Background: VCA in WASP proteins binds Arp2/3 complex and actin via a variable number of WH2 domains.
Results: WH2 repeats in V, VC, and VCA of N-WASP bind actin in 1:2 complexes; VC-actin crystallizes as a 1:1 complex, and the VCA·actin·Arp2/3 complex has 1:1:1 stoichiometry.
Conclusion: Data reconcile the studies of VCA-actin-Arp2/3.
Significance: Data provide insight in filament branching.
Keywords: Actin, Protein Complexes, Protein Domains, Protein Self-assembly, Protein Structure, Protein-Protein Interactions, Arp2/3 Complex, N-WASP, WASP Homology 2 Domain, filament Branching
Abstract
Wiskott-Aldrich syndrome proteins (WASP) are a family of proteins that all catalyze actin filament branching with the Arp2/3 complex in a variety of actin-based motile processes. The constitutively active C-terminal domain, called VCA, harbors one or more WASP homology 2 (WH2) domains that bind G-actin, whereas the CA extension binds the Arp2/3 complex. The VCA·actin·Arp2/3 entity associates with a mother filament to form a branched junction from which a daughter filament is initiated. The number and function of WH2-bound actin(s) in the branching process are not known, and the stoichiometry of the VCA·actin·Arp2/3 complex is debated. We have expressed the tandem WH2 repeats of N-WASP, either alone (V) or associated with the C (VC) and CA (VCA) extensions. We analyzed the structure of actin in complex with V, VC, and VCA using protein crystallography and hydrodynamic and spectrofluorimetric methods. The partial crystal structure of the VC·actin 1:1 complex shows two actins in the asymmetric unit with extensive actin-actin contacts. In solution, each of the two WH2 domains in V, VC, and VCA binds G-actin in 1:2 complexes that participate in barbed end assembly. V, VC, and VCA enhance barbed end depolymerization like profilin but neither nucleate nor sever filaments, in contrast with other WH2 repeats. VCA binds the Arp2/3 complex in a 1:1 complex even in the presence of a large excess of VCA. VCA·Arp2/3 binds one actin in a latrunculin A-sensitive fashion, in a 1:1:1 complex, indicating that binding of the second actin to VCA is weakened in the ternary complex.
Introduction
The WASP2/Scar/WAVE family of proteins counts many members, all of which stimulate actin polymerization in a dendritic array of actin filaments that deform membranes in a large number of motile processes (1–3). These processes include the extension of the lamellipodium of migrating cells, as well as phagocytosis, dendritic spine activity, immune synapse formation, spatial organization of the Golgi, and endosomal vesicle scission (4–12). WASP proteins work as membrane-bound, stimulus-responsive enzymes that catalyze cycles of filament branching and share the Arp2/3 complex as a substrate (13, 14).
All proteins of the WASP family use the same molecular mechanism to multiply filaments by branching, albeit with variable efficiencies (15–17). The conserved, constitutively active C-terminal moiety of the WASP enzymes, conventionally called VCA, is made of one or two WH2 (WASP homology 2) domains (“V” referring to either a single or multiple WH2 domains in Fig. 1), followed by a connector domain (“C”) and an acidic short extension (“A”). In the VCA region, the WH2 domain(s) bind G-actin, whereas the CA domain binds Arp2/3 complex. The resulting VCA·actin·Arp2/3 ternary complex (in which the shortened notation Arp2/3 refers to the Arp2/3 complex) associates with a filament to form the branched junction. Binding of G-actin to VCA is generally required for the branching reaction (18), although a basic branching activity of yeast Arp2/3 complex alone has been detected (19). The Arp2/3 complex is incorporated at the branched junction at each catalytic cycle and thus penetrates the network at the same pace as actin, by insertional polymerization (20–22).
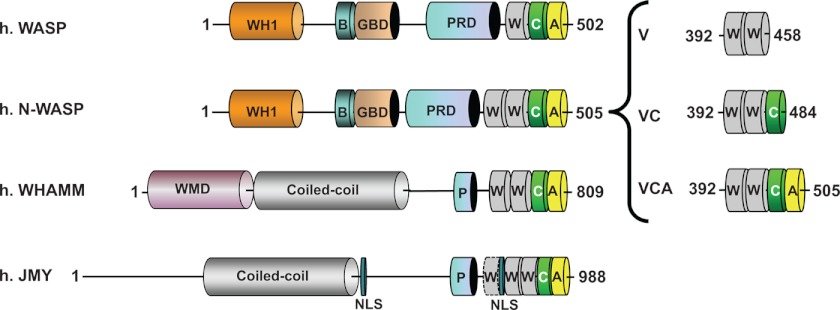
FIGURE 1.
Domain organization of WASP family proteins and design of N-WASP peptides. Modular structures found in WASP, N-WASP, WHAMM, and JMY (17) contain the following: WH1, WASP homology region 1; B, basic region; GBD, GTPase-binding domain; NLS, nuclear localization sequence; PRD/P, pro-rich domain; WMD, WHAMM membrane interaction domain; W, WH2 domain; C, connector region; A, acidic domain. His6-tagged peptides including both WH2 domains (WW, referred as V), VC, and VCA peptides were designed.
How the morphology of the dendritic array develops and perpetuates at the leading edge and how protrusive force is produced against the membrane tightly depend on the molecular mechanism of filament branching. However, despite extensive structural and biochemical studies, this issue remains elusive. The structural organization and stoichiometry of the active VCA·actin·Arp2/3 complex (called “branching complex”), its mode of binding to a filament, the nature of elementary steps and transients in the branching process, the fate of all protein partners in the branching reaction, and the possible requirement of additional G-actin molecules to nucleate a daughter filament are missing pieces of the puzzle (23–28). Among these questions, the functional role of the actin-WH2 interaction(s) in the enzymatic branching activity is not understood.
WH2 domains (first found in WASP/WAVE proteins) are small ubiquitous actin-binding modules found as single or repeated domains in many signaling modular proteins (29). They fold like thymosin β4 upon binding to actin. As single domains, they either sequester G-actin or act like profilin, i.e. make a complex with ATP·G-actin that participates in filament elongation at barbed ends exclusively (30, 31). As WH2 tandem repeats and/or in association with adjacent motifs/domains interacting with actin or actin-binding proteins, they display multifunctional regulation of actin assembly (29, 31, 32). In contrast to other WASP/SCAR/WAVE family members, the VCA domains of N-WASP, WHAMM, and JMY contain tandem repeats of WH2 domains (3). The VCA domain of N-WASP displays a higher efficiency of branching than WASP or WAVE VCA (15). The WH2 repeats of JMY confer to the protein an additional nucleating activity (17). The structures of the complexes of actin with the larger VC and VCA domains of N-WASP are not known. The isolated C domain of SCAR VCA binds G-actin like a WH2 domain using its N-terminal α-helix (33). The VC domain of N-WASP binds the Arp2/3 complex via the C domain and branches filaments with a lower efficiency than VCA (13). The CA domain binds the Arp2/3 complex at two sites of different affinities (27), and two Alexa 488-labeled VCAs co-sediment with Arp2/3 in analytical centrifugation studies (26).
In a first step toward understanding the general molecular mechanism for filament branching by WASP proteins, in particular the functional role of the association of actin to their WH2 domain(s) in the branching reaction, we describe here the crystal structure of the VC moiety of N-WASP in complex with ATP-G-actin, in the absence of either DNase I, gelsolin, or latrunculin A. In parallel, we analyze the binding of V, VC, and VCA to G-actin in solution and compare the functional properties of the complexes in filament assembly. Although only one of the two WH2 domains of N-WASP binds actin in the crystal structure of VC-actin, in the isolated V, VC, and VCA domains of N-WASP these two WH2 domains form 1:1 (VA) and 1:2 (VA2) complexes with G-actin (A) in solution, which participate in barbed end assembly like profilin-actin. Despite their binding two actin molecules, V, VC and VCA do not nucleate nor sever filaments. Arp2/3 complex binds VCA in a 1:1 complex and forms with VCA and actin a single 1:1:1 ternary complex. Thus, binding of the second actin to VCA is abolished in the ternary VCA·actin·Arp2/3 complex.
EXPERIMENTAL PROCEDURES
Proteins and Peptides
Actin was purified from rabbit skeletal muscle and was isolated as a monomeric CaATP-G-actin form by gel filtration in G buffer (5 mm Tris/HCl, pH 7.8, 1 mm dithiothreitol, 0.2 mm ATP, 0.1 mm CaCl2, and 0.01% NaN3). Actin was pyrenyl-labeled. Spectrin-actin seeds were prepared from human erythrocytes (34). The Arp2/3 complex from bovine brain, full-length recombinant human gelsolin, and the VCA domain of human neural Wiskott-Aldrich syndrome protein (N-WASP) were purified as described previously (35).
From a DNA fragment encoding human N-WASP VCA(389–505), two C-terminal His6 recombinant peptides VC(392–484) and V(392–458) were cloned and expressed as follows. The peptides were amplified by PCR using nucleotides carrying adaptors including either NdeI or XmaI cutting sites, respectively, VC-XmaI-3′ (5′-tccccccgggTCAAGAATGAATGGCTTTGCTCCT-3′) and VC-NdeI-5′ (5′-ggaattccatatgcatcatcatcatcatcattatccaagatcttacCCTTCTGATGGGGACCATCAGG-3′); V1-NdeI-5′ and V2-XmaI-3′ (5′-tccccccgggtcatggtgtagactcttggccatc-3′). After restriction enzyme digestion, each gene was cloned under the control of an arabinose-inducible promoter in the expression vector pBAD33-GFP (36). Plasmids were expressed in Escherichia coli strain BL21. Cultures were grown at 310 K in LB medium containing 25 μg·ml−1 chloramphenicol until A600 nm = 1.2. Cells were induced by the addition of 10 mm arabinose and harvested 1 h post-induction. They were centrifuged twice for 20 min at 10,000 × g and frozen at 193 K. The cell pellet was resuspended in 20 mm Tris/HCl, pH 8.0, 200 mm NaCl, 5 mm EDTA, 1 mm PMSF. Cells were lysed with a French press at 69 megapascals. His6-tagged peptides were isolated on 15 ml of nickel-Sepharose 6 Fast Flow (Amersham Biosciences) (loading solution: 90 ml of 100 mm Tris/HCl, pH 8.9, 1 m NaCl, 5 mm imidazole, 0.1% N-lauryl sarcosyl), washed with 60 ml of solution A (20 mm Tris/HCl, pH 8.0, 1 m NaCl, 5 mm imidazole), and eluted by a 100-ml linear gradient of imidazole (50–400 mm) at the flow rate of 2 ml·min−1. The fractions containing the peptide were pulled and exchanged into C buffer (25 mm NaOH/HOAc, pH 4.3, 1 mm EDTA, 25 mm NaCl, 5 mm DTT). Peptide solution was loaded on a Source 15S (Amersham Biosciences) and eluted by a linear gradient of NaCl (5 mm to 1 m). Purity of peptides was checked by SDS-PAGE and isoelectric focusing.
Molar Extinction Coefficient Determination
The 280-nm molar extinction coefficient of the peptides VC and V was measured by the titration of the Cys-431 free sulfhydryl group by Ellman's reagent (Pierce) (E280 = 3180 m−1·cm−1) and from lyophilization of 3.79 mg of VC (E280 = 2830 m−1·cm−1). These values are in reasonable agreement with the value of E280 = 2980 m−1·cm−1 calculated according to Pace et al. (37).
Analytical Size-exclusion Chromatography
Size-exclusion chromatography of VC- or VCA-incubated mixtures with G-actin was carried out using a Superdex 200 10/300 GL column (Fig. 1A). The column was calibrated using standard globular proteins. Isocratic elution was carried out at 0.3 ml/min in (5 mm Tris, pH 7.5, 100 mm NaCl, 0.1 mm CaCl2, 0.1 mm ATP, 1 mm DTT). Elutions were monitored by absorbance at 280 nm and analyzed by densitometry on SDS-PAGE.
Crystallization and Data Collection
Solutions of CaATP-G-actin and N-WASP-peptide (in a 1:2 actin/peptide molar ratio) were concentrated to 180 μm of actin in 5 mm Tris/HCl, pH 7.4, 0.2 mm ATP, 20 μm CaCl2, 20 mm tris(2-carboxyethyl)phosphine, 0.01% NaN3.
Crystals were obtained by the vapor-diffusion method at 278 K, under argon, in 3-μl drops containing a 2:1 mixture of protein solution and precipitant, 11.5% (w/v) polyethylene glycol 8000 (Sigma), 8.5% TacsimateTM (Hampton Research), 100 mm Hepes, pH 7.0. After 3 weeks of growth, the reservoirs were changed to 13.8% (w/v) PEG 8000, 12.5% TacsimateTM, 100 mm Hepes, pH 7.0. Crystals were soaked in 16% (w/v) PEG 8000, 12,5% TacsimateTM, 100 mm Hepes, pH 7.0, 15% ethylene glycol, before flash freezing in liquid nitrogen. Crystals with similar shapes were also obtained using either 150 mm sodium citrate and 9% PEG 8000, 500 mm sodium formate and 11% PEG 8000, 200 mm sodium malonate and 12% PEG 8000, or 400 mm sodium tartrate and 10.5% PEG 8000.
Structure Determination and Refinement
Data were collected at 100 K at beamline ID14-4 and ID23-2 at the European Synchrotron Radiation Facility in Grenoble, France. We obtained diffraction patterns of the two complexes VC·G-actin and V·G-actin. All of them are isomorphous, but only the first one was of high enough quality to collect data. Data were indexed and scaled in the programs MOSFLM (38) and SCALA (39). The apparent point group was P622, but according to Padilla and Yeates (40), we detected a merohedral twin with a twinning factor of 0.432. Molecular replacement using AMoRe (41) was carried out with the starting model 2A3Z of actin alone (42) in all P622 enantiomorph groups. Two sets of two highly contrasted solutions of rotation and of translation functions were obtained in the P6122 group. They correspond to two molecules per asymmetric unit and to the same solution rotated along the twin law.
These solutions were also obtained using PHASER (43) but turning off the crash test. Indeed, the twin resulted in a clash between molecules related by the 2-fold axis perpendicular to the crystallographic 6-fold axis in the P6122 super group. Therefore, we concluded that the true space group was P61 with a twinning axis in the 110 direction and two molecules A and B per asymmetric unit. The solvent content is 76%, and the Matthews coefficient is evaluated to 5.2 Å3/dalton. Molecules A and B are related to the other by a noncrystallographic true 2-fold axis perpendicular to the c axis and at an angle of −24.4° with the a axis in the a, b plane. In addition to this contact, analysis of the crystal packing showed that molecules A and B are each organized along the 61 axis in a similar manner. The contacts of each molecule of the asymmetric unit with their neighbors along the 61 axis are identical; consequently, the structure can be refined with strict noncrystallographic symmetry constraints.
The model of actin was refined in CNS (44) with the hemihedral twinning procedure. We applied first a rigid-body refinement with eight bodies corresponding to the four subdomains of each actin molecule of the asymmetric unit (twinned Rfact = 0.386 twinned Rfree = 0.390). It was followed by simulated annealing and by energy minimization applying a strict NCS constraint. Manual rebuilding was facilitated because the region linked to molecule A by the twin law contained only solvent. So the electron map was unusually clear for such a twinned structure. We had to rebuild essentially three zones as follows: first, the 10 C-terminal missing residues at the C terminus part of the protein, which were absent in the initial model; second, the D-loop, and third, the VC peptide. Bulk solvent correction was applied, and two restrained and grouped B-factors per residue were refined. Final statistics are twinned Rfact = 0.275 and twinned Rfree = 0.331.
Analysis of the Fluorescence Data for Binding of V and VC to AEDANS-Actin
The observed fluorescence of AEDANS-actin was expressed as the sum of the contributions of all species of AEDANS-actin in free and liganded states, as shown in Equations 1 and 2.
where f0, f1, and f2 represent the intrinsic fluorescence of free AEDANS·actin (A), of VA and VA2 complex, respectively (f2 refers to the intrinsic fluorescence of actin averaged over the two G-actin molecules in VA2), and [Atot] represents the total concentration of actin. The saturation of actin by V or VC, Y, was expressed by the relative extent of fluorescence change shown in Equation 3,
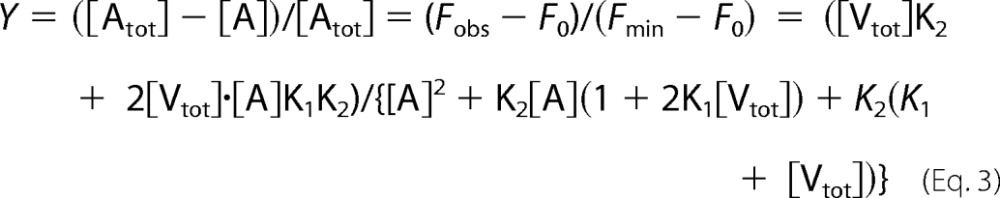 |
where [Vtot] = [V] + [VA] + [VA2].
The fluorescence anisotropy of AEDANS-actin in the presence of V or VC was expressed as the sum of the contributions of A, VA and VA2 species as in Equation 4.
in which r0, r1, and r2 represent the anisotropies of AEDANS bound to free actin and to actin in VA and VA2 complexes, respectively. Because of the cubic nature of the equations expressing the saturation of actin by V and VC, the values of K1 and K2 were determined by manual adjustment of theoretical curves that were generated as follows using the Excel software. Values of [V] were incremented and used to calculate the corresponding values of [A], [VA], and [VA2], derived as shown in Equations 5 and 6.
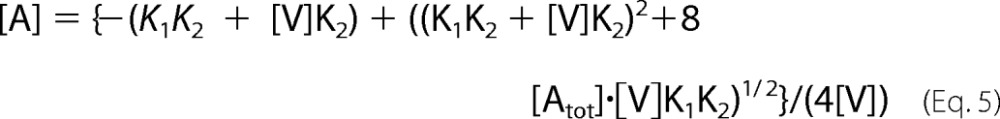 |
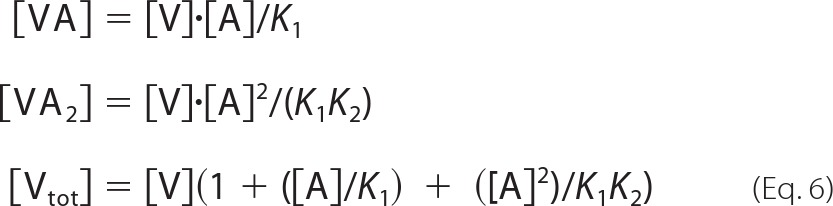 |
The fluorescence intensity data and the anisotropy data (Fig. 2) were analyzed in a global fashion, to find a set of values of K1 and K2 that best fitted the data.
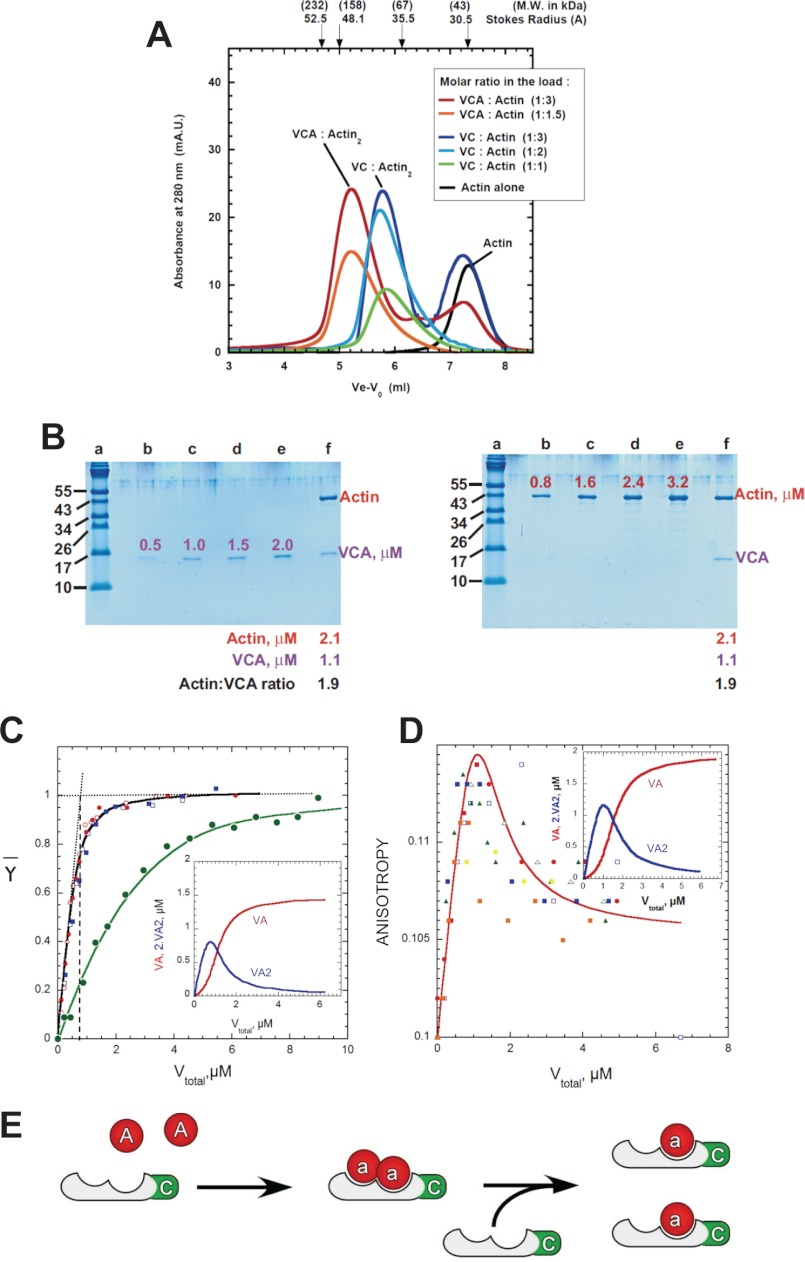
FIGURE 2.
Actin binds to each of the two WH2 domains of N-WASP in the isolated V, VC, and VCA fragments. A, analysis of the complexes of VC or VCA with G-actin on size-exclusion chromatography and SDS-PAGE. Samples at indicated molar ratios containing 7 μm VC or VCA with either 21 (red, blue), 14 (cyan), 11 (orange), or 7 μm (green) G-actin were loaded and eluted on a Superdex 200 10 × 300 column at near-physiological ionic strength. The elution of G-actin alone (5 μm) is shown in black. B, SDS-PAGE analysis of the saturated eluted complex of VCA and G-actin (red curve in A). The peak fraction (lanes f) was co-electrophoresed with protein markers (lanes a), VCA, and actin standards (lanes b–e). The densitometric analysis indicates a VCA/actin molar ratio of 1:2 in the peak fraction. A similar 1:2 VC/actin molar ratio is obtained in the peak fraction of the saturated eluted VC·actin complex (data not shown). C, fluorescence titration curves of AEDANS-actin by the V, VC. and VCA fragments of N-WASP. AEDANS-G-actin (1.5 μm) was titrated by V (blue), VC (red), or VCA (green closed circles). The resulting quenching of AEDANS fluorescence is normalized to represent the saturation of actin by the peptides. The curves are calculated assuming the formation of VA and VA2 complexes (see text) with values of 0.02 and 0.3 μm for K1 and K2, and a K1 value of 1.2 μm for a putative single 1:1 complex of VCA with actin. The equivalence point (black dashed line) at ∼0.75 μm V for 1.5 μm AEDANS-G-actin indicates a 1:2 VC/A stoichiometry. Inset, fraction of actin in VA (red) and VA2 (blue) complexes in the conditions of the main frame. D, fluorescence anisotropy titration curves of AEDANS-actin (1.5 μm) by V (yellow, orange, and green closed symbols) and VC (red and blue closed symbols and blue and green open symbols). Independent experiments are superimposed. The continuous red line is calculated according to Equation 4 and using anisotropy values of 0.1, 0.106, and 0.117 for G-actin, VA, and VA2 complexes respectively, and values of K1 and K2 as in C. Inset, fraction of actin in VA (red) and VA2 (blue) complexes. E, scheme for binding of the V, VC, and VCA fragments of N-WASP to G-actin.
Analysis of the Seeded Filament Growth Rates
In the presence of V or VC, if only free actin participates in filament elongation (pointed end growth), the rate of filament elongation from seeds is expressed as shown in Equations 7 and 8,
In the absence of V or VC,
where [Atot] is the total concentration of G-actin in the growth assay; k+P the on rate constant for G-actin to the filament pointed end and ACP is the critical concentration for actin assembly at pointed ends. The data were expressed as normalized rates of elongation as in Equation 9.
The iteration method described above was used to calculate [V0], [A], and Vel and to compute simulated curves. The best fit to the experimental curves was searched by manual adjustment of K1 and K2.
If VA and VA2 complexes participate in barbed end growth and V caps barbed ends with an equilibrium dissociation constant KF, then the normalized elongation rate was described by Equation 10 (in which the critical concentrations for barbed end assembly of VA and VA2 have been neglected).
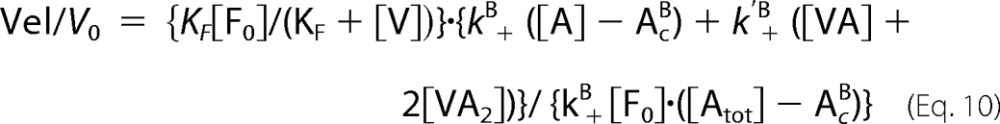 |
As described above, the values of [V] were incremented, leading to calculated values of [A], [VA], [VA2], and [V0]. Theoretical curves of Vel/V0 were generated. The values of the rate constants for association of VA or VA2 to barbed ends (k′B) were assumed to be identical. Both k+′B and KF were adjusted manually for the best fit to the data.
RESULTS
Design and Production of the V and VC Fragments of N-WASP
Two His6-tagged constructions of fragments were derived from the VCA domain of human N-WASP (Fig. 1) (35). A V construct included the two WH2 domains starting from residue 392 to 458. The VC construct (residues 392–484) included the two WH2 domains and the C region.
Binding of G-actin to the V, VC, and VCA Regions of N-WASP
The complexes of V, VC, and VCA constructs with G-actin were analyzed using hydrodynamic and spectroscopic methods. Gel filtration analysis, at protein concentrations in the 10−5–10−6 m range and near physiological ionic strength, provided evidence for the binding of two G-actin molecules to either V, VC, or VCA domains of N-WASP (Fig. 2A). The same complex of Stokes radius of ∼42 Å was eluted from the column when the loaded sample contained 7 μm VC and either 21, 14, or 7 μm G-actin. Similarly, the VCA·actin complex eluted with a Stokes radius of ∼47 Å. The Stokes radii of these complexes are larger than the Stokes radius of actin (30 Å) or of known 1:1 WH2·ATP·G-actin complexes obtained with Cobl constructs of 8–8.9 kDa containing a single WH2 domain (34–35 Å), indicating that two actin molecules are bound to V, VC, and VCA. This stoichiometry was confirmed by densitometric analysis of the peak fractions on SDS-PAGE (Fig. 2B).
The interaction between G-actin and the WH2 domains of N-WASP was then monitored using the fluorescence of AEDANS-actin as a probe. The fluorescence of AEDANS-actin was decreased by 39 ± 2% upon binding either V or VC at low ionic strength (G-buffer), as was observed upon binding of thymosin β4 (45), the WH2 domains of Spire (46) and Cordon-Bleu (47). This result is consistent with the similar binding of the N-terminal amphipathic α-helix of β-thymosin and WH2 domains to the shear zone between actin subdomains 1 and 3 at the barbed face of the actin monomer. Identical fluorescence titration curves were obtained with V and VC (Fig. 2C). The binding was tight enough to exclude the binding of a single actin molecule. Actually all data points fall well above the titration curve expected for a tight 1:1 complex, and up to 80% of the saturation level are on a straight line corresponding to a 1:2 V·actin complex. Therefore, actin binds to each of the two WH2 domains of V and VC in a VA2 complex, consistent with the gel filtration results. The superimposable binding curves obtained with V and VC indicate that in VC the C region does not bind a third G-actin molecule. In contrast, the isolated C region of human Scar has been reported to bind G-actin (33).
The stable fluorescence plateau reached at saturation of AEDANS-actin by V or VC suggests that the fluorescence of AEDANS-actin is quenched identically in possible 1:1 and 1:2 complexes of V or VC with actin. Therefore, the anisotropy of fluorescence of AEDANS-actin, which is expected to differ in VA and VA2 complexes due to their different sizes, was used to provide complementary information on the distribution of actin in various complexes. Fluorescence anisotropy titrations of AEDANS-actin by either V or VC show a biphasic shape (Fig. 2D). Upon addition of increasing amounts of V or VC, the anisotropy of AEDANS-actin first increased from 0.100 to 0.115 ± 0.001 and then decreased to a final lower value of 0.106 ± 0.001. These data indicate that a large VA2 complex, characterized by a high anisotropy value (r = 0.117), is formed at V/actin ratios lower than 1:2, although when V or VC are in excess over G-actin, the smaller 1:1 VA complex (r = 0.106), predominates. Data in Fig. 2, C and D, were analyzed as described in under “Experimental Procedures” (Equations 3–10), using the following simple (sequential) binding scheme which implicitly assumes that actin binds identically (K1) to either WH2 domain in VA complex and with a lower affinity (K2) to the available WH2 domain in VA, giving VA2. (Note that three equilibrium dissociation constants would be required if a square isoenergetic binding scheme was used.)
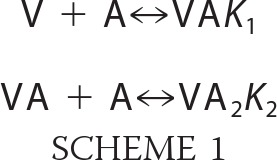 |
The global analysis of the fluorescence intensity and anisotropy binding curves in Fig. 1 was consistent with values of 0.02 ± 0.02 and 0.3 ± 0.1 μm for K1 and K2, respectively, in G-buffer. Note that these two values are low enough to account for the apparent cooperative binding recorded in gel filtration experiments performed at higher protein concentrations. The distribution of actin in VA and VA2 complexes as a function of the total concentration of V is also shown in Fig. 2, C and D. The curve in Fig. 2D was calculated using the values of K1 and K2 derived from Fig. 2C, and values of 0.117 and 0.106 for the anisotropies of VA2 and VA complexes. The generated curve fits reasonably well the experimental data, which show appreciable scatter.
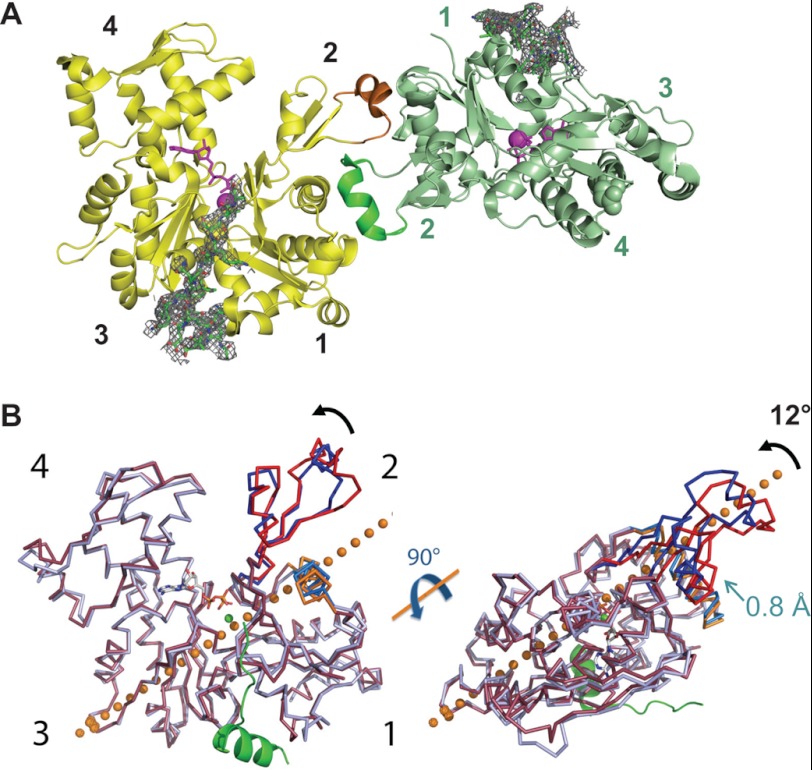
FIGURE 3.
Crystal structure of the 1:1 VC·actin complex. A, representation of the Ca-ATP·G-actin·N-WASP WH2 complex asymmetric unit. The actin molecules A and B of the asymmetric unit (yellow and pale green) are related by a noncrystallographic 2-fold axis, nearly normal to the figure. The D-loops (green and brick) are folded in α-helices stabilized by extensive contacts with the symmetric molecule. VC peptides (ball and stick) are shown in 2Fo − Fc electron density map (1,2 σ). B, Ca-ADP·G-actin·rhodamine structure (1J6Z, blue) is superimposed onto the molecule A (red) of the asymmetric unit (root mean square fit on the 5–31 and 73–372 main chain). A ribbon representation of the VC peptide (green) is shown. As compared with ADP-actin, the subdomain 2 rotates forward by 12° around the hinge axis (orange spheres). The α-helix Asn-78 to Arg-95 of subdomain 3 is translated by 0.8 Å along its axis (orange and marine blue).
To evaluate the robustness of the relative values of K1 and K2, the fit to the anisotropy curves was performed assuming different values of the anisotropy of VA and VA2. A good fit can also be obtained with K1 = 0.08 μm and K2 = 0.3 μm, imposing anisotropy values of 0.106 and 0.125 for VA and VA2 respectively. Overall the two sets of values are in reasonable agreement. As a matter of comparison, anisotropy values of 0.157 and 0.125 were found, respectively, for complexes of AEDANS·actin with Spire, in which four actins are bound to the four WH2 repeats (46) and with a fragment of Cordon-Bleu, in which two actins are bound to two WH2 repeats (47).
Identical titration curves and values of binding constants were obtained under physiological ionic conditions (MgATP-actin in 0.1 m KCl, 1 mm MgCl2, and 0.2 mm EGTA). Hence, within experimental error, the VA and VA2 complexes are as stable in polymerization buffer as in G buffer. The presence of 25 μm latrunculin A only very slightly decreased the stability of the complexes (data not shown), as observed previously on Spire·actin complex (46).
The fluorescence of AEDANS-actin was quenched by 37 ± 2% upon binding VCA, but the titration curves were very different from those obtained with V and VC and showed no straightforward evidence for a 2:1 molar ratio in the complex, due to an overall low affinity of actin to VCA. (Fig. 2C). No overshoot in fluorescence anisotropy of AEDANS-actin was detectable either in the range of concentrations of VCA lower than AEDANS-actin. Consistent with fluorescence intensity measurements, the anisotropy increased slightly upon addition of VCA from 0.100 ± 0.001 up to a single higher limit value of 0.105 ± 0.002, indicating a small change in mobility of actin. The resulting large scatter of the data did not allow us to derive a significant equilibrium constant (data not shown). Either actin binds more weakly to the two WH2 domains when the acidic region was present or actin binds to one WH2 and its binding to the second WH2 on VCA was too weak to be detected. In conclusion, the additional C-terminal acidic region (A) of N-WASP has a strong influence on the affinity of actin for the WH2 domains; however, a major 1:2 complex is formed with unlabeled actin at concentrations above micromolar (Fig. 2A). The actin binding modes of the WH2 domains of V, VC, and VCA of N-WASP are summarized in Fig. 2E.
Crystal Structure of VC·Actin Complex
Crystals of Ca-ATP·G-actin·N-WASP peptide complexes were obtained only with a 2-fold excess of peptide over G-actin, to avoid actin polymerization induced by the high concentrations of protein and salts used in crystallization solutions. Both V and VC fragments of N-WASP co-crystallized in a 1:1 complex with Ca-ATP·G-actin in identical conditions and gave isomorphous crystals, but only the VC·actin complex yielded data of high enough quality at 3.2 Å resolution. The crystals were merohedrally twinned; nevertheless, the structure was solved by molecular replacement starting from the crystal structure of actin·DNase I·WASP WH2 (PDB code 2A3Z) and was refined using CNS with the hemihedral twinning procedure (twinned Rfact = 0.275 and twinned Rfree = 0.331) (Table 1).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P61 |
| Wavelength | 0.97551 Å |
| Resolution | 48.3 to 3.2 Å (3.37 to 3.20 Å)a |
| Unit cell parameters | a = b = 136.24 c = 205.41 Å |
| Unique reflections | 35,056 |
| Redundancy | 3.5 (2.3) |
| Completeness | 98.5% (95.0%) |
| Average I/σ(I) | 8.8 (2.3) |
| Rmergeb | 12.0% (40.0%) |
| Yeates 〈|L|〉 and 〈|L2|〉c | 0.458 and 0.284 |
| Estimated Twin fraction (α) | 0.432 |
| Refinement | |
| Resolution range | 20.0 to 3.2 Å (3.31–3.20 Å) |
| Unique reflections | 32,407 (2,338) |
| No. of protein atoms | 6070 |
| No. of heteroatoms | 86 |
| No. of solvent atoms | 0 |
| Root mean square bond deviation | 0.011 Å |
| Root mean square angle deviation | 1.614° |
| Twinned Rworkd | 27.5% (29.6%) |
| Twinned Rfreee | 33.1% (32.5%) |
| Mean temperature factor | 28.6 Å2 |
| Ramachandran plot | |
| Favored | 81.6% |
| Allowed | 18.0% |
| Generous | 0.3% |
| Disallowed | 0.1% |
a Figures in parentheses apply to the highest resolution shell.
b Rmerge = ΣhΣi |I(h,i) − 〈I(h)〉|/ΣhΣiI(h,i), where I(h,i) is the intensity of the ith observation of reflection h, and 〈I(h)〉 is the average intensity of redundant measurements of reflection h.
c Yeates 〈|L|〉untwinned = 0.500 and perfectly twinned = 0.375 and 〈L2〉 untwinned = 0.333 and perfectly twinned = 0.200.
d Twinned Rwork = Σh|Fobs −((1 − α)Icalc(h1) + α Icalc(h2))1/2|/Σh Fobs for F ≥ 3σ(F) with I(h1) and I(h2) reflections related by the twinning operation but not by crystallographic symmetry.
e Twinned Rfree = Σh|Fobs − ((1 − α)Icalc(h1) + α Icalc(h2))1/2|/Σh Fobs for F ≥3σ(F) for 5% of the reserved reflections.
The VC·actin complex crystallizes with two molecules per asymmetric unit (Fig. 3A), which are related to each other by a true 2-fold noncrystallographic axis. The main surface contact lies near the D-loop, which is buried at the contact of residues 87–99 and 127 on subdomain I of the second molecule of the asymmetric unit. The total accessible surface area (ASA) buried at the interface between the two actin molecules of the asymmetric unit is 1835 Å2 for the two actin molecules together (48). This value lies within the limits of the “standard size” (1600 ± 400 Å2) of total buried ASA observed in protein-protein recognition sites (49). Qualitatively, the buried ASA includes hydrogen bonds and hydrophobic interactions with two hydrophobic residues Val-45 and Tyr-91, which are almost entirely buried in the interface. For comparison, the other ASA buried between symmetry-related molecules in the crystal packing are composed between 280 and 665 Å2 for each pair contact.
The N-WASP VC peptide is tightly bound to actin. Whether actin binds the first or the second of the two consecutive WH2 domains of N-WASP, which share 13 identical residues among 18, cannot be discriminated on the 3.2 Å electron density map. However, the distance between the N and C termini of the nearest symmetry-related WH2 is larger than 48 Å, which excludes the possibility that the two WH2 bound to the two actins in the asymmetric unit represent the consecutive (10 residue apart) WH2 domains of a single VC. Hence, the crystal structure corresponds to VC·actin 1:1 complex. The sequence of the second WH2 domain (residues 433–451) fits the noncrystallographic symmetry averaged electronic density better for the long side chain of residues Arg-434, Asp-435, and Ile-445 and for the short side chain of residues Ser-449 and Ala-451 than the corresponding sequence 405–423 in the first WH2 domain. The sequence 433–451 was therefore refined on both molecules A and B, leaving the connector (C) domain and the first WH2 domain disordered.
In the VC·actin complex, VC shares the fold of WH2 domains of WASP/WIP/WAVE/MIM in their complexes with actin·DNase I (42, 50); the N-terminal part of WH2 forms an amphiphilic α-helix with its hydrophobic side buried in the groove between subdomains 1 and 3 of actin. The C terminus moiety (Gly-444–Ala-451) is in an extended conformation. In the LKKT-related sequence, Lys-445 is in the vicinity of but does not interact directly with Asp-24 of actin. Similarly, Lys-445 and -446 in WASP and at homologous positions in WIP and MIM do not interact with actin directly. A total ASA of 1640 Å2 is buried at the contact between actin and the second WH2 domain. The calculated free energy of solvation is −12.4 kcal/mol (48). The hydrophobic residues Leu-434, Leu-435, Ile-438, and Leu-444 are entirely buried, and Ile-442 and Val-447 are partially buried.
Two main differences with the structure of actin·DNase I·WASP WH2 complex (Protein Data Bank code 2A3Z) are noticed. First, the actin D-loop (residues 41–54) adopts an α-helix conformation in the VC·actin complex, as observed in the ADP-TMR-actin structure (Protein Data Bank code 1J6Z) (51). Because ATP is seen bound to actin in the VC·actin complex, we conclude that the D-loop can adopt several conformations (α-helix, antiparallel β-turn, extended, or unstructured loop), independently of the nature of the bound nucleotide, as pointed out by Rould et al. (52). The helix of the D-loop is further stabilized by contacts with the second molecule of the asymmetric unit, suggesting that the conformation of the D-loop is governed by crystal packing. As compared with the ADP-TMR-actin structure, however, in VC·actin the residues of the subdomain 2 are rotated by 12° in a rigid-body motion (Fig. 3B) (DynDom) (53). The rotation of subdomain 2 thus appears independent of the D-loop secondary structure and of the neighboring contacts and could correlate with Pi release (51), which would promote a shift of the Asn-12–Val-17 hairpin toward the γ-phosphate position in ADP-actin as compared with the present ATP-bound actin·WH2 structure. The main consequences are a shift of the hydrogen-bonded main chain Val-30,-Ser-33 and the reorganization of the “sensor loop” Ile-71–His-73. The subdomain 2 (Ser-33–Glu-72) rotates around the axis represented in Fig. 3B, and the α-helix Asn-78–Arg-95 is translated by 0.8 Å along its axis toward the N terminus. The second difference between VC·actin and actin-DNase I·WASP WH2 concerns the structure of the 10 C-terminal residues of actin, which are disordered in actin-DNase I·WASP WH2, although they appear clearly defined in the Fourier difference map of the VC·actin structure. The refined structure of the C terminus converged to the amphiphilic helix observed in the majority of actin structures.
Functional Analysis of the V and VC Regions of N-WASp in Actin Assembly
The complexes of V, VC, and VCA with G-actin behaved very similarly in kinetic and steady state measurements of actin assembly in F-buffer. Their functional properties were investigated in detail because they are likely involved in the overall mechanism of filament branching with Arp2/3 complex.
Preliminary assays showed that like profilin both V and VC inhibited spontaneous actin assembly even at substoichiometric amounts, conditions favoring formation of VA2 complexes. Hence, in contrast with the tandem repeats of WH2 domains of JMY (17), the two adjacent WH2 domains of N-WASP bind two actins but do not nucleate actin assembly.
The effect of V and VC on F-actin assembled at steady state was then analyzed (Fig. 4A). Superimposable curves were obtained with V and VC. The slopes of the linear declines in the amount of F-actin as a function of the total amount of V or VC, indicating sequestration of G-actin, were much steeper when barbed ends were capped, whereas insignificant depolymerization was observed when barbed ends were free. In the simple case of a canonical sequestering agent S making a nonpolymerizable 1:1 complex with G-actin, like Tβ4, the relative values of these slopes are determined by the values of the critical concentrations for actin assembly in filaments (AC), with free and capped barbed ends, respectively, and the value of the equilibrium dissociation constant KS for the complex SA of actin with the sequestering agent, as described by Equation 11,
The situation is more complex with V and VC because both make VA and VA2 complexes. The possible sequestration of actin in these complexes therefore was expressed as shown in Equations 12–14.
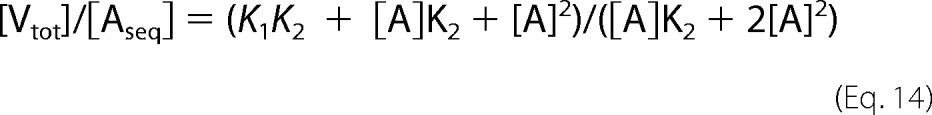 |
The concentration of sequestered actin at a given total concentration of V or VC, [Aseq], was experimentally determined when barbed ends were free or gelsolin-capped and was incorporated in Equation 14, using values of [A] equal to the values of the critical concentrations at either the barbed end (0.1 μm) or the pointed end (0.6 μm). This procedure generated a unique set of values for K1 (3.4 μm) and K2 (0.18 μm) that could satisfy the data at both barbed and pointed ends in Fig. 4A in terms of sequestration of actin in VA and VA2 complexes. These values of K1 and K2 are significantly higher than and incompatible with those derived from titration curves in G- or F-buffers (0.02 and 0.3 μm, respectively). Thus, a pure sequestration activity of V and VC is inconsistent with the data. One should note that VA and VA2 are not equally represented at steady state when barbed ends are free or capped, because the ratio [VA2]/[VA] = [A]/K2 depends on the critical concentration [A]. Using values of 0.1 and 0.6 μm for [A] at steady state when barbed ends are free and capped, respectively, one finds that VA2 is 6-fold more abundant than VA when barbed ends are capped than when barbed ends are free. In summary, steady state measurements strongly suggest that like profilin, V and VC do not behave as bona fide sequesterers when barbed ends are free.
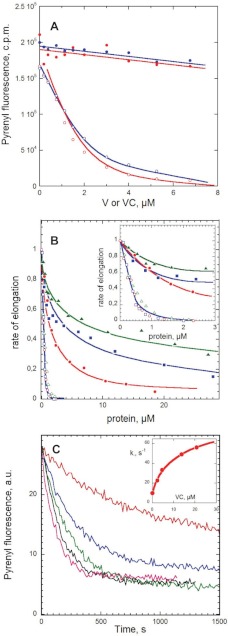
FIGURE 4.
Functional properties of V, VC, and VCA in actin assembly. A, effects of V and VC on the amount of F-actin assembled at steady state. Actin (10% pyrenyl-labeled, 3.8 μm) was polymerized with (open symbols) or without (closed symbols) 13 nm gelsolin and incubated overnight in the presence of V (blue curves) or VC (red curves) at the indicated concentrations. Note that when barbed ends are capped by gelsolin, F-actin is totally depolymerized by addition of less than 1 molar eq V or VC, consistent with sequestration of actin in high affinity VA2 complexes. In contrast, V and VC fail to significantly depolymerize F-actin when barbed ends are free. B, effects of V, VC, and VCA on barbed and pointed end growth rates. Initial rates of barbed end (closed symbols) and pointed end (open symbols) growth was measured in the presence of 2.5 μm MgATP-G-actin and the indicated amounts of V (blue curves), VC (red curves), and VCA (green curves). Normalized values of the growth rates are shown. Inset, expanded graph in the 0–3 μm peptide range. C, increase in barbed end depolymerization rate by VC. Shown are time courses of depolymerization of pre-assembled F-actin (2.5 μm) induced by 40-fold dilution in polymerization buffer containing VC at the following concentrations: 0 (red), 2.3 μm (blue), 4.6 μm (green), 13.8 μm (black), and 20.8 μm (magenta). Inset, plot of the initial depolymerization rate constant versus VC concentration.
To understand how V and VC affect barbed end and pointed end assembly dynamics, the effect of V and VC on the rate of barbed end or pointed end growth was examined (Fig. 4B). Both V and VC inhibited pointed end growth very efficiently. Total inhibition was reached at a substoichiometric molar ratio of V or VC with respect to G-actin, consistent with the sequestration of G-actin in a tight predominant VA2 complex. The predominance of VA2 in this concentration range was confirmed by calculating the distribution of VA and VA2 at different total concentrations of V or VC, using values of binding constants derived from the titration assays.
The effects of V and VC on barbed end growth rate were more complex. The rate of barbed end growth at 2.5 μm MgATP-G-actin decreased in a biphasic fashion with increasing concentrations of V or VC. A steep initial 40% decrease in rate was observed in the range of low concentrations of V or VC (0 to 3 μm) in which total inhibition of growth at pointed ends was recorded (Fig. 4B, inset). The partial inhibition of barbed end growth was followed, at a higher concentration of V or VC (5–50 μm), by a much less steep decrease in rate. These data indicate that VA and VA2 complexes can productively associate with barbed ends like profilin-actin; however, our data do not allow accurate determination of the kinetic parameters for barbed end assembly from VA and VA2 specifically. The slower decline in rate at a high concentration of V or VC, although the calculated concentration of VA complex remains constant, suggests that V and VC may also cap barbed ends with very low affinity to prevent their growth fed by VA. The same behavior at barbed ends and the same inhibition at very high concentrations were observed with VCA (green curve in Fig. 4B). The experimental data were well accounted for by a model (Equation 10 under “Experimental Procedures”) in which VA and VA2 support barbed end growth with on rate constants (k+B) of 5.5, 4, and 6.5 μm−1·s−1 for V, VC, and VCA, respectively, and V, VC, and VCA cap barbed ends with low affinity (KF = 15, 5, and 20 μm, respectively).
The evidence for barbed end growth from the complexes of actin with V, VC, and VCA implies that these WH2 domains interact with barbed ends, at least transiently. To further address this point, the effect of V, VC, and VCA on dilution-induced depolymerization of actin filaments was assayed (Fig. 4C). For V, VC, and VCA, all three increased the rate of barbed end dissociation up to a maximum of 7-fold, in a concentration-dependent behavior consistent with V and VC binding to barbed ends with a Kd of 5 ± 2 μm. In contrast, neither V nor VC affected the rate of pointed end depolymerization (data not shown). Hence, the increase in barbed end depolymerization rate is not due to severing of the filaments, but it results from specific binding of the WH2 domains of V or VC to barbed ends, causing faster release of the V·actin complex than actin. A similar effect has been reported for profilin (54, 55) and for the WH2 domains of Cordon-Bleu, which exhibit functional similarity to profilin (47).
Analysis of the Complexes Formed between VCA, Actin, and Arp2/3 Complex
Branching of filaments by VCA occurs by association of a branching complex of VCA with actin and Arp2/3 complex with a filament. The Arp2/3/VCA/actin stoichiometry of the branching complex (in which the shortened notation Arp2/3 refers to Arp2/3 complex) is not well known and has been debated recently (25–27). In small angle x-ray scattering solution studies of Arp2/3 in complex with a covalent (N-WASP)VCA·actin 1:1 complex, the V region binds actin; the C region binds the barbed face of Arp2 (25); and the A region binds the small subunit p21 of the Arp2/3 complex (56). However, recent reports proposed that Arp2/3 complex was active when bound to two WASP VCA domains. One study (26) showed binding of two Alexa 488-labeled VCAs to Arp2/3 complex, and the other study (27) demonstrated that the Arp2/3 complex made a 1:2 complex with two molecules of the isolated CA domain. Our data show that the WH2 domains of N-WASP interact with G-actin with an affinity that appears weakened by the acidic (A) C-terminal region, in the isolated VCA state. Upon interaction with Arp2/3 complex, the contacts made by the acidic region with subunits of the Arp2/3 complex may neutralize this region and restore enhanced strength of binding of the two WH2 domains of VCA in an unknown fashion. In summary, if VCA can bind two actin molecules and if two VCA molecules bind to Arp2/3, the Arp2/3·VCA·actin may potentially display up to 1:2:4 stoichiometry.
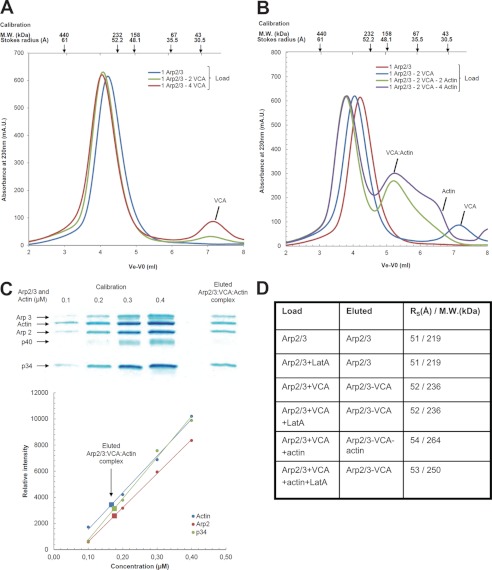
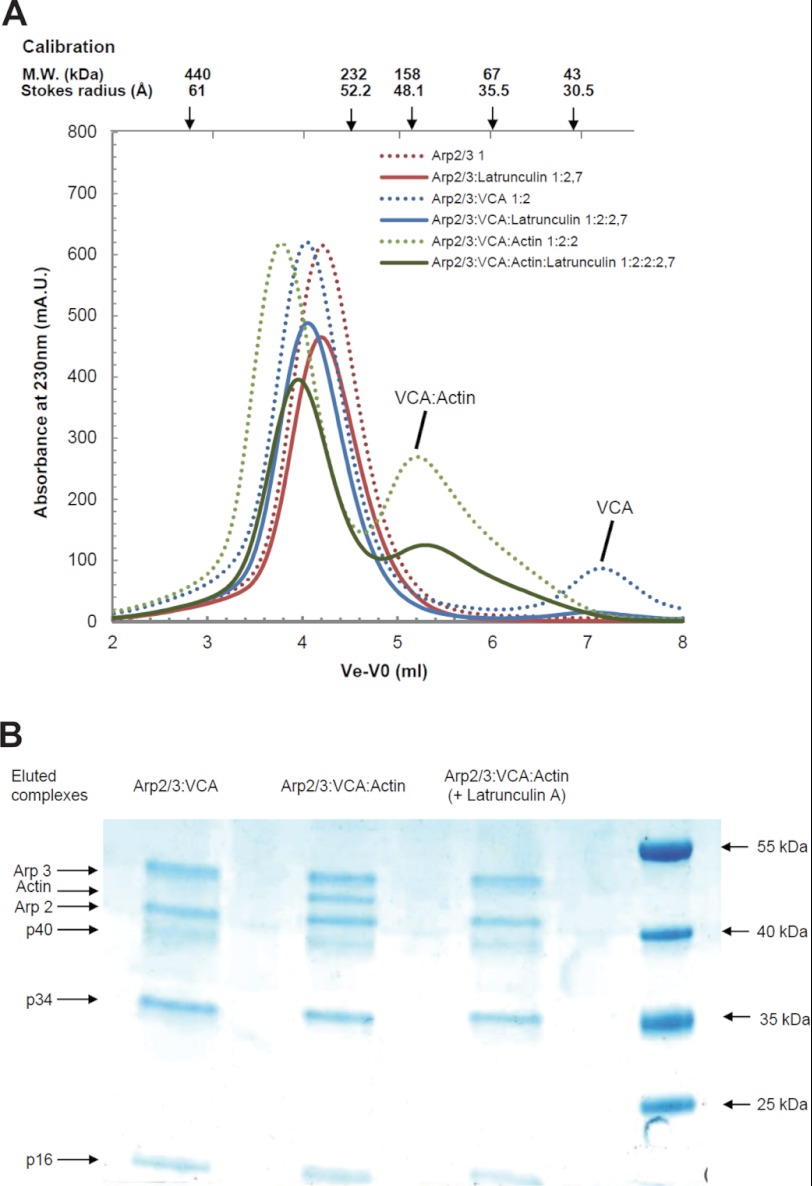
The noncovalent complexes of Arp2/3 complex with unlabeled VCA and actin were characterized by size-exclusion chromatography (Fig. 5). The Stokes radius of the Arp2/3 complex increased from 51 Å to a maximum of 52 Å upon addition of VCA, at various VCA·Arp2/3 complex molar ratios in the load. A peak corresponding to free VCA appeared as soon as the VCA·Arp2/3 complex molar ratio in the load was higher than 1, indicating that a single 1:1 VCA·Arp2/3 complex species was formed (Fig. 5A). Further addition of G-actin in increasing amounts in the load led to a single faster migrating species of Stokes radius at 54 Å, consistent with a 1:1:1 VCA·actin·Arp2/3 complex. The theoretical molecular mass of 266 kDa, assuming a globular shape, is reasonably consistent with the calculated molecular mass of 275 kDa for a 1:1:1 complex. The molar ratio was confirmed both by SDS-PAGE analysis of the content in actin and the Arp2/3 complex in the fractions of the eluted complex, and by the appearance of an actin peak at actin·Arp2/3 complex molar ratios higher than 1 in the loaded samples (Fig. 5, B–D). No evidence was observed for a second binding site of VCA to Arp2/3 complex in these experiments nor for binding of two actins to the V moiety of VCA when the CA region was engaged in contacts with Arp2/3 complex. When the chromatography of samples of Arp2/3, actin and VCA was performed in the presence of latrunculin A, the elution profiles of Arp2/3 complex and of VCA·Arp2/3 complex were unchanged, but a complex could no longer be detected between actin and VCA·Arp2/3 complex (Fig. 6). Because latrunculin A does not prevent binding of VCA to actin, this result indicates that the pointed face of VCA-bound actin in the ternary complex interacts with components of Arp2/3 complex in a manner incompatible with latrunculin A binding.
FIGURE 5.
Gel filtration analysis of the complexes of Arp2/3 complex with VCA and actin. A, samples containing Arp2/3 complex alone (3 μm) or supplemented with VCA at indicated molar ratios were loaded and eluted on a Superdex 200 10 × 300 column at physiological ionic strength. B, elution patterns of samples containing Arp2/3 complex alone or supplemented with VCA and actin at indicated ratios. Conditions as in A. C, SDS-PAGE analysis of the eluted VCA·actin·Arp2/3 ternary complex and standards consisting of pre-mixed actin and Arp2/3 in a 1:1 molar ratio and at the indicated concentrations, from which the molar ratio of actin Arp2 and p34 in the eluted complex was derived. Standards and samples were electrophoresed on the same gel to establish the molar composition of VCA·actin·Arp2/3 complex rigorously. D, hydrodynamic radius and apparent molecular weight of the eluted complexes derived from the size-exclusion chromatography calibration with known protein standards. Stokes radius values are within 10% standard deviation.
FIGURE 6.
Gel filtration analysis of the complexes of Arp2/3 complex with VCA and actin with latrunculin A. A, samples containing Arp2/3 complex alone or supplemented with VCA and actin at the indicated ratios and with latrunculin A were loaded and eluted on a Superdex 200 10 × 300 column at physiological ionic strength. The dotted lines refer to elution patterns observed without latrunculin A (reproduced from Fig. 5). B, SDS-PAGE analysis of the VCA·Arp2/3 complex and VCA·actin·Arp2/3 ternary complex eluted with latrunculin A. (Note absence of actin in eluted complex.) For the electrophoretic positions of actin, some subunits of the Arp2/3 complex and protein molecular weight standards are indicated.
DISCUSSION
The analysis of the structural/functional interaction of the two adjacent WH2 domains of N-WASP in the V, VC, and VCA domains and the characterization of the ternary VCA·actin·Arp2/3 complex provides new insight into the multifunctionality of WH2 repeats and its use in the filament branching reaction.
Each of the two WH2 domains of N-WASP binds one G-actin molecule, with an affinity that is lowered by the presence of the acidic C-terminal extension (VCA). Both VA and VA2 complexes are formed from V, VC, and VCA, with affinities in the 107 m−1 range, and are observed in solution in proportions depending on the concentrations of actin and V or VC or VCA. The fact that the affinity of actin is lowered by the acidic region A in VCA may have an interesting physiological implication, a weak affinity facilitating the dissociation of actin from VCA in the branching process.
In the crystal structure of VC·actin 1:1 complex, two VC-actin molecules form the asymmetric unit. The D-loop adopts an α-helix conformation and forms the major part of the interface between the two actins. This contact differs from the actin-actin contacts in the filament. The apolar buried surface, calculated according to Bahadur and Chakrabarti (57) was 989 Å2, and the fraction of buried atoms was 0.366. Despite the low free energy of solvation, estimated to −4.4 kcal/mol, and of the low resolution and high value of the B factor of residues in the D loop, these figures are suggestive of possible functional interactions (48, 57). Whether similar contacts might occur in nature, for instance in the structural organization of the branch junction, in which VCA transiently binds G-actin, the Arp2 and Arp3 subunits of the Arp2/3 complex and the mother filament, is an issue raised by these data.
The present results reinforce the view that the protein environment of the WH2 domains plays a determinant role in their multifunctionality. The two WH2 domains of N-WASP are not nucleators of actin filaments, even when they are present in polymerizing actin solutions at concentrations such that VA2 is the dominant complex. Therefore, two adjacent actin-bound WH2 domains do not necessarily generate a nucleation activity. This conclusion is in full agreement with the observation that the two adjacent WH2 domains of Cordon-Bleu do not nucleate actin in the absence of the N-terminally adjacent lysine-rich K region (47). Conversely, filament nucleation by WH2 domains does not require two adjacent WH2 domains, because the first WH2 domain of Cordon-Bleu, flanked by the K region nucleates as well as two consecutive domains. Similarly, the C-terminal dimerization domain of the pathogen protein VopL weakly nucleates actin, and nucleation is enhanced by additional WH2 domains, which by themselves do not nucleate (58, 59). A general mechanism of “facilitated spontaneous nucleation” of actin filaments by WH2 domains, differing from the initial template mechanism (60–62), has been proposed, in which binding of a WH2 domain to an actin monomer enhances its electrostatic interaction with another actin monomer to form a pre-nucleus actin dimer (47, 63). Thus, WH2 domains that harbor appropriate sequence elements to decrease the electronegativity of G-actin lower the free energy of nucleation associated with elementary steps of the regular actin nucleation pathway. The WH2 domains of N-WASP do not harbor this peculiar property; therefore, in the absence of the Arp2/3 complex they do not nucleate actin either in isolated state or in the context of the activated full-length N-WASP protein in which the VCA C-terminal domain is exposed. Binding of the Arp2/3 complex is not sufficient to confer a nucleating activity to VCA, because the association with a mother filament is required for nucleation of a daughter filament (64, 65). However, binding of putative ligands to N-WASP or to WAVE might confer an actin-nucleating activity to the WH2 domain by modifying its electrostatic environment. The nucleating activity then might synergize with the Arp2/3 complex-dependent branching activity and enhance the potency of the WASP protein to generate new barbed ends.
In contrast with other WH2 tandem repeats like Spire or Cordon-Bleu (46, 47), V and VC do not sever filaments. The absence of severing activity correlates with the absence of nucleating activity.
V, VC, and VCA share two properties with profilin and a few other WH2/β-thymosin domains. First, their complexes with MgATP-G-actin participate in barbed end growth (Table 2). Second, in dilution-induced depolymerization assays, in which the core ADP-F-actin subunits dissociate endwise, their interaction with the barbed ends enhances by 7-fold the dissociation of MgADP-F-actin subunits. These data bring growing evidence for a variety of functional interactions of WH2 domains with barbed ends, including the capture of barbed ends by the WH2 domain of N-WASP (66), the capping of barbed ends by Spire WH2 domains (46, 67), and the tracking of barbed ends by the WH2 domain of VASP (68, 69). Table 3 summarizes the diverse multifunctionality patterns reported on various WH2 repeats. Domain swapping and engineering of chimeric or mutated WH2 repeats will allow us to design customized WH2 domains and to modulate at will the overall biological function of the proteins in which they are inserted.
TABLE 2.
Characteristic parameters of the V, VC and VCA fragments of N-WASP for G-actin binding and filament barbed end assembly
TABLE 3.
Compared multifunctionality of various WH2 domains in single module or tandem repeats in actin assembly
Symbols used are as follows: +, −, and ? indicate that the property has been observed, not observed, or not tested, respectively.
| Protein (WH2 no.) | G-actin (ATP, ADP) sequestration | Barbed end elongation | Filament nucleation | Filament severing | Barbed end capping | Barbed end tracking |
|---|---|---|---|---|---|---|
| N-WASP (2) | − | + | − | − | − | + |
| ActA (1) | − | + | − | − | − | − |
| VASP (1) | − | + | + | − | − | + |
| INF2 (1 + FH2) | − | ? | + | + | ? | ? |
| Spire (4) | + (ATP, ADP) | − | + | + | + | − |
| Cordon-Bleu (3) | + (ADP) | + | + | + | − | − |
Filament branching using the Arp2/3 complex is a property shared by all WASP family proteins despite their variable number of WH2 domains, indicating that one actin bound to one WH2 domain interacts with the Arp2/3 complex in a functionally conserved fashion in the branching process. In the (N-WASP)VCA·Arp2/3·actin complex, G-actin is probably bound to the second WH2 domain of N-WASP, in a position homolog to WASP WH2. A supplementary WH2 domain of N-WASP was reported to increase the efficiency of branching (15). Assuming that the WH2 domain adjacent to CA binds actin in VCA·Arp2/3·actin complex, binding of the first (N-terminal) WH2 domain of (N-WASP)VCA to another actin molecule in the branching process might account for the enhanced branching efficiency. Such a barbed end tracking function, comforted by other data (66), might participate in the protrusive activity of dendritic actin arrays.
Only a 1:1 complex of VCA with the Arp2/3 complex is isolated using unmodified proteins in a wide range of molar ratios. This result contrasts with the reported association of two Alexa 488-labeled VCA molecules to the Arp2/3 complex (26) and with the binding of fission yeast CA to two sites on the Arp2/3 complex, albeit with very different affinities (27). The present data do not favor the view that branching of filaments requires the binding of two WASP molecules to the Arp2/3 complex. The difference between the various results may be due to the labeling of VCA or to the difference in reactivity of CA versus VCA and of yeast versus bovine proteins. However, the present evidence for single noncovalent 1:1 VCA·Arp2/3 complex and 1:1:1 VCA·actin·Arp2/3 complexes is in agreement with the small angle x-ray scattering solution structure of VCA·actin·Arp2/3 in which VCA and actin were covalently linked (25). Although two actin molecules bind to the VCA domain of N-WASP irrespective of the presence of latrunculin A, actin does not bind to the 1:1 VCA·Arp2/3 complex in the presence of latrunculin A, suggesting that new latrunculin-sensitive contacts are established between actin and Arp2 in the ternary branching complex, also consistent with solution studies (25). The fact that one single branching enzyme (VCA) is bound to its substrates Arp2/3 complex and actin simplifies the possible molecular mechanisms for filament branching and the possible binding modes of the VCA·actin·Arp2/3 branching complex with a filament. What minimum size F-actin unit can functionally interact with the VCA·actin·Arp2/3 complex is the next challenging issue.
The atomic coordinates and structure factors (code 2VCP) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- WASP
- Wiskott-Aldrich syndrome protein
- Arp2/3 complex
- actin-related proteins 2 and 3 containing complex
- WH2
- WASP homology 2
- SCAR
- suppressor of c-AMP receptor
- WAVE
- WASP family Verprolin homologous protein
- AEDANS
- acetyl-N′-(5 sulfo-1-naphthyl)ethylenediamine
- PEG
- polyethylene glycol; C and A refer to the connector and acidic regions of N-WASP
- ASA
- accessible surface area.
REFERENCES
- 1. Rottner K., Hänisch J., Campellone K. G. (2010) WASH, WHAMM, and JMY. Regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20, 650–661 [DOI] [PubMed] [Google Scholar]
- 2. Veltman D. M., Insall R. H. (2010) WASP family proteins. Their evolution and its physiological implications. Mol. Biol. Cell 21, 2880–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kollmar M., Lbik D., Enge S. (2012) Evolution of the eukaryotic ARP2/3 activators of the WASP family. WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res. Notes 5, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Svitkina T. M., Borisy G. G. (1999) Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takenawa T., Suetsugu S. (2007) The WASP-WAVE protein network. Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 6. Tsuboi S., Meerloo J. (2007) Wiskott-Aldrich syndrome protein is a key regulator of the phagocytic cup formation in macrophages. J. Biol. Chem. 282, 34194–34203 [DOI] [PubMed] [Google Scholar]
- 7. Huang Y., Burkhardt J. K. (2007) T-cell receptor-dependent actin regulatory mechanisms. J. Cell Sci. 120, 723–730 [DOI] [PubMed] [Google Scholar]
- 8. Wegner A. M., Nebhan C. A., Hu L., Majumdar D., Meier K. M., Weaver A. M., Webb D. J. (2008) N-WASP and the Arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J. Biol. Chem. 283, 15912–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campellone K. G., Webb N. J., Znameroski E. A., Welch M. D. (2008) WHAMM is an Arp2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 134, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Derivery E., Sousa C., Gautier J. J., Lombard B., Loew D., Gautreau A. (2009) The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev. Cell. 17, 712–723 [DOI] [PubMed] [Google Scholar]
- 11. Jia D., Gomez T. S., Metlagel Z., Umetani J., Otwinowski Z., Rosen M. K., Billadeau D. D. (2010) WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci. U.S.A. 107, 10442–10447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suraneni P., Rubinstein B., Unruh J. R., Durnin M., Hanein D., Li R. (2012) The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J. Cell Biol. 197, 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delatour V., Helfer E., Didry D., Lê K. H., Gaucher J. F., Carlier M. F., Romet-Lemonne G. (2008) Arp2/3 controls the motile behavior of N-WASP-functionalized GUVs and modulates N-WASP surface distribution by mediating transient links with actin filaments. Biophys. J. 94, 4890–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Weisswange I., Newsome T. P., Schleich S., Way M. (2009) The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature 458, 87–91 [DOI] [PubMed] [Google Scholar]
- 15. Yamaguchi H., Miki H., Suetsugu S., Ma L., Kirschner M. W., Takenawa T. (2000) Two tandem verprolin homology domains are necessary for a strong activation of Arp2/3 complex-induced actin polymerization and induction of microspike formation by N-WASP. Proc. Natl. Acad. Sci. U.S.A. 91, 12631–12636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zalevsky J., Lempert L., Kranitz H., Mullins R. D. (2001) Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr. Biol. 11, 1903–1913 [DOI] [PubMed] [Google Scholar]
- 17. Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B., Mullins R. D. (2009) p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 11, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higgs H. N., Pollard T. D. (2001) Regulation of actin filament network formation through ARP2/3 complex. Activation by a diverse array of proteins. Annu. Rev. Biochem. 70, 649–676 [DOI] [PubMed] [Google Scholar]
- 19. Wen K. K., Rubenstein P. A. (2005) Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require and Arp2/3 complex activating protein. J. Biol. Chem. 280, 24168–24174 [DOI] [PubMed] [Google Scholar]
- 20. Wiesner S., Helfer E., Didry D., Ducouret G., Lafuma F., Carlier M. F., Pantaloni D. (2003) A biomimetic motility assay provides insight into the mechanism of actin-based motility. J. Cell Biol. 160, 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iwasa J. H., Mullins R. D. (2007) Spatial and temporal relationships between actin filament nucleation, capping, and disassembly. Curr. Biol. 17, 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lai F. P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H. G., Stradal T. E., Dunn G. A., Small J. V., Rottner K. (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rouiller I., Xu X. P., Amann K. J., Egile C., Nickell S., Nicastro D., Li R., Pollard T. D., Volkmann N., Hanein D. (2008) The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beltzner C. C., Pollard T. D. (2008) Pathway of actin filament branch formation by Arp2/3 complex. J. Biol. Chem. 283, 7135–7144 [DOI] [PubMed] [Google Scholar]
- 25. Boczkowska M., Rebowski G., Petoukhov M. V., Hayes D. B., Svergun D. I., Dominguez R. (2008) X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure 16, 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Padrick S. B., Doolittle L. K., Brautigam C. A., King D. S., Rosen M. K. (2011) Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci. U.S.A. 108, E472–E479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ti S. C., Jurgenson C. T., Nolen B. J., Pollard T. D. (2011) Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 108, E463–E471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu X. P., Rouiller I., Slaughter B. D., Egile C., Kim E., Unruh J. R., Fan X., Pollard T. D., Li R., Hanein D., Volkmann N. (2012) Three-dimensional reconstructions of Arp2/3 complex with bound nucleation promoting factors. EMBO J. 31, 236–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dominguez R. (2009) Actin filament nucleation and elongation factors. Structure-function relationships. Crit. Rev. Biochem. Mol. Biol. 44, 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hertzog M., van Heijenoort C., Didry D., Gaudier M., Coutant J., Gigant B., Didelot G., Préat T., Knossow M., Guittet E., Carlier M. F. (2004) The β-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117, 611–623 [DOI] [PubMed] [Google Scholar]
- 31. Renault L., Bugyi B., Carlier M. F. (2008) Spire and Cordon-bleu. Multifunctional regulators of actin dynamics. Trends Cell Biol. 18, 494–504 [DOI] [PubMed] [Google Scholar]
- 32. Carlier M. F., Husson C., Renault L., Didry D. (2011) Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int. Rev. Cell Mol. Biol. 290, 55–85 [DOI] [PubMed] [Google Scholar]
- 33. Kelly A. E., Kranitz H., Dötsch V., Mullins R. D. (2006) Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J. Biol. Chem. 281, 10589–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Casella J. F., Barron-Casella E. A., Torres M. A. (1995) Quantitation of Cap Z in conventional actin preparations and methods for further purification of actin. Cell Motil. Cytoskeleton 30, 164–170 [DOI] [PubMed] [Google Scholar]
- 35. Egile C., Loisel T. P., Laurent V., Li R., Pantaloni D., Sansonetti P. J., Carlier M. F. (1999) Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin-based motility. J. Cell Biol. 146, 1319–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. (1995) How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4, 2411–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leslie A. G., Powell H. R. (2007) in Evolving Methods for Macromolecular Crystallography (Read R. J., Sussman J. L., eds) Vol. 245, pp. 41–45, Springer NATO Science Series, New York [Google Scholar]
- 39. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 40. Padilla J. E., Yeates T. O. (2003) A statistic for local intensity differences. Robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 59, 1124–1130 [DOI] [PubMed] [Google Scholar]
- 41. Navaza J. (1994) AMoRe. An automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 [Google Scholar]
- 42. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR System. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 45. De La Cruz E. M., Ostap E. M., Brundage R. A., Reddy K. S., Sweeney H. L., Safer D. (2000) Thymosin-β(4) changes the conformation and dynamics of actin monomers. Biophys J. 78, 2516–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., Carlier M. F. (2007) Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- 47. Husson C., Renault L., Didry D., Pantaloni D., Carlier M. F. (2011) Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 43, 464–477 [DOI] [PubMed] [Google Scholar]
- 48. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 49. Janin J., Chothia C. (1990) The structure of protein-protein recognition sites. J. Biol. Chem. 265, 16027–16030 [PubMed] [Google Scholar]
- 50. Lee S. H., Kerff F., Chereau D., Ferron F., Klug A., Dominguez R. (2007) Structural basis for the actin-binding function of missing-in-metastasis. Structure 15, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otterbein L. R., Graceffa P., Dominguez R. (2001) The crystal structure of uncomplexed actin in the ADP state. Science 293, 708–711 [DOI] [PubMed] [Google Scholar]
- 52. Rould M. A., Wan Q., Joel P. B., Lowey S., Trybus K. M. (2006) Crystal structures of expressed nonpolymerizable monomeric actin in the ADP and ATP states. J. Biol. Chem. 281, 31909–31919 [DOI] [PubMed] [Google Scholar]
- 53. Hayward S., Berendsen H. J. (1998) Systematic analysis of domain motions in proteins from conformational change. New results on citrate synthase and T4 lysozyme. Proteins 30, 144–154 [PubMed] [Google Scholar]
- 54. Kinosian H. J., Selden L. A., Gershman L. C., Estes J. E. (2002) Actin filament barbed end elongation with nonmuscle MgATP-actin and MgADP-actin in the presence of profilin. Biochemistry 41, 6734–6743 [DOI] [PubMed] [Google Scholar]
- 55. Bubb M. R., Yarmola E. G., Gibson B. G., Southwick F. S. (2003) Depolymerization of actin filaments by profilin. Effects of profilin on capping protein function. J. Biol. Chem. 278, 24629–24635 [DOI] [PubMed] [Google Scholar]
- 56. Kreishman-Deitrick M., Goley E. D., Burdine L., Denison C., Egile C., Li R., Murali N., Kodadek T. J., Welch M. D., Rosen M. K. (2005) NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry 44, 15247–15256 [DOI] [PubMed] [Google Scholar]
- 57. Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2004) A dissection of specific and nonspecific protein-protein interfaces. J. Mol. Biol. 336, 943–955 [DOI] [PubMed] [Google Scholar]
- 58. Yu B., Cheng H. C., Brautigam C. A., Tomchick D. R., Rosen M. K. (2011) Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 18, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Namgoong S., Boczkowska M., Glista M. J., Winkelman J. D., Rebowski G., Kovar D. R., Dominguez R. (2011) Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 18, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. (2005) Drosophila Spire is an actin nucleation factor. Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- 61. Rebowski G., Boczkowska M., Hayes D. B., Guo L., Irving T. C., Dominguez R. (2008) X-ray scattering study of actin polymerization nuclei assembled by tandem W domains. Proc. Natl. Acad. Sci. U.S.A. 105, 10785–10790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qualmann B., Kessels M. M. (2009) New players in actin polymerization. WH2-domain-containing actin nucleators. Trends Cell Biol. 19, 276–285 [DOI] [PubMed] [Google Scholar]
- 63. Carlier M. F. (2011) A new twist in actin filament nucleation. Nat. Struct. Mol. Biol. 18, 967–969 [DOI] [PubMed] [Google Scholar]
- 64. Machesky L. M., Mullins R. D., Higgs H. N., Kaiser D. A., Blanchoin L., May R. C., Hall M. E., Pollard T. D. (1999) Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 96, 3739–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pantaloni D., Boujemaa R., Didry D., Gounon P., Carlier M. F. (2000) The Arp2/3 complex branches filament barbed ends. Functional antagonism with capping proteins. Nat. Cell Biol. 2, 385–391 [DOI] [PubMed] [Google Scholar]
- 66. Co C., Wong D. T., Gierke S., Chang V., Taunton J. (2007) Mechanism of actin network attachment to moving membranes. Barbed end capture by N-WASP WH2 domains. Cell 128, 901–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ito T., Narita A., Hirayama T., Taki M., Iyoshi S., Yamamoto Y., Maéda Y., Oda T. (2011) Human Spire interacts with the barbed end of the actin filament. J. Mol. Biol. 408, 18–25 [DOI] [PubMed] [Google Scholar]
- 68. Hansen S. D., Mullins R. D. (2010) VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191, 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Breitsprecher D., Kiesewetter A. K., Linkner J., Vinzenz M., Stradal T. E., Small J. V., Curth U., Dickinson R. B., Faix J. (2011) Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30, 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gutsche-Perelroizen I., Lepault J., Ott A., Carlier M. F. (1999) Filament assembly from profilin-actin. J. Biol. Chem. 274, 6234–6243 [DOI] [PubMed] [Google Scholar]
- 71. Jégou A., Niedermayer T., Orbán J., Didry D., Lipowsky R., Carlier M. F., Romet-Lemonne G. (2011) Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PloS Biol. e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]