Background: Mitochondrial rhomboid protease PARL mediates the cleavage of PINK1 in healthy mitochondria.
Results: PARL mediates the cleavage of PGAM5 in damaged mitochondria.
Conclusion: PARL regulates differential cleavage of PINK1 and PGAM5 depending on the health status of mitochondria.
Significance: This is the first implication of stress-dependent regulation of PARL-mediated RIP.
Keywords: Intramembrane Proteolysis, Mitochondria, Phosphatase, Rhomboid Protease, Stress Response
Abstract
Regulated intramembrane proteolysis is a widely conserved mechanism for controlling diverse biological processes. Considering that proteolysis is irreversible, it must be precisely regulated in a context-dependent manner. Here, we show that phosphoglycerate mutase 5 (PGAM5), a mitochondrial Ser/Thr protein phosphatase, is cleaved in its N-terminal transmembrane domain in response to mitochondrial membrane potential (ΔΨm) loss. This ΔΨm loss-dependent cleavage of PGAM5 was mediated by presenilin-associated rhomboid-like (PARL). PARL is a mitochondrial resident rhomboid serine protease and has recently been reported to mediate the cleavage of PINK1, a mitochondrial Ser/Thr protein kinase, in healthy mitochondria with intact ΔΨm. Intriguingly, we found that PARL dissociated from PINK1 and reciprocally associated with PGAM5 in response to ΔΨm loss. These results suggest that PARL mediates differential cleavage of PINK1 and PGAM5 depending on the health status of mitochondria. Our data provide a prototypical example of stress-dependent regulation of PARL-mediated regulated intramembrane proteolysis.
Introduction
Intramembrane cleaving proteases (I-Clips)2 are an evolutionarily conserved group of multipass membrane proteins that catalyze the cleavage of transmembrane (TM) domains within lipid bilayers (1). Intramembrane proteolysis controls many biological processes by liberating the functional domains of substrates; thus, it is referred to as regulated intramembrane proteolysis (RIP). Rhomboid proteases, which belong to the I-Clips family of proteins, are serine proteases that were originally identified as regulators of epidermal growth factor receptor signaling in Drosophila (2, 3). At least five active rhomboids have been found in the mammalian genome, including presenilin-associated rhomboid-like (PARL) (4). PARL has been shown to be targeted to the inner mitochondrial membrane (IMM) (5, 6). PARL knock-out (KO) mice, whose primary defects in mitochondria appeared to be impaired cristae junctional sealing resulting from the reduced processing of OPA1, a component of the mitochondrial fusion machinery, die post-natally because of massive apoptosis (6, 7). However, several lines of evidence suggest that PARL is only partly responsible for the cleavage of OPA1, at least in mammals (8, 9), and other PARL substrates, such as Omi/HtrA2, have been identified (10), suggesting that unidentified PARL substrates still exist in mitochondria.
Phosphoglycerate mutase 5 (PGAM5) belongs to the PGAM family, an evolutionarily conserved enzyme family, the prototype of which converts 3-phosphoglycerate to 2-phosphoglycerate during glycolysis (11). Although all the amino acid residues composing the catalytic core of the other family members are conserved in PGAM5, we have previously reported that PGAM5 lacks mutase activity and instead acts as a Ser/Thr protein phosphatase that activates the stress-activated MAP kinase pathways (12). PGAM5 has also been reported to be targeted to mitochondria through its N-terminal TM domain (13), indicating that PGAM5 is a unique mitochondrial resident phosphatase involved in cellular stress responses. It was recently reported that PGAM5 plays a critical role in necrosis induction by facilitating mitochondrial fragmentation (14).
In this study we show that PARL mediates the cleavage of PGAM5 and that this is induced in response to mitochondrial membrane potential (ΔΨm) loss. Interestingly, recent reports have suggested that, in healthy mitochondria that maintain ΔΨm, PARL mediates the cleavage of PINK1, a mitochondrial Ser/Thr protein kinase known as a gene product responsible for early-onset autosomal recessive Parkinson disease (15–17). Taken together with this finding, our data suggest that the PARL regulates differential cleavage of PINK1 and PGAM5 depending on the health status of mitochondria. Our data provide a prototypical example of stress-dependent regulation of PARL-mediated RIP.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
Cells were cultured as previously described (12). The transfection of expression plasmids was performed using FuGENE6 (Roche Applied Science) or PEI-Max (Polysciences). For RNAi, cells were transfected with the following siRNAs (Invitrogen) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacture's instructions: human PARL (#1), 5′-CCCAGAAGGGAGGCUUGCCAUUAUU-3′; human PARL (#2), 5′-CCUAUCCUAUAAGGAGUCUCAUAAA-3′; mouse PARL (#1), 5′-AAAGACUGUUUCUUCCAGAGGCGGG-3′; mouse PARL (#2), 5′-AAACAAGUUAAACCUGCGUCCAAGC-3′; human OMA1, 5′-CAGCAGUCCCUAGUCUGUCAGUAUU-3′. Stealth RNAi Negative CTL Medium GC Duplex #2 was used as a control.
Quantitative Reverse Transcription (RT)-PCR
Total RNA was isolated from siRNA-treated HEK293A cells using Isogen (Wako) and reverse-transcribed with the QuantiTect Reverse Transcription kit (Qiagen). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Roche Applied Science) on an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). To normalize the relative expression of PARL and OMA1 to the S18 control, a standard curve was prepared for S18, PARL, and OMA1.
Expression Plasmids and Mutagenesis
cDNAs for human PARL and PINK1 were cloned by RT-PCR using total RNA isolated from HEK293A cells and subcloned into the mammalian expression plasmid pcDNA3 (Invitrogen). Site-directed mutagenesis of PARL (S277G, H335A) and PGAM5 (S24F) was performed using the QuikChange site-directed mutagenesis kit (Stratagene) with the following primers and their respective complementary sequence primers: PARL (S277G), 5′-CCATCACTTGGTGCAGGTGGTGCCATCATGACA-3′; PARL (H335A), 5′-TTTGATCATGCGGCAGCTCTTGGGGGAGCTCTT-3′; PGAM5 (S24F), 5′-GCCGCCGTGCTCTTCTTTGCCGTGGCGGTAGGG-3′. The generation of AIF-FLAG, FLAG-Fis1, and DRP1 K38A-GFP mammalian expression vectors was described previously (18–20).
Antibodies
The generation of the rabbit polyclonal anti-PGAM5 antibody (RTL) was described previously (12). The rat monoclonal anti-PGAM5 antibody (K1B6) was raised against purified GST-tagged PGAM5 lacking the TM domain (amino acids 30–289) as previously described (21). To generate the rabbit polyclonal anti-cleaved PGAM5 antibody (RB2), antisera raised against the synthetic peptide NH2-AVAVGKPR-COOH were affinity-purified with the same peptide. The purified fraction was then negatively purified using a peptide mimicking the uncleaved protein (N-terminally acetylated AVAVGKPR). Mouse polyclonal anti-cleaved PGAM5 antibody (K15) was generated using the same peptide as an immunogen (22). A pool of hybridomas-secreting antibodies that specifically recognized the non-acetylated peptide was selected by screening with enzyme-linked immunosorbent assay. The following antibodies were also used in this study: FLAG (D8, Santa Cruz; M2, Sigma; 1E6, Wako), HA (3F10, Roche Applied Science), OPA1 (BD Transduction Laboratories), Complex III subunit CORE 1 (Invitrogen), voltage-dependent anion channel protein (VDAC; Abcam), GRIM19 (Mito Sciences), PINK1 (Novus Biologicals), cytochrome c (BD Pharmingen), and actin (Sigma). The generation of Tom70, Mitofilin, and Tim23 antibodies was described previously (23–25).
Immunoblotting and Immunocytochemistry
Immunoblotting was performed as previously described (12). For immunocytochemistry, cultured cells were fixed with 2% formaldehyde in PBS and permeabilized with 0.2% Triton X-100. After 30 min of blocking with 2% BSA in PBS, cells were stained with the respective antibodies. ΔΨm was monitored by staining with 50 nm MitoTracker Red CMXRos (Molecular Probes) for 15 min before fixation. Cells were imaged using an inverted microscope (LSM510 Meta; Carl Zeiss, Inc.).
Generation of Recombinant PGAM5
pGEX-4T (GE Healthcare)-based bacterial expression plasmids encoding PGAM5 (amino acids 24–289, 25–289, and 26–289) in which the Factor Xa recognition sequence (Ile-Glu-Gly-Arg) was inserted immediately N-terminal to PGAM5 were constructed and expressed in Escherichia coli strain BL21. Purified GST-IEGR-PGAM5 proteins on a glutathione-Sepharose column were cleaved with 0.04 unit/mg of protein Factor Xa (GE Healthcare) in a reaction buffer (1 mm CaCl2, 100 mm NaCl, and 50 mm Tris-HCl, pH 8.0) for 30 min at 4 °C. Cleaved products were eluted with the same reaction buffer.
Assays of Isolated Mitochondria
For trypsin digestion, isolated mitochondria were resuspended in the following buffers with various concentrations of trypsin (Sigma) for 30 min on ice: isotonic buffer (10 mm HEPES-KOH, pH 7.4, 0.22 m mannitol, and 0.07 m sucrose), hypotonic buffer (10 mm HEPES-KOH, pH 7.4), or Triton X-100 buffer (1% Triton X-100, 10 mm HEPES-KOH, pH 7.4, 0.22 m mannitol, and 0.07 m sucrose). For sucrose density gradient centrifugation, an isolated mitochondrial membrane fraction was layered over a linear gradient of sucrose from 0.6 to 1.6 m and centrifuged in a SW 41 Ti rotor (Beckman Coulter) at 100,000 × g for 15 h at 4 °C. For alkaline extraction, crude mitochondria were treated with a hypotonic buffer containing 10 mm Tris-HCl (pH 7.4 or pH 11; adjusted with Na2CO3), 50 mm NaCl, and 1 mm EDTA for 5 min on ice. Membranes and supernatants were isolated by centrifugation at 8000 × g for 10 min at 4 °C. For protease inhibitor assay, isolated mitochondria were incubated with each protease inhibitor for 5 min on ice followed by stimulation with 20 μm carbonyl cyanide m-chlorophenyl hydrazone (CCCP) for 1 h at 30 °C in vitro. The following protease inhibitors purchased from Sigma were used: 1 mm o-phenanthroline, 20 μm E64d, 5 μm pepstatin A, 100 μm dichloroisocoumarin (DCI), 100 μm N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK), and 1 mm phenylmethylsulfonyl fluoride (PMSF).
Quantification of Cellular ATP Level
Cellular ATP was quantified using the ATP Determination kit (Invitrogen) according to the manufacturer's instructions.
Generation of HeLa-PGAM5-FLAG Cells
HeLa cells stably expressing PGAM5 C-terminally tagged with FLAG (HeLa-PGAM5-FLAG cells) were established by infecting cells with recombinant retroviruses. Retrovirus packaging PLAT-E cells (provided by T. Kitamura, University of Tokyo) (26) were transfected with a pMXs-puro vector encoding PGAM5-FLAG and were cultured for 48 h. The viral supernatant was collected and used for infection. Before infection, HeLa cells were transfected with an expression vector encoding murine cationic amino acid transporter 1 (mCAT; provided by N. Fujita and T. Yoshimori, Osaka University), which mediates ecotropic retrovirus infection. HeLa cells were plated onto a 6-well plate before infection, and the medium was replaced with the viral supernatant including 10 μg/ml Polybrene (Sigma). Two days later infected cells were selected in medium containing 10 μg/ml puromycin.
Electron Microscopy and Immunoelectron Microscopy Using Ultrathin Cryosections
Cells grown on gridded glass were fixed in 2% glutaraldehyde, 2% paraformaldehyde (PA) buffered with 0.1 m phosphate buffer, pH 7.2, for ordinary electron microscopy and in 4% PA buffered with phosphate buffer. pH 7.2, for immunoelectron microscopy (27). In the former procedure the cells were post-fixed with 1% OsO4 and embedded in Epon 812 after dehydration. Ultrathin sections, 60 nm, were cut with an ultramicrotome (UC6, Leica) and stained with uranium acetate and lead citrate. In the latter procedure, fixed cells were rotated in 2.3 m sucrose in phosphate buffer overnight and plunged into liquid nitrogen. Sections ∼60-nm thick were cut with a cryo-ultramicrotome (UC7/FC7, Leica) and reacted overnight at 4 °C with rabbit anti-FLAG (Santa Cruz; 1:10) and then for 1 h with goat anti-rabbit IgG conjugated with 10-nm colloidal gold particles (British Biocell International). The specimens were examined with a Hitachi H-7100 electron microscope.
RESULTS
PGAM5 Is an IMM-resident Protein Phosphatase
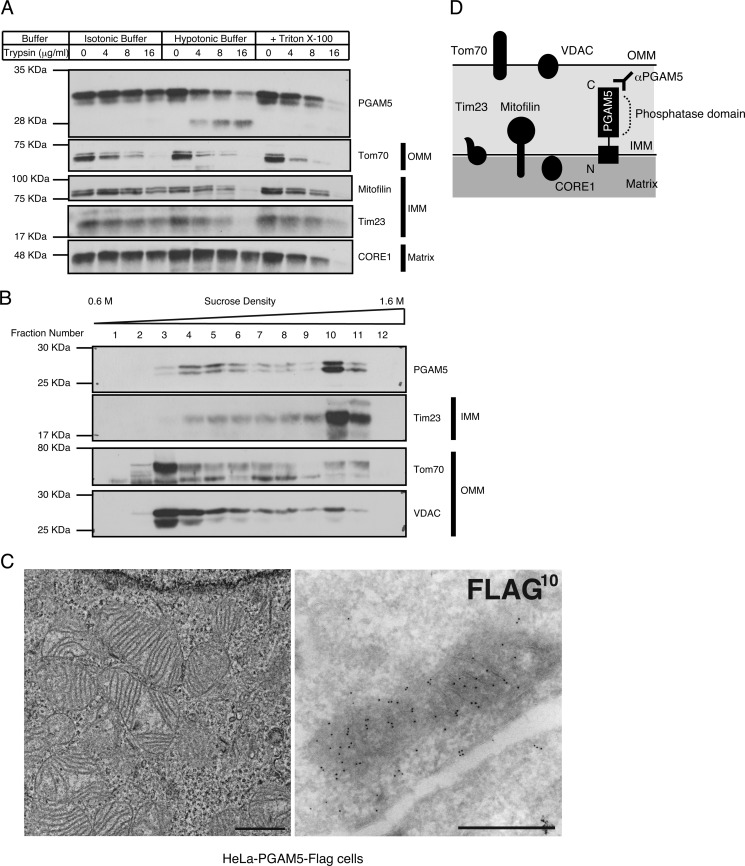
PGAM5 has been reported to be targeted to mitochondria through its N-terminal TM domain (13). To characterize the localization and membrane topology of PGAM5 in mitochondria, we treated the isolated mitochondria with trypsin under isotonic or hypotonic conditions (Fig. 1A). The IMM-resident proteins, Mitofilin and Tim23, were digested more efficiently under hypotonic conditions than under isotonic conditions, indicating that mitochondrial integrity was preserved during isolation and subsequent trypsin digestion of mitochondria. Similar to the IMM-resident proteins examined, PGAM5 was digested more efficiently under hypotonic conditions than under isotonic conditions, suggesting that PGAM5 is an IMM-resident protein (Fig. 1A). In addition, because the antibody used in this experiment specifically recognizes the C terminus of PGAM5, the C-terminal phosphatase domain appears to face the intermembrane space. The fractionation of mitochondrial membranes also revealed that PGAM5 was mainly co-fractionated with Tim23 but not with outer mitochondrial membrane (OMM)-resident proteins Tom70 and voltage-dependent anion channel protein (VDAC; Fig. 1B). To strengthen these biochemical results, we further examined the localization of PGAM5 in mitochondria in HeLa cells stably expressing PGAM5 (HeLa-PGAM5-FLAG cells) by immunoelectron microscopy analysis. In these cells mitochondrial cristae structure was well organized within each mitochondrion (Fig. 1C, left panel), and gold particles representing PGAM5 accumulated inside mitochondria, mostly along cristae (Fig. 1C, right panel). Although PGAM5 was previously described as an OMM-resident protein (13), our results suggest that PGAM5 is mainly targeted to the IMM with the phosphatase domain facing the intermembrane space (Fig. 1D).
FIGURE 1.
PGAM5 is an IMM-resident protein phosphatase. A and B, biochemical characterization of PGAM5 in mitochondria is shown. Isolated mitochondria from HEK293A cells treated with the indicated concentrations of trypsin in the indicated buffer conditions were solubilized and subjected to immunoblotting (IB) (A). The isolated membrane fraction from mouse liver mitochondria was subjected to sucrose density gradient centrifugation followed by IB (B). C, ultrastructural characterization of the localization of PGAM5 in mitochondria is shown. HeLa-PGAM5-FLAG cells were subjected to ordinary electron microscopy (left panel) and immunoelectron microscopy using ultrathin cryosections (right panel). Most of the 10-nm gold particles representing PGAM5-FLAG are mostly localized along mitochondrial cristae. Scale bars: 1 μm (left panel) and 0.5 μm (right panel). D, shown is a schematic view of the localization of the mitochondrial proteins examined in A and B. PGAM5 is mainly localized in the IMM with the phosphatase domain facing the intermembrane space.
PGAM5 Is Cleaved in Its TM Domain in Response to ΔΨm Loss
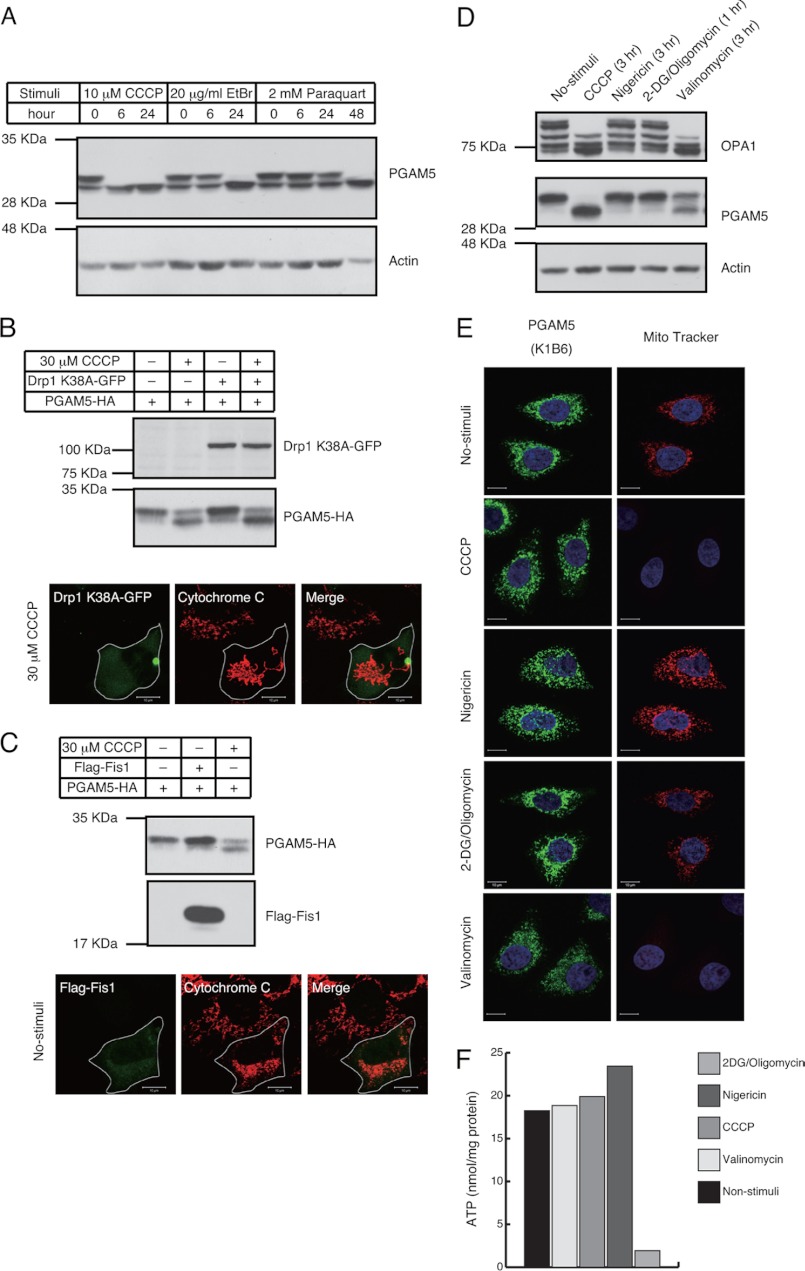
From the above finding that PGAM5 is an IMM-resident protein together with our previous finding that PGAM5 regulates the stress-activated signaling pathways (12), we wondered whether PGAM5 might respond to stresses that damage mitochondria. Among various mitochondrial stress inducers, we tested CCCP, which causes depolarization of mitochondria, ethidium bromide, which preferentially impairs replication and transcription of the mitochondrial genome, and paraquat, which promotes accumulation of reactive oxygen species. Interestingly, a doublet band of PGAM5 observed in unstimulated cells by SDS-PAGE converged into a lower single band when cells were stimulated with these reagents (Fig. 2A). This suggested that the modification of PGAM5 is commonly induced by various forms of mitochondrial damage.
FIGURE 2.
ΔΨm loss induces the dynamic changes in the band pattern of PGAM5. A, mitochondria-damaging agents induce changes in the band pattern of PGAM5. HEK293A cells treated with 10 μMCCCP, 20 μg/ml ethidium bromide (EtBr), or 2 mm paraquat for indicated time periods were subjected to IB. B, mitochondrial fragmentation is dispensable for the changes in the band pattern of PGAM5. 48 h after co-transfection of Drp1 K38A-GFP and PGAM5-HA in HEK293A cells, cells were untreated or treated with 30 μm CCCP for 3 h and subjected to IB (upper panels) or immunocytochemistry (lower panels). Scale bar = 10 μm. C, mitochondrial fragmentation is not sufficient to change the band pattern of PGAM5. 24 h after co-transfection of FLAG-Fis1 and PGAM5-HA in HEK293A cells, cells were untreated or treated with 30 μm CCCP for 3 h and subjected to IB (upper panels) or immunocytochemistry (lower panels). Scale bar = 10 μm. D–F, the changes in the band pattern of PGAM5 are induced by the ΔΨm loss-inducing agents. HeLa cells were treated with 20 μm CCCP, 20 μm nigericin, 20 μm valinomycin, or 10 mm 2-deoxy-d-glucose (2DG)/10 μm oligomycin for the indicated time periods. The treatment with 2-deoxy-d-glucose/oligomycin was performed in glucose-free medium. After each stimulus, cells were lysed and subjected to IB (D). ΔΨm in D was detected with the ΔΨm-dependent dye, MitoTracker red CMXRos. Scale bar = 10 μm (E). Cellular ATP levels in D were quantified using the ATP detection kit (F).
Because depolarization of mitochondria is known to induce their fragmentation (19), we examined whether mitochondrial fragmentation triggers the modification of PGAM5. Mitochondrial fragmentation induced by CCCP is known to be suppressed by overexpressing Drp1 K38A, a dominant-negative mutant that inhibits the mitochondrial fission machinery (19). Although the mitochondria in Drp1 K38A-expressing cells failed to fragment even in the presence of CCCP (Fig. 2B, lower panels), CCCP-dependent changes in the band pattern of PGAM5 were not disrupted (Fig. 2B, upper panels). In contrast, overexpression of a component of the mitochondrial fission machinery, Fis1, which caused mitochondrial fragmentation with minimal perturbation of ΔΨm (18) (Fig. 2C, lower panels), was not sufficient to change the band pattern of PGAM5 (Fig. 2C, upper panels). These results suggest that mitochondrial damage-induced changes in the post-translational modifications of PGAM5 do not depend on the mitochondrial fragmentation.
CCCP, acting as a uniporter for H+, dissipates both ΔpH and ΔΨm across the IMM and, therefore, inhibits ATP production. To determine which of these factors triggers the post-translational modifications of PGAM5, we tested the effects of a couple of ionophores, the H+/K+ antiporter nigericin, which lowers ΔpH but not ΔΨm, and the K+ uniporter valinomycin, which dissipates ΔΨm but not ΔpH, as well as the effect of glucose-free medium containing inhibitors of glycolysis and the ATP synthase, 2-deoxy-d-glucose and oligomycin, respectively, as previously described (28). We found that treatment of cells with valinomycin but not nigericin or 2-DG/oligomycin induced the modification change of PGAM5, although less effectively than that with CCCP (Fig. 2, D–F). These results suggest that the modification of PGAM5 depends mainly on ΔΨm loss rather than on ΔpH and ATP loss.
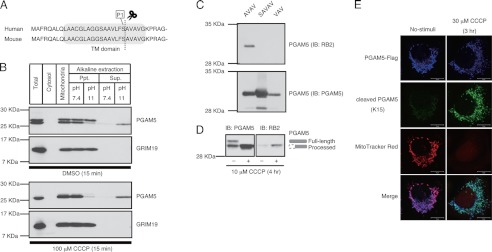
Because similar changes in the band pattern of OPA1 were demonstrated to be the result of ΔΨm loss-dependent proteolytic cleavage (8, 29–31), we hypothesized the possibility that PGAM5 might also be cleaved by proteases. Because the CCCP-induced lower band of PGAM5 was detected by an antibody that recognizes the C terminus of PGAM5 (Fig. 2A), we speculated that the cleavage site was located in the N-terminal region. Therefore, we performed Edman degradation analysis of the lower band of PGAM5 and determined that its N-terminal sequence was Ala-Val-Ala-Val-Gly (amino acids 25–29). This suggested that the lower band of PGAM5 was a processed form and that the cleavage site was located in the TM domain (Fig. 3A). When isolated mitochondria were subjected to alkaline extraction, the membrane-anchoring efficiency of the processed form of PGAM5 was lower than that of full-length PGAM5 (Fig. 3B), supporting the above result that the processed form of PGAM5 has lost most of the N-terminal region of the TM domain. Nevertheless, cleaved PGAM5 was still mostly localized in the IMM when we examined the localization of cleaved PGAM5 in CCCP-treated HEK293A and HeLa-PGAM5-FLAG cells by trypsin protection assay and immunoelectron microscopy analysis, respectively (supplemental Fig. S1, A–C).
FIGURE 3.
PGAM5 is cleaved in its N-terminal TM domain in response to ΔΨm loss. A, the identified cleavage site is located in the TM domain of PGAM5. B, the cleaved form of PGAM5 is released from mitochondria upon alkaline extraction. Mitochondria isolated from MEFs treated with DMSO or 100 μm CCCP for 15 min were subjected to alkaline extraction with the indicated pH buffers. After centrifugation, pellet (Ppt.) and supernatant (Sup.) were collected and subjected to IB. C, cleaved PGAM5 antibody (RB2) specifically recognizes proteins with the identified cleaved end. Recombinant PGAM5 proteins with the indicated cleaved ends were subjected to IB using the antibody specific for cleaved PGAM5 (RB2). D, CCCP-induced lower band of PGAM5 is detected by cleaved PGAM5 antibody (RB2). HEK293A cells treated with CCCP were subjected to IB using the antibody RB2. E, the staining with cleaved PGAM5 antibody (K15) is detected only in mitochondria lacking ΔΨm. HeLa cells transfected with PGAM5-FLAG were treated with 10 μm CCCP for 4 h and subjected to immunocytochemistry using the antibody specific for cleaved PGAM5 (K15). Mitochondria with intact ΔΨm were detected with the ΔΨm-dependent dye, MitoTracker red CMXRos. Scale bar = 10 μm.
To confirm the cleavage site, we generated an antibody against a synthetic peptide starting with the identified cleavage site (amino acids 25–32 of PGAM5) that does not cross-react with the N-terminally acetylated peptide that mimics the uncleaved protein. The antibody generated against cleaved PGAM5, named RB2, specifically recognized PGAM5 with the identified cleaved end (AVAV) but did not recognize PGAM5 with the cleaved end one amino acid forward (SAVAV) or backward (VAV), demonstrating the specificity of the antibody (Fig. 3C). Importantly, this antibody specifically recognized the lower band of PGAM5 that was increased after CCCP treatment but not the upper band (Fig. 3D). When we used MitoTracker red CMXRos, which accumulates in mitochondria with an intact ΔΨm and thus discriminates healthy mitochondria from damaged mitochondria lacking ΔΨm, staining with another cleaved PGAM5 antibody (K15) was detected only in damaged mitochondria with much lower MitoTracker staining (Fig. 3E). These results indicate that PGAM5 is cleaved in its TM domain in response to ΔΨm loss.
PARL Mediates the Cleavage of PGAM5
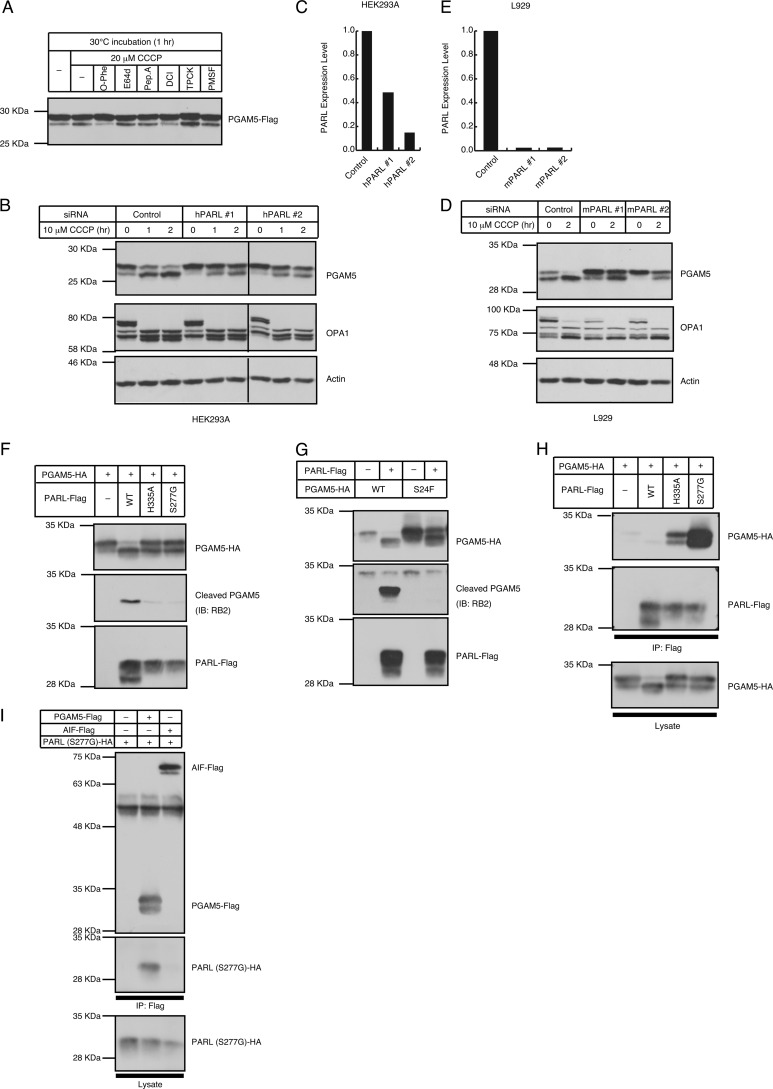
Next, we sought to identify the protease that mediates the cleavage of PGAM5. Because the cleavage site is located in the TM domain, we speculated that PGAM5 might be cleaved by I-Clips. Currently known I-Clips are divided into three classes of proteases depending on their catalytic residues: the metalloproteases (site 2 proteases), the aspartyl proteases (presenilin and signal peptide peptidase), and the serine proteases (rhomboid proteases) (1). To determine which class of I-Clips is responsible for the cleavage of PGAM5, mitochondria isolated from cells expressing PGAM5 were treated with several protease inhibitors under CCCP stimuli. As shown in Fig. 4A, CCCP-induced cleavage of PGAM5 was inhibited by the metalloprotease inhibitor o-phenanthroline (o-Phe) and the serine protease inhibitor dichloroisocoumarin (DCI), whereas it was insensitive to other serine protease inhibitors, such as TPCK and PMSF.
FIGURE 4.
PARL mediates ΔΨm loss-induced cleavage of PGAM5. A, sensitivity of CCCP-induced cleavage of PGAM5 to various protease inhibitors is shown. 24 h after transfection of PGAM5-FLAG in HEK293A cells, mitochondria were isolated from the cells and pretreated with or without the indicated protease inhibitors followed by stimulation with or without 20 μm CCCP in vitro. B–E, PARL is required for CCCP-induced cleavage of PGAM5. 48 h after transfection with siRNAs for human PARL (hPARL), HEK293A cells were treated with 10 μm CCCP for the indicated time periods and subjected to IB (B). In mouse L929 cells, knockdown of PARL using siRNAs for mouse PARL (mPARL) also suppressed the CCCP-induced cleavage of PGAM5 (D). Knockdown efficiency of siRNAs used in (B and D) was evaluated using quantitative RT-PCR (C and E). F, co-expressed PARL, but not inactive mutants of PARL, cleaves PGAM5. PGAM5 was co-expressed with C-terminally FLAG-tagged PARL (PARL-WT-FLAG) or the inactive mutants of PARL (PARL-S277G-FLAG and PARL-H335A-FLAG) in HEK293A cells. After 48 h, cells were lysed and subjected to IB. G, mutating the helix-breaking residue in P1 position disturbs the cleavage of PGAM5 at the correct site. HEK293A cells were transfected with PARL-WT-FLAG and PGAM5-WT-HA or a mutant of the P1 position, PGAM5-S24F-HA. After 48 h, cells were lysed and subjected to IB. H and I, PARL interacts with PGAM5. HEK293A cells were transfected with the indicated expression plasmids. After 48 h, cells were lysed, and PARL (H) and PGAM5 and AIF (I) were immunoprecipitated (IP) using a FLAG antibody. All the immunoprecipitates (top and middle panels) and of the total cell lysate (bottom panels) were subjected to IB. Based on the quantification of the band intensity using the ImageJ software, ∼5 and 1% of total PGAM5 appeared to bind to PARL (S277G) and PARL (H335A), respectively (H).
The protease inhibition profile was interesting because dichloroisocoumarin is a unique serine protease inhibitor that inhibits a broad spectrum of rhomboid-mediated intramembrane proteolysis (32). We thus examined the possibility that the rhomboid proteases regulate the cleavage of PGAM5, although o-phenanthroline-sensitive processes may also be involved in PGAM5 cleavage (see “Discussion”). A comprehensive genome sequence analysis of the eukaryotic rhomboid protease family revealed that there are at least five genes encoding putative rhomboid proteases in mammals: RHBDL1, RHBDL2, RHBDL3, RHBDD1, and PARL (4). Because PARL has only been reported to be localized in mitochondria, we knocked down PARL using three siRNAs that target different regions in the PARL gene. As shown in Fig. 4, B and C, CCCP-induced cleavage of PGAM5, but not of OPA1, was severely impaired by suppressing PARL expression in HEK293A cells, suggesting that PARL is required for CCCP-induced cleavage of PGAM5. To negate that this was cell- and/or species-specific result, we also used mouse L929 cells (Fig. 4, D and E). In these cells we found the requirement of PARL for CCCP-induced cleavage of PGAM5, suggesting that PARL-mediated cleavage of PGAM5 is conserved at least in mammals. Consistent with this, co-expression of PGAM5 with PARL strongly induced the processing of PGAM5 even in the absence of CCCP (Fig. 4F, top panel, second lane), whereas the co-expression of the protease-inactive PARL mutants, in which the active site Ser-277 or His-335 was substituted with Gly or Ala, respectively, failed to process PGAM5 (Fig. 4F, top panel, third and fourth lanes). Moreover, the processed form of PGAM5 induced by co-expression with PARL was specifically detected by the cleaved PGAM5 antibody (RB2) (Fig. 4F, second 2nd panel), indicating that PARL mediates the cleavage of PGAM5 at the same site as is induced by CCCP.
Recently, it has been reported that rhomboids recognize a specific amino acid sequence surrounding the cleavage site of their substrates (33). Among these, the helix-destabilizing residues, such as Ala, Gly, Cys, and Ser, in the P1 position (indicated in Fig. 3A) are important because they have been shown to facilitate local helix unfolding and allow the efficient cleavage within a lipid bilayer. To examine the requirement of this motif for PARL-mediated cleavage of PGAM5, we generated the PGAM5 S24F mutant in which Ser-24 at the P1 position was substituted by a bulky amino acid, Phe. As shown in Fig. 4G, co-expression with PARL still induced the band shift of PGAM5 S24F to some extent; however, the lower band was no longer detected by the cleaved PGAM5 antibody (RB2), suggesting that the cleavage of PGAM5 S24F at least between Phe-24 and Ala-25 was completely blocked. The residual band shift of PGAM5 S24F may represent cleavage at other sites, because many helix-breaking residues exist in the TM domain of PGAM5 (Fig. 3A).
Consistent with our data showing the localization of PGAM5 in the IMM and the previous reports that PARL is also located in the IMM (5, 6), PGAM5 interacted with the two different inactive mutants of PARL (Fig. 4H). It was unlikely that the PARL mutants used above tended to non-specifically bind to IMM-localized proteins, because another IMM-localized protein, AIF, did not interact with PARL (Fig. 4I). The binding efficiency of PGAM5 and inactive PARL was higher than that of PGAM5 and wild-type PARL, suggesting that wild-type PARL but not the inactive PARL is rapidly dissociated from PGAM5 after the cleavage. These results, taken together strongly suggest that the ΔΨm loss-induced cleavage of PGAM5 is mediated by PARL.
PARL-mediated cleavage of PINK1 and PGAM5 Is Reciprocally Regulated Depending on the ΔΨm Status
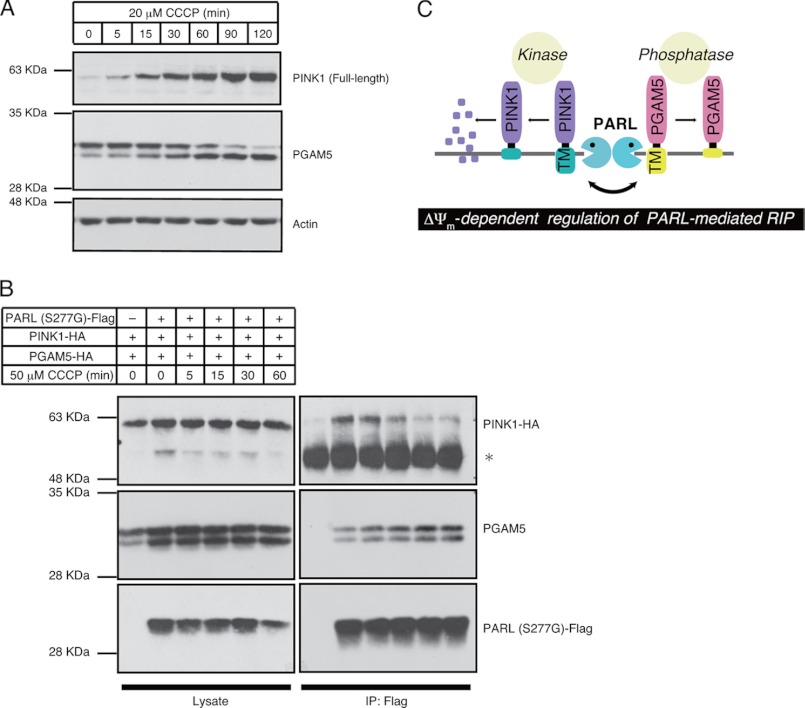
Recently, PINK1 has been reported to be rapidly and constitutively degraded under steady-state conditions but stabilized in response to ΔΨm loss (34, 35). Stabilized PINK1 acts as a marker of damaged mitochondria and triggers mitophagy, a system for selective elimination of damaged mitochondria by autophagy (36). More recently, it has been reported that the proteolytic destabilization of PINK1 in healthy mitochondria is mediated by PARL (15). Drosophila genetics also provides evidence that PINK1 is a substrate of PARL (37). From these findings, we speculated that PARL could change its substrate from PINK1 to PGAM5 in response to ΔΨm loss. We found that the time-course of CCCP-induced stabilization of endogenous PINK1 was coincident with that of CCCP-induced cleavage of endogenous PGAM5 (Fig. 5A). Moreover, PINK1 bound to the inactive mutant of PARL, PARL (S277G), under unstimulated conditions and dissociated from PARL in a time-dependent manner in response to CCCP, whereas the PGAM5-PARL interaction increased in a manner inversely correlated with the PINK1-PARL interaction (Fig. 5B). Similar results were obtained when another inactive mutant of PARL, PARL (H335A), was used as bait (supplemental Fig. S2). These results suggest that the protease activity of PARL is maintained regardless of the health status of mitochondria but that PARL mediates differential cleavage of PINK1 and PGAM5 depending on ΔΨm.
FIGURE 5.
PARL-mediated cleavage of PINK1 and PGAM5 are reciprocally regulated in a ΔΨm-dependent manner. A, time-course of PINK1 stabilization and PGAM5 cleavage in response to CCCP is shown. HEK293A cells treated with 20 μm CCCP for indicated time periods were lysed and subjected to IB. B, CCCP-dependent reciprocal binding of PINK1 and PGAM5 to PARL is shown. HEK293A cells were transfected with the indicated plasmids. After 48 h, cells were treated with 50 μm CCCP for indicated time periods. After stimuli, cells were lysed, and PARL was immunoprecipitated (IP) using a FLAG antibody. Immunoprecipitates and cell lysates were subjected to IB. *, indicates IgG band. C, a proposed model of ΔΨm-dependent regulation of PARL-mediated cleavage of PINK1 and PGAM5 is shown.
DISCUSSION
In this study we revealed that PARL mediates the cleavage of PGAM5 in response to ΔΨm loss. In addition, PARL-mediated cleavage of PINK1 and PGAM5 is reciprocally regulated depending on the ΔΨm status (Fig. 5C).
The interaction between inactive mutants of PARL and PGAM5 was stronger than that between wild type PARL and PGAM5 (Fig. 4H). In addition, the helix-breaking residue at the P1 position, which is known to be critical for recognition by rhomboids, appeared to also be critical in the case of recognition of PGAM5 by PARL (Fig. 4G). These results suggest that PGAM5 is directly cleaved by PARL. Currently, however, it is difficult to exclude the possibility that PARL indirectly regulates the cleavage of PGAM5 via unknown direct substrates of PARL. The establishment of an in vitro reconstitution assay system of PARL-mediated cleavage will enable us to address this important question.
We found that ΔΨm status is a critical determinant of PARL-mediated cleavage of PGAM5 (Fig. 2, A–E). Recently, it has been reported that the cleavage of several mitochondrial resident proteins, such as OPA1 and PINK1, is regulated depending on ΔΨm (36, 38). Whereas the cleavage of OPA1 is induced by ΔΨm loss, the cleavage of PINK1 is inhibited by ΔΨm loss, resulting in the accumulation of full-length PINK1 on the OMM. It has also been reported that the cleavage of a zinc metalloprotease, OMA1, is inhibited by ΔΨm loss, and the resulting stabilized OMA1 is in turn responsible for ΔΨm loss-dependent cleavage of OPA1 (30). These findings indicate that the ΔΨm loss-dependent cleavage of PGAM5 is a hot example of these complicated proteolysis regulations in mitochondria.
Because the cleavage of PGAM5 was inhibited by the metalloprotease inhibitor, o-phenanthroline, as by the serine protease inhibitor, dichloroisocoumarin (DCI; Fig. 4A), we examined the involvement of OMA1 in the cleavage of PGAM5 (supplemental Fig. S3A). Whereas OMA1 knockdown impaired CCCP-induced cleavage of OPA1 as previously reported (30, 31), we observed a marginal, if any, impairment of the cleavage of PGAM5. In contrast, the attenuation of the cleavage of PGAM5 by PARL knockdown was more significant. These results suggest that the contribution of PARL to the cleavage of PGAM5 is higher than that of OMA1 at least under cellular conditions examined here. Elucidation of the relationship between OMA1 and PARL in the cleavage of PGAM5 (e.g. independent or co-operative) will uncover the precise molecular mechanism how ΔΨm-dependent cleavage of PGAM5 is regulated.
What are the underlying molecular mechanisms that regulate the ΔΨm-dependent change of substrates for PARL? Considering that the transport of PINK1 to the IMM is blocked by ΔΨm loss (15), it was possible that the disappearance of PINK1 from the IMM allowed the PARL-mediated cleavage of PGAM5. However, we found that the cleavage of PGAM5 was not induced by siRNA-mediated suppression of PINK1 expression (data not shown). Thus, the PARL-mediated cleavage of PINK1 and PGAM5 appears not to be regulated in a competitive manner with each other. Moreover, because the affinity of PGAM5 for PARL increased in response to ΔΨm loss (Fig. 5B and supplemental Fig. S2), it is rather likely that some changes in PGAM5 and/or PARL induced by ΔΨm loss facilitate proteolysis of PGAM5 by PARL. Although the regulatory mechanisms of rhomboid-mediated proteolysis are not well known in mammals, some models, especially those of how the substrates are presented to rhomboids, have been proposed in other organisms. For example, in Drosophila, a regulated compartmentalization mechanism has been well characterized; Rhomboid-1 processes its substrate Spitz in Golgi, before which a membrane protein, Star, escorts Spitz from the endoplasmic reticulum to Golgi and, therefore, is required for the efficient processing of Spitz (2). Although this compartmentalization model operates at the inter-organelle level, a similar model, but at the inner-organelle level, might apply to PGAM5; it would be possible that some Star-like proteins assist the translocation of PGAM5 to the PARL-resident compartment in the IMM in response to ΔΨm loss. Another possibility is that the scissile region within the TM domain of PGAM5, which may be concealed in healthy mitochondria, is presented to PARL in response to ΔΨm loss. In yeast it has been reported that IMM-resident motor proteins mediate the correct positioning of the TM domains of substrate proteins within lipid bilayers, facilitating the presentation of their scissile regions to Rbd1/Pcp1, a yeast ortholog of PARL (39, 40).
At present, the physiological role of ΔΨm loss-dependent cleavage of PGAM5 is not known. ΔΨm loss-dependent cleavage of OPA1 is thought to be a mechanism that prevents the re-fusion of damaged mitochondria to healthy mitochondria, because the mitochondrial fusion activity of OPA1 is inhibited by its cleavage (38). On the other hand, ΔΨm loss-dependent stabilization and accumulation of PINK1 on the OMM triggers mitophagy, one of the mitochondrial quality control systems (36). These findings indicate that ΔΨm-dependent proteolysis, including that of PGAM5, might be utilized as a general tool for the stress response of mitochondria.
It has been reported that PARL KO mice display defects in postnatal growth and lifespan because of increased sensitivity to cell death-inducing stimuli (6, 7), indicating the anti-apoptotic roles of PARL. Interestingly, recent reports have suggested that PGAM5 is involved in multiple cell death pathways in a context-dependent manner. For example, we have recently reported that flies deficient in the Drosophila ortholog of mammalian PGAM5 (dPGAM5) are vulnerable to heat shock stress mainly because of increased apoptosis in neurons in the mushroom body, suggesting the role for PGAM5 in apoptosis regulation (41). On the other hand, dPGAM5 has recently been shown to act as an exacerbating factor for mitochondrial degeneration and dopaminergic neuronal cell death in the model of Parkinson disease induced by mutation of the Drosophila PINK1 gene (42). In addition, it has more recently been reported that PGAM5 is activated downstream of necrosis promoting kinases, RIP1 and RIP3, and subsequently induces necrosis via activating mitochondrial fission factor, Drp1 (14). Thus, it is important to determine whether or not the PARL-mediated cleavage of PGAM5 plays a critical role in the regulation of cell death.
In conclusion, we presented here a prototypical example of stress-induced regulation of PARL-mediated RIP. Importantly, PARL appears to mediate differential cleavage of kinases and phosphatases depending on mitochondrial conditions and might thus indirectly regulate the phosphorylation status of their common and/or respective substrates. The elucidation of the detailed mechanisms and roles of this model may provide important insights into the mitochondrial and cellular stress responses.
Acknowledgments
We are grateful to T. Kitamura, N. Fujita, and T. Yoshimori for providing materials. We thank T. Natsume, S. Iemura, and T. Tomita for valuable discussions.
This work was supported by the Japan Science and Technology Agency PRESTO program, KAKENHI from the Japan Society for the Promotion of Science and Ministry of Education, Culture, Sports, Science, and Technology of Japan, Global Center of Education and Research for Chemical Biology of the Diseases, the “Understanding of molecular and environmental bases for brain health” conducted under the Strategic Research Program for Brain Sciences by Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Naito Foundation Natural Science Scholarship, the Cosmetology Research Foundation, and the Tokyo Biochemical Research Foundation.

This article contains supplemental Figs. S1–S3.
- I-Clip
- intramembrane cleaving protease
- IMM
- inner mitochondrial membrane
- OMM
- outer mitochondrial membrane
- OPA1
- optic atrophy 1
- PINK1
- PTEN-induced kinase 1
- RIP
- regulated intramembrane proteolysis
- PARL
- presenilin-associated rhomboid-like
- TM
- transmembrane
- PGAM5
- phosphoglycerate mutase 5
- TPCK
- N-p-tosyl-l-phenylalanine chloromethyl ketone
- PA
- paraformaldehyde
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- IB
- immunoblot
REFERENCES
- 1. Urban S., Freeman M. (2002) Intramembrane proteolysis controls diverse signaling pathways throughout evolution. Curr. Opin. Genet. Dev. 12, 512–518 [DOI] [PubMed] [Google Scholar]
- 2. Lee J. R., Urban S., Garvey C. F., Freeman M. (2001) Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell 107, 161–171 [DOI] [PubMed] [Google Scholar]
- 3. Urban S., Lee J. R., Freeman M. (2001) Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell 107, 173–182 [DOI] [PubMed] [Google Scholar]
- 4. Lemberg M. K., Freeman M. (2007) Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17, 1634–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pellegrini L., Scorrano L. (2007) A cut short to death. Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 14, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 6. Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., Derks C., Dejaegere T., Pellegrini L., D'Hooge R., Scorrano L., De Strooper B. (2006) Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell 126, 163–175 [DOI] [PubMed] [Google Scholar]
- 7. Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G. V., Rudka T., Bartoli D., Polishuck R. S., Danial N. N., De Strooper B., Scorrano L. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 [DOI] [PubMed] [Google Scholar]
- 8. Ishihara N., Fujita Y., Oka T., Mihara K. (2006) Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25, 2966–2977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duvezin-Caubet S., Koppen M., Wagener J., Zick M., Israel L., Bernacchia A., Jagasia R., Rugarli E. I., Imhof A., Neupert W., Langer T., Reichert A. S. (2007) OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol. Biol. Cell 18, 3582–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chao J. R., Parganas E., Boyd K., Hong C. Y., Opferman J. T., Ihle J. N. (2008) Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 452, 98–102 [DOI] [PubMed] [Google Scholar]
- 11. Jedrzejas M. J. (2000) Structure, function, and evolution of phosphoglycerate mutases. Comparison with fructose-2,6-bisphosphatase, acid phosphatase, and alkaline phosphatase. Prog. Biophys. Mol. Biol. 73, 263–287 [DOI] [PubMed] [Google Scholar]
- 12. Takeda K., Komuro Y., Hayakawa T., Oguchi H., Ishida Y., Murakami S., Noguchi T., Kinoshita H., Sekine Y., Iemura S., Natsume T., Ichijo H. (2009) Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc. Natl. Acad. Sci. U.S.A. 106, 12301–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lo S. C., Hannink M. (2008) PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp. Cell Res. 314, 1789–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang Z., Jiang H., Chen S., Du F., Wang X. (2012) The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 [DOI] [PubMed] [Google Scholar]
- 15. Jin S. M., Lazarou M., Wang C., Kane L. A., Narendra D. P., Youle R. J. (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 191, 933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deas E., Plun-Favreau H., Gandhi S., Desmond H., Kjaer S., Loh S. H., Renton A. E., Harvey R. J., Whitworth A. J., Martins L. M., Abramov A. Y., Wood N. W. (2011) PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum. Mol. Genet. 20, 867–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shi G., Lee J. R., Grimes D. A., Racacho L., Ye D., Yang H., Ross O. A., Farrer M., McQuibban G. A., Bulman D. E. (2011) Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson disease. Hum. Mol. Genet. 20, 1966–1974 [DOI] [PubMed] [Google Scholar]
- 18. Jofuku A., Ishihara N., Mihara K. (2005) Analysis of functional domains of rat mitochondrial Fis1, the mitochondrial fission-stimulating protein. Biochem. Biophys. Res. Commun. 333, 650–659 [DOI] [PubMed] [Google Scholar]
- 19. Ishihara N., Jofuku A., Eura Y., Mihara K. (2003) Regulation of mitochondrial morphology by membrane potential and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 301, 891–898 [DOI] [PubMed] [Google Scholar]
- 20. Otera H., Ohsakaya S., Nagaura Z., Ishihara N., Mihara K. (2005) Export of mitochondrial AIF in response to proapoptotic stimuli depends on processing at the intermembrane space. EMBO J. 24, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takeda K., Shimozono R., Noguchi T., Umeda T., Morimoto Y., Naguro I., Tobiume K., Saitoh M., Matsuzawa A., Ichijo H. (2007) Apoptosis signal-regulating kinase 2 (ASK2) functions as a mitogen-activated protein kinase kinase kinase in a heteromeric complex with ASK1. J. Biol. Chem. 282, 7522–7531 [DOI] [PubMed] [Google Scholar]
- 22. Sado Y., Inoue S., Tomono Y., Omori H. (2006) Lymphocytes from enlarged iliac lymph nodes as fusion partners for the production of monoclonal antibodies after a single tail base immunization attempt. Acta Histochem. Cytochem. 39, 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki H., Maeda M., Mihara K. (2002) Characterization of rat TOM70 as a receptor of the preprotein translocase of the mitochondrial outer membrane. J. Cell Sci. 115, 1895–1905 [DOI] [PubMed] [Google Scholar]
- 24. Eura Y., Ishihara N., Oka T., Mihara K. (2006) Identification of a novel protein that regulates mitochondrial fusion by modulating mitofusin (Mfn) protein function. J. Cell Sci. 119, 4913–4925 [DOI] [PubMed] [Google Scholar]
- 25. Ishihara N., Mihara K. (1998) Identification of the protein import components of the rat mitochondrial inner membrane, rTIM17, rTIM23, and rTIM44. J. Biochem. 123, 722–732 [DOI] [PubMed] [Google Scholar]
- 26. Morita S., Kojima T., Kitamura T. (2000) Plat-E. An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 27. Mori Y., Koike M., Moriishi E., Kawabata A., Tang H., Oyaizu H., Uchiyama Y., Yamanishi K. (2008) Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9, 1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2009) Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy 5, 706–708 [DOI] [PubMed] [Google Scholar]
- 29. Song Z., Chen H., Fiket M., Alexander C., Chan D. C. (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Head B., Griparic L., Amiri M., Gandre-Babbe S., van der Bliek A. M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D., Martinou J. C., Westermann B., Rugarli E. I., Langer T. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Urban S., Wolfe M. S. (2005) Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc. Natl. Acad. Sci. U.S.A. 102, 1883–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Urban S., Freeman M. (2003) Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 34. Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C. A., Sou Y. S., Saiki S., Kawajiri S., Sato F., Kimura M., Komatsu M., Hattori N., Tanaka K. (2010) PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanaka A. (2010) Parkin-mediated selective mitochondrial autophagy, mitophagy. Parkin purges damaged organelles from the vital mitochondrial network. FEBS Lett. 584, 1386–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whitworth A. J., Lee J. R., Ho V. M., Flick R., Chowdhury R., McQuibban G. A. (2008) Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis. Model Mech. 1, 168–174; discussion 173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Detmer S. A., Chan D. C. (2007) Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 [DOI] [PubMed] [Google Scholar]
- 39. Herlan M., Bornhövd C., Hell K., Neupert W., Reichert A. S. (2004) Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J. Cell Biol. 165, 167–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 26, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ishida Y., Sekine Y., Oguchi H., Chihara T., Miura M., Ichijo H., Takeda K. (2012) Prevention of apoptosis by mitochondrial phosphatase PGAM5 in the mushroom body is crucial for heat shock resistance in Drosophila melanogaster. PLoS One 7, e30265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Imai Y., Kanao T., Sawada T., Kobayashi Y., Moriwaki Y., Ishida Y., Takeda K., Ichijo H., Lu B., Takahashi R. (2010) The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 6, e1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]