes and colocalized only with FA-localized GIT1 (Fig. 4A, top), whereas PIX colocalized with both GIT1 S46A and S46D (Fig. 4A, bottom). GIT1 cytoplasmic complexes have been reported to be involved in paxillin trafficking (10). We therefore analyzed MCF7 cells expressing GFP-GIT1 S46D in more detail by time lapse video microscopy (Fig. 4B and supplemental movie). Pictures of cells were captured every 40 s for the duration of 20 min and the movement of individual GFP-GIT1 S46D complexes was analyzed. Fig. 4B shows for a chosen GIT1 S46D-positive complex (arrow) the movement toward to the cell periphery. It thus appears that phosphorylation at serine 46 shifts the localization of GIT1 from focal adhesions to highly motile, paxillin-transporting cytoplasmic complexes.
FIGURE 4.
GIT1 S46D cytoplasmic complexes are motile and contain betaPIX. A, MCF7 cells were plated onto collagen-coated coverslips and transiently co-transfected with expression vectors encoding GFP-GIT1 S46A or S46D and either Myc-tagged PAK1 or Flag-tagged betaPIX. The next day, cells were fixed and stained with Myc- or Flag-specific primary and Alexa Fluor 546-conjugated secondary antibodies (red). The images shown are projections of several confocal sections. Scale bar, 20 μm. B, MCF7 cells stably expressing GFP-GIT1 S46D were plated at low density onto collagen-coated glass bottom cell culture dishes, and fluorescence images of cells were captured using a video microscope every 40 s for a duration of 20 min. Representative photos (time points as indicated) of the movement of a chosen complex (arrow) to the cell periphery are shown. Scale bar, 20 μm.
GIT1 Phosphorylation on Serine 46 Increases Spreading, Protrusive Activity, and Migration of MCF7 Cells
GIT1 is known to be important for cell spreading and migration, processes which rely on paxillin trafficking and focal adhesion turnover. We therefore investigated whether GIT1 phosphorylation on serine 46 modulates these biological activities using MCF7 cells stably expressing GIT1 wt, S46D, and S46A. Importantly, phosphorylation of GIT1 on serine 46 also depends on PKD3 in MCF7 cells (supplemental Fig. S4). To analyze initial cell adhesion and spreading, cells were plated at low density onto collagen-coated microtiter plates arrayed with electrodes (E-plates) and impedance (cell index) was measured using an xCELLigence device. The cell index is proportional to the surface area covered by the cells and therefore serves as a measure for cell adhesion and spreading. Compared with cells expressing GIT1 wild type, the cell index of MCF7 cells stably expressing the phosphomimetic mutant S46D was higher, whereas that of cells expressing the GIT1 S46A mutant was lower during the first hour of cell plating (Fig. 5A), indicating that GIT1 phosphorylation on serine 46 positively affects its ability to promote cell adhesion and spreading.
FIGURE 5.
Phosphorylation of GIT1 on Ser-46 increases spreading, protrusive activity and migration of MCF7 cells. A–D, MCF7 cells stably expressing GFP-GIT1 wt, S46A, and S46D were plated (A) into collagen-coated E-plates (96-well plate format, 10,000 cells), and the impedance (cell index) of adhering cells was measured every 5 min using a xCELLigence device, starting 25 min after plating. The mean cell index is plotted (n = 4). Error bars represent S.D. In B, cells were plated at low density onto collagen-coated glass bottom plates. The next day, single cells were video-imaged by microscopy (GFP and phase contrast) every 30 min. Selected GFP-positive cells were outlined at the beginning and the end of a selected 30-min time interval, the images were overlaid, and the change in cell area (in %) was calculated using Image J software. Data show the mean of at least 25 independent single cells. Error bars represent S.E., **, p ≤ 0.01. In C, 50,000 cells were seeded in medium containing 0.5% FCS into the upper chamber of a transwell coated with collagen on the underside. The lower well contained medium supplemented with 10% FCS. Cells that had migrated across the filter after overnight incubation were fixed and stained. The number of migrated cells was determined by counting five independent microscopic fields (20-fold magnification) per filter. Data show the mean of four independent experiments performed with duplicate filters, the wt was set as 1. Error bars represent S.E., ***, p ≤ 0.001. D, transwell migration was analyzed as described in C in the presence of 50 nm PDBu in upper and lower chambers. Data show the mean of two independent experiments performed with duplicate filters; unstimulated cells were set as 1. Error bars represent S.E.
We next examined if phosphorylation of GIT1 on serine 46 influences the overall protrusive activity of cells. Live cell images of MCF7 cells plated on collagen-coated glass dishes were taken every 30 min and the percentage of change in total cell area after a selected 30 min interval was determined using Image J software (Fig. 5B, outline). Based on this analysis, cells expressing the phosphomimetic S46D mutant showed significantly more protrusive activity than the wild type control cells, whereas cells expressing GIT1 S46A showed less protrusive activity (Fig. 5B). Finally, in Transwell migration assays, MCF7 cells expressing the GIT1 S46D protein displayed significantly increased cell motility after overnight incubation compared with those expressing the wild type protein, whereas those expressing GIT1 S46A displayed reduced cell motility, in accordance with the results obtained in the other biological assays (Fig. 5C). In the presence of PDBu, the migration of cells expressing GIT1 wt was increased by 1.5-fold, whereas that of cells expressing GIT1 S46D was only increased by 1.2-fold in comparison to the untreated controls (Fig. 5D). This indicates that PDBu-induced phosphorylation of wild type GIT1 contributes to enhanced cell migration, whereas in cells expressing the phosphomimetic GIT1 S46D variant cell migration is already enhanced (Fig. 5C) and cannot be stimulated much further. In summary, we identified a novel phosphorylation site in GIT1 on serine 46 that is specifically phosphorylated by PKD3 and is associated with increased cellular protrusive activity and motility.
DISCUSSION
In this study we identify and characterize the function of a novel phosphorylation site within GIT1 on serine 46. Previous analysis of GIT1 by mass spectrometry did not identify serine 46 to be phosphorylated (16), which is likely due to the stimulation of cells with PDBu in our study to specifically activate the PKD pathway. In addition to serine 46, five further novel phosphosites were mapped and several others confirmed (see supplemental Table S1). Our findings suggest that the phosphorylation state of GIT1 on serine 46 regulates paxillin trafficking and cellular protrusive activity by modulating GIT1 subcellular localization. In MCF7 cells, wild type GIT1 localized mainly to FAs. By contrast, MCF7 cells expressing a GIT1 S46D phosphomimetic mutant contained a high number of cytoplasmic paxillin-positive complexes, which were absent in cells expressing GIT1 S46A. We further noticed that the GIT1 S46A mutant was less soluble and could only be efficiently extracted from cells using an SDS-containing lysis buffer, indicating that the dephosphorylated protein associates more strongly with cytoskeletal structures. PBDu stimulation of cells shifted the balance toward a higher number of cytoplasmic complexes, providing further evidence that serine 46 phosphorylation regulates GIT1 subcellular localization.
GIT1 localization to adhesions and the leading edge was reported to depend on the PBS whereas targeting to cytoplasmic complexes required the central region that contains the ankyrin repeats and PIX binding domain (10). Opposed to the observation by Manabe et al., in MCF7 cells PAK colocalized with GIT1 only in FAs and was not contained in GIT1 S46D complexes. GIT1 phosphorylation at serine 46 may thus alter protein conformation, modulating the interaction with specific protein partners and thereby affecting localization. Serine 46 is located within the GAP domain of GIT1 and could thus potentially regulate the GAP function of the protein, but Arf6 activity was not significantly altered in MCF7 cells stably expressing the GIT1 phosphorylation site mutants as judged by GGA3 pulldowns (data not shown). Serine 46 phosphorylation thus appears to decrease the focal adhesion localization of GIT1 independently of its GAP function. We therefore propose that the enhanced cell spreading and motility observed in GIT1 S46D-expressing cells arises from increased focal adhesion turnover through the continuous trafficking of paxillin to membrane protrusions. By contrast, cells expressing GIT1 S46A are less motile due to the stabilization of focal adhesions. Finally, the PKD phosphorylation site at serine 46 is conserved in GIT2, raising the possibility that GIT2 is regulated in a similar manner, although it should be noted that GIT1 and GIT2 are not exchangeable as they also possess non-redundant functions (2).
The external cues that activate PKD3 to trigger GIT1 phosphorylation remain to be identified. Our results show that the pharmacological inhibition of novel PKCs by Gö6983 does not block GIT1 phosphorylation on serine 46. Phosphorylation of PKD by novel PKCs in the activation loop is generally considered as a prerequisite for activation. However, a PKC-independent role for PKD3 has been suggested in basal glucose uptake of skeletal muscle cells (35) and more recent studies demonstrated that the rapid PKC-dependent PKD activation is followed by a sustained, PKC-independent phase of catalytic activation (36).
Emerging evidence suggests that the nature of the PKD isoform activated determines whether cell migration is regulated in a positive or negative manner. PKD1/2-mediated phosphorylation of substrates such as SSH1L and RIN1 leads to the inhibition of cell migration (21, 23, 25, 37) and reduced PKD1 expression levels were observed by immunohistological analyses of invasive breast cancer samples (38), suggesting that the loss of PKD1 may be associated with the development of metastasis. By contrast, PKD3 was found to be up-regulated in primary prostate cancers and prostate cancer cell lines to stimulate prosurvival pathways (39), indicating a positive correlation between PKD3 expression and cancer progression. This implies that PKD3 regulates a distinct set of substrates to fulfill such functions, which is in accordance with our findings that PKD3 selectively phosphorylates GIT1 to enhance cell spreading and motility. In future studies, it will be interesting to determine how PKD substrate specificity is achieved as the kinases phosphorylate the same consensus sequence in vitro. Indeed, GIT1 was also phosphorylated by PKD1 in an in vitro kinase assay (data not shown), indicating that in a cellular context specificity is achieved by different spatial localization and/or the binding of specific protein cofactors. For example, the amino terminal region of nonpolar amino acids and PDZ-binding motif contained in PKD1 and PKD2 are absent in PKD3 (20), providing a means for the formation of distinct protein complexes.
While this study focuses on the function of GIT1 in epithelial cells, GIT1 has also been extensively studied in neuronal cells where it regulates neurite extension, dendritic spine morphogenesis, and synapse formation. Interestingly, PKD3 is strongly expressed in neuronal cells in the developing embryo (40), making it tempting to speculate that PKD3 may also control GIT1-mediated cytoskeletal rearrangements in the central nervous system.
Acknowledgment
We thank Gisela Link for excellent technical support.
This work was funded by the German Cancer Aid 109583.

This article contains supplemental Figs. S1–S4, Table S1, and movie.
- GIT1
- G-protein-coupled receptor kinase-interacting protein 1
- ca
- catalytically active
- FA
- focal adhesion
- FAK
- focal adhesion kinase
- GAP
- GTPase-activating protein
- kd
- kinase dead
- PAK
- p21-activated kinase
- PBS
- paxillin-binding site
- PDBu
- phorbol-12,13-dibutyrate
- PKD
- protein kinase D.
REFERENCES
- 1. Frank S. R., Hansen S. H. (2008) The PIX-GIT complex: a G protein signaling cassette in control of cell shape. Semin Cell Dev. Biol. 19, 234–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Randazzo P. A., Inoue H., Bharti S. (2007) Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583–600 [DOI] [PubMed] [Google Scholar]
- 3. Hoefen R. J., Berk B. C. (2006) The multifunctional GIT family of proteins. J. Cell Sci. 119, 1469–1475 [DOI] [PubMed] [Google Scholar]
- 4. Deakin N. O., Turner C. E. (2008) Paxillin comes of age. J. Cell Sci. 121, 2435–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. West K. A., Zhang H., Brown M. C., Nikolopoulos S. N., Riedy M. C., Horwitz A. F., Turner C. E. (2001) The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL). J. Cell Biol. 154, 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nishiya N., Kiosses W. B., Han J., Ginsberg M. H. (2005) An alpha4 integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat. Cell Biol. 7, 343–352 [DOI] [PubMed] [Google Scholar]
- 7. Brown M. C., Turner C. E. (2004) Paxillin: adapting to change. Physiol. Rev. 84, 1315–1339 [DOI] [PubMed] [Google Scholar]
- 8. Webb D. J., Donais K., Whitmore L. A., Thomas S. M., Turner C. E., Parsons J. T., Horwitz A. F. (2004) FAK-Src signaling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 6, 154–161 [DOI] [PubMed] [Google Scholar]
- 9. Zhao Z. S., Manser E., Loo T. H., Lim L. (2000) Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell. Biol. 20, 6354–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manabe R., Kovalenko M., Webb D. J., Horwitz A. R. (2002) GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 115, 1497–1510 [DOI] [PubMed] [Google Scholar]
- 11. Loo T. H., Ng Y. W., Lim L., Manser E. (2004) GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol. Cell Biol. 24, 3849–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown M. C., West K. A., Turner C. E. (2002) Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol. Biol. Cell 13, 1550–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Cesare A., Paris S., Albertinazzi C., Dariozzi S., Andersen J., Mann M., Longhi R., de Curtis I. (2000) p95-APP1 links membrane transport to Rac-mediated reorganization of actin. Nat. Cell Biol. 2, 521–530 [DOI] [PubMed] [Google Scholar]
- 14. Matafora V., Paris S., Dariozzi S., de Curtis I. (2001) Molecular mechanisms regulating the subcellular localization of p95-APP1 between the endosomal recycling compartment and sites of actin organization at the cell surface. J. Cell Sci. 114, 4509–4520 [DOI] [PubMed] [Google Scholar]
- 15. Albertinazzi C., Za L., Paris S., de Curtis I. (2003) ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol. Biol. Cell 14, 1295–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Webb D. J., Mayhew M. W., Kovalenko M., Schroeder M. J., Jeffery E. D., Whitmore L., Shabanowitz J., Hunt D. F., Horwitz A. F. (2006) Identification of phosphorylation sites in GIT1. J. Cell Sci. 119, 2847–2850 [DOI] [PubMed] [Google Scholar]
- 17. Ren Y., Yu L., Fan J., Rui Z., Hua Z., Zhang Z., Zhang N., Yin G. (2012) Phosphorylation of GIT1 tyrosine 321 is required for association with FAK at focal adhesions and for PDGF-activated migration of osteoblasts. Mol Cell Biochem. 365, 109–118 [DOI] [PubMed] [Google Scholar]
- 18. Webb D. J., Kovalenko M., Whitmore L., Horwitz A. F. (2006) Phosphorylation of serine 709 in GIT1 regulates protrusive activity in cells. Biochem. Biophys. Res. Commun. 346, 1284–1288 [DOI] [PubMed] [Google Scholar]
- 19. Fu Y., Rubin C. S. (2011) Protein kinase D: coupling extracellular stimuli to the regulation of cell physiology. EMBO Rep. 12, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Q. J. (2006) PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol. Sci. 27, 317–323 [DOI] [PubMed] [Google Scholar]
- 21. Eiseler T., Döppler H., Yan I. K., Kitatani K., Mizuno K., Storz P. (2009) Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat. Cell Biol. 11, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scholz R. P., Gustafsson J. O., Hoffmann P., Jaiswal M., Ahmadian M. R., Eisler S. A., Erlmann P., Schmid S., Hausser A., Olayioye M. A. (2011) The tumor suppressor protein DLC1 is regulated by PKD-mediated GAP domain phosphorylation. Exp. Cell Res. 317, 496–503 [DOI] [PubMed] [Google Scholar]
- 23. Ziegler S., Eiseler T., Scholz R. P., Beck A., Link G., Hausser A. (2011) A novel protein kinase D phosphorylation site in the tumor suppressor Rab interactor 1 is critical for coordination of cell migration. Mol. Biol. Cell 22, 570–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eiseler T., Hausser A., De Kimpe L., Van Lint J., Pfizenmaier K. (2010) Protein kinase D controls actin polymerization and cell motility through phosphorylation of cortactin. J. Biol. Chem. 285, 18672–18683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peterburs P., Heering J., Link G., Pfizenmaier K., Olayioye M. A., Hausser A. (2009) Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 69, 5634–5638 [DOI] [PubMed] [Google Scholar]
- 26. Spratley S. J., Bastea L. I., Döppler H., Mizuno K., Storz P. (2011) Protein kinase D regulates cofilin activity through p21-activated kinase 4. J. Biol. Chem. 286, 34254–34261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayhew M. W., Webb D. J., Kovalenko M., Whitmore L., Fox J. W., Horwitz A. F. (2006) Identification of protein networks associated with the PAK1-betaPIX-GIT1-paxillin signaling complex by mass spectrometry. J. Proteome Res. 5, 2417–2423 [DOI] [PubMed] [Google Scholar]
- 28. Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., Sommer R. J., Macek B. (2010) Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Olsen J. V., Macek B. (2009) High accuracy mass spectrometry in large-scale analysis of protein phosphorylation. Methods Mol. Biol. 492, 131–142 [DOI] [PubMed] [Google Scholar]
- 30. Koch A., Krug K., Pengelley S., Macek B., Hauf S. (2011) Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci. Signal 4, rs6. [DOI] [PubMed] [Google Scholar]
- 31. Cox J., Matic I., Hilger M., Nagaraj N., Selbach M., Olsen J. V., Mann M. (2009) A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 4, 698–705 [DOI] [PubMed] [Google Scholar]
- 32. Döppler H., Storz P., Li J., Comb M. J., Toker A. (2005) A phosphorylation state-specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J. Biol. Chem. 280, 15013–15019 [DOI] [PubMed] [Google Scholar]
- 33. Fugmann T., Hausser A., Schöffler P., Schmid S., Pfizenmaier K., Olayioye M. A. (2007) Regulation of secretory transport by protein kinase D-mediated phosphorylation of the ceramide transfer protein. J. Cell Biol. 178, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu G., Chen J., Espinoza L. A., Garfield S., Toshiyuki S., Akiko H., Huppler A., Wang Q. J. (2007) Protein kinase D 3 is localized in vesicular structures and interacts with vesicle-associated membrane protein 2. Cell Signal 19, 867–879 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Lu G., Wang Q. J. (2005) Protein kinase C-independent effects of protein kinase D3 in glucose transport in L6 myotubes. Mol. Pharmacol. 67, 152–162 [DOI] [PubMed] [Google Scholar]
- 36. Jacamo R., Sinnett-Smith J., Rey O., Waldron R. T., Rozengurt E. (2008) Sequential protein kinase C (PKC)-dependent and PKC-independent protein kinase D catalytic activation via Gq-coupled receptors: differential regulation of activation loop Ser(744) and Ser(748) phosphorylation. J. Biol. Chem. 283, 12877–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eiseler T., Schmid M. A., Topbas F., Pfizenmaier K., Hausser A. (2007) PKD is recruited to sites of actin remodeling at the leading edge and negatively regulates cell migration. FEBS Lett. 581, 4279–4287 [DOI] [PubMed] [Google Scholar]
- 38. Eiseler T., Döppler H., Yan I. K., Goodison S., Storz P. (2009) Protein kinase D1 regulates matrix metalloproteinase expression and inhibits breast cancer cell invasion. Breast Cancer Res. 11, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen J., Deng F., Singh S. V., Wang Q. J. (2008) Protein kinase D3 (PKD3) contributes to prostate cancer cell growth and survival through a PKCϵ/PKD3 pathway downstream of Akt and ERK 1/2. Cancer Res. 68, 3844–3853 [DOI] [PubMed] [Google Scholar]
- 40. Ellwanger K., Pfizenmaier K., Lutz S., Hausser A. (2008) Expression patterns of protein kinase D 3 during mouse development. BMC Dev. Biol. 8, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]