Background: Aggregation of a protein, huntingtin, is a pathological hallmark of Huntington disease.
Results: Oxidation of a methionine in huntingtin occurs only after aggregation and alters the aggregate morphologies.
Conclusion: Properties of protein aggregates can be controlled by a “post-aggregation” modification.
Significance: Learning factors that modulate properties of protein aggregates is relevant to understanding the molecular pathomechanism of neurodegenerative diseases.
Keywords: Methionine, Neurodegenerative Diseases, Polyglutamine, Post-translational Modification, Protein Aggregation
Abstract
Aggregation of protein molecules is a pathological hallmark of many neurodegenerative diseases. Abnormal modifications have often been observed in the aggregated proteins, supporting the aggregation mechanism regulated by post-translational modifications on proteins. Modifications are in general assumed to occur in soluble proteins before aggregation, but actually it remains quite obscure when proteins are modified in the course of the aggregation. Here we focus upon aggregation of huntingtin (HTT), which causes a neurodegenerative disorder, Huntington disease, and we show that oxidation of a methionine residue in HTT occurs in vitro and also in vivo. Copper ions as well as added hydrogen peroxide are found to oxidize the methionine residue, but notably, this oxidative modification occurs only in the aggregated HTT but not in the soluble state. Furthermore, the methionine oxidation creates additional interactions among HTT aggregates and alters overall morphologies of the aggregates. We thus reveal that protein aggregates can be a target of oxidative modifications and propose that such a “post-aggregation” modification is a relevant factor to regulate properties of protein aggregates.
Introduction
Misfolding of protein molecules often results in the formation of insoluble aggregates, which are major pathological products observed in many neurodegenerative diseases (1). In proteins composing the aggregates, abnormal post-translational modifications have been often identified, and misregulation of the protein modifications is hence considered to occur under pathogenic conditions and to play roles in the formation of aggregates (2–4).
Post-translational modifications are well known to affect the structural stability of soluble proteins and also the protein-protein interactions (5). Any abnormalities in the modifications hence potentially destabilize the native/functional structure of a protein molecule, which would then facilitate or even trigger the misfolding and aggregation. Abnormally modified proteins are, however, often isolated from insoluble aggregates but not from a soluble fraction of the affected tissues (2, 4), implying that some modifications occur after the formation of insoluble aggregates. Given that insoluble aggregates tend to be regarded as final products with inert structures, the protein modifications after aggregation (i.e., “post-aggregation modifications”) appear to be underrepresented.
In this study, we have illuminated such post-aggregation modifications by focusing on huntingtin (HTT)2 as a model protein. HTT is a large protein with approximately 350 kDa of the molecular mass, in which a polyglutamine (polyQ) tract follows the N-terminal 17 amino acids (6). Although a physiological function of HTT is still controversial (7), a normal HTT possesses a polyQ tract with 6–35 consecutive glutamines and localizes at the cytoplasm as a soluble protein (8). In contrast, HTT forms insoluble aggregates in the nucleus as well as the cytoplasm, when the length of a polyQ tract becomes abnormally expanded (>36) (8). Such aggregation of mutant HTT is a pathological hallmark of a neurodegenerative disease, Huntington disease (HD) (9). In the course of HD, oxidative stress has been reported to significantly increase (10); therefore, the aggregates of mutant HTT are supposed to be susceptible to oxidative modifications.
For example, in HD model mice expressing an N-terminal fragment of mutant HTT, abnormal accumulation of copper ions in striatum and cortex, which are the most affected areas of HD, has been observed at a later stage of the disease (11). A copper ion functions as a pro-oxidant and is suggested to oxidize cysteine residues to form disulfide-linked HTT oligomers (12). Given that almost all of mutant HTT proteins are already aggregated at the disease end stage, however, the aggregation model triggered by oxidative modification on soluble HTT proteins seems questionable. Indeed, at the preclinical stage of the disease, where the inclusions are almost absent, a copper concentration in striatum and cortex is not different between a HD model mouse and its wild-type littermate (11). Recently, furthermore, an aggregation process of mutant HTT has been shown to associate with the production of hydrogen peroxide albeit via an as yet unknown mechanism (13). Compared with soluble HTT proteins, therefore, HTT aggregates are considered to be placed in more oxidative intracellular environments, but it remains unknown whether oxidative modifications occur on HTT aggregates.
We have shown here that Met8 in HTT is oxidized with copper ions as well as added hydrogen peroxide and found that this Met oxidation occurs only when HTT is in the insoluble aggregated state. A soluble form of HTT is not oxidized even by exogenous addition of copper ions and hydrogen peroxide. We have, furthermore, found that Met oxidation creates additional interactions among HTT aggregates to form the aggregate tangles with increased overall size. We thus propose that a protein remains susceptible for oxidation even in the aggregated state and that properties of protein aggregates can be modulated by such post-aggregation modifications.
EXPERIMENTAL PROCEDURES
Expression and Purification of HTTEX1 Proteins
A human HTTEX1 cDNA was cloned in an expression vector, pGEX6P-2 (GE Healthcare), using BamHI and XhoI sites; thereby a GST protein was fused at the N terminus of HTTEX1 (14). The length of a polyQ tract in human HTTEX1 used in this study is either 18 or 42 for an in vitro model of normal and mutant HTT, respectively. Furthermore, the six consecutive His residues were directly fused to the C terminus of HTTEX1 for facilitation of protein purification. Mutations were introduced by an inverse PCR method using KOD DNA polymerase (TOYOBO), and all constructs used in this study were confirmed by DNA sequencing. Escherichia coli, Rosetta(DE3) (Novagen), was transformed with a plasmid, and the expression of HTTEX1 proteins was induced with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside at 20 °C at 200 rpm for 48 h.
The cells were lysed with three cycles of freeze and thaw and resuspended in PBS containing 2% Triton X-100, 420 μm MgCl2, and DNaseI. A soluble supernatant was obtained by centrifugation at 14,000 rpm for 10 min, filtrated using a 0.2-μm filter and then mixed with His-SELECT nickel affinity gel (Sigma) (2 ml/40 ml of supernatant). HTTEX1 proteins bound on the gel were washed with a buffer at pH 8.0 containing 50 mm Tris, 500 mm NaCl, and 10 mm imidazole and eluted by increasing the concentration of imidazole to 250 mm in the buffer solution. Eluted proteins were further purified by incubating with glutathione-Sepharose high performance (GE Healthcare) on ice for 2 h. After washed with a GST wash buffer (50 mm Tris, 200 mm NaCl, pH 8.0), HTTEX1 proteins were collected with a GST elution buffer (50 mm Tris, 200 mm NaCl, 10 mm reduced GSH, pH 8.0). Protein samples were concentrated and ultracentrifuged at 110,000 × g for 15 min. The resultant supernatant was subsequently used or stored at −80 °C for further experiments. Concentration of a GST-fused HTTEX1 with a His tag was determined from the absorption at 280 nm with an extinction coefficient of 42,860 cm−1 m−1.
Aggregation of Purified HTTEX1 Proteins in Vitro
After removal of any insoluble materials by ultracentrifugation, 150 μl of 20 μm GST-HTTEX1-His was prepared in a GST elution buffer. The sample was set in a well of a 96-well plate and mixed with 2 units of a HRV3C protease (Novagen), which specifically cleaves the site between GST and HTTEX1. In this experimental condition, we have confirmed almost completion of the cleavage reactions within 30 min by SDS-PAGE analysis (supplemental Fig. S1). HTTEX1(42Q) liberated from a GST tag has been known to start aggregation, which was monitored by increase in the solution turbidity (the absorbance at 350 nm) using a plate reader (Epoch; BioTek). If necessary, 50 μm bathocuproine disulfonate (BCS), a specific chelator for Cu(I) ions, was added before addition of HRV3C but had no effects on the cleavage reactions by HRV3C protease.
Western Blotting Analysis
After aggregation reactions, 150 μl of samples were fractionated into soluble supernatant and insoluble pellets with ultracentrifugation at 110,000 × g for 15 min. Insoluble HTTEX1 pellets were washed three times with a buffer containing 50 mm Tris and 200 mm NaCl at pH 8.0 and resolubilized by incubation in 50 μl of TFA at 37 °C for 1 h. TFA was then removed by a centrifugal evaporator (SpeedVac), and the resultant protein pellets were dissolved in 150 μl of a buffer containing 500 mm Tris and 5% SDS at pH 8.0. Soluble supernatant and redissolved pellets were mixed with an SDS-PAGE sample buffer containing 10% β-mercaptoethanol, boiled, and then loaded on a 12.5% SDS-polyacrylamide gel. After electrophoresis, proteins were blotted on a 0.2-μm PVDF membrane and probed with mouse anti-polyglutamine (1C2; Merck Millipore; 1:5000), mouse anti-huntingtin (EM48; Merck Millipore; 1:3000), and mouse anti-GST (WAKO; 1:3000) as primary antibodies. Goat anti-mouse conjugated with HRP (Pierce; 1:3000) was then used as a secondary antibody.
MALDI-TOF Mass Analysis on HTTEX1 in Vitro
To analyze a soluble form of HTTEX1, 6 m guanidine HCl and 0.1% TFA were added to the solution, and then the protein was desalted using ZipTip C18 (Millipore). For the mass analysis of HTTEX1 protein in the aggregated state, HTTEX1 aggregates (40 μg) were first resolubilized with 50 μl of TFA and then dried up by centrifugal evaporator. The protein pellets were then dissolved in 50 μl of 6 m guanidine HCl, acidified by addition of 0.1% TFA, and desalted with ZipTip C18. Sinapic acid was chosen as a matrix, and MALDI-TOF mass spectra were acquired using UltraflexTM (Bruker) in a reflectron mode. Mass spectra were calibrated using insulin (Nacalai; m/z 5,734.1), horse heart cytochrome c (Nacalai; m/z 12,361), and equine heart myoglobin (Sigma; m/z 16,951.5).
To analyze the core regions of HTTEX1 aggregates, the aggregates collected by ultracentrifugation were resuspended in a GST elution buffer. After ultrasonication, the aggregates (30 μg, ∼40 μl) were mixed with 150 μl of 50 mm Tris, 100 mm NaCl, 5 mm CaCl2, pH 8.0, and then incubated with 5 μg of Pronase at 37 °C for 12 h. Insoluble pellets were collected, washed twice with a GST wash buffer by ultracentrifugation, and dissolved in 50 μl of TFA with ultrasonication and incubation at 37 °C for 1 h. After removal of TFA with a centrifugal evaporator, the pellets were redissolved in 50 μl of 6 m guanidine HCl with 0.1% TFA, desalted with ZipTip C18, and analyzed with MALDI-TOF mass spectrometry using sinapic acid as a matrix. Mass spectra were further calibrated with insulin (m/z 5,734.1) and horse heart cytochrome c (m/z 12,361), and identification of peptides based upon the observed m/z values was performed using PAWS software (ProteoMetrics).
MALDI-TOF Mass Analysis on HTTEX1 Aggregates in Vivo
The mouse experiments were approved by the animal experiment committee of the RIKEN Brain Science Institute. A whole brain except cerebellum of an R6/2 mouse (15) at either 8, 12, or 14 weeks of age was homogenized with 3 ml of PBS containing 0.1% Triton X-100, 5 mm EDTA, 0.8 m NaCl, 10% sucrose, and protease inhibitor mixture (Complete-EDTA; Roche Applied Science) using a Teflon homogenizer. After centrifugation at 20,000 × g, 4 °C for 20 min, 1% Sarkosyl was added to the supernatant, which was further incubated at room temperature for 2 h. The sample was ultracentrifuged at 110,000 × g, 4 °C for 20 min, and the resultant pellets were washed once with 500 μl of PBS containing 5 mm EDTA, 0.8 m NaCl, and 10% sucrose and then once with 500 μl of PBS. The pellets were resuspended with ultrasonication in 100 μl of PBS containing 5 mm EDTA, 0.8 m NaCl, and 50% sucrose and incubated at room temperature for 30 min. After ultracentrifugation at 110,000 × g at 4 °C for 20 min, the supernatant was collected, and then the concentration of sucrose was decreased to 10% by adding PBS with 5 mm EDTA and 0.8 m NaCl. Again, the sample was ultracentrifuged; thereby, the pellets were collected as HTTEX1 aggregates in vivo and resolubilized with the incubation in 100 μl of TFA at 37 °C for 1 h. Successful recovery of HTTEX1 was confirmed by Western blotting analysis using an anti-polyglutamine antibody (1C2). Resolubilized HTTEX1 aggregates in vivo were then dissolved in a buffer (150 μl, pH 8.0) containing 50 mm Tris, 100 mm NaCl, 5 mm CaCl2, and 1 m urea, and further incubated with 2 μg of an endoproteinase Glu-C (Roche Applied Science) at 25 °C for 3 days. After digestion, 80 mg of guanidine HCl and 0.1% (final concentration) of TFA was added, and the supernatant was obtained by ultracentrifugation, desalted with ZipTip C18, and then analyzed by MALDI-TOF mass spectrometry in linear mode using sinapic acid as a matrix. Mass spectra were calibrated by human angiotensin I (Calbiochem; m/z 1,046.2), human ACTH(18–39) (WAKO; m/z 4,930.4), and bombesin (LKT Laboratories; m/z 1619.86).
Electron Microscopy
HTTEX1 aggregates were adsorbed on STEM100Cu grids coated by elastic carbon (Okenshoji), washed with water, and then negatively stained with 2% phosphotungstic acid. Images were obtained using an electron microscope (TecnaiTM Spirit, FEI).
Static Laser Light Scattering
HTTEX1(42Q) aggregates prepared in the presence and absence of BCS were analyzed by using SALD-7100H equipped with SALD-71HC (Shimadzu). The data were analyzed by the WingSALD II software (Shimadzu).
Gel Filtration Chromatography
HTTN16(WT and M8A) peptides were chemically synthesized (Biologica) and dissolved in pure water. 100 μl of 10 μm HTTN16 peptides were loaded on Nanofilm SEC-150 (Sepax Technologies) fitted to a LaChrom Elite (Hitachi) HPLC system. The column was pre-equilibrated with a buffer containing 50 mm Na-Pi and 100 mm NaCl at pH 7.4, and the flow rate was 0.35 ml/min. The chromatogram was obtained by monitoring the absorbance at 214 nm and calibrated using 100 μl of 0.05 mg/ml human ACTH(18–39) (4,930.4 Da), 0.1 mg/ml human angiotensin I (1,296.5 Da), and 0.05 mg/ml bovine milk α-lactoalbumin (Sigma; 14,200 Da).
RESULTS
Methionine Oxidation in Mutant HTT Occurs after the Formation of Insoluble Aggregates
Full-length HTT is a large protein with ∼350 kDa, but the N-terminal fragment corresponding to the first exon in HTT gene (HTTEX1) is sufficient for causing HD-like symptoms and neuropathological changes in animal models (15). HTTEX1, which is composed of N-terminal 17 amino acids followed by a polyQ tract and a Pro-rich region, was purified as a fusion protein with GST and then liberated from the GST tag by proteolytic cleavage using a specific protease, HRV3C (16). To evaluate the aggregation kinetics of HTTEX1 proteins, we have set 0 min at the time when HRV3C was introduced, although almost (∼90%) completion of this proteolytic process required 30 min (supplemental Fig. S1). As shown in Fig. 1A, 20 μm HTTEX1 with a pathogenic length of a polyQ tract (42Q) increased its solution turbidity, whereas no changes were observed when the length of a polyQ tract is normal (18Q). This reproduces previous findings that aggregation of HTTEX1 displays a threshold for polyQ length (17).
FIGURE 1.
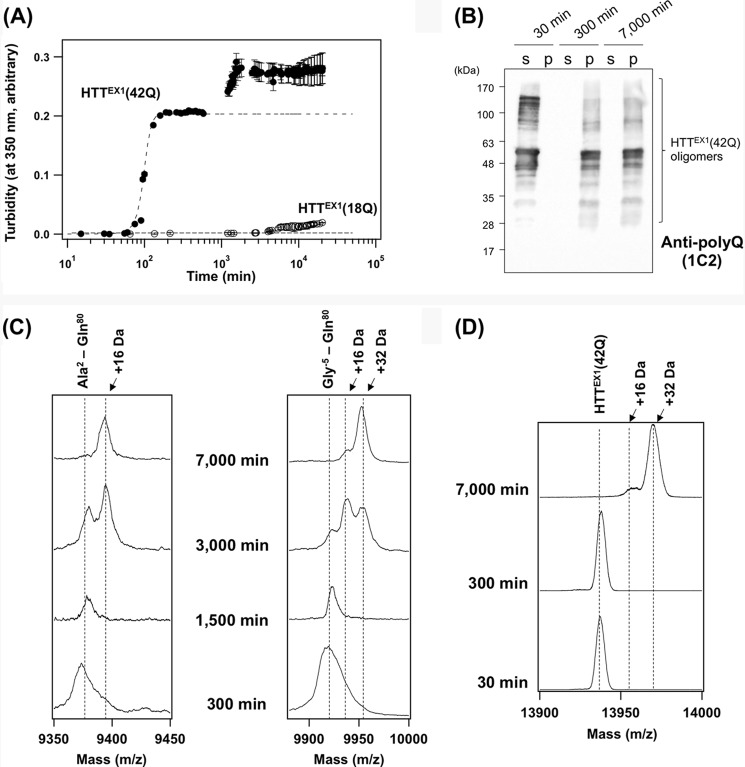
Oxidative modification occurs on HTTEX1 after the formation of insoluble aggregates. A, 20 μm GST-fused HTTEX1 proteins in a GST elution buffer were incubated at 25 °C with 2 units of HRV3C protease to liberate a HTTEX1 fragment, which triggered an aggregation reaction. Increase of the solution turbidity at 350 nm in HTTEX1(42Q) (filled circles) until 500 min was fit with a sigmoidal function (broken curve). In contrast, HTTEX1(18Q) (open circles) did not change its solution turbidity, suggesting no aggregation. B, after addition of an HRV3C protease, HTTEX1(42Q) proteins incubated for 30, 300, and 7,000 min were fractionated by ultracentrifugation into soluble supernatant (s) and insoluble pellets (p). Insoluble pellets were further treated with TFA to resolubilize HTTEX1(42Q) proteins (see “Experimental Procedures”). Both fractions were loaded on a 12.5% SDS-polyacrylamide gel and analyzed by Western blotting with anti-polyQ antibody (1C2). Because of the high aggregation propensity of polyQ-containing proteins, HTTEX1(42Q) proteins were detected as multiple bands in the higher molecular weight region. C, pronase-resistant insoluble pellets of HTTEX1(42Q) aggregates at 300, 1,500, 3,000, and 7,000 min were resolubilized with TFA and analyzed by MALDI-TOF mass spectrometry in linear mode. Two representative mass peaks were shown corresponding to Ala2–Gln80 (left panel) and Gly−5–Gln80 (right panel). Other mass peaks were summarized in supplemental Fig. S3. D, MALDI-TOF mass analysis (reflectron mode) on the soluble HTTEX1(42Q) before aggregation (30 min) and the insoluble HTTEX1(42Q) aggregates formed at 300 and 7,000 min after incubation. The observed mass of HTTEX1(42Q) coincides with the calculated one (13,937 Da).
Consistent with the nucleation-dependent aggregation model (17), the solution turbidity of HTTEX1(42Q) exhibited a sigmoidal change and reached a plateau at 300 min after the addition of HRV3C, where the aggregation appeared to complete (Fig. 1A). Formation of insoluble aggregates was also examined by Western blotting after ultracentrifugal fractionation of the samples into soluble supernatant and insoluble pellets (Fig. 1B and supplemental Fig. S2B). At 30 min after addition of HRV3C, multiple bands immunoreactive to anti-polyQ antibody (1C2) were observed only in soluble supernatant but not in the pellet fraction (Fig. 1B). These bands were also detected with anti-HTT antibody (EM48) but not with anti-GST antibody (supplemental Fig. S2, A and B). Given that an SDS-PAGE analysis on our starting GST-HTTEX1(42Q) sample has shown a major protein band at 45 kDa with small amounts of impurities (0 min in supplemental Fig. S1), the high molecular weight multiple bands in Fig. 1B and supplemental Fig. S2B represent the SDS-resistant soluble oligomers of HTTEX1(42Q). Indeed, multiple bands in high molecular weight regions have been previously observed in HTTEX1 with an elongated polyQ tract in the electrophoretic analysis (14, 16). Notably, at 300 min after the addition of HRV3C, Western blotting analysis using both 1C2 and EM48 antibodies have shown that all of HTTEX1(42Q) molecules are converted from soluble species into insoluble pellets (Fig. 1B and supplemental Fig. S2B), supporting the completion of aggregation at 300 min.
Surprisingly, when we further left the solution containing HTTEX1(42Q), the solution turbidity was found to again start to rise (Fig. 1A). In our experimental conditions, the time when this second phase of the turbidity increase starts was found to be significantly varied among the sample preparations; however, at 300 min, the first phase of the turbidity increase always finished, and the second phase did not start yet. At 7,000 min, we have consistently confirmed that the second phase of the turbidity increase is almost or completely finished. Furthermore, the second phase of the turbidity change did not simply mean the progression of HTTEX1(42Q) aggregation, because there were no differences in the amounts of pelletable HTTEX1(42Q) between 300 and 7,000 min (Fig. 1B and supplemental Fig. S2B). We have also confirmed that all HTTEX1(18Q) molecules remain soluble until 7,000 min after the addition of HRV3C (supplemental Fig. S2, C and D). Given that the solution turbidity depends upon shapes of the scattering particles (or aggregates) (18, 19), we initially suspected that HTTEX1(42Q) aggregates change its molecular structure during the second phase of turbidity changes.
Protein aggregates are generally composed of protease-resistant core and the associated “fuzzy coat” that is a susceptible region for proteolysis (20). To get insight into the structural core of the HTTEX1 aggregates before (300 min) and after (7,000 min) the second phase of the turbidity increase, the fuzzy coat regions in the aggregates were removed by treatment with a nonspecific protease, Pronase (21). The resultant Pronase-resistant materials, which were collected as insoluble pellets by ultracentrifugation, were resolubilized with TFA and analyzed by MALDI-TOF mass spectrometry. All of the identified peptides were found to cover a polyQ tract, consistent with its primary role in the formation of aggregates (supplemental Fig. S3). We also found that the set of peptides constituting the Pronase-resistant cores of HTTEX1(42Q) aggregates was unchanged during the second phase of the turbidity increase (from 300 to 7,000 min in supplemental Fig. S3). In contrast to our initial expectation, therefore, this result suggests no drastic changes in the core structure of HTTEX1(42Q) aggregates during the post-aggregation process.
Despite this, we have noticed that several Pronase-resistant peptides (No. 8–12 mass peaks in supplemental Fig. S3) exhibit the increase of a 16-Da mass unit during the post-aggregation process. For example, during the second phase of turbidity changes (from 300 to 7,000 min), the mass of the Ala2–Gln80 peptide increased by 16 Da (Fig. 1C, left panel), and the 16- and 32-Da (16 Da × 2) increases of the mass were observed in the Gly−5–Gln80 peptide (Fig. 1C, right panel). Such increase in the mass was also detected when the analysis was performed on HTTEX1(42Q) (Fig. 1D) but not on HTTEX1(18Q) (supplemental Fig. S4). Notably, the 16-Da mass increase was observed in the Met8–Gln80 peptide but not in the Lys9–Gln80 peptide (supplemental Fig. S3); therefore, we suspected that the mass increase of 16 Da represents the oxidation of a Met residue to methionine sulfoxide.
Met8 in HTTEX1 Is Oxidized during the Post-aggregation Increase in the Solution Turbidity
HTTEX1 possesses two Met residues at positions 1 and 8, but the initial Met residue has been known to be removed soon after translation in vivo (22). Our HTTEX1 proteins in vitro was, in contrast, prepared by proteolytic cleavage of an N-terminal GST tag, which leaves Met1 with the tag remnant five amino acids (Gly-Pro-Leu-Gly-Ser) at the N-terminal region (supplemental Fig. S3B). We have thus introduced M1A mutation in our in vitro HTTEX1(42Q) protein but still observed a second phase of the turbidity change (Fig. 2A, black) with a 16-Da increase of its mass (Fig. 2B, left panel). These results are consistent with the idea that the oxidation of Met8 contributes to a second phase of the turbidity increase. Indeed, M8A mutation in HTTEX1(42Q) abolished the post-aggregation increase of solution turbidity (Fig. 2A, red), although the mass increase of 16 Da was still observed (Fig. 2B, middle panel). In HTTEX1(42Q) with M1A/M8A double mutations, neither turbidity increase nor mass increase was observed as the post-aggregation process (Fig. 2, A, blue, and B, right panel). It is also notable that a mutation at Met residue significantly delays the aggregation kinetics of HTTEX1(42Q) (Fig. 2A). The N-terminal region of HTTEX1 has been shown to adopt an amphipathic helix structure, of which the hydrophobic surface plays a role in accelerating the aggregation of HTTEX1 with an elongated polyQ tract (23). Met8 constitutes a hydrophobic surface of the helix (24); therefore, its mutation to Ala will perturb the interactions for aggregation and is thus considered to decelerate the aggregation reaction. In HTTEX1(42Q) with M8A mutation, furthermore, such an increased lag phase (∼500 min) before the initial increase of the turbidity might delay the appearance of the second phase; however, we could not observe significant changes in the solution turbidity after 7,000 min at least until 20,000 min (Fig. 2A). Based upon these results, therefore, both Met1 and Met8 in the aggregated state of HTTEX1 in vitro are susceptible for oxidation, but only the oxidation of Met8 leads to the second phase of the turbidity increase.
FIGURE 2.
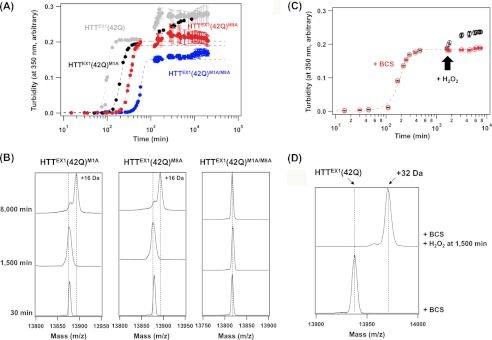
Post-aggregation oxidation at Met8 in HTTEX1(42Q) causes further increase in solution turbidity. A, aggregation reactions of HTTEX1(42Q) with M1A mutation (black), M8A mutation (red), and M1A/M8A mutations (blue) were examined under the same experimental conditions with those in Fig. 1A. For comparison, the aggregation kinetics of HTTEX1(42Q) shown in Fig. 1A was also overlaid (gray). Broken curves represent the sigmoidal fits to the turbidity changes. Note that the second phase of turbidity increase was evident in M1A but not in M8A and M1A/M8A mutant proteins. B, MALDI-TOF mass analysis (reflecton mode) on HTTEX1(42Q) proteins with Met mutations was performed before (30 min) and after (1,500 and 8,000 min) aggregation. The observed mass coincides with the calculated one in all samples examined as follows; 13,877 Da (HTTEX1(42Q)M1A and HTTEX1(42Q)M8A) and 13,817 Da (HTTEX1(42Q)M1A/M8A). C, aggregation kinetics of 20 μm HTTEX1(42Q) in a GST elution buffer in the presence of 50 μm BCS was monitored by the increase of solution turbidity (shown in red). Further addition of 0.1 mm H2O2 at 1,500 min (indicated with an arrow) triggered the second phase of turbidity increase (shown in black). D, MALDI-TOF mass analysis (reflectron mode) confirms that HTTEX1(42Q) aggregates are not oxidized in the presence of BCS even after 7,000 min incubation (a lower trace). In contrast, after H2O2 was added at 1,500 min in the presence of BCS (see C), the aggregates were collected at 7,000 min and analyzed by MALDI-TOF mass spectrometry (an upper trace), showing the 32-Da mass increase.
Oxidation of Met residues is often catalyzed by heavy metal ions (25), and we thus supposed that a trace amount of copper ions often contaminated in chemical reagents and water was responsible for Met1/8 oxidation in HTTEX1 aggregates. Indeed, when a copper-specific chelator, BCS, was included, the second phase of the turbidity increase completely disappeared in the aggregation of HTTEX1(42Q) (Fig. 2C, red) without any changes in the mass until 7,000 min (Fig. 2D). In contrast, even in the presence of BCS, the addition of an oxidant, H2O2, at 1,500 min after incubation started to further increase the solution turbidity (Fig. 2C, black) and resulted in a mass increase of 32 (16 × 2) Da (Fig. 2D). A soluble form of HTTEX1(42Q) before aggregation (at 30 min in Fig. 1A) was not oxidized by incubation with 0.1 mm H2O2 for 30 min (supplemental Fig. S5A). Only when HTTEX1(42Q) aggregates (at 300 min in Fig. 1A) were incubated with 0.1 mm H2O2 for 30 min was mass increase by 32 Da caused by Met oxidation confirmed (supplemental Fig. S5B). Aggregation of HTTEX1 would be thus associated with such conformational changes that render Met8 prone to oxidation. As another negative control, HTTEX1(18Q) was not oxidized by incubating with either copper sulfate or H2O2 for 7,000 min (supplemental Fig. S4). Taken together, we have successfully identified the oxidation of Met8 in HTTEX1 that specifically occurs as the post-aggregation process. Our next question is how such a post-aggregation oxidation of Met residues leads to the turbidity increase.
Post-aggregation Control of Interactions between HTTEX1 Molecules by Met8 Oxidation
As mentioned above, factors affecting turbidity include shape, size, and number of particles (aggregates) in solution (18, 19). No changes were confirmed in the amounts of HTTEX1 aggregates before and after the oxidation of Met8 (Fig. 1B); furthermore, the protease-resistant core structures of HTTEX1 aggregates appeared not to be affected by the oxidation (supplemental Fig. S3). We thus expect that the oxidation of Met8 somehow increases the overall size of HTTEX1 aggregates.
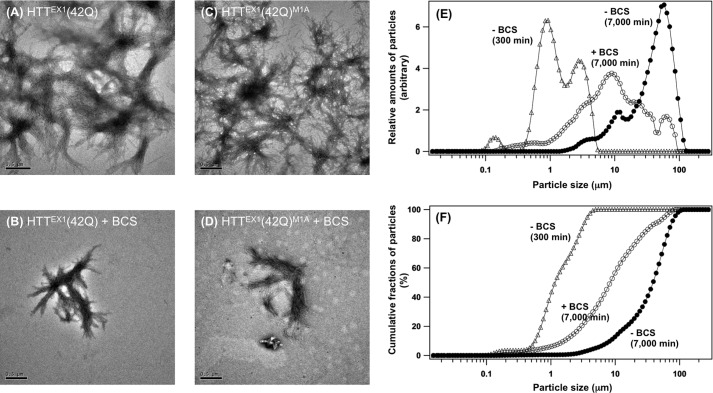
To examine effects of the Met oxidation on the size of HTTEX1 aggregates, we first prepared aggregates without and with the oxidation of Met8 by incubating HTTEX1(42Q) for 7,000 min in the presence and absence of 50 μm BCS, respectively; then these aggregates were directly observed by a transmission electron microscope. Aggregates of HTTEX1 with an elongated polyQ tract have been well known to possess fibrillar morphologies (26), which was also confirmed irrespective of the oxidation state at Met8 in this study (Fig. 3, A and B). Instead, when the Met residues were oxidized, fibrils were found to more extensively interact with each other, and overall appearance of HTTEX1(42Q) aggregates was changed from the isolated and relatively small tangles (Fig. 3B) to the intricate and extended weblike structures (Fig. 3A). HTTEX1(42Q) aggregates before oxidation at Met8 were also examined by incubation for 300 min in the absence of BCS (Fig. 1), and the appearance of those aggregates was confirmed to be similar to the ones prepared by incubation for 7,000 min in the presence of BCS (supplemental Fig. S6A). Morphological differences of HTTEX1 aggregates before and after oxidation at a Met residue were also observed in HTTEX1(42Q) with M1A mutation (Fig. 3, C and D); in contrast, the aggregates of HTTEX1(42Q) with M8A mutation remained relatively small and isolated even after incubation for 7,000 min in the absence of BCS (supplemental Fig. S6B). These results thus support our idea that the post-aggregation oxidation at Met8 adds extended interactions between fibrils.
FIGURE 3.
Post-aggregation oxidation at Met8 facilitates the interactions between HTTEX1(42Q) fibrils to increase overall size of the aggregates. A–D, electron micrograms of HTTEX1(42Q) aggregates (A and B) and HTTEX1(42Q)M1A (C and D) aggregates were shown. The aggregates were prepared by incubation of GST-HTTEX1(42Q) proteins with HRV3C in a GST elution buffer for 7,000 min, and 50 μm BCS was further included to inhibit the oxidation of Met residues (B and D). A bar at the lower left corner of each panel represents 0.5 μm. E and F, size of HTTEX1(42Q) aggregates was analyzed by static laser light scattering. Size distribution of HTTEX1(42Q) aggregates, which formed at 7,000 min in the absence (filled circles) and presence (open circles) of BCS, was shown in E. The HTTEX1(42Q) aggregates formed at 300 min after the addition of HRV3C in the absence of BCS were also examined (open triangles). A cumulative distribution of the aggregate size was calculated and plotted in F: the aggregates at 7,000 min in the presence (open circles) and absence (filled circles) of BCS and the aggregates formed at 300 min in the absence of BCS (open triangles).
We have further performed quantitative analysis on the size of HTTEX1(42Q) aggregates by static laser light scattering. As shown in Fig. 3 (E and F), the size of Met-oxidized HTTEX1(42Q) aggregates, which formed at 7,000 min in the absence of BCS, was mainly populated at ∼60 μm with a minor fraction of the aggregates smaller than 20 μm (filled circles). In contrast, in the HTTEX1(42Q) aggregates formed at 7,000 min in the presence of BCS, where Met8 remains in the thioether state, significantly more fractions of aggregates exhibited the size smaller than 20 μm (Fig. 3, E and F, open circles). Furthermore, the aggregates formed at 300 min in the absence of BCS, where Met8 also remains in the thioether state (Fig. 1A), were found to be much smaller in size and populated between 0.5 and 5 μm (Fig. 3, E and F, open triangles). Oxidation at Met8 thus appears not to be an only factor determining the overall size of HTTEX1(42Q) aggregates, but these quantitative results are consistent with our qualitative transmission electron microscope observations on the morphologies of HTTEX1(42Q) aggregates, and the oxidation of Met8 was confirmed to increase the size of aggregates.
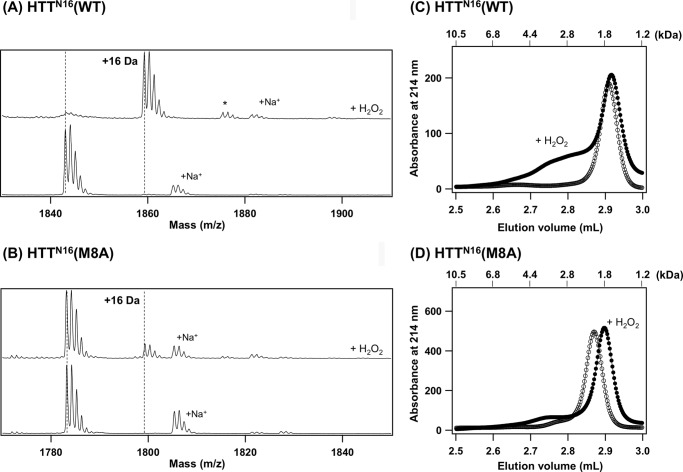
To reveal how the interfibril interactions are augmented by Met oxidation, we focused upon the association state of the N-terminal 16 amino acids (from Ala2 to Phe17, HTTN16) adjacent to the polyQ tract in HTT. HTTN16 was synthesized as a peptide, and its Met8-oxidized form was prepared by incubation with 100 mm H2O2. Successful oxidation was confirmed by MALDI-TOF mass spectrometry; the H2O2-treated HTTN16 peptide increased its mass by as much as 16 Da (Fig. 4A). Besides, these changes were dramatically suppressed in the HTTN16 peptide with M8A mutation (Fig. 4B), indicating that Met8 is the most susceptible site for oxidation in HTTN16. Using these HTTN16 peptides, the association states were analyzed by gel filtration chromatography. As shown in Fig. 4C, HTTN16 was eluted as a single peak corresponding to the monomer size (open circles), whereas the oxidation at Met8 notably associated with the broad peak eluted earlier than the monomer (filled circles). HTTN16 peptides were thus considered to become easily associated with each other upon the oxidation at Met8. Although it remains obscure why a slight difference in the retention time of HTTN16(M8A) peptide was observed upon the addition of H2O2, a broad peak corresponding to the oligomers of HTTN16 was not observed in H2O2-treated HTTN16 with M8A mutation (Fig. 4D). Based upon these results, therefore, the post-aggregation oxidation at Met8 is considered to promote the interactions between the N-terminal regions of HTTEX1; thereby, the individual fibrils tangle to form the weblike structures, resulting in an increased overall size for aggregates.
FIGURE 4.
An N-terminal region of HTTEX1 (HTTN16) forms oligomers upon oxidation at Met8. A and B, oxidation of HTTN16 peptide with hydrogen peroxide was confirmed by MALDI-TOF mass spectrometry in reflectron mode. A, HTTN16(WT) (monoisotopic mass, 1842.0) exhibited a mass peak at m/z 1843.1, which almost completely shifted to the peak at m/z 1859.3 (+16 Da) after treatment with H2O2. A minor peak at m/z 1875.5 (+32 Da, indicated with an asterisk) also appeared, suggesting that the Met oxidation proceeds into the sulfone state in a small fraction of the HTTN16 peptide. B, in contrast, a mass peak of HTTN16(M8A) (monoisotopic mass, 1782.0) was unchanged at m/z 1783.2 even after the addition of H2O2, although a minor peak at m/z 1799.3 (+16 Da) was observed. Mass peaks indicated with +Na+ are due to the Na+ adducts of the peptides. C and D, the gel filtration chromatograms of HTTN16(WT) (C) and HTTN16(M8A) (D). Open and filled circles represent the chromatograms of the peptides without and with the addition of H2O2, respectively. Molecular mass (kDa) calculated by calibration of the column is also shown on top of the figure.
Met8 of HTT Is Oxidized in the Pathological Aggregates of HD Model Mice
Expression of human HTTEX1 with an elongated polyQ tract in mice has been known to well reproduce fibrillar aggregation of mutant HTTEX1 (15). As performed in the aggregates in vitro (Fig. 1D), therefore, we attempted to find the 16-Da mass increase of mutant HTTEX1 purified from transgenic mice (called R6/2 mice (15)), but it failed to obtain a mass peak corresponding to mutant HTTEX1 even after resolubilization with TFA. This is probably because the length of a polyQ tract is relatively long (∼130 Q) in mutant HTTEX1 expressed in an R6/2 mouse; that is, increasing length of a polyQ tract would make it more difficult to keep HTTEX1 soluble and less tolerant for the MALDI-TOF mass analysis. We have thus attempted to perform a mass analysis on soluble fragments that can be produced through a limited proteolysis of mutant HTTEX1.
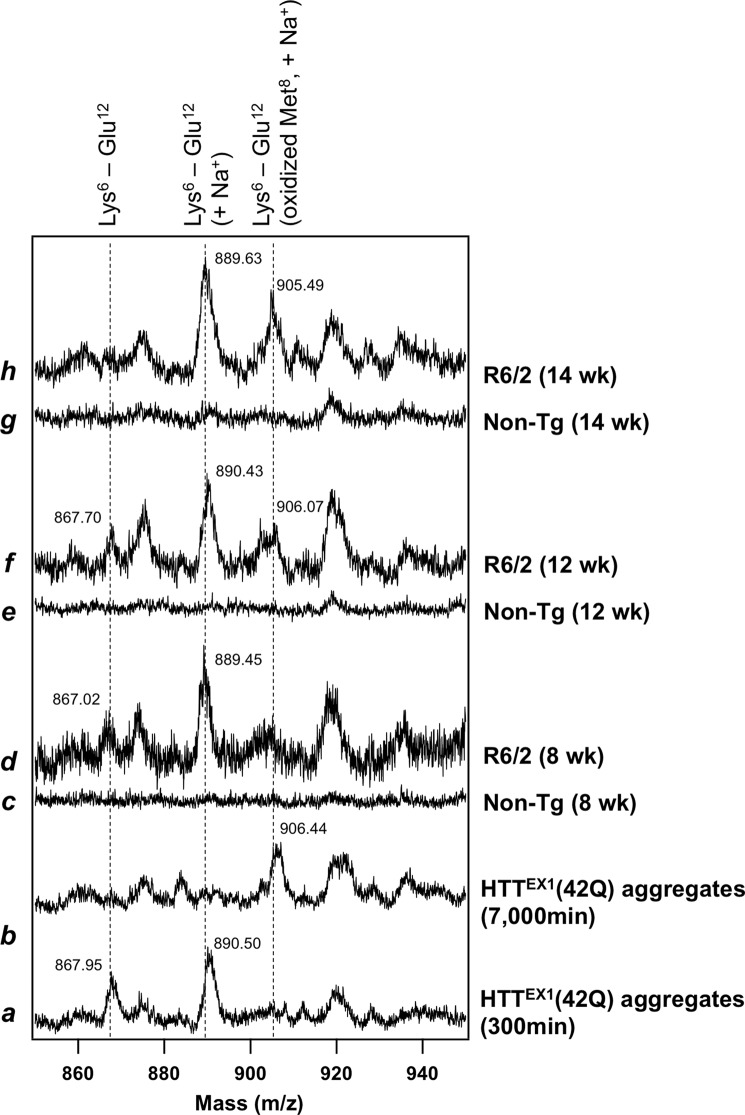
For that purpose, an endoproteinase, Glu-C, was used for a limited proteolysis of HTTEX1, which can produce a relatively small proteolytic fragment (Lys6–Glu12) that contains Met8 but not a polyQ tract. To test whether digestion of HTTEX1 with Glu-C produces a Lys6–Glu12 peptide, in vitro aggregates of HTTEX1(42Q) before Met8 oxidation were resolubilized with TFA, incubated with Glu-C and then analyzed with MALDI-TOF mass spectrometry (see “Experimental Procedures”). As shown in Fig. 5 (trace a), mass peaks were observed at m/z 867.95 and 890.50, which correspond to a Lys6–Glu12 peptide and its sodium adduct, respectively. Furthermore, when the Glu-C treatment was performed on in vitro aggregates after Met8 oxidation, a mass peak was observed at m/z 906.44, which corresponds to a sodium adduct of the Lys6–Glu12 peptide with oxidized Met8 (Fig. 5, trace b). Oxidation of Met8 in HTT aggregates can thus be examined by mass analysis of the Lys6–Glu12 peptide that is produced by the Glu-C treatment.
FIGURE 5.
Proteolytic analysis supports the oxidation of Met8 in HTTEX1 aggregates reproduced in the HD model mouse. Aggregates of HTTEX1(42Q) in vitro (traces a and b) and HTTEX1 purified from R6/2 mice (traces d, f, and h; 8, 12, and 14 weeks of age; see on the right of figure) were first redissolved in TFA and treated with Glu-C protease. The resultant proteolytic fragments were then analyzed with MALDI-TOF mass spectrometry in linear mode. A Lys6-Glu12 peptide (calculated m/z 866.09) exhibited a mass peak at m/z 866–867, and the mass peak of its sodium adduct (+23 Da) was also observed at m/z 889–890. Furthermore, the mass peak observed at m/z 905–906 represents a sodium adduct of a Lys6–Glu12 peptide with oxidized Met8. The observed m/z values were also shown at mass peaks in the figure. These mass peaks were not obtained when nontransgenic mice were analyzed instead of R6/2 mice (traces c, e, and g), supporting that the mass peaks were assigned to the proteolytic fragments of HTTEX1.
Based upon the in vitro results above, further in vivo testing of the Met8 oxidation was performed by purification of HTTEX1 aggregates from brain tissues of R6/2 mice (see “Experimental Procedures”). After resolubilized with TFA, the purified in vivo aggregates were also treated with Glu-C protease and analyzed by MALDI-TOF mass spectrometry. As shown in Fig. 5 (traces d, f, and h), mass peaks of a Lys6–Glu12 peptide (m/z 867) and its sodium adduct (m/z 889–890) were observed, and more importantly, we found the mass peak at m/z 905–906, which corresponds to the peptide with oxidized Met8. All of these mass peaks were not observed from insoluble materials isolated from nontransgenic littermates (traces c, e, and g). It is also notable that the mass peak representing the oxidized Met8 becomes more evident with aging of R6/2 mice. Although MALDI-TOF mass spectrometry is generally not suitable for quantitative analysis, these in vivo observations are consistent with our in vitro results showing the oxidative modifications on HTT aggregates as a post-aggregation process.
DISCUSSION
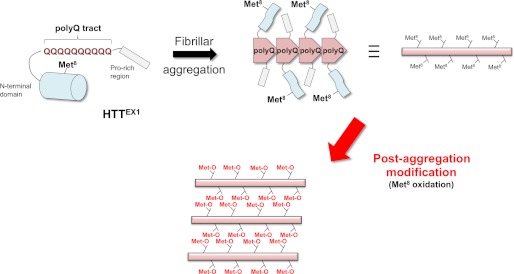
Formation of HTT-immunoreactive inclusions in neurons is a pathological hallmark of HD (9). In various rodent models established so far, expression of HTTEX1 with an elongated polyQ tract is sufficient for reproduction of the disease pathologies including the formation of inclusions (27). Although the abnormal expansion of polyQ is a primary factor to trigger the aggregation, recent studies have supported modulatory roles of post-translational modifications at the flanking sequence (i.e., the regions other than polyQ in HTTEX1) in the aggregation of mutant HTT. In particular, modifications in HTTN16 such as phosphorylation at Ser13/16 (28) and sumoylation/ubiquitylation at Lys6/9/15 (29) have been shown to affect the aggregation kinetics in vitro and in vivo and even the HD-like neuropathologies in model animals. Now, as reported here, we add the oxidation at Met8 as a novel modification in the HTTN16 region in vitro and in vivo, and interestingly, this Met oxidation appears to occur only in the aggregated state of HTTEX1 and to regulate the overall morphologies of aggregates (Fig. 6).
FIGURE 6.
Schematic representation of our proposed mechanism on a post-aggregation modification in mutant HTT proteins. HTTEX1 is composed of an N-terminal region followed by a polyQ tract and a Pro-rich region. In a soluble state shown on the upper left, an N-terminal region of HTTEX1 has been proposed to adopt an α-helical structure, which is represented as a cylinder. When a polyQ tract is abnormally expanded, HTTEX1 forms fibrillar aggregates, in which the elongated polyQ constitutes a structural core (shown on the upper right). Upon fibrillar aggregation, the N-terminal region would be structurally disturbed, which is further supposed to render Met8 susceptible for oxidation. Oxidation at Met8 then introduces the interactions between fibrils via N-terminal regions of HTTEX1, resulting in a size increase for HTTEX1 aggregates.
In general, Met oxidation is not a rare post-translational modification in proteins; rather, Met belongs to the most easily oxidizable amino acids and forms the sulfoxide even under physiologically mild conditions (30). Notably, the increased oxidative stress has been observed in neurodegenerative diseases including HD (10); therefore, Met residues in proteins are supposed to have an increased chance to be oxidized under pathogenic/pathological environment. Indeed, for example, oxidation of a Met residue has been reported in an Aβ peptide forming senile plaques of Alzheimer's diseases (31) and also in infectious forms of PrP proteins in prion diseases (32). Furthermore, the effects of Met oxidation on the kinetics of fibrillar aggregation in vitro have been examined in several disease-related proteins such as α-synuclein (33), PrP (34), and ApoA-I (35). In these previous studies, oxidative modifications have been assumed to precede the aggregation of protein molecules and thereby affect its formation kinetics; however, it remains obscure when proteins are oxidized during the formation of pathological inclusions in vivo. Given that aggregates generally adopt tightly packed structures (36), small oxidants could not attack the amino acid residues in the core but are expected to be accessible to the residues in solvent-exposed regions of aggregates. It is hence possible that some of the protein oxidations occur at the solvent-exposed surface of pathological inclusions even after aggregation. Indeed, we have found here that Met8 is not a constituent of the protease-resistant core in HTTEX1 aggregates and is susceptible to post-aggregation oxidation (Fig. 6).
In soluble HTTEX1 with nonpathogenic length of a polyQ tract, HTTN16 region adopts an amphipathic α-helical structure (37), where Met8 occupies the central area of the nonpolar surface. Met8 is thus supposed to be buried in hydrophobic cores formed by HTTEX1 proteins (23) and protected from the attack by oxidants such as hydrogen peroxide; indeed, computer simulations have supported the compact conformation of a HTT fragment comprising HTTN16 and a polyQ tract (38), where hydrophobic residues in HTTN16 are sequestered in the interface between HTTN16 and a polyQ tract. In contrast, Met8 became oxidized in the aggregated state of HTTEX1 (Figs. 1 and 2), implying conformational changes of HTTN16 upon aggregation. In several studies, HTTN16 has been proposed to be incorporated into the core structures of fibrillar HTTEX1 aggregates (23, 39), but we have found that a polyQ tract but not the HTTN16 region constitutes a protease-resistant core in the aggregates (supplemental Fig. S3). Notably, our results are consistent with recent solid state NMR studies showing that HTTN16 region remains α-helical even in the aggregates but display increased dynamics and water exposure (40). As summarized in Fig. 6, therefore, the hydrophobic surface of α-helical HTTN16 is covered by a polyQ tract in the soluble state, but upon aggregation, polyQ undergoes significant structural changes to constitute a core of fibrils, which would abrogate the interaction between HTTN16 and a polyQ tract. These structural changes associated with aggregation are then considered to increase the exposure of hydrophobic surface of HTTN16 and facilitate the oxidation of Met8.
In our in vitro experiments, Met oxidation appears to create new interactions between HTTEX1 fibrils and thus increases the overall size of aggregates (Fig. 3). Although a molecular mechanism describing the oxidation-dependent increase in size of aggregates remains to be established, a Met residue has been known to become less hydrophobic or even hydrophilic upon its oxidation from thioether into sulfoxide form (41). Such drastic changes in chemical properties of Met would contribute to the oxidation-induced formation of oligomeric structures in a HTTN16 peptide (Fig. 4). As schematically shown in Fig. 6, we thus speculate that the HTTN16 region behaves like an oxidation-induced “zipper” to connect the individual HTTEX1 fibrils, increasing the overall size of aggregates.
Understanding roles of our proposing post-aggregation oxidation in the disease pathomechanism will require further investigations. Although both toxic and beneficial roles of protein aggregation in the pathomechanism of HD have been suggested (15, 42), distinct conformations of HTTEX1 fibrils have been found to exhibit different cytotoxicity toward cultured neuronal cells (43). Although the relation between fibril conformations and toxicity still remains unclear, it is possible that the potential cytotoxicity of HTT aggregates is modulated through those morphological changes upon the Met oxidation. Interestingly, an age-dependent increase in the size of the cytoplasmic HTT aggregates has been observed in biochemical as well as histological examinations on R6/2 HD model mice (44, 45), in which we have confirmed that pathological HTTEX1 proteins are oxidized at its Met8 residue (Fig. 5). Furthermore, the neuropil aggregates in the HD patients have been reported to increase in size as the disease progresses (46). Nonetheless, the age-dependent increase in the aggregate size can also be interpreted as the simple accumulation of mutant HTT in surviving neurons; therefore, further investigation is also required to reveal pathological roles of Met oxidation as a zipper to increase the size of aggregates in HD.
In conclusion, we propose a mechanism describing the post-aggregation modification on protein aggregates, where structural changes caused by aggregation play critical roles. Significant changes in structures upon aggregation have not been limited to HTT but so far suggested in many other pathogenic proteins (21, 47–49). Post-aggregation modifications are thus supposed to describe pathological changes such as oxidation and phosphorylation detected in inclusions. Although exact roles of post-aggregation modifications in disease pathomechanisms remain to be established, insoluble aggregates are not considered to be inert products of protein misfolding but are able to change its morphologies and structures by post-aggregation modifications. Increasing evidence has recently supported the idea that different structures/morphologies of protein aggregates are related to distinct phenotypes (43, 50, 51); therefore, a possible outcome of post-aggregation modifications may be to modulate disease phenotypes/symptoms even after the onset of disease.
Acknowledgments
We thank Prof. Shuji Akiyama (Institute for Molecular Science, Okazaki, Japan) for fruitful discussion on light scattering and Drs. Mitsuru Maruyama and Atsushi Masumoto (Shimadzu, Japan) for the particle analysis using static laser light scattering.
This work was supported by Grants-in-Aid 24111542 (to Y. F.) and 22110004 (to N. N.) for Scientific Research on Innovative Areas and Grant-in-Aid 22770162 for Young Scientists (B) (to Y. F.) and Strategic Research Program for Brain Sciences (to N. N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, funds from CREST from Japan Science and Technology Agency (to N. N.), a grant-in-aid from the Research Committee for Ataxic Diseases from MHLW (to N. N.), and a grant from the Naito Foundation (to Y. F.).

This article contains supplemental Figs. S1–S6.
- HTT
- huntingtin
- HD
- Huntington disease
- HTTEX1
- the N-terminal fragment corresponding to the first exon in HTT gene
- HTTN16
- the N-terminal 16 amino acids adjacent to the polyQ tract in HTT
- polyQ
- polyglutamine
- BCS
- bathocuproine disulfonate.
REFERENCES
- 1. Ross C. A., Poirier M. A. (2004) Protein aggregation and neurodegenerative disease. Nat. Med. 10, (Suppl.) S10–S17 [DOI] [PubMed] [Google Scholar]
- 2. Furukawa Y., Fu R., Deng H. X., Siddique T., O'Halloran T. V. (2006) Disulfide cross-linked protein represents a significant fraction of ALS-associated Cu,Zn-superoxide dismutase aggregates in spinal cords of model mice. Proc. Natl. Acad. Sci. U.S.A. 103, 7148–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grundke-Iqbal I., Iqbal K., Tung Y. C., Quinlan M., Wisniewski H. M., Binder L. I. (1986) Abnormal phosphorylation of the microtubule-associated protein Tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 5. Seet B. T., Dikic I., Zhou M. M., Pawson T. (2006) Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7, 473–483 [DOI] [PubMed] [Google Scholar]
- 6. Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 7. Cattaneo E., Zuccato C., Tartari M. (2005) Normal huntingtin function. An alternative approach to Huntington's disease. Nat. Rev. Neurosci. 6, 919–930 [DOI] [PubMed] [Google Scholar]
- 8. DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J. P., Carraway R., Reeves S. A. (1995) Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14, 1075–1081 [DOI] [PubMed] [Google Scholar]
- 9. Zoghbi H. Y., Orr H. T. (2000) Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 10. Browne S. E., Bowling A. C., MacGarvey U., Baik M. J., Berger S. C., Muqit M. M., Bird E. D., Beal M. F. (1997) Oxidative damage and metabolic dysfunction in Huntington's disease. Selective vulnerability of the basal ganglia. Ann. Neurol. 41, 646–653 [DOI] [PubMed] [Google Scholar]
- 11. Fox J. H., Kama J. A., Lieberman G., Chopra R., Dorsey K., Chopra V., Volitakis I., Cherny R. A., Bush A. I., Hersch S. (2007) Mechanisms of copper ion mediated Huntington's disease progression. PLoS One 2, e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fox J. H., Connor T., Stiles M., Kama J., Lu Z., Dorsey K., Liebermann G., Sapp E., Cherny R. A., Banks M., Volitakis I., DiFiglia M., Berezovska O., Bush A. I., Hersch S. M. (2011) Cysteine oxidation within N-terminal mutant huntingtin promotes oligomerization and delays clearance of soluble protein. J. Biol. Chem. 286, 18320–18330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hands S., Sajjad M. U., Newton M. J., Wyttenbach A. (2011) In vitro and in vivo aggregation of a fragment of huntingtin protein directly causes free radical production. J. Biol. Chem. 286, 44512–44520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. (2009) Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 29, 5153–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 16. Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 17. Wetzel R. (2012) Physical chemistry of polyglutamine. Intriguing tales of a monotonous sequence. J. Mol. Biol. 421, 466–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer R. A., Brunsting A. (1975) Light scattering from nucleated biological cells. Biophys. J. 15, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akiyama S. (2010) Quality control of protein standards for molecular mass determinations by small-angle X-ray scattering. J. Appl. Crystallogr. 43, 237–243 [Google Scholar]
- 20. Wischik C. M., Novak M., Thøgersen H. C., Edwards P. C., Runswick M. J., Jakes R., Walker J. E., Milstein C., Roth M., Klug A. (1988) Isolation of a fragment of tau derived from the core of the paired helical filament of Alzheimer disease. Proc. Natl. Acad. Sci. U.S.A. 85, 4506–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furukawa Y., Kaneko K., Yamanaka K., Nukina N. (2010) Mutation-dependent polymorphism of Cu,Zn-superoxide dismutase aggregates in the familial form of amyotrophic lateral sclerosis. J. Biol. Chem. 285, 22221–22231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aiken C. T., Steffan J. S., Guerrero C. M., Khashwji H., Lukacsovich T., Simmons D., Purcell J. M., Menhaji K., Zhu Y. Z., Green K., Laferla F., Huang L., Thompson L. M., Marsh J. L. (2009) Phosphorylation of threonine 3. Implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 284, 29427–29436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tam S., Spiess C., Auyeung W., Joachimiak L., Chen B., Poirier M. A., Frydman J. (2009) The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat. Struct. Mol. Biol. 16, 1279–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stadtman E. R. (1990) Metal ion-catalyzed oxidation of proteins. Biochemical mechanism and biological consequences. Free Radic Biol. Med. 9, 315–325 [DOI] [PubMed] [Google Scholar]
- 26. Díaz-Hernández M., Moreno-Herrero F., Gómez-Ramos P., Morán M. A., Ferrer I., Baró A. M., Avila J., Hernández F., Lucas J. J. (2004) Biochemical, ultrastructural, and reversibility studies on huntingtin filaments isolated from mouse and human brain. J. Neurosci. 24, 9361–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beal M. F., Ferrante R. J. (2004) Experimental therapeutics in transgenic mouse models of Huntington's disease. Nat. Rev. Neurosci. 5, 373–384 [DOI] [PubMed] [Google Scholar]
- 28. Gu X., Greiner E. R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J. S., Thompson L. M., Wetzel R., Yang X. W. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron 64, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., Marsh J. L. (2004) SUMO modification of Huntingtin and Huntington's disease pathology. Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 30. Vogt W. (1995) Oxidation of methionyl residues in proteins. Tools, targets, and reversal. Free Radic Biol. Med. 18, 93–105 [DOI] [PubMed] [Google Scholar]
- 31. Dong J., Atwood C. S., Anderson V. E., Siedlak S. L., Smith M. A., Perry G., Carey P. R. (2003) Metal binding and oxidation of amyloid-beta within isolated senile plaque cores. Raman microscopic evidence. Biochemistry 42, 2768–2773 [DOI] [PubMed] [Google Scholar]
- 32. Canello T., Engelstein R., Moshel O., Xanthopoulos K., Juanes M. E., Langeveld J., Sklaviadis T., Gasset M., Gabizon R. (2008) Methionine sulfoxides on PrPSc. A prion-specific covalent signature. Biochemistry 47, 8866–8873 [DOI] [PubMed] [Google Scholar]
- 33. Glaser C. B., Yamin G., Uversky V. N., Fink A. L. (2005) Methionine oxidation, alpha-synuclein and Parkinson's disease. Biochim. Biophys. Acta 1703, 157–169 [DOI] [PubMed] [Google Scholar]
- 34. Wolschner C., Giese A., Kretzschmar H. A., Huber R., Moroder L., Budisa N. (2009) Design of anti- and pro-aggregation variants to assess the effects of methionine oxidation in human prion protein. Proc. Natl. Acad. Sci. U.S.A. 106, 7756–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wong Y. Q., Binger K. J., Howlett G. J., Griffin M. D. (2010) Methionine oxidation induces amyloid fibril formation by full-length apolipoprotein A-I. Proc. Natl. Acad. Sci. U.S.A. 107, 1977–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sawaya M. R., Sambashivan S., Nelson R., Ivanova M. I., Sievers S. A., Apostol M. I., Thompson M. J., Balbirnie M., Wiltzius J. J., McFarlane H. T., Madsen A. Ø., Riekel C., Eisenberg D. (2007) Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 [DOI] [PubMed] [Google Scholar]
- 37. Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Secondary structure of Huntingtin amino-terminal region. Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williamson T. E., Vitalis A., Crick S. L., Pappu R. V. (2010) Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J. Mol. Biol. 396, 1295–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lakhani V. V., Ding F., Dokholyan N. V. (2010) Polyglutamine induced misfolding of huntingtin exon1 is modulated by the flanking sequences. PLoS Comput. Biol. 6, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sivanandam V. N., Jayaraman M., Hoop C. L., Kodali R., Wetzel R., van der Wel P. C. (2011) The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 133, 4558–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gellman S. H. (1991) On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry 30, 6633–6636 [DOI] [PubMed] [Google Scholar]
- 42. Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 43. Nekooki-Machida Y., Kurosawa M., Nukina N., Ito K., Oda T., Tanaka M. (2009) Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 9679–9684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stack E. C., Kubilus J. K., Smith K., Cormier K., Del Signore S. J., Guelin E., Ryu H., Hersch S. M., Ferrante R. J. (2005) Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. J. Comp. Neurol. 490, 354–370 [DOI] [PubMed] [Google Scholar]
- 45. Weiss A., Klein C., Woodman B., Sathasivam K., Bibel M., Régulier E., Bates G. P., Paganetti P. (2008) Sensitive biochemical aggregate detection reveals aggregation onset before symptom development in cellular and murine models of Huntington's disease. J. Neurochem. 104, 846–858 [DOI] [PubMed] [Google Scholar]
- 46. Gutekunst C. A., Li S. H., Yi H., Mulroy J. S., Kuemmerle S., Jones R., Rye D., Ferrante R. J., Hersch S. M., Li X. J. (1999) Nuclear and neuropil aggregates in Huntington's disease. Relationship to neuropathology. J. Neurosci. 19, 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding F., Furukawa Y., Nukina N., Dokholyan N. V. (2012) Local unfolding of Cu,Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates. J. Mol. Biol. 421, 548–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laidman J., Forse G. J., Yeates T. O. (2006) Conformational change and assembly through edge beta strands in transthyretin and other amyloid proteins. Acc. Chem. Res. 39, 576–583 [DOI] [PubMed] [Google Scholar]
- 49. Liu C., Sawaya M. R., Eisenberg D. (2011) β-microglobulin forms three-dimensional domain-swapped amyloid fibrils with disulfide linkages. Nat. Struct. Mol. Biol. 18, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Angers R. C., Kang H. E., Napier D., Browning S., Seward T., Mathiason C., Balachandran A., McKenzie D., Castilla J., Soto C., Jewell J., Graham C., Hoover E. A., Telling G. C. (2010) Prion strain mutation determined by prion protein conformational compatibility and primary structure. Science 328, 1154–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tanaka M., Chien P., Naber N., Cooke R., Weissman J. S. (2004) Conformational variations in an infectious protein determine prion strain differences. Nature 428, 323–328 [DOI] [PubMed] [Google Scholar]