Background: Mammalian complex I spontaneously forms a deactive state when it is not turning over.
Results: Deactive complex I is a Na+/H+ antiporter.
Conclusion: In deactive complex I the ion transfer domain is functionally disconnected from the redox domain; the antiporter-like subunits assert their independent function.
Significance: Na+/H+ exchange by deactive complex I may influence the outcome of ischemia-reperfusion.
Keywords: Bioenergetics, Enzyme Mechanisms, Mitochondria, Proton Transport, Respiratory Chain, Sodium Proton Exchange, Complex I, Mrp Antiporter, NADH:Ubiquinone Oxidoreductase
Abstract
In mitochondria, complex I (NADH:ubiquinone oxidoreductase) uses the redox potential energy from NADH oxidation by ubiquinone to transport protons across the inner membrane, contributing to the proton-motive force. However, in some prokaryotes, complex I may transport sodium ions instead, and three subunits in the membrane domain of complex I are closely related to subunits from the Mrp family of Na+/H+ antiporters. Here, we define the relationship between complex I from Bos taurus heart mitochondria, a close model for the human enzyme, and sodium ion transport across the mitochondrial inner membrane. In accord with current consensus, we exclude the possibility of redox-coupled Na+ transport by B. taurus complex I. Instead, we show that the “deactive” form of complex I, which is formed spontaneously when enzyme turnover is precluded by lack of substrates, is a Na+/H+ antiporter. The antiporter activity is abolished upon reactivation by the addition of substrates and by the complex I inhibitor rotenone. It is specific for Na+ over K+, and it is not exhibited by complex I from the yeast Yarrowia lipolytica, which thus has a less extensive deactive transition. We propose that the functional connection between the redox and transporter modules of complex I is broken in the deactive state, allowing the transport module to assert its independent properties. The deactive state of complex I is formed during hypoxia, when respiratory chain turnover is slowed, and may contribute to determining the outcome of ischemia-reperfusion injury.
Introduction
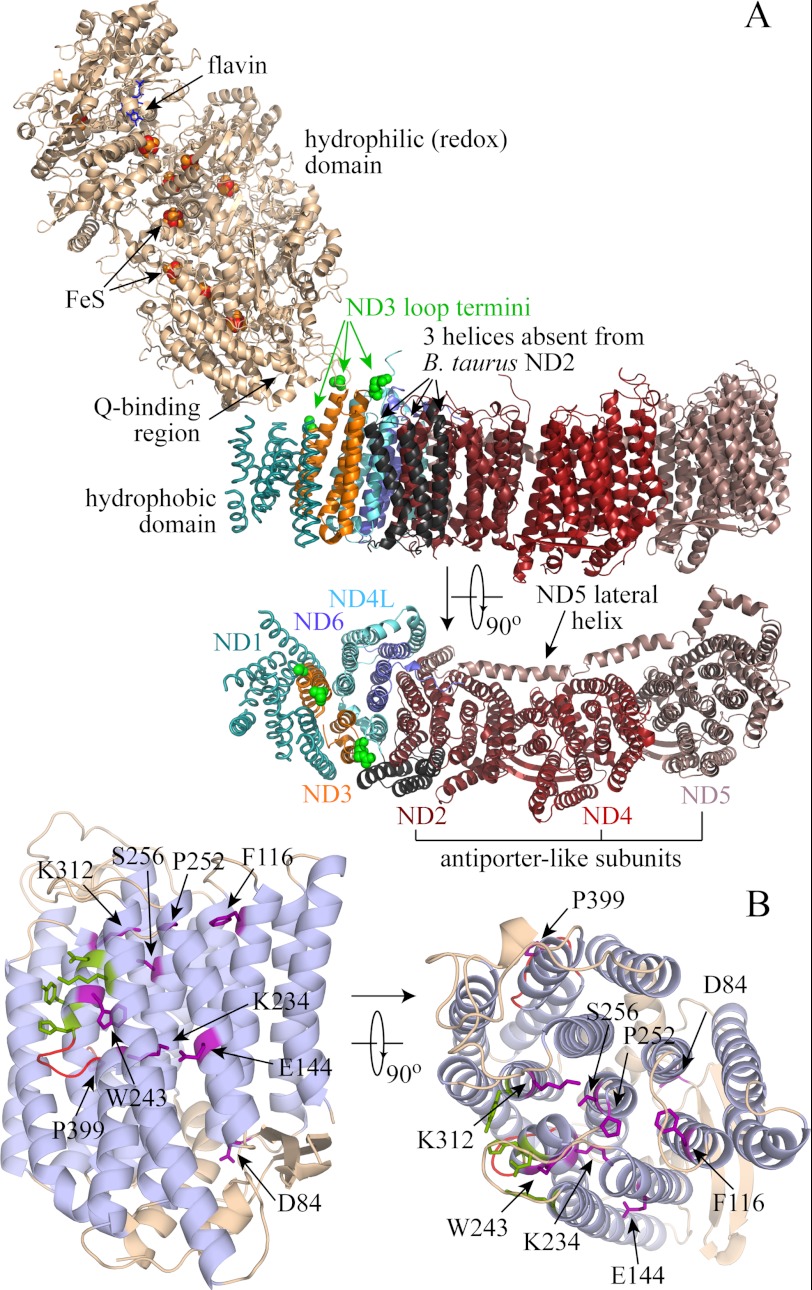
In mitochondria, complex I (NADH:ubiquinone oxidoreductase) (1) uses potential energy from the oxidation of NADH by ubiquinone (ΔE) to translocate protons across the mitochondrial inner membrane, contributing to the proton-motive force (Δp) that is used for ATP synthesis and transport processes. Complex I has an unusual L-shaped structure (Fig. 1A); the redox reaction is contained in the hydrophilic domain that extends into the mitochondrial matrix, and proton translocation in the membrane-bound domain. Recent structural data from two prokaryotic complexes I (2) has revealed that the membrane-bound ND2, ND4, and ND5 subunits form three related structural units linked by a lateral helix from ND5; together they form the part of the membrane domain farthest from the hydrophilic domain. ND2, ND4, and ND5 are related to two subunits from the Mrp family of Na+/H+ antiporters (3, 4) (see supplemental Fig. 1 and Fig. 1B), suggesting that they share a common ancestor and that their mechanisms of ion translocation are related. Together, these observations suggest that, for energy transduction by complex I, redox-coupled conformational changes drive proton transfer through the three antiporter-like subunits.
FIGURE 1.
Structural information about complex I, taken from 3M9S.pdb (Thermus thermophilus complex I), 3I9V.pdb (T. thermophilus hydrophilic domain), and 3RKO.pdb (six subunits of the E. coli hydrophobic domain) (2, 57). A, complex I is an L-shaped enzyme, with a large hydrophilic domain containing the flavin mononucleotide, iron-sulfur (FeS) clusters, and the binding site for the ubiquinone headgroup. The membrane domain contains seven subunits; the structure of subunit ND1 has been determined only at low resolution, and two possible positions for helix I of ND3 are shown, from the low resolution structure of the intact enzyme and the E. coli membrane domain. The three helices absent from the mammalian ND2 subunit are shown in black (51), and the ends of the cysteine-containing loop of ND3, which is not resolved in the structure, are highlighted in green (the ends of ND3 helices I and II). The membrane domain is also shown from the top, with the hydrophilic domain removed, and labeled with the names of the B. taurus subunits. The antiporter-like subunits, ND2, ND4, and ND5, are indicated. B, the structure of E. coli NuoM (ND4; the 14 core helices of ND2 and ND5 have the same structure) (2). The transmembrane helices are in light blue; fully conserved residues between all of the antiporter-like subunits of B. taurus and E. coli and MrpA and MrpD from B. subtilis are shown in purple (see supplemental Fig. 1). Contact points with the lateral helix of ND5 are highlighted in green, and the loops in the two broken helices in red. On the right, the same structure is shown from the top.
Proton transfer is firmly established as the basis for energy conversion in mitochondria, but some bacteria use sodium ion transfer instead. For example, some organisms use Na+-coupled ATP synthases, and only minor adjustments to the residues of the ion-binding site are required to alter the specificity (5). Redox-driven Na+ translocation has been proposed also for complex I. The complexes I from Klebsiella pneumoniae (6) and Escherichia coli (7) have been proposed to be primary Na+ pumps (transport Na+ out of the cytoplasm), those from Rhodothermus marinus and Escherichia coli have been proposed to transport protons out of and Na+ into the cytoplasm together (4, 8, 9), and E. coli complex I has also been reported to be a simple H+-coupled enzyme (10).
Here, we use three preparations of tightly coupled membrane vesicles to define the ion translocation reactions of mitochondrial complex I (CI):2 proteoliposomes (PLs) containing CI from Bos taurus (Bt) or the yeast Yarrowia lipolytica (Yl) and submitochondrial particles (SMPs) from B. taurus heart. PLs are simpler than SMPs because they contain only CI, but SMPs are more versatile because other respiratory enzymes can be used to set Δp and reduce the quinone pool. In accord with the well established consensus, our results exclude the possibility of redox-driven sodium ion translocation by mitochondrial complex I. However, they reveal an additional ion transfer process in preparations containing the “deactive” form of BtCI. In the absence of substrates, BtCI converts slowly but spontaneously to a deactive form, which is readily reactivated by turnover (NADH:ubiquinone oxidoreduction) (11, 12). The “deactive transition” has been observed in SMPs, permeabilized mitochondria (13), cultured cells (14), and heart tissue (15), and it may be important during ischemia (14). Here, we show that the deactive form of BtCI is a Na+/H+ antiporter. Our results provide new information about the mechanism of coupled ion translocation in complex I, and may be relevant to the role of complex I in ischemia-reperfusion injury.
EXPERIMENTAL PROCEDURES
Preparation of CI, PLs, and SMPs
BtCI and YlCI were prepared as described previously (16–19) in 20 mm Tris-Cl (pH 7.4), 150 mm NaCl, 10% (v/v) ethylene glycol, 0.02% (w/v) n-dodecyl-β-d-maltoside (Anatrace), and 20 mm Na-MOPS (pH 7.2), 150 mm NaCl, 0.05% (w/v) n-dodecyl-β-d-maltoside, respectively. To prepare BtPLs, a 1:2:1 (w/w) mixture of bovine heart phosphatidylcholine, phosphatidylethanolamine, and cardiolipin (Avanti Polar Lipids Inc.) in chloroform was dried under N2 and then homogenized to 10 mg ml−1 in 10 mm Tris-SO4, pH 7.5, 50 mm NaCl (unless otherwise stated) and extruded through a 0.1-μm Whatman Nuclepore track-etched membrane. The preformed liposomes were partially solubilized (20) using 1.2% n-octyl-β-d-glucoside (Anatrace), and the solution was sonicated to clarify it. Then, 0.15–0.25 mg ml−1 BtCI (10–15 mg ml−1 stock solution) was added, the mixture was incubated for 10 min on ice, and prewashed SM-2 Bio-Beads (Bio-Rad) were added to remove the detergents (21) (50 μl of wet Bio-Beads per ml every 30 min for 4 h). The Bio-Beads were removed, and the BtPLs were collected by centrifugation (70,000 × g for 30 min) and resuspended. YlPLs were prepared similarly using soybean asolectin (Sigma-Aldrich) (18), with enzyme that had not undergone the final gel filtration step of the purification (this enzyme contains low levels of cytochrome c oxidase). Following reconstitution, the complex I in PLs was predominantly deactive (11, 12); when required, PLs (∼20 mg ml−1) were activated by incubating them on ice in 800 μm NADH and 800 μm decylubiquinone (DQ) for 10 min (until the NADH was consumed), and then the active PLs were collected by centrifugation, resuspended, and used immediately. For CI in PLs, the deactive status was confirmed by treating the PLs with 1 mm N-ethylmaleimide (NEM) for 25 min; following treatment, the deactive CI is catalytically inactive, whereas catalysis by the active CI is unaffected by the NEM (22).
BtSMPs were prepared as described previously, in 10 mm Tris-SO4 (pH 7.5) and 250 mm sucrose (23), except the NADH incubation step was omitted. To deactivate the complex I, they were incubated at 37 °C for 20 min; to activate it, ∼10 mg ml−1 BtSMPs were incubated in 1 mm NADH at 4 °C for 20 min and used immediately (11, 12). The active/deactive status of BtSMPs was confirmed by measuring the reverse electron transfer activity; deactive BtSMPs exhibit less than 10% of the reverse electron transfer activity of active BtSMPs.
Spectroscopic Assays
Unless otherwise stated, SMP assays were in 10 mm Tris-SO4 (pH 7.5) and 250 mm sucrose, and PL assays were in 5 mm Tris-SO4 (pH 7.5) and 20 mm KCl, at 32 °C. NADH:O2 and NADH:DQ oxidoreduction were followed at 340–380 nm (ϵNADH = 4.81 mm−1 cm−1), and succinate:O2 oxidoreduction (in 20 mm succinate) was followed with a Clark electrode. Reverse electron transfer (succinate:NAD+ oxidoreduction driven by the Δp from ATP hydrolysis) by SMPs was measured in 10 mm succinate, 1 mm NAD+, 1 mm ATP-MgSO4, and 400 μm KCN (23). 10 μg ml−1 gramicidin (a mixture of A, B, C, and D, Sigma-Aldrich) or 25 μg ml−1 alamethicin (Trichoderma viride, Sigma-Aldrich) was used to dissipate Δp. Complex I inhibitors (24, 25) were added at concentrations sufficient for >95% inhibition of the NADH:O2 or NADH:DQ activity: 50 μm capsaicin, 100 nm 2-decyl-4-quinazolinyl amine (from Dr. V. Zickermann, Frankfurt, Germany) (26), 30 μm ethyl-isopropyl amiloride (EIPA) (10, 27–29), 150 nm fenpyroximate (30), 30 μm Δlac-acetogenin (from Prof. H. Miyoshi, Kyoto, Japan, compound 7 in Ref. 31), 5 mm 1-methyl-4-phenyl-pyridinium, 25 μm palmitic acid (32), 1 μm piericidin A, 200 μm ranolazine (33), 200 nm rotenone, and 250 nm stigmatellin (34). To expose PLs and SMPs to NEM, they were incubated for 25 min on ice in 1 mm NEM (added from a 500 mm stock solution in DMSO) and then washed and collected by centrifugation and resuspended as above. Cytochrome c oxidase was inhibited by 400 μm KCN.
The fluorescent dye 9-amino-6-chloro-2-methoxyacridine (ACMA, 0.25 μm, Invitrogen Molecular Probes) was used to monitor ΔpH formation across the vesicular membranes (excitation at 419 nm, emission at 483 nm) (23, 35). In BtSMPs, the Δp from ATP hydrolysis was determined, with KCN to inhibit cytochrome c oxidase, by balancing the ΔE for NADH:fumarate oxidoreduction against Δp (23).
Measurements of the Physical Properties of the Vesicles
“Oriented complex I” is complex I with its NADH-binding site exposed to the external solution; for PLs, the fractions of oriented complex I were determined from catalytic rates in the presence of gramicidin, measured in the presence and absence of 25 μg ml−1 T. viride alamethicin, a pore-forming antibiotic that allows NADH to enter the vesicular lumen (36). The hydrodynamic radii of the PLs and SMPs were determined by dynamic light scattering, using a Wyatt Technology Corp. DynaPro Titan or a Malvern Zetasizer Nano S, according to the Raleigh spheres model (37); each value is the average of three measurements, each consisting of 10 readings. The total internal volumes of the PL preparations were determined by including iron nitrate (10 μg liter−1 of iron, prepared from the Sigma-Aldrich iron AA/ICP calibration standard) in the reconstitution. The PLs were washed three times by centrifugation and resuspension in iron-free buffer, and then the total internal volumes were determined by measuring the iron contents (19) of matched pairs of samples prepared with and without additional iron.
RESULTS
Characterization of the PLs and SMPs
Comprehensive characterizations of the three preparations of coupled vesicles used here (BtPLs, YlPLs and BtSMPs) are summarized in Table 1. Each preparation displays high catalytic activity (NADH:DQ oxidoreduction for PLs (16, 38) or NADH:O2 oxidoreduction for SMPs (involving complexes I, III, and IV of the respiratory chain, with endogenous ubiquinone-10 as an intermediate)) that is inhibited by piericidin A, a canonical complex I inhibitor. Gramicidin, an ionophore that collapses Δp, stimulates catalysis significantly, showing that Δp is substantial (see below also). Finally, average hydrodynamic radii and total internal volume measurements showed that each particle contains at least 100 correctly oriented enzyme molecules, allowing them to be treated as homogeneous populations.
TABLE 1.
Characterization of the PL and SMP preparations
The values reported in rows 1, 2, and 4 are in μmol NADH min−1 (mg of protein)−1. The respiratory control ratio (RCR, row 3) is a qualitative measure of how well the particles are coupled, but it depends also on the catalytic activity, the enzyme concentration in the membrane, and the ability of an enzyme to work against Δp. In independent measurements on SMPs, ATP hydrolysis displayed an RCR value of only 1.6 but was shown to support a Δp of more than 0.15 V (23). “Oriented complex I” is complex I with its NADH-binding site exposed to the external solution. The values reported in row 7 are in μmol NADH min−1 (mg of complex I)−1). Comparison of the values for BtPLs and BtSMPs shows that the CI in SMPs is around twice as active as that in PLs. However, the activity in BtPLs is commensurate with values from the most active preparations of BtCI (16,58). The difference probably arises from the nonphysiological quinone analogue required for the BtPLs and from the harsh procedure for preparing BtPLs (solubilization and chromatography in detergent-containing solutions and reconstitution into an artificial membrane) relative to BtSMPs (sonication and centrifugation steps that retain the native membrane). Our SMPs are comparable in size with those of Li et al. (59) (∼100 nm), and the value given for their total internal volume (in μl (mg of protein)−1) is a representative value based on that of Sorgato et al. (60) (1.3 μl (mg of protein)−1) and an equivalent value determined for this preparation using the same protocol (0.83 ± 0.12 μl (mg of protein)−1); the value is similar to values determined for inverted bacterial membrane vesicles using an EPR spin probe (1 μl (mg of protein)−1) (8, 9). The numbers of oriented complex I per particle are average values calculated from rows 5, 6, 8, and 9 by considering a uniform set of spherical particles, membrane thickness of 5 nm, and a uniform distribution of complex I.
| PLs |
SMPs (B. taurus) | ||
|---|---|---|---|
| B. taurus | Y. lipolytica | ||
| NADH:DQ or O2 oxidoreduction | 0.90 ± 0.04 | 0.46 ± 0.03 | 0.25 ± 0.01 (23) |
| Row 1 with gramicidin | 2.39 ± 0.19 | 1.70 ± 0.04 | 0.74 ± 0.02 (23) |
| RCR value (row 2/row 1) | 2.66 ± 0.24 | 3.70 ± 0.26 | 3.02 ± 0.15 |
| Row 2 with piericidin A | 0.13 ± 0.03 | 0.055 ± 0.01 | 0.05 ± 0.02 |
| Fraction of oriented complex I | 0.55 ± 0.03 | 0.48 ± 0.03 | 0.81 ± 0.05 (23) |
| Fraction of protein that is complex I | 0.95 ± 0.01 | ∼1 | ∼0.096 (23) |
| Row 2 corrected for rows 6 and 5 | 4.57 ± 0.48 | 3.54 ± 0.24 | 9.55 ± 0.70 |
| Particle radius (nm) | 143 ± 12 | 129 ± 4 | 163 ± 25 |
| Total internal volume | 22.8 ± 4.4 | 21.0 ± 6.2 | ∼1.0 |
| Oriented complex I per particle | 140 | 100 | 700 |
Confirmation That Mitochondrial CI Catalyzes Only Redox-driven Proton Translocation
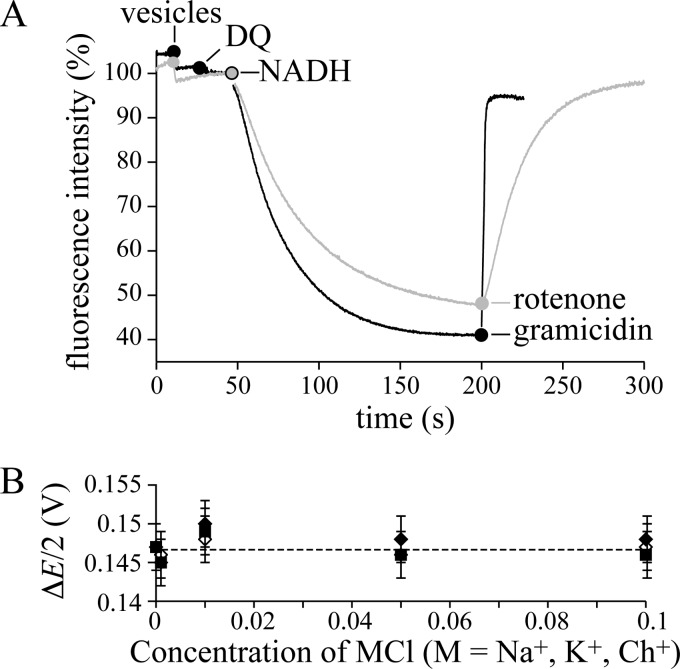
ACMA is a fluorescent dye that is used widely to demonstrate ΔpH formation across vesicular membranes; when ACMA is taken up by activated vesicles its fluorescence is quenched, most likely by dimerization and excimer formation (39). Fig. 2A shows that adding NADH and DQ to BtPLs, and NADH to BtSMPs, rapidly quenches the ACMA fluorescence; the effect is reversed when ΔpH is dissipated by gramicidin or when proton translocation is inhibited by rotenone. Note that a membrane-permeant charge-compensating ion is required to dissipate Δψ (so that Δψ → 0 and ΔpH → Δp). Thus, for BtSMPs, chloride or nitrate was added to the external solution (the membrane is readily permeable to both (23, 35, 40)), and for the PL experiment of Fig. 2A, 25 mm K+ and valinomycin (a K+ uniporter) were present. The data in Fig. 2A are fully consistent with redox-driven proton translocation by mitochondrial CI, but do not exclude additional ion translocation events. Two strategies, one kinetic and one thermodynamic, were used to investigate this possibility.
FIGURE 2.
Mitochondrial complex I is a redox-coupled proton pump. A, quenching of the ACMA fluorescence demonstrates ΔpH formation in BtPLs and BtSMPs. Black: BtPLs (1.3 μg-protein ml−1), 200 μm DQ, and 200 μm NADH; 2 μg ml−1 gramicidin abolishes Δp. Conditions are: 10 mm Tris-SO4, pH 7.5, 125 mm sucrose, 25 mm KCl, 2 μg ml−1 valinomycin, 20 °C. Gray: BtSMPs (3.8 μg-protein ml−1), and 200 μm NADH; 200 nm rotenone stops proton translocation, and Δp dissipates. Conditions are: 10 mm Tris-SO4, pH 7.5, 125 mm sucrose, 80 mm KCl, 20 °C. B, the effect of Na+, K+ and Ch+ on the apparent Δp (ΔE/2), generated by ATP hydrolysis by SMPs. The rate of NADH:fumarate oxidoreduction was recorded as a function of ΔE, where ΔE (V) = −0.315 − RT/2F.ln{([NADH][fumarate])/([NAD+][succinate])} using 0.1 mm [NADH], 1 mm [NAD+], 0.5 mm [succinate], 0.025–40 mm [fumarate]). When the net rate is zero, ΔE/2 = Δp (assuming 4H+ per NADH). Conditions are: 10 mm Tris-SO4, pH 7.5, 32 °C, 0 to 100 mm MCl (M, Na+, K+, Ch+) with variable sucrose concentration (250 to 50 mm) to maintain the osmolarity. Error bars indicate S.D.
First, the rates of NADH:DQ oxidoreduction by BtCI or YlCI were measured as a function of the Na+, K+, and [(CH3)3N(CH2)2OH]+ (choline, Ch+) concentrations (0–100 mm); Ch+ is a monovalent cation that is clearly distinct from Na+ and K+. Some cation-dependent variation was evident (for BtCI, Na+ stimulates the rate by up to 40% at intermediate concentrations, K+ stimulates less and at lower concentrations, and Ch+ does not stimulate at all, whereas for YlCI, the effects of K+ and Na+ are more similar). However, the limited magnitude of these effects indicates a secondary effect, not tightly coupled Na+ or K+ translocation; we note that catalysis by E. coli complex I has a similar Na+ concentration dependence (10). Second, a “thermodynamic balance” experiment was used previously to quantify the Δp produced by ATP hydrolysis in BtSMPs (23); when the net rate of NADH:fumarate oxidoreduction (catalyzed by complexes I and II) is zero, the redox potential (ΔE) is balanced exactly by Δp (at 2ΔE = 4Δp, assuming the reaction has a 2e− to 4H+ stoichiometry). Here, we asked whether there is a thermodynamic contribution from coupled Na+ or K+ transport that affects the point of balance. For example, if Na+ are co-translocated with the H+, then adding Na+ to the external solution would create a ΔNa+ that favors NADH oxidation over NAD+ reduction (an apparent uncoupling) and the apparent Δp would decrease. Fig. 2B shows that the external Na+, K+, or Ch+ concentration does not affect the apparent Δp (the rate of ATP hydrolysis was not affected), so mitochondrial CI catalyzes only redox-coupled H+ transfer.
ACMA Fluorescence Experiments Indicate That Deactive BtCI Catalyzes Na+/H+ Exchange
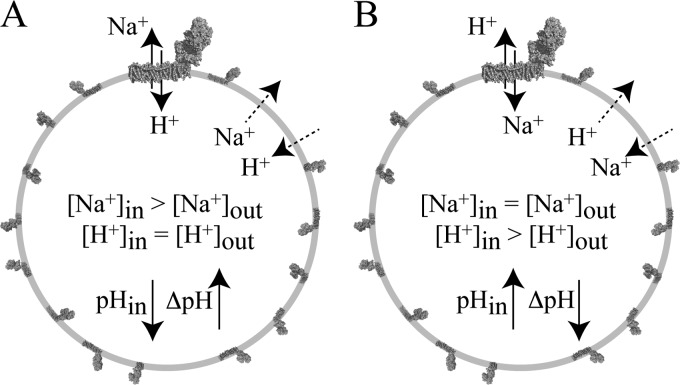
In Experiment A (Fig. 3), PLs loaded with Na+ (formed in high [Na+] buffer) were placed into low Na+ buffer (with the same pH and ionic strength, balanced with Ch+). If CI acts as an antiporter then Na+ from inside are exchanged for H+ from outside, and the intravesicular pH decreases (ΔNa+ is converted to ΔpH). It is not possible to control the Na+ concentration in the lumen of SMPs (see supplemental Information), so Experiment A is only possible for PLs. In Experiment B, SMPs were loaded with protons by succinate:O2 oxidoreduction (independently of CI) to drive proton uptake. Then, succinate oxidation was inhibited, and Na+ was added to the external solution. If CI acts as an antiporter, then H+ from inside are exchanged for Na+ from outside, and the intravesicular pH rises (ΔpH is converted to ΔNa+). It is not possible to load the lumen of PLs with sufficient H+ for this experiment without using CI itself, so Experiment B is only possible for SMPs. Ideally, both the intravesicular H+ and Na+ concentrations should be monitored during these experiments. However, despite extensive trials (see supplemental Information), we were unable to incorporate H+- and Na+-specific probes into the lumenal volumes of either PLs or SMPs while retaining the catalytic activity of CI. Therefore, we used ACMA to monitor the formation or dissipation of ΔpH (with Δψ collapsed to zero using a permeant ion).
FIGURE 3.
Schematic representation of two experiments that reveal the Na+/H+ antiporter activity of complex I. The diagrams represent a typical complex I PL (see data in Table 1); the complex at the top has been magnified. A, Na+-loaded vesicles are placed into a low [Na+] buffer. If complex I is a Na+/H+ antiporter, then Na+ efflux drives H+ uptake and the pH of the lumen rapidly decreases, forming ΔpH. B, the vesicles have been loaded with protons to create ΔpH. The driving force for H+ efflux is high, but uncatalyzed H+ efflux is slow; rapid H+ efflux is observed if it is coupled to Na+ uptake by complex I (the pH of the lumen rapidly increases, and ΔpH collapses).
During initial trials with BtPLs and YlPLs in Experiment A, and BtSMPs in Experiment B, it was noted that the YlPLs behaved more reproducibly than the BtPLs or BtSMPs; for BtCI, different preparations presented very different levels of apparent antiporter activity. Subsequent investigation revealed that BtCI, in both PLs and SMPs, was present as a mixture of deactive and active forms (11, 12), whereas YlPLs were essentially all deactive (as judged by their NEM sensitivity) (41). Therefore, in all the experiments described below, the deactive/active status of each sample was defined as exactly as possible (see “Experimental Procedures”).
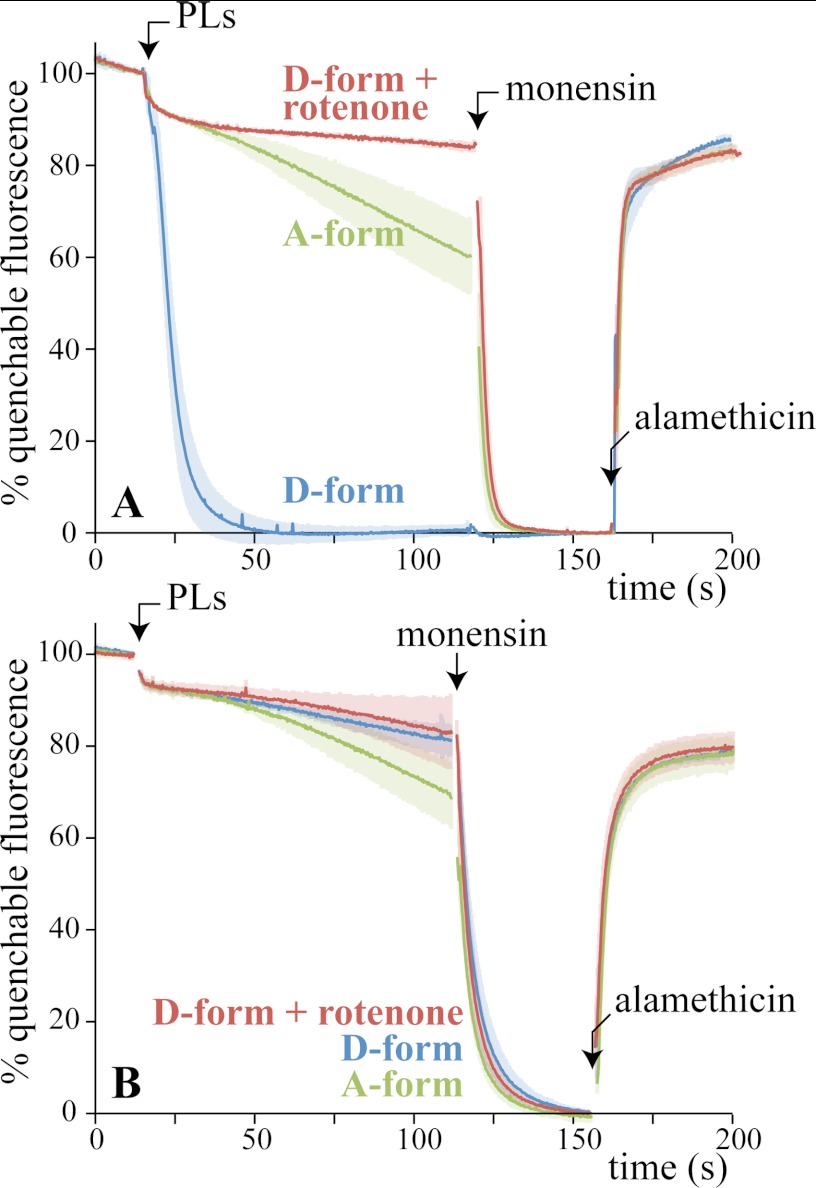
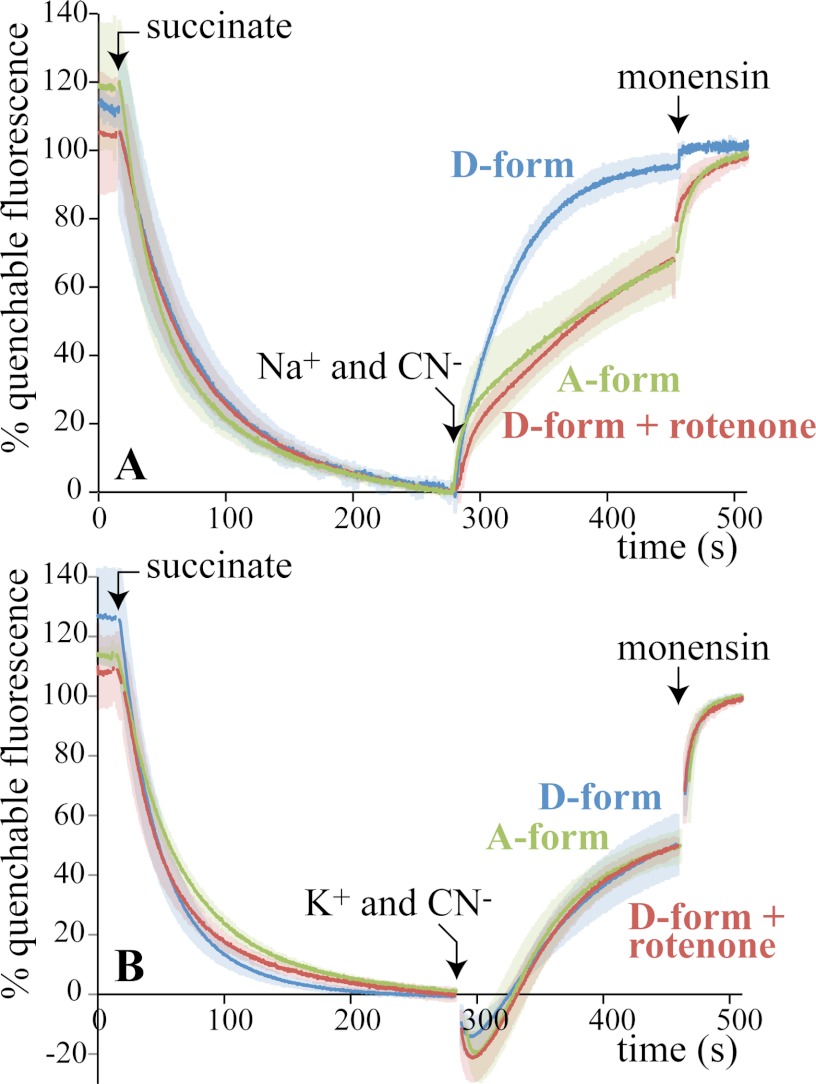
Fig. 4A shows how the fluorescence of ACMA responds when BtPLs containing 50 mm NaCl are placed into buffer containing 5 mm NaCl and 45 mm ChCl. Deactive BtPLs immediately quench the fluorescence, as Na+ efflux drives H+ uptake, creating ΔpH. Importantly, no significant quenching occurred with rotenone present or when the internal and external Na+ concentrations were equal. Furthermore, the effect of adding active BtPLs was much slower and more limited than for deactive BtPLs. In each case, monensin, a small-molecule Na+/H+ exchanger, takes the fluorescence to the same end-point level, as ΔNa+ and ΔpH equalize; the deactive BtPLs have already reached this level. Finally, the pore-forming antibiotic alamethicin was added to dissipate both ΔNa+ and ΔpH; the fluorescence rapidly recovered its starting value. The results in Fig. 4A suggest strongly that deactive BtCI (but not active BtCI) is a Na+/H+ antiporter that can be inhibited by rotenone. Fig. 4B shows equivalent experiments using K+ instead of Na+, showing that the activity is specific to Na+.
FIGURE 4.
Sodium ion efflux from PLs drives proton uptake by deactive BtCI (Experiment A). A, BtPLs were prepared in 5 mm Tris-SO4, 50 mm NaCl, pH 7.4, and then placed into buffer containing 5 mm Tris-SO4, 5 mm NaCl, 45 mm ChCl, pH 7.4, and 0.25 μm ACMA. Their active/deactive status (A- or D-form) was set beforehand (see “Experimental Procedures”). Each trace is the average from at least three experiments, with the standard deviations indicated by shading. 200 nm rotenone was included in the assay buffer as indicated, and 2 μg ml−1 monensin and 2 μg ml−1 alamethicin were added as indicated. The signal has been normalized to 100% when the BtPLs were added and to 0% in the presence of monensin; ∼70% of the total fluorescence was quenchable. Note that the fluorescence is quenched slightly by the ethanol used for monensin and alamethicin. B, same as A except that the PLs contained 50 mm KCl and were placed into buffer containing 5 mm KCl and 45 mm ChCl. Conditions are: 21 °C, ∼20 μg ml−1 BtCI.
For Experiment B (Fig. 5A), succinate was added to BtSMPs, to use succinate:O2 oxidoreduction to create ΔpH (the protons are translocated by complexes III and IV) and quench the ACMA fluorescence (Cl−, a permeant anion, ensures that Δψ is converted to ΔpH). Then, cyanide was added (to inhibit catalysis by complex IV and so stop proton translocation), along with 10 mm Na+ (ΔpH collapses when an antiporter is present as H+ efflux is coupled to Na+ uptake). For deactive BtSMPs, the fluorescence recovers immediately (as ΔpH collapses); it recovers much more slowly in the presence of rotenone, or for active BtSMPs. Note that deactive and active BtSMPs exhibit the same respiratory control ratio for succinate:O2 oxidoreduction (1.6 ± 0.25), so the deactive particles are not uncoupled. In addition, when cycles of activation and deactivation were applied to BtSMPs, the results from Experiment B alternated accordingly, although accompanied by a gradual loss of succinate:O2 oxidoreduction activity and/or coupling. Fig. 5B shows equivalent experiments using K+ instead of Na+; the data are consistent with the antiporter activity being specific to Na+. Finally, the small apparent “burst” of recovery in Fig. 5A upon the addition of Na+ and the “dip” in Fig. 5B upon the addition of K+ are experiment-independent, ion-dependent artifacts of the ACMA system. In fact, otherwise identical experiments with Li+, Na+, K+, or Rb+ revealed an individual “signature” for each ion; we are currently unable to explain these effects.
FIGURE 5.
Proton efflux from BtSMPs drives sodium ion uptake by deactive BtCI (Experiment B). A, BtSMPs were placed into buffer containing 10 mm Tris-SO4, 80 mm ChCl, 250 mm sucrose, pH 7.5, and 0.25 μm ACMA. The active/deactive status of complex I (A- or D-form) was set beforehand (see “Experimental Procedures”). Each trace is the average from at least three experiments, with the standard deviations indicated by shading. 1 mm succinate, 5 mm Na2SO4, 0.5 mm KCN, and 2 μg ml−1 monensin were added as indicated. 200 nm rotenone was added along with the Na2SO4 and KCN where indicated. The signal has been normalized to 100% at the end of the experiment (to compare the traces during the recovery phase) and to 0% before the addition of M+ and cyanide (where M+ = Li+, Na+, K+, or Rb+); ∼60% of the total fluorescence was quenchable. B, same as A except with 5 mm K2SO4 instead of 5 mm Na2SO4. Conditions are: 21 °C, ∼30 μg ml−1 protein.
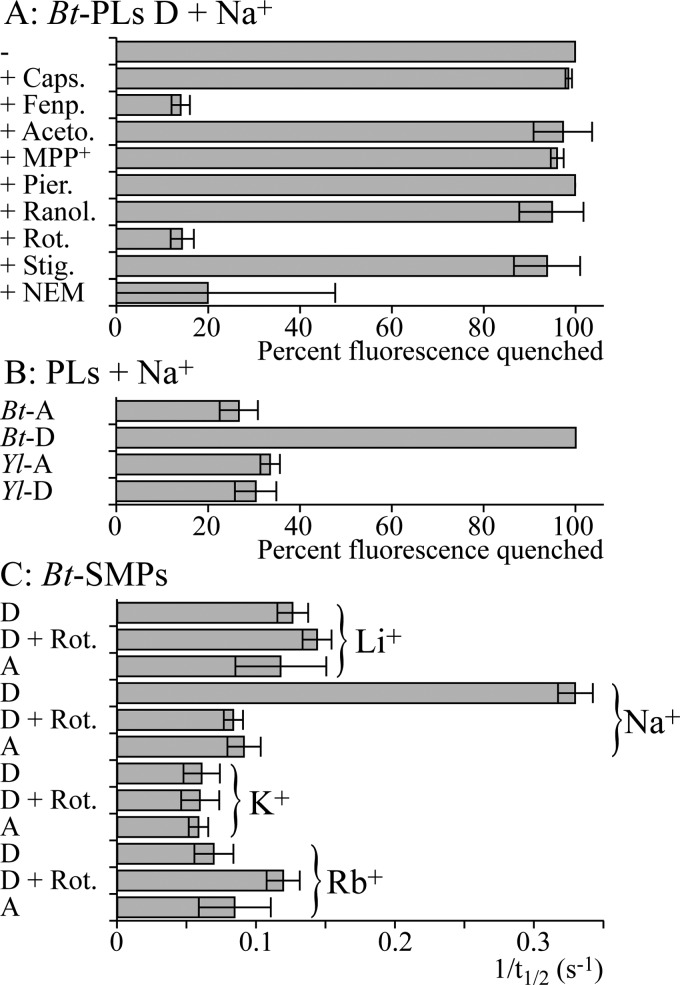
Fig. 6 shows a selection of results that have been quantified using the normalized fluorescence intensity 75 s after the PLs were added (Fig. 4, Experiment A, expressed as the percentage of the normalized fluorescence that was quenched) or the time taken for the normalized fluorescence intensity to recover to 50% (t½) (Fig. 5, Experiment B, expressed as 1/t½). In each case, high values indicate significant antiporter activity. Fig. 6A shows the results from testing several complex I inhibitors in Experiment A (see “Experimental Procedures”). Capsaicin, the Δlac-acetogenin, 1-methyl-4-phenyl-pyridinium, piericidin A, ranolazine, and stigmatellin had no effect on the antiporter activity, whereas fenpyroximate (as well as rotenone) inhibited it. However, fenpyroximate was atypical because the slow initial rate of quenching eventually begins to increase; this was not observed for rotenone. EIPA is a canonical Na+/H+ antiporter inhibitor (42) that has been used to study proton translocation by complex I (10, 27–29), and palmitic acid has been linked to the deactive-active transition in complex I (32). Both inhibitors abolished ACMA fluorescence quenching by deactive BtPLs, but no quenching was observed when monensin was added, and further investigation revealed that they are both uncouplers (see also Ref. 43). It was not possible to identify any concentrations (from 5 pm to 5 mm) that inhibited but did not uncouple; EIPA and palmitic acid should not be used to study ion translocation by complex I. Similarly, 2-decyl-4-quinazolinyl amine is both a complex I inhibitor (26) and an uncoupler. In Experiment B, inhibition and uncoupling oppose each other, and inhibition of the antiporter activity by 2-decyl-4-quinazolinyl amine could be observed at low concentrations (100 nm).
FIGURE 6.
Characteristics of the antiporter reactions in PLs and BtSMPs. A, BtPLs, with deactive CI, were tested using Experiment A in the presence of various inhibitors. A high value for the normalized fluorescence quench (100% fluorescence quench = fluorescence in the presence of monensin) 75 s after the PLs were added indicates high antiporter activity (Fig. 4). Caps.: 50 μm capsaicin; Fenp.: 150 nm fenpyroximate; Aceto.: 30 μm Δlac-acetogenin; MPP+, 5 mm 1-methyl-4-phenyl-pyridinium; Pier.: 1 μm piericidin A; Ranol.: 200 μm ranolazine; Rot.: 200 nm rotenone; Stig.: 250 nm stigmatellin. NEM indicates that the BtPLs were preincubated in 1 mm NEM for 25 min and then washed and collected by centrifugation. B, BtPLs and YlPLs were tested using Experiment A following activation (-A) or deactivation (-D). C, BtSMPs, with active or deactive CI, in the presence or absence of 200 nm rotenone (Rot.), were tested for M+/H+ antiporter activity (M+ = Li+, Na+, K+, or Rb+). A high 1/t½ (where t½ is the time taken for the fluorescence to recover to half its full value upon the addition of M+) indicates significant antiporter activity (Fig. 5).
Fig. 6A shows that treating deactive BtPLs with NEM, as required to derivatize the widely conserved cysteine in ND3 that is accessible only in the deactive enzyme (22) (Fig. 1A), inhibits the antiporter activity. Note that it was necessary to remove the NEM before the assay to avoid artifacts in the ACMA measurement; this extra experimental step, and the effects of small amounts of DMSO, resulted in greater variation in the results. Qualitatively similar results were obtained from SMPs, except that side reactions of NEM with complex II compromised the formation of ΔpH, decreasing the intensity of the ACMA response. In separate experiments, ATP hydrolysis was used to create ΔpH and inhibited by adenosine 5′-(β,γ-imido)triphosphate (44); ATP hydrolysis was also affected by NEM, but a large enough ΔpH could be formed to confirm that the antiporter activity of complex I is sensitive to NEM. Fig. 6B shows that YlPLs do not display any Na+/H+ antiporter activity, irrespective of attempts to convert the Y. lipolytica enzyme between the deactive and active forms. Finally, Fig. 6C compares the results from Experiment B with Li+, Na+, K+, and Rb+, confirming that antiporter activity is Na+-specific.
Vesicle-swelling Experiments Confirm That Deactive BtCI Catalyzes Na+/H+ Exchange
Mitochondria-swelling experiments in isotonic media have been used to characterize ion movement across the inner mitochondrial membrane (45–47); ions accumulating in the matrix cause water uptake (both a cation and an anion must be taken up to balance the charge), and the mitochondria swell as the inner membrane unfolds. Here, we used an analogous approach to monitor ion uptake by BtSMPs; changes in the hydrodynamic radii (the “radii”) were monitored by dynamic light scattering. Thus, sodium acetate was added to the external buffer solution; when an antiporter is present, Na+ uptake coupled to H+ efflux creates an “inverted” ΔpH (alkali inside) that drives the uptake of neutral acetic acid and the net uptake of sodium acetate, driving water uptake and swelling.
Deactive BtSMPs were placed into 250 mm sucrose and 10 mm Tris-SO4, pH 7.5, and their average radius was measured to be 145 ± 3 nm. When 250 mm sodium acetate was added to the external buffer (at constant sucrose and buffer concentrations), the radius increased significantly to 190 ± 15 nm; measurements were taken over 10 min from the addition of the sodium acetate (as the BtSMPs continue to swell until they burst). In equivalent experiments with active BtSMPs or with lithium acetate or potassium acetate, no significant changes in radius were observed (the variation was <5 nm). The results are consistent with Na+/H+ exchange by deactive complex I only. However, experiments to test the effects of rotenone were unsuccessful because rotenone alone caused a significant (20 ± 5 nm) increase in the radius of both deactive and active BtSMPs.
DISCUSSION
Monitoring Redox-driven Ion Translocation by Complex I
In our initial experiments, we used the ΔpH-sensitive dye ACMA to reconfirm that mitochondrial complex I catalyzes redox-linked H+ translocation, and a thermodynamic balance experiment to exclude the possibility of additional redox-linked Na+ translocation. To monitor H+ and Na+ transfer directly, in real time, we investigated several pH- and Na+-sensitive dyes, but were unable to identify any that did not inhibit catalysis and that could be incorporated into the lumenal volumes of coupled vesicles (see supplemental Information). Thus, to monitor Na+/H+ exchange, we designed experiments that relied upon the formation or dissipation of ΔpH monitored by using ACMA. ACMA was compatible with both PLs and SMPs, but its response is only semiquantitative; its fluorescence is affected by many molecules used here (notably DQ, ethanol, and M+ (where M+ = Li+, Na+, K+, or Rb+)), the level of quenching observed is subject to the size and concentration of the vesicles, the enzyme activity/concentration, and the lipid composition, and equal ΔpH values created differently (using different pH buffers, different NaCl concentrations with monensin or valinomycin + carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone) (FCCP) give different responses. Therefore, we compared only “like-with-like,” employed exhaustive sets of controls, and supported our conclusions by vesicle-swelling experiments, a completely different measure of ion transport. We note that Na+ translocation by several prokaryotic complexes I has been studied previously by using radioactive 22Na, atomic absorption spectroscopy, or 23Na NMR with a shift reagent in the external solution to follow Na+ uptake into vesicles directly (6–9). However, these are not real-time methods (so short-lived ion gradients established during catalysis may relax before the measurement). Furthermore, the conclusions of studies of redox-driven ion translocation by E. coli complex I using these methods have disagreed completely; they have proposed that Na+ transfer occurs in opposite directions (7, 9).
The Na+/H+ Antiporter Activity of Deactive B. taurus Complex I
Our results show that deactive BtCI is a Na+/H+ antiporter; it exchanges Na+ and H+ across a membrane to balance the ion- and proton-motive forces. Importantly, Na+/H+ exchange is independent of energy transduction; it halts when the enzyme is reactivated by redox-linked H+ translocation. Together with the sequence similarity of the complex I ND2, ND4, and ND5 subunits with the Mrp antiporter subunits MrpA and MrpD (see supplemental Fig. 1 and Fig. 1B), the ability of the complex I ND2, ND4, and ND5 subunits to rescue Mrp antiporter subunit knockouts in Bacillus subtilis (3, 4), and the Na+/H+ antiporter activity of an overexpressed truncated form of ND5 (27), our results suggest that one or more of ND2, ND4, and ND5 retain an inherent antiporter capability that is exhibited in the deactive enzyme. We propose that in deactive BtCI, the functional connection between the redox and antiporter modules (Fig. 1) is broken; energy-transducing catalysis abolishes the antiporter activity by initiating their reconnection, whereupon the redox domain “harnesses” the antiporter domain for proton translocation. It is further possible that the H+ transfer-ready states created by the redox reaction and by Na+ transfer resemble one another. Interestingly, rotenone binds more strongly to active BtCI than to the deactive enzyme, and it partially prevents and reverses deactivation (48). Thus, rotenone does not inhibit the antiporter activity of deactive complex I directly; it abolishes it by functionally reconnecting the antiporter domain to the redox domain. The antiporter domain is brought under control and can no longer freewheel. Our proposal is consistent with the location of the rotenone-binding site in complex I, at the base of the redox domain, not in the antiporter-like subunits (49).
Antiporter activity was observed here only in deactive BtCI, and not in YlCI, despite the fact that the ND3 cysteine in “as prepared” YlCI could be derivatized by NEM, a key diagnostic criterion for the deactive enzyme. Derivatizing the ND3 cysteine with NEM in BtCI inhibits the antiporter activity, further confirming the link between the active/deactive status of the enzyme and the antiporter activity. Clear differences between the active/deactive transitions in BtCI and YlCI have been noted previously; for example, the apparent active/deactive transition is much less temperature-dependent in YlCI (50). Thus, we propose that the deactive and active states in BtCI differ more extensively than they do in YlCI, with only the transition in BtCI progressing to the functional disconnection of the redox and transport domains. It is possible that the truncation of ND2 in BtCI (Fig. 1A) is one determinant of the extent of the transition (51); the widely conserved cysteine in ND3 that is accessible to NEM only in the deactive enzyme is in a loop close to the three (truncated) helices (2, 22). Finally, we note that increased ACMA quenching was observed during catalysis by E. coli complex I PLs upon the addition of either extra NaCl or ETH-157, a sodium ionophore (although no antiporter activity was observed in an experiment similar to that shown in Fig. 4) (10). This result was used to suggest “secondary antiporter activity,” but no readily permeant ion was present, so it may arise from enhanced ion transfer that relaxes Δψ and facilitates ΔpH formation instead.
A Physiological Role for the Antiporter Activity of Deactive Complex I?
The deactive transition of complex I may contribute to determining the outcome of ischemia-reperfusion. Partially inhibiting the respiratory chain during ischemia, for example by inhibiting complex I, protects against mitochondrial damage and myocardial injury (52). Decreased respiratory chain turnover also favors the deactivation of complex I, making it susceptible to thiol reagents, such as S-nitrosothiols, that “lock” it in the deactive state (14) until the thiol pools recover. Thus, mitochondrially targeted S-nitrosothiols protect against ischemia-reperfusion injury (53). Our observation that deactive complex I is a Na+/H+ antiporter suggests an additional role for it during ischemia-reperfusion. Under normal conditions, transport cycles in the mitochondrial inner membrane, comprising Na+/H+ exchange, Na+/Ca2+ exchange, and Ca2+ uniport, contribute to cellular Ca2+ homeostasis (54). During ischemia, the intracellular Ca2+, Na+, and H+ concentrations rise, and Ca2+ accumulates in the mitochondrial matrix, contributing to opening the permeability transition pore (54, 55). It is possible that Na+/H+ exchange by deactive complex I influences Ca2+ accumulation by altering the ion transport properties of the inner membrane. Furthermore, Na+/H+ exchange across the inner membrane of mammalian mitochondria was identified more than 40 years ago by mitochondria-swelling experiments (45–47), but the protein responsible has not been identified (56). In our experiments, complex I is the dominant Na+/H+ exchanger, and a clear preference for Na+ over K+ was also observed in nonenergized mitochondria, consistent with a significant contribution from deactive complex I under these conditions. Finally, inhibiting Na+/H+ exchange across the plasma cell membrane, to attenuate the rise in intracellular Na+, is a well developed strategy for limiting ischemia-reperfusion injury (55). EIPA is one of the 5-N-substituted amiloride family of Na+/H+ exchange inhibitors, and ranolazine, in clinical use for angina, is proposed to be a Na+/H+ exchange inhibitor and also a complex I inhibitor (33). Here, we found no evidence that either compound inhibits Na+/H+ exchange by deactive complex I (although EIPA is an efficient uncoupler of Δp). Thus, the response of deactive complex I to pharmacological agents is clearly distinct from that of the plasma membrane exchangers, inviting the prospect of targeting them individually.
Acknowledgments
We thank Kenneth R. Pryde (MRC) for characterizing the SMP preparation and Dr. Volker Zickermann (Frankfurt, Germany) and Professor Hideto Miyoshi (Kyoto, Japan) for complex I inhibitors.
This work was supported by the Medical Research Council.

This article contains supplemental Information and Fig. 1.
- CI
- complex I
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- Bt
- B. taurus
- Ch+
- choline
- DQ
- decylubiquinone
- EIPA
- ethyl-isopropyl amiloride
- NEM
- N-ethyl maleimide
- PL
- proteoliposome
- SMP
- submitochondrial particle
- Yl
- Y. lipolytica.
REFERENCES
- 1. Hirst J. (2010) Toward the molecular mechanism of respiratory complex I. Biochem. J. 425, 327–339 [DOI] [PubMed] [Google Scholar]
- 2. Efremov R. G., Sazanov L. A. (2011) Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 [DOI] [PubMed] [Google Scholar]
- 3. Mathiesen C., Hägerhäll C. (2002) Transmembrane topology of the NuoL, M, and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochim. Biophys. Acta 1556, 121–132 [DOI] [PubMed] [Google Scholar]
- 4. Moparthi V. K., Kumar B., Mathiesen C., Hägerhäll C. (2011) Homologous protein subunits from Escherichia coli NADH:quinone oxidoreductase can functionally replace MrpA and MrpD in Bacillus subtilis. Biochim. Biophys. Acta 1807, 427–436 [DOI] [PubMed] [Google Scholar]
- 5. Krah A., Pogoryelov D., Langer J. D., Bond P. J., Meier T., Faraldo-Gómez J. D. (2010) Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase F0 rotors. Biochim. Biophys. Acta 1797, 763–772 [DOI] [PubMed] [Google Scholar]
- 6. Gemperli A. C., Dimroth P., Steuber J. (2003) Sodium ion cycling mediates energy coupling between complex I and ATP synthase. Proc. Natl. Acad. Sci. U.S.A. 100, 839–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Steuber J., Schmid C., Rufibach M., Dimroth P. (2000) Na+ translocation by complex I (NADH:quinone oxidoreductase) of Escherichia coli. Mol. Microbiol. 35, 428–434 [DOI] [PubMed] [Google Scholar]
- 8. Batista A. P., Fernandes A. S., Louro R. O., Steuber J., Pereira M. M. (2010) Energy conservation by Rhodothermus marinus respiratory complex I. Biochim. Biophys. Acta 1797, 509–515 [DOI] [PubMed] [Google Scholar]
- 9. Batista A. P., Pereira M. M. (2011) Sodium influence on energy transduction by complexes I from Escherichia coli and Paracoccus denitrificans. Biochim. Biophys. Acta 1807, 286–292 [DOI] [PubMed] [Google Scholar]
- 10. Stolpe S., Friedrich T. (2004) The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J. Biol. Chem. 279, 18377–18383 [DOI] [PubMed] [Google Scholar]
- 11. Vinogradov A. D., Grivennikova V. G. (2001) The mitochondrial complex I: progress in understanding of catalytic properties. IUBMB Life 52, 129–134 [DOI] [PubMed] [Google Scholar]
- 12. Kotlyar A. B., Vinogradov A. D. (1990) Slow active/inactive transition of the mitochondrial NADH-ubiquinone reductase. Biochim. Biophys. Acta 1019, 151–158 [DOI] [PubMed] [Google Scholar]
- 13. Grivennikova V. G., Kapustin A. N., Vinogradov A. D. (2001) Catalytic activity of NADH-ubiquinone oxidoreductase (complex I) in intact mitochondria. Evidence for the slow active/inactive transition. J. Biol. Chem. 276, 9038–9044 [DOI] [PubMed] [Google Scholar]
- 14. Galkin A., Abramov A. Y., Frakich N., Duchen M. R., Moncada S. (2009) Lack of oxygen deactivates mitochondrial complex I: implications for ischemic injury? J. Biol. Chem. 284, 36055–36061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maklashina E., Sher Y., Zhou H. Z., Gray M. O., Karliner J. S., Cecchini G. (2002) Effect of anoxia/reperfusion on the reversible active/deactive transition of NADH-ubiquinone oxidoreductase (complex I) in rat heart. Biochim. Biophys. Acta 1556, 6–12 [DOI] [PubMed] [Google Scholar]
- 16. Sharpley M. S., Shannon R. J., Draghi F., Hirst J. (2006) Interactions between phospholipids and NADH:ubiquinone oxidoreductase (complex I) from bovine mitochondria. Biochemistry 45, 241–248 [DOI] [PubMed] [Google Scholar]
- 17. Sherwood S., Hirst J. (2006) Investigation of the mechanism of proton translocation by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria: does the enzyme operate by a Q-cycle mechanism? Biochem. J. 400, 541–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kashani-Poor N., Kerscher S., Zickermann V., Brandt U. (2001) Efficient large scale purification of His-tagged proton translocating NADH:ubiquinone oxidoreductase (complex I) from the strictly aerobic yeast Yarrowia lipolytica. Biochim. Biophys. Acta 1504, 363–370 [DOI] [PubMed] [Google Scholar]
- 19. Bridges H. R., Grgic L., Harbour M. E., Hirst J. (2009) The respiratory complexes I from the mitochondria of two Pichia species. Biochem. J. 422, 151–159 [DOI] [PubMed] [Google Scholar]
- 20. Rigaud J. L., Pitard B., Levy D. (1995) Reconstitution of membrane proteins into liposomes: application to energy-transducing membrane proteins. Biochim. Biophys. Acta 1231, 223–246 [DOI] [PubMed] [Google Scholar]
- 21. Rigaud J. L., Levy D., Mosser G., Lambert O. (1998) Detergent removal by nonpolar polystyrene beads: applications to membrane protein reconstitution and two-dimensional crystallization. Eur. Biophys. J. 27, 305–319 [Google Scholar]
- 22. Galkin A., Meyer B., Wittig I., Karas M., Schägger H., Vinogradov A., Brandt U. (2008) Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 283, 20907–20913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pryde K. R., Hirst J. (2011) Superoxide is produced by the reduced flavin in mitochondrial complex I: a single, unified mechanism that applies during both forward and reverse electron transfer. J. Biol. Chem. 286, 18056–18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Degli Esposti M. (1998) Inhibitors of NADH-ubiquinone reductase: an overview. Biochim. Biophys. Acta 1364, 222–235 [DOI] [PubMed] [Google Scholar]
- 25. Miyoshi H. (1998) Structure-activity relationships of some complex I inhibitors. Biochim. Biophys. Acta 1364, 236–244 [DOI] [PubMed] [Google Scholar]
- 26. Okun J. G., Lümmen P., Brandt U. (1999) Three classes of inhibitors share a common binding domain in mitochondrial complex I (NADH:ubiquinone oxidoreductase). J. Biol. Chem. 274, 2625–2630 [DOI] [PubMed] [Google Scholar]
- 27. Steuber J. (2003) The C-terminally truncated NuoL subunit (ND5 homologue) of the Na+-dependent complex I from Escherichia coli transports Na+. J. Biol. Chem. 278, 26817–26822 [DOI] [PubMed] [Google Scholar]
- 28. Nakamaru-Ogiso E., Kao M. C., Chen H., Sinha S. C., Yagi T., Ohnishi T. (2010) The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of Escherichia coli complex I. J. Biol. Chem. 285, 39070–39078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Batista A. P., Marreiros B. C., Pereira M. M. (2011) Decoupling of the catalytic and transport activities of complex I from Rhodothermus marinus by sodium/proton antiporter inhibitor. ACS Chem. Biol. 6, 477–483 [DOI] [PubMed] [Google Scholar]
- 30. Shiraishi Y., Murai M., Sakiyama N., Ifuku K., Miyoshi H. (2012) Fenpyroximate binds to the interface between PSST and 49-kDa subunits in mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry 51, 1953–1963 [DOI] [PubMed] [Google Scholar]
- 31. Ichimaru N., Murai M., Abe M., Hamada T., Yamada Y., Makino S., Nishioka T., Makabe H., Makino A., Kobayashi T., Miyoshi H. (2005) Synthesis and inhibition mechanism of Δlac-acetogenins, a novel type of inhibitor of bovine heart mitochondrial complex I. Biochemistry 44, 816–825 [DOI] [PubMed] [Google Scholar]
- 32. Loskovich M. V., Grivennikova V. G., Cecchini G., Vinogradov A. D. (2005) Inhibitory effect of palmitate on the mitochondrial NADH:ubiquinone oxidoreductase (complex I) as related to the active/deactive enzyme transition. Biochem. J. 387, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aldakkak M., Camara A. K., Heisner J. S., Yang M., Stowe D. F. (2011) Ranolazine reduces Ca2+ overload and oxidative stress and improves mitochondrial integrity to protect against ischemia-reperfusion injury in isolated hearts. Pharmacol. Res. 64, 381–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Degli Esposti M., Ghelli A., Crimi M., Estornell E., Fato R., Lenaz G. (1993) Complex I and complex III of mitochondria have common inhibitors acting as ubiquinone antagonists. Biochem. Biophys. Res. Commun. 190, 1090–1096 [DOI] [PubMed] [Google Scholar]
- 35. Rottenberg H., Lee C. P. (1975) Energy-dependent hydrogen ion accumulation in submitochondrial particles. Biochemistry 14, 2675–2680 [DOI] [PubMed] [Google Scholar]
- 36. Gostimskaya I. S., Grivennikova V. G., Zharova T. V., Bakeeva L. E., Vinogradov A. D. (2003) In situ assay of the intramitochondrial enzymes: use of alamethicin for permeabilization of mitochondria. Anal. Biochem. 313, 46–52 [DOI] [PubMed] [Google Scholar]
- 37. Pecora R. E. (1985) Dynamic Light Scattering: Applications of Photocorrelation Spectroscopy, Plenum Press, New York [Google Scholar]
- 38. King M. S., Sharpley M. S., Hirst J. (2009) Reduction of hydrophilic ubiquinones by the flavin in mitochondrial NADH:ubiquinone oxidoreductase (complex I) and production of reactive oxygen species. Biochemistry 48, 2053–2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grzesiek S., Otto H., Dencher N. A. (1989) ΔpH-induced fluorescence quenching of 9-aminoacridine in lipid vesicles is due to excimer formation at the membrane. Biophys. J. 55, 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bashford C. L., Thayer W. S. (1977) Thermodynamics of the electrochemical proton gradient in bovine heart submitochondrial particles. J. Biol. Chem. 252, 8459–8463 [PubMed] [Google Scholar]
- 41. Galkin A., Moncada S. (2007) S-nitrosation of mitochondrial complex I depends on its structural conformation. J. Biol. Chem. 282, 37448–37453 [DOI] [PubMed] [Google Scholar]
- 42. Vigne P., Frelin C., Cragoe E. J., Jr., Lazdunski M. (1983) Ethylisopropyl-amiloride: a new and highly potent derivative of amiloride for the inhibition of the Na+/H+ exchange system in various cell types. Biochem. Biophys. Res. Commun. 116, 86–90 [DOI] [PubMed] [Google Scholar]
- 43. Davies K., Solioz M. (1992) Assessment of uncoupling by amiloride analogues. Biochemistry 31, 8055–8058 [DOI] [PubMed] [Google Scholar]
- 44. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 45. Blondin G. A., Vail W. J., Green D. E. (1969) The mechanism of mitochondrial swelling. II. Pseudoenergized swelling in the presence of alkali metal salts. Arch. Biochem. Biophys. 129, 158–172 [DOI] [PubMed] [Google Scholar]
- 46. Douglas M. G., Cockrell R. S. (1974) Mitochondrial cation-hydrogen ion exchange: sodium selective transport by mitochondria and submitochondrial particles. J. Biol. Chem. 249, 5464–5471 [PubMed] [Google Scholar]
- 47. Brierley G. P., Jurkowitz M., Jung D. W. (1978) Osmotic swelling of heart mitochondria in acetate and chloride salts: evidence for two pathways for cation uptake. Arch. Biochem. Biophys. 190, 181–192 [DOI] [PubMed] [Google Scholar]
- 48. Grivennikova V. G., Maklashina E. O., Gavrikova E. V., Vinogradov A. D. (1997) Interaction of the mitochondrial NADH-ubiquinone reductase with rotenone as related to the enzyme active/inactive transition. Biochim. Biophys. Acta 1319, 223–232 [DOI] [PubMed] [Google Scholar]
- 49. Tocilescu M. A., Fendel U., Zwicker K., Kerscher S., Brandt U. (2007) Exploring the ubiquinone-binding cavity of respiratory complex I. J. Biol. Chem. 282, 29514–29520 [DOI] [PubMed] [Google Scholar]
- 50. Maklashina E., Kotlyar A. B., Cecchini G. (2003) Active/deactive transition of respiratory complex I in bacteria, fungi, and animals. Biochim. Biophys. Acta 1606, 95–103 [DOI] [PubMed] [Google Scholar]
- 51. Birrell J. A., Hirst J. (2010) Truncation of subunit ND2 disrupts the threefold symmetry of the antiporter-like subunits in complex I from higher metazoans. FEBS Lett. 584, 4247–4252 [DOI] [PubMed] [Google Scholar]
- 52. Chen Q., Camara A. K., Stowe D. F., Hoppel C. L., Lesnefsky E. J. (2007) Modulation of electron transport protects cardiac mitochondria and decreases myocardial injury during ischemia and reperfusion. Am. J. Physiol. Cell. Physiol. 292, C137–C147 [DOI] [PubMed] [Google Scholar]
- 53. Prime T. A., Blaikie F. H., Evans C., Nadtochiy S. M., James A. M., Dahm C. C., Vitturi D. A., Patel R. P., Hiley C. R., Abakumova I., Requejo R., Chouchani E. T., Hurd T. R., Garvey J. F., Taylor C. T., Brookes P. S., Smith R. A., Murphy M. P. (2009) A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proc. Natl. Acad. Sci. U.S.A. 106, 10764–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Crompton M. (1999) The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 341, 233–249 [PMC free article] [PubMed] [Google Scholar]
- 55. Murphy E., Eisner D. A. (2009) Regulation of intracellular and mitochondrial sodium in health and disease. Circ. Res. 104, 292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bernardi P. (1999) Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol. Rev. 79, 1127–1155 [DOI] [PubMed] [Google Scholar]
- 57. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 58. Shinzawa-Itoh K., Seiyama J., Terada H., Nakatsubo R., Naoki K., Nakashima Y., Yoshikawa S. (2010) Bovine heart NADH-ubiquinone oxidoreductase contains one molecule of ubiquinone with ten isoprene units as one of the cofactors. Biochemistry 49, 487–492 [DOI] [PubMed] [Google Scholar]
- 59. Li W., Aurora T. S., Haines T. H., Cummins H. Z. (1986) Elasticity of synthetic phospholipid vesicles and submitochondrial particles during osmotic swelling. Biochemistry 25, 8220–8229 [DOI] [PubMed] [Google Scholar]
- 60. Sorgato M. C., Ferguson S. J., Kell D. B., John P. (1978) The protonmotive force in bovine heart submitochondrial particles: magnitude, sites of generation, and comparison with the phosphorylation potential. Biochem. J. 174, 237–256 [DOI] [PMC free article] [PubMed] [Google Scholar]