Background: Blo t 21 is a paralogue of the group 5 allergen with comparable allergenicity.
Results: Blo t 21 has an additional conformational IgE epitope and only a low to moderate cross-reactivity with Blo t 5.
Conclusion: Blo t 21 represents a novel group of major allergen in the dust mite Blomia tropicalis.
Significance: The structure and IgE epitopes of Blo t 21 allow design of hypoallergen for immunotherapy.
Keywords: Allergen, Immunotherapy, Mutagenesis Site-specific, Nuclear Magnetic Resonance, Structural Biology, Blomia tropicalis, Cross-reactivity, IgE Epitopes
Abstract
Blo t 21 is a paralogue of the group 5 allergen, Blo t 5, a major allergen from the dust mite Blomia tropicalis. Blo t 21 has moderate sequence identity (40.7%) to Blo t 5 and low to moderate cross-reactivity to Blo t 5. In B. tropicalis, the most prevalent and allergenic allergens are in the order of Blo t 21, Blo t 5, and Blo t 7. Here, we determined the NMR solution structure of Blo t 21, which represents the first structure of the group 21 dust mite allergen. The structure of Blo t 21 closely resembles the structures of Blo t 5 and Der p 5, comprising three anti-parallel α-helices arranged in a helical bundle. Using site-directed mutagenesis and specific IgE binding ELISA, Blo t 21 was found to contain both conserved and unique charged IgE epitope residues at the L2 loop region and on helix α3. Cross-inhibition assays confirmed that Blo t 21 has a low to moderate cross-reactivity with Blo t 5 and Der p 5 and represents a novel group of major allergen in B. tropicalis. In addition to group 5 allergens, Blo t 21 has also a low to moderate cross-reactivity with group 21 allergens from Dermatophagoides mites, confirming that B. tropicalis is a major and distinct source of dust mite allergens.
Introduction
House dust mites are recognized as one of the major sources of allergen in house dust, causing allergic asthma worldwide. Approximately 10–30% of the general population and 90% of individuals with allergic asthma have house dust mite allergy. In tropical regions, sensitization to the dust mite Blomia tropicalis is highly prevalent in terms of occurrence when compared with Dermatophagoides mites and is strongly associated with allergic diseases (1, 2). Among the seven allergens isolated from B. tropicalis, the group 5 allergen, Blo t 5, was shown to be the most important allergenic component, with 43% of patients showing a positive reaction (3, 4). Recently, the group 21 allergen was identified in B. tropicalis (Blo t 21) (5), Dermatophagoides pteronyssinus (Der p 21) (6), and Dermatophagoides farinae (Der f 21) (GenBank code EF027123).2 Blo t 21 shares a low to moderate sequence identity with Blo t 5 (40.7%), Der p 5 (28.3%), Der p 21 (38.1%), and Der f 21 (38.9%) and is a paralogue of the group 5 dust mite allergen (5). Blo t 21 is able to elicit a positive allergic response in 57.9% (286 of 494) of atopic patients attending outpatient allergy clinics over 1.5 years (5). Following this, the patterns of mite component-specific IgE were investigated in a total of 253 atopic children (7). The results revealed that the most common mite allergens recognized were Der p 2 (71%), Der p 1 (64%), Blo t 21 (56%), Blo t 5 (45%), and Blo t 7 (44%) (7). Quantitatively, Blo t 21, Blo t 5, and Blo t 7 represented, on average, 36.2, 21.0, and 18.1% of the total specific IgE against B. tropicalis allergens, respectively (7). These results suggest that Blo t 21 is a group of major allergens that is equally or even more important than Blo t 5 in B. tropicalis. In contrast, Der p 1 and 2 represented ∼46 and 29% of the total specific IgE against D. pteronyssinus allergens, respectively. IgE binding to mid-range allergens, such as Der p 21, Der p 7, and Der p 5, generally occurs only in individuals with high titer specific IgE to D. pteronyssinus (7).
The NMR structure of Blo t 5 was determined as a helical bundle consisting of three anti-parallel α-helices (8, 9). A linear epitope comprising four charged residues surrounding the turn region connecting α2 and α3 has been identified to be involved in IgE binding (8). Subsequently, the crystal structure of the homologous group 5 allergen, Der p 5, was determined (10). The exact IgE-binding epitopes on Der p 5, however, were not reported. Furthermore, neither the NMR nor crystal structure of the group 21 allergen is currently available. Based on the protein sequence alignment between Blo t 21 and Blo t 5, all the putative IgE epitopes identified in Blo t 5 are also present in Blo t 21. However, even though the majority (>75%) of the sensitized individuals showed co-sensitization to both Blo t 5 and Blo t 21, these two allergens demonstrated only a low to moderate degree of cross-reactivity (5). A low cross-reactivity between Der p 21 and the group 5 allergens was also observed through IgE and IgG reactivity data and cross-inhibition studies (6). These findings suggest that Blo t 21 represents a novel group of allergens in B. tropicalis and may contain unique IgE epitopes not present in Blo t 5. In addition, it has long been known that the same group of allergens from B. tropicalis and D. pteronyssinus usually displays only a low to moderate degree of cross-reactivity (11). For instance, Blo t 5 exhibits low levels of IgE cross-reactivity with the homologous Der p 5 allergen (12, 13), and there is a lack of human IgE cross-reactivity between Blo t 1 and Der p 1 (14). It would be of interest to determine the cross-reactivity between Blo t 21 and the homologous group 21 allergen from D. pteronyssinus or D. farinae.
In this study, we have determined the NMR structure of Blo t 21 and mapped its IgE-binding epitopes almost to the same region as Blo t 5, but with an additional conformational epitope. The cross-reactivity of Blo t 21 with Blo t 5, Der p 5, and Der f 21 was low; this suggests that Blo t 21 represents a novel group of major allergens in B. tropicalis and should be included in further diagnostic and therapeutic studies of B. tropicalis-based allergies.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Blo t 21
Modified cDNA of Blo t 21 was subcloned into pET-M vector (modified from pET-32a; Merck) for expression of protein for NMR experiments and pGEX-4T-1 vector (GE Healthcare) for expression of proteins for ELISA experiments. The expression vectors with Blo t 21 insert were subsequently transformed into Escherichia coli BL21 cells for expression. Overnight bacterial cultures were grown at 37 °C in 2 liters of LB broth culture medium until A600 reached 0.6. Protein expression was then induced with 0.3 mm of isopropyl-β-d-thiogalactopyranoside at 20 °C overnight. The cell suspension was harvested and resuspended in nickel binding buffer (20 mm Tris-HCl, pH 7.9, 0.5 m NaCl, and 5 mm imidazole). Sonication was used to lyse the cell suspension, and the lysate was later purified by using nickel-nitrilotriacetic acid affinity column (Qiagen). The N-terminal His tag was cleaved from the purified protein using thrombin (3 unit/mg of protein). Untagged Blo t 21 was further purified using a HiLoad 16/60 Superdex 75-pg gel filtration column (GE Healthcare). Purified Blo t 21 was dialyzed against 50 mm phosphate buffer at pH 7.0 and concentrated to ∼0.8 mm. 15N- and 13C-labeled protein was obtained by growing the bacteria in M9 minimal medium; 15N ammonium chloride was used as a nitrogen source, and 13C glucose was used as the sole carbon source.
CD Spectropolarimetry
CD experiments were conducted with 15 μm of protein in 50 mm phosphate buffer at room temperature. CD spectra were acquired with a Jasco J-810 spectropolarimeter using a Hellma quartz cuvette with a 0.1-cm path length. The spectra were recorded at a wavelength range of 190–260 nm with 0.1 nm resolution using a scan speed of 50 nm/min and averaged for eight scans. Each spectrum was corrected by subtracting buffer signal and normalized to mdeg = 0 at wavelength 260 nm. For thermal denaturation, far UV CD signal at 222 nm was followed with a temperature slope of 1 °C/min and a resolution of 0.1 °C over the temperature range from 25 to 80 °C in PBS.
NMR Experiments
All NMR experiments were carried out in Bruker 800 NMR spectrometer equipped with a cryo-probe. The data were processed using NMRPipe (15) and analyzed with NMRDraw and Sparky software (16). The experiments were performed at 298 K using 0.8 mm protein solution in 50 mm phosphate buffer at pH 7.0. Amide proton and 15N chemical shifts were obtained using 1H-15N heteronuclear single quantum coherence. Backbone and side chain 13C chemical shift assignments were carried out using standard triple resonance techniques: HNCACB (17), CBCA(CO)NH (18), and (H)CC(CO)NH (19) experiments. Side chain proton chemical shifts were obtained with H(CCO)NH (19) and HCCH-TOCSY3 (20). Aromatic side chain residue assignment was carried out using two-dimensional TOCSY, two-dimensional NOESY, and double quantum filtered COSY. Proton distance constraints were obtained from three-dimensional 15N-edited NOESY and 13C-edited NOESY experiments with a 100 ms mixing time. HNCACB, CBCA(CO)NH, (H)CC(CO)NH, H(CCO)NH, and 15N-edited NOESY experiments were carried out using buffers containing 5% D2O; HCCH-TOCSY, two-dimensional TOCSY, two-dimensional NOESY, double quantum filtered COSY, and 13C NOESY were carried out using buffers in 100% D2O.
Structural Calculations
NOE distance constraints were derived from three-dimensional 15N-edited NOESY and 13C-edited NOESY experiments. Secondary structure calculations were performed using the chemical shift index of Cα and Cβ-13C chemical shift data (21). Torsion angle constraints were derived from the TALOS (Torsion Angle Likelihood Obtained from Shifts and sequence similarity) program (22). NOE assignments and structure calculations were performed using CNS software (23). The ensemble of structures was subsequently refined using RECOORD standard protocol by CNS in a hydrated environment (24, 25). Twenty of the lowest energy structures of 100 calculated structures were used for analysis. PROCHECK NMR (26) and WHATCHECK (27) software were used to validate the final structure.
Side-directed Mutagenesis
The cDNA of Blo t 21 was subcloned into PGEX-4T-1 expression vector and transformed into E. coli DH5α cells. Eighteen pairs of oligonucleotide primers (1st Base, Singapore) containing mismatch were designed specifically to create single point mutations using PCR-based site-directed mutagenesis. Using the modified SDM kit method, only a single round of PCR using high fidelity Pfu DNA polymerase (Promega) was required. The elongation temperature was extended to 6 min for the amplification of the whole plasmid using the mismatched primers. After 15 cycles, the PCR products were purified using spin column (Qiagen). The methylated, wild-type plasmid in the purified PCR product was digested with DpnI fast digest restriction enzyme (Fermentas). The digested mixture was further purified with the same procedure, prior to the addition of T4 DNA ligase (Fermentas). The mutated plasmids were transformed into E. coli BL21 (DE3) cells and plated on LB broth agar with ampicillin (100 μg/ml) for at least 16 h at 37 °C. Several colonies were selected for sequencing to confirm the mutations. Multiple mutants were generated using the mutated plasmids as template.
Specific IgE Binding ELISA Experiment
All of the ELISA experiments were conducted using GST-tagged fusion proteins for enhanced binding on ELISA plates. All of the patient sera were preadsorbed with 5 mg/ml of GST protein before use. The sera used in this study were prescreened for IgE responses to Blo t 21 in our previous study (7). Wild-type Blo t 21, GST, and mutant proteins were diluted in PBS to 20 μg/ml. Wild-type Blo t 21, GST, or mutants at a concentration of 1 μg/ml was coated onto Maxisorp plates (Nunc) and incubated overnight at 4 °C. The plates were blocked, washed, and incubated with 50 μl of diluted sera from patients at room temperature for 3.5 h. After washing, biotin-conjugated anti-human IgE monoclonal antibody (BD-Pharmingen), diluted 1:250 in PBS-T (1× phosphate-buffered saline with 0.05% Tween 20) 0.05%, was coated into the wells. After 2 h, avidin-conjugated HRP (BD-Pharmingen), diluted 1:1000 in PBS-T 0.05%, was added and incubated for 30 min. The plates were washed thoroughly with PBS-T 0.05% before the addition of 3,3′,5,5′-tetramethylbenzidine substrate. The reaction was detected through absorbance measurements at 655 nm using an ELISA plate reader. Readings obtained from a GST sample were used as a negative control for a base line of IgE binding. All of the experiments were carried out in duplicate, and the results were reported as mean values with standard deviations.
End Point Inhibition and Cross-inhibition Assay
For the end point inhibition assay, 1 μg of GST-tagged protein in a 100 μl volume was coated onto a Maxisorp ELISA plate (Nunc) overnight at 4 °C. The sera were preadsorbed with 5 mg/ml of GST protein and 100 μg/ml of GST-tagged protein (inhibitor). For the cross-inhibition assay, three different concentrations (0.1, 1, and 100 μg/ml) of inhibitors were used. After overnight incubation at 4 °C, plates were washed with PBS-T and blocked with 4% skim milk in PBS with 0.1% Tween 20 for 30 min at room temperature. The plates were washed again with PBS-T and incubated with sera preadsorbed with inhibitor for 2.5 h at room temperature. Subsequent antibody incubation and colorimetric development were performed as described in specific IgE binding ELISA experiment. The results are presented as the mean values from duplicates with standard deviations. The percentage of inhibition was calculated using the following formula.
 |
Protein Data Bank and Biological Magnetic Resonance Bank Deposition
The NMR structure of Blo t 21 has been deposited at the Protein Data Bank with accession code of 2LM9. The NMR chemical shift assignment of Blo t 21 has been deposited at the Biological Magnetic Resonance Bank database with accession code 18107.
RESULTS
NMR Solution Structure of Blo t 21
The cDNA of Blo t 21 was modified by removing the N-terminal 17 residues from Leu-1 to Val-17 (Fig. 1). Based on the sequence alignment with Blo t 5, this region is highly unstructured, and a significant improvement in the 15N heteronuclear single quantum coherence spectrum was achieved by excluding it from the construct of Blo t 21. For NMR sample preparation, Blo t 21 (from residues Asn-18 to Glu-113) was expressed at a high level in E. coli BL21 (DE3) cells as a soluble His-tagged fusion protein at about 30 mg/liter in M9 minimal medium. The N-terminal His tag was removed by thrombin digestion, and the protein was purified to homogeneity using gel filtration chromatography. The purified protein was dialyzed against 50 mm phosphate buffer at pH 7.0 and remained soluble at a concentration of 0.8 mm at 25 °C without precipitation or degradation.
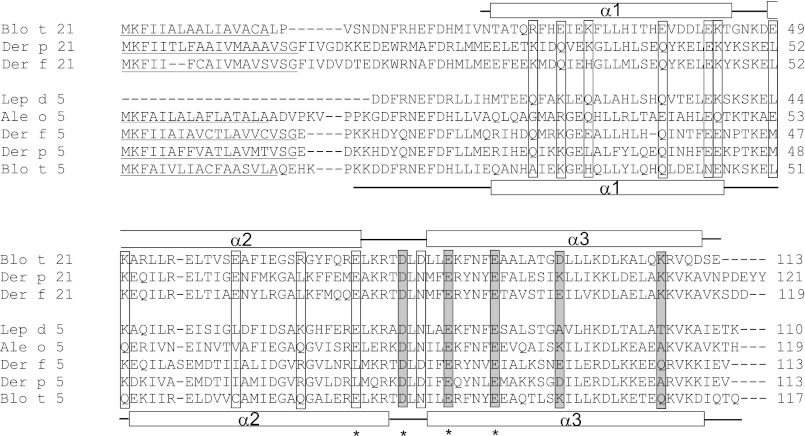
FIGURE 1.
Sequence alignment of Blo t 21 with group 21 and 5 allergens from other house dust mites (Der p 21, Der f 21, Blo t 5, Der p 5, and Der f 5) and less common storage product mites (Ale o 5 from Aleuroglyphus ovatus and Lep d 5 from Lepidoglyphus destructor). Seventeen charged residues (boxed) were chosen for site-directed mutagenesis. Between Blo t 21 and Blo t 5, 13 of the charged residues showed distinct properties, whereas four of the charged residues (marked by asterisks) were previously identified as IgE epitopes in Blo t 5, corresponding to residues Glu-74, Asp-79, Glu-84, and Glu-89 in Blo t 21. Mutation of the residues (gray shaded boxes) were found to cause a significant (>20%) drop in IgE binding in 6 of the 12 allergic patients. The signal peptide regions predicted by SIG-Pred are underlined. The boundaries of the secondary structures from the NMR structure of Blo t 21 and Blo t 5 are shown above and below the sequences, respectively. Note that the N-terminal 17 residues of Blo t 21 are highly unstructured and are not included in sample preparation and structure determination.
The structure of Blo t 21 was determined by employing the angle and distance restraints derived from a series of heteronuclear multidimensional NMR experiments, as described under “Experimental Procedures.” Backbone amides of all but residues Thr-21 and His-27 were assigned, and 97% of the nonlabile and nonaromatic protons were assigned. A total of 1,729 NOE restraints and 171 TALOS angle restraints were used to calculate the three-dimensional structure using RECOORD by CNS in a hydrated environment; more detailed statistics of the solution structure can be seen in Table 1. The final structure is represented as a 20-conformer ensemble with a well ordered backbone and all atoms with a root mean square of 0.87 ± 0.16 and 1.73 ± 0.12 Å, respectively (Fig. 2A). As shown in Fig. 2B, Blo t 21 consists of three anti-parallel α-helices forming an elongated helix bundle. The three α-helices, denoted α1, α2, and α3, are formed by residues Thr-19 to Thr-44, Glu-49 to Glu-74, and Leu-82 to Asp-111, respectively, separated by two loops, L1 and L2. Similar to most of the other inhalant allergens, Blo t 21 contains a high percentage of basic (15.0%) and acidic (20.4%) residues. This high percentage of charged residues is also observed in Blo t 5 (15.4% basic and 22.2% acidic), Der p 5 (18.6% basic and 23.9% acidic), Der p 21 (19.0% basic and 22.3% acidic), and Der f 21 (16.0% basic and 22.7% acidic). The surface of the protein is highly charged, with an even distribution of exposed basic and acidic residues (Fig. 2C).
TABLE 1.
Structural statistics for the NMR structure of Blo t 21
| Data collection | Statistics |
|---|---|
| Restraints used for calculation | |
| Total NOE restrains | 1729 |
| Intraresidue | 523 |
| Sequential (|i − j| = 1) | 454 |
| Medium range (1 < |i − j| < 5) | 416 |
| Long range (|i − j| ≥ 5) | 216 |
| Hydrogen bond restraints | 120 |
| Dihedral angle restraints (ϕ, ψ) | 171 |
| Violations (means ± S.D.) | |
| Distance constraints (Å) | 0.03 ± 0.01 |
| Dihedral angle constraints (°) | 0.66 ± 0.07 |
| Deviations from idealized covalent geometry | |
| Bond length (Å) | 0.01 ± 2.72e−4 |
| Bond angles (°) | 1.35 ± 0.03 |
| Impropers (°) | 1.34 ± 0.06 |
| Average pairwise root mean square deviation | |
| All backbone atoms (Å) | 1.08 ± 0.87 |
| Well ordered backbone atoms (Å) | 0.87 ± 0.16 |
| All heavy atoms (Å) | 1.94 ± 0.17 |
| Well ordered heavy atoms (Å) | 1.73 ± 0.12 |
| Ramachandran plot statistics | |
| Most favored (%) | 94.6 ± 1.2 |
| Additionally allowed (%) | 4.2 ± 1.2 |
| Generously allowed (%) | 0.6 ± 0.8 |
| Disallowed (%) | 0.7 ± 0.7 |
| Energies | |
| NOE (kcal/mol) | 1.1 ± 0.9 |
| Dihedrals (kcal/mol) | 9.3 ± 2.0 |
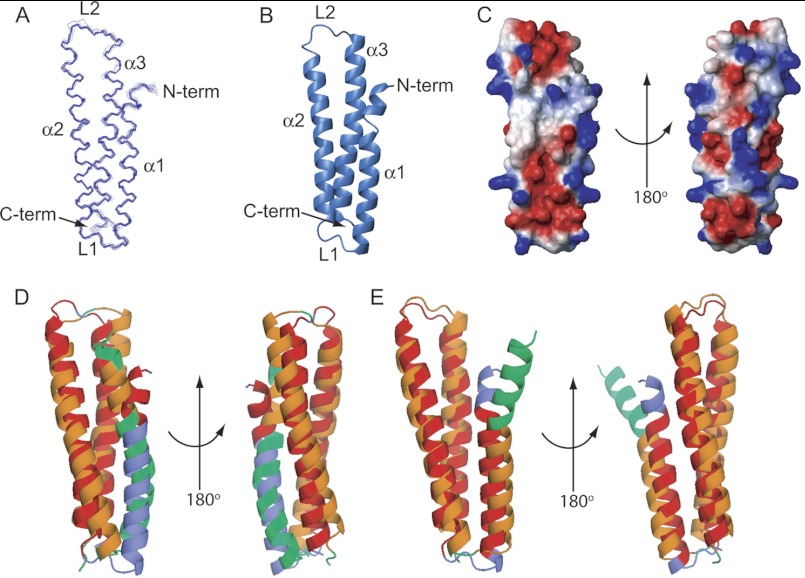
FIGURE 2.
The NMR solution structure of Blo t 21. A, an ensemble of 20 of the best structures of Blo t 21 solved by NMR. The N terminus and helices α1, α2, and α3 are labeled in the diagram. B, a ribbon diagram of the lowest energy conformer of Blo t 21. C, two orientations of the surface diagram of Blo t 21 with electrostatic potential as calculated by the program MOLMOL (32). D and E, superposition of Blo t 21 with Blo t 5 (D, Protein Data Bank code 2JMH) and Der p 5 (E, Protein Data Bank code 3MQ1) prepared using the TopMatch web service (28, 29). The query structure (Blo t 21) is colored blue, and the target sequences (Blo t 5 or Der p 5) are colored green. Pairs of structurally equivalent residues are colored orange (Blo t 21) and red (Blo t 5 or Der p 5). The figures were generated by the program PyMOL (33). N-term, N-terminal; C-term, C-terminal.
To make comparisons between the structures of Blo t 21, Blo t 5, and Der p 5, structure superposition was performed using the TopMatch (28, 29) web service. Blo t 21 superimposed with Blo t 5 (30) with a root mean square deviation of 2.8 Å for Cα atoms of 89 structurally equivalent residues; the main differences were found at the N-terminal region of helix α1 (Fig. 2D). For Der p 5 (10), Blo t 21 superimposed with a root mean square deviation of 2.4 Å for Cα atoms of 91 structurally equivalent residues; the main differences were also found at the extreme N-terminal region of helix α1 (Fig. 2E). Overall, the structures of Blo t 21, Blo t 5, and Der p 5 are highly similar, with small discrepancies at the N-terminal helix.
Site-directed Mutagenesis and IgE Epitope Mapping of Blo t 21
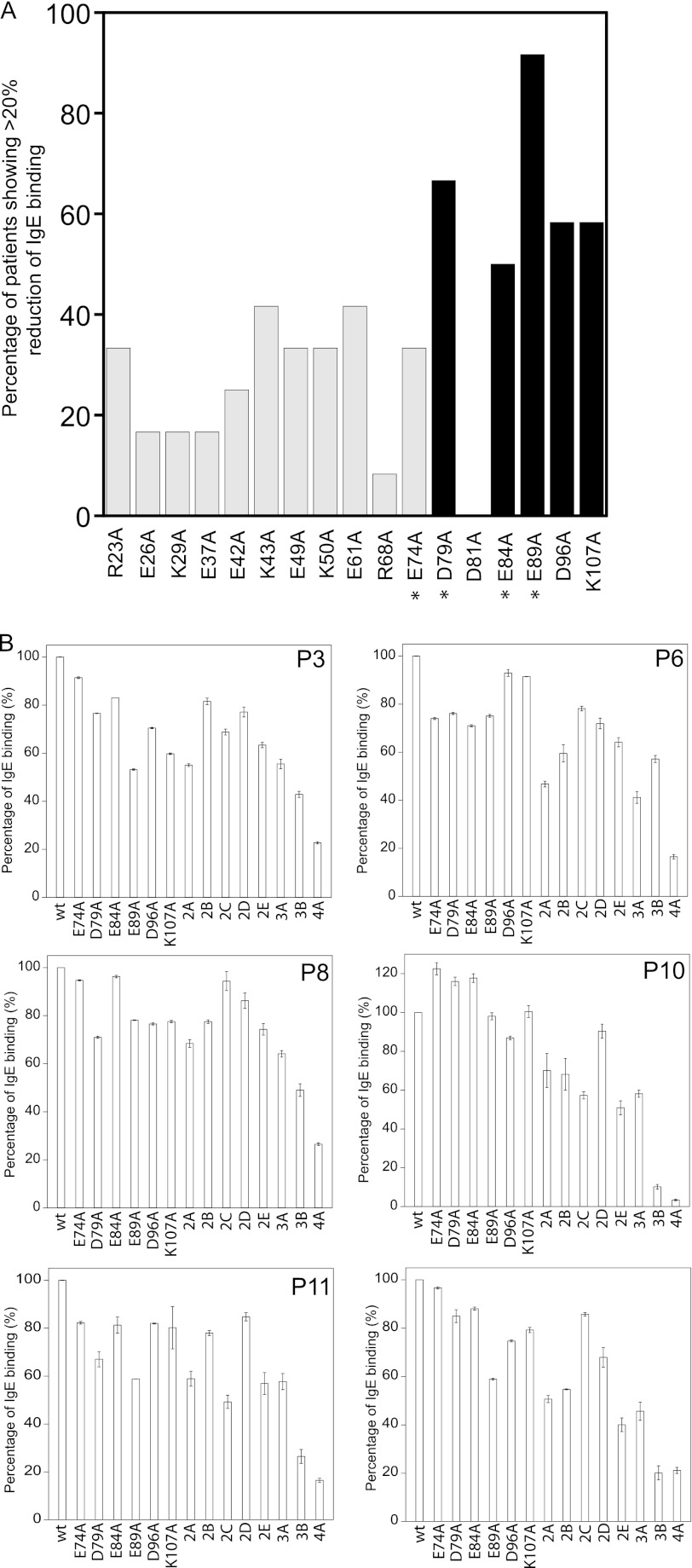
To identify novel IgE epitopes in Blo t 21, 13 charged residues with distinct properties as compared with the residues in Blo t 5 were selected for site-directed mutagenesis using alanine substitution (Fig. 1). In addition, the four previously identified IgE epitope residues of Blo t 5 (corresponding to Glu-74, Asp-79, Glu-84, and Glu-89 of Blo t 21) that are conserved in Blo t 21 were also targeted. Each of these Blo t 21 mutants were then tested for their IgE binding capabilities. All of the ELISA experiments were conducted using sera from 12 allergic patients, labeled as P1–P12. Mutations were scored as significant if they caused more than a 20% reduction in IgE binding as compared with the wild-type Blo t 21. As shown in Fig. 3A, of the 17 residues mutated, five caused a significant reduction in IgE binding in 50% or more of the patients' sera. Mutation of residue Glu-89 to alanine (E89A) conferred the most significant IgE reduction, as demonstrated in 11 of the 12 (91.7%) serum samples tested; this was followed by 8 of 12 (66.7%) for mutant D79A, 7 of 12 (58.3%) for mutants D96A and K107A, and 6 of 12 (50%) for mutant E84A. Among these five residues, the corresponding residues of Asp-79, Glu-84, and Glu-89 in Blo t 5 were also identified as three of the four essential IgE-binding epitopes in Blo t 5 (Fig. 3A) (8). These data suggest that the main IgE-binding epitope of Blo t 21 is located in similar region to that observed in Blo t 5 and that these two allergens could share at least some common IgE epitopes.
FIGURE 3.
Site-directed mutagenesis of Blo t 21 for determination of IgE-binding epitopes. A, prevalence of IgE binding reduction as induced by single site mutations on Blo t 21. The percentage of occurrence of significant reduction (>20%) in IgE binding caused by each of the 17 mutations among a total of 12 patients is determined. The five Blo t 21 mutants D79A, E84A, E89A, D96A, and K107A that caused a significant reduction in IgE binding in ≥50% of patients are highlighted in black. The Blo t 21 mutants that contain a single mutation at the previously identified IgE-binding epitope residue of Blo t 5 are marked with asterisks. B, percentage of IgE binding by wild-type and mutant Blo t 21 using sera from individual patients. As compared with the single mutants of Blo t 21, there was an overall further reduction of IgE binding with double mutants (2A, E74A/D79A; 2B, E74A/E84A; 2C, E74A/E89A; 2D, D79A/E89A; and 2E, E89A/D96A), triple mutants (3A, E74A/D79A/E89A; and 3B, D79A/E89A/D96A), and the quadruple mutant (4A, E74A/D79A/E89A/D96A). In all of the graphs, the averages of three independent experiments with duplicates are plotted with their standard deviations.
There are, however, some differences in the IgE epitope residues observed between Blo t 5 and Blo t 21. In Blo t 5, alanine substitution of the residue corresponding to Asp-79 demonstrated the most drastic reduction in IgE binding (8), whereas in Blo t 21, residue Glu-89 seemed to play a more important role in IgE binding. We found that the four conserved residues, i.e., Glu-74, Asp-79, Glu-84, and Glu-89, in Blo t 21, and the corresponding residues in Blo t 5 were oriented differently (Fig. 4, A and B), suggesting that these residues may play different roles in IgE binding in these proteins.
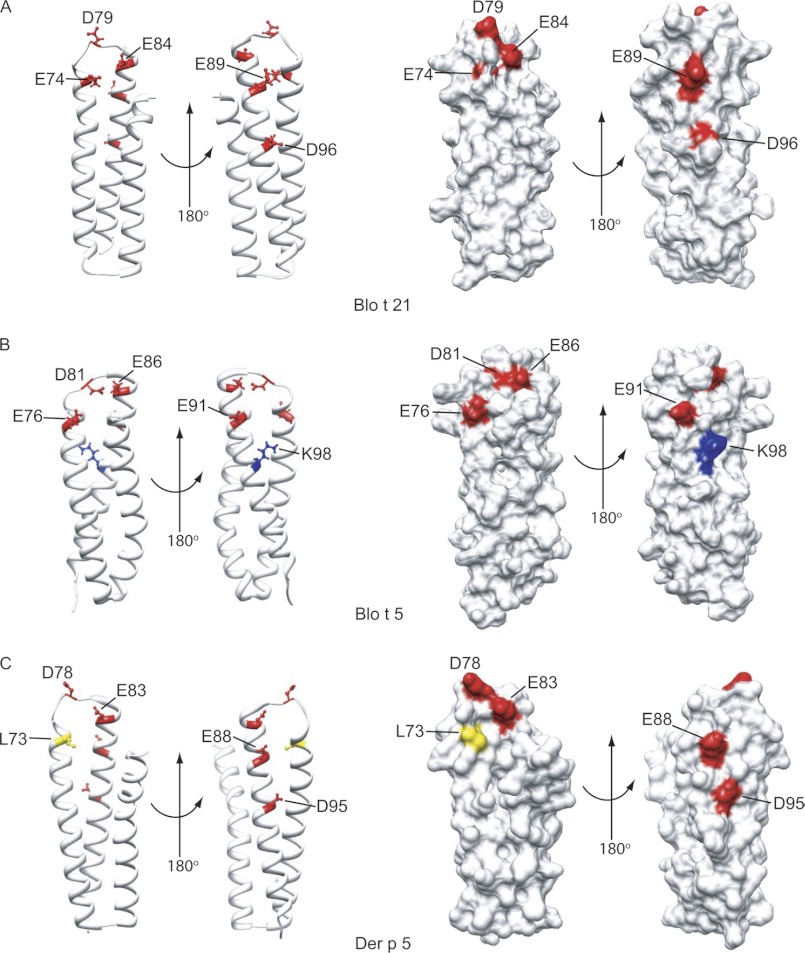
FIGURE 4.
Corresponding locations of the IgE-binding epitopes of Blo t 21 on the structures of Blo t 21, Blo t 5, and Der p 5. A, the mapped IgE-binding epitope residues of Blo t 21 are showed as ball-and-stick models on the ribbon diagram of Blo t 21, in two different orientations. The surface diagram of Blo t 21 shows that residues Glu-74, Asp-79, and Glu-84 form a surface cluster on one side of Blo t 21, whereas residues Glu-89 and Asp-96 form another cluster on the opposite side. B and C, the corresponding locations of the mapped IgE-binding epitope residues of Blo t 21 on the ribbon and surface diagrams of Blo t 5 and Der p 5, respectively. Positively charged, negatively charged, and hydrophobic residues are colored in blue, red, and yellow, respectively. Note that Asp-96 of Blo t 21 is replaced with Lys-98 in Blo t 5, whereas Glu-74 of Blo t 21 is replaced with Leu-73 in Der p 5. The figures were generated by the program Chimera (34).
Asp-96 is a novel IgE-binding epitope residue, and it is located in the vicinity of the previously identified IgE-binding epitopes of Blo t 5 (Fig. 4A). In Blo t 5, the corresponding residue of Asp-96 is replaced with an oppositely charged residue, Lys-98 (Fig. 4B). Based on the structure of Blo t 21, Asp-96 could form a conformational IgE-binding epitope with Glu-89; therefore, it was selected for preparation of mutants together with other IgE-binding epitope residues of Blo t 21. Mutation of Lys-107, on the other hand, caused a reduction in IgE binding for a similar number of patient sera as Asp-96; however, this residue is isolated and located further away from the other IgE-binding epitopes of Blo t 21, so further site-directed mutagenesis experiments were performed without Lys-107.
A Novel Conformational Epitope in Blo t 21 That Is Absent in Blo t 5
In Blo t 21, the four identified IgE epitope residues (Asp-79, Glu-84, Glu-89, and Asp-96) and Glu-74 (one of the identified IgE epitope of Blo t 5) are arranged in two separate patches, with Glu-74, Asp-79, and Glu-84 on one side and Glu-89 and Asp-96 on the other side of the protein (Fig. 4A). In Blo t 5, residues corresponding to Glu-74, Asp-79, Glu-84, and Glu-89 of Blo t 21 were arranged as a linear epitope and in close proximity to each other. Asp-96 is a unique IgE epitope in Blo t 21 that is replaced with Lys-98 in Blo t 5 (Fig. 4B). In Der p 5, a hydrophobic Leu-73 residue corresponds with the Glu-74 in Blo t 21 (Fig. 4C). To investigate the effect of these residues on IgE binding, various double, triple, and quadruple point mutants, encompassing these five charged residues, were generated in Blo t 21. Far UV CD spectra were acquired for each mutant to ensure that the loss of IgE binding was due to the removal of the IgE epitope rather than changes in the structure of the protein. Only mutants that gave CD spectra closely resembling that of the wild-type Blo t 21 will be included in the IgE binding assay (supplemental Fig. S1). Five double mutants (2A, E74A/D79A; 2B, E74A/E84A; 2C, E74A/E89A; 2D, D79A/E89A; and 2E, E89A/D96A), two triple mutants (3A, E74A/D79A/E89A; and 3B, D79A/E89A/D96A), and one quadruple mutant (4A, E74A/D79A/E89A/D96A) were able to be used. Thermal denaturation of the triple mutants 3A and 3B and the quadruple mutant 4A showed that these mutants had similar Tm as the wild-type Blo t 21 (supplemental Fig. S2). However, the CD spectra of Blo t 21 mutants D79A/E84A, E74A/D79A/E84A/D96A, and E74A/E84A/E89A/D96A were found to be significantly different from that of the wild-type Blo t 21 and were excluded from the IgE binding assay (supplemental Fig. S1). In addition, other mutants including E84A/E89A, E74A/D79A/E84A/E89A, and E74A/D79A/E84A/E89A/D96A were found to be unstable during purification and were also not included in the IgE binding assay (data not shown).
The suitable mutants of Blo t 21 were tested for their IgE binding reactivity using sera from six patients with significantly higher IgE titers. As shown in Fig. 3B, double mutants 2A, 2B, and 2E showed a further reduction in IgE binding reactivity as compared with those of the single mutants. In general, the degree of reduction in IgE binding was higher for 2A as compared with 2B and 2E. When compared with the double mutants, further reductions in IgE binding were demonstrated by both triple mutants, with mutant 3B showing a higher reduction than 3A; this suggests that Asp-96 is more important in the IgE-binding epitope than Glu-74 in Blo t 21. As expected, the quadruple mutant, 4A, was able to even further reduce the IgE binding as compared with the triple mutants (Fig. 3B). This result indicates that residues Glu-74, Asp-79, Glu-89, and Asp-96 account for most of the IgE binding in Blo t 21. The IgE reactivity of mutant 4A ranged from 3.4 to 26.5% of the wild-type Blo t 21, with an average of 17.8% for these six patients. We expect that this reduction of IgE binding could be further enhanced by including a Glu-84 mutation. However, it seems that including this mutation renders the protein unstable, and thus, data for the E74A/D79A/E84A/E89A/D96A mutant are not available.
Low to Moderate Cross-reactivity between Blo t 21, Blo t 5, and Der p 5
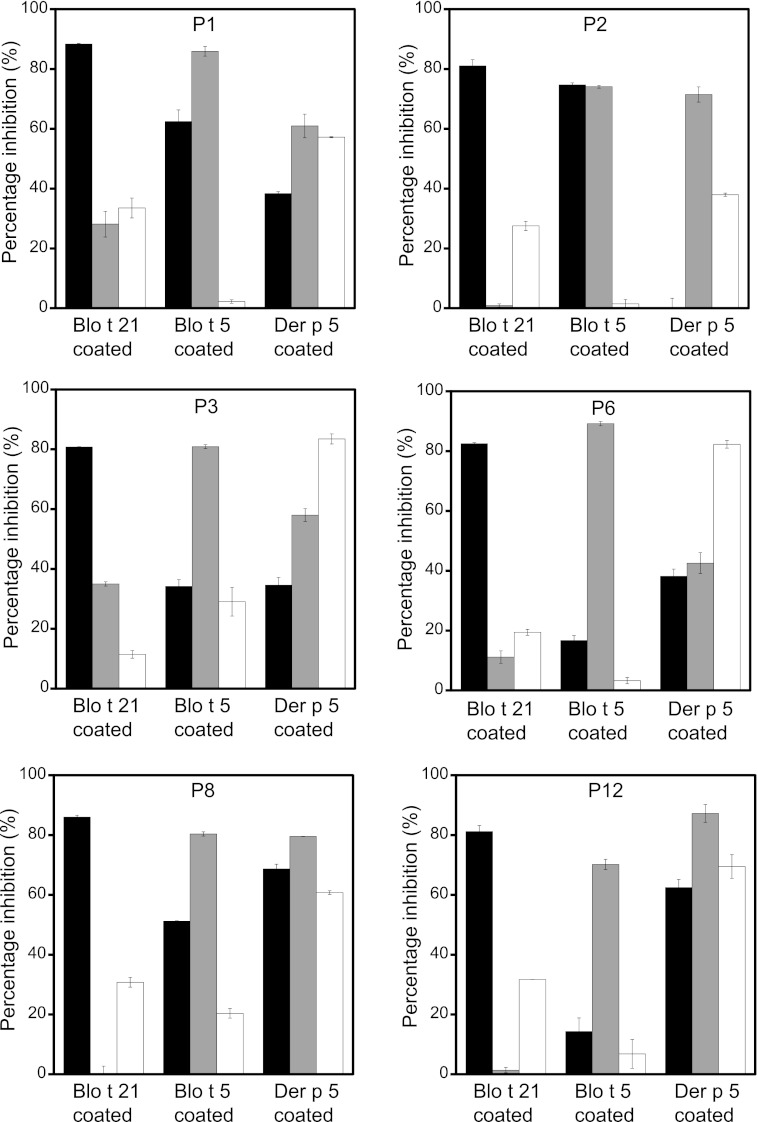
To confirm that Blo t 21 indeed represents a novel group of dust mite allergens in B. tropicalis that is different from the group 5 allergens, we investigated the cross-reactivity of Blo t 21 with Blo t 5 and Der p 5 by cross-inhibition IgE binding assay using sera from six allergic patients. Fig. 5 shows that Blo t 21 inhibited, on average, 83.3% (range, 80.8–88.3%) of IgE binding to itself but inhibited 42.3% (range, 14.3–74.7%) of IgE binding to Blo t 5. In contrast, Blo t 5 inhibited an average of 80.1% (range, 70.2–89.2%) of IgE binding to itself, whereas it could only inhibit 12.8% (range, 0.1–35%) of IgE binding to Blo t 21. These results suggest that Blo t 21 can moderately inhibit IgE binding to Blo t 5 but that Blo t 5 can only inhibit IgE binding to Blo t 21 to a very low degree. Our results are consistent with previous work showing that Blo t 21 and Blo t 5 had only a low to moderate degree of cross-reactivity (5).
FIGURE 5.
Cross-inhibition of IgE binding among Blo t 21, Blo t 5, and Der p 5. End point cross-inhibition experiments of IgE binding were conducted using sera from six patients preadsorbed with 100 μg/ml of inhibitor protein. The bars represent percentages of inhibition of IgE binding. Black, gray, and white bars indicate the serum preadsorbed with GST-tagged Blo t 21, Blo t 5, and Der p 5, respectively. Specific IgE binding experiments were performed with plates coated with Blo t 21, Blo t 5, or Der p 5 using the sera preadsorbed with different inhibitor proteins. The averages of two independent experiments with duplicates were plotted with their standard deviations.
A similar trend of cross-reactivity was also observed between Blo t 21 and Der p 5. Blo t 21 inhibited, on average, 40.4% (range, 0–68.7%) of IgE binding to Der p 5, but Der p 5 could only weakly inhibit 25.7% (range, 11.4–33.5%) of IgE binding to Blo t 21, even though it could inhibit an average of 65.2% (range, 38–83.5%) of IgE binding to itself (Fig. 5). These data suggest that Blo t 21 has only a low to moderate degree of cross-reactivity to Der p 5 and confirms that Blo t 21 represents a novel group of allergens that is distinct from group 5 allergens from both B. tropicalis (Blo t 5) and D. pteronyssinus (Der p 5).
Low to Moderate Cross-reactivity between Blo t 21 and Der f 21
Blo t 5 and Der p 5 are group 5 allergens from different species of mites that have a relatively low sequence identity of 44.2%. Furthermore, they only cross-inhibit at a low or moderate level, as determined by extensive in vitro and in vivo studies (11–13, 31). Here, we confirm that Blo t 5 can inhibit moderately, with an average of 66.7% (range, 42.6–87.2%) of IgE binding to Der p 5; by contrast, Der p 5 can only inhibit very weakly, with an average of 10.6% (range, 1.5–29%) of IgE binding to Blo t 5 (Fig. 5). These data agree with the low to moderate cross-reactivity reported for these allergens. Because the sequence identity between Blo t 21 and Der f 21 (sequence identity of 70.6% to Der p 21) is also relatively low at 38.1%, we sought to investigate the degree of cross-reactivity for group 21 allergens across different mite species.
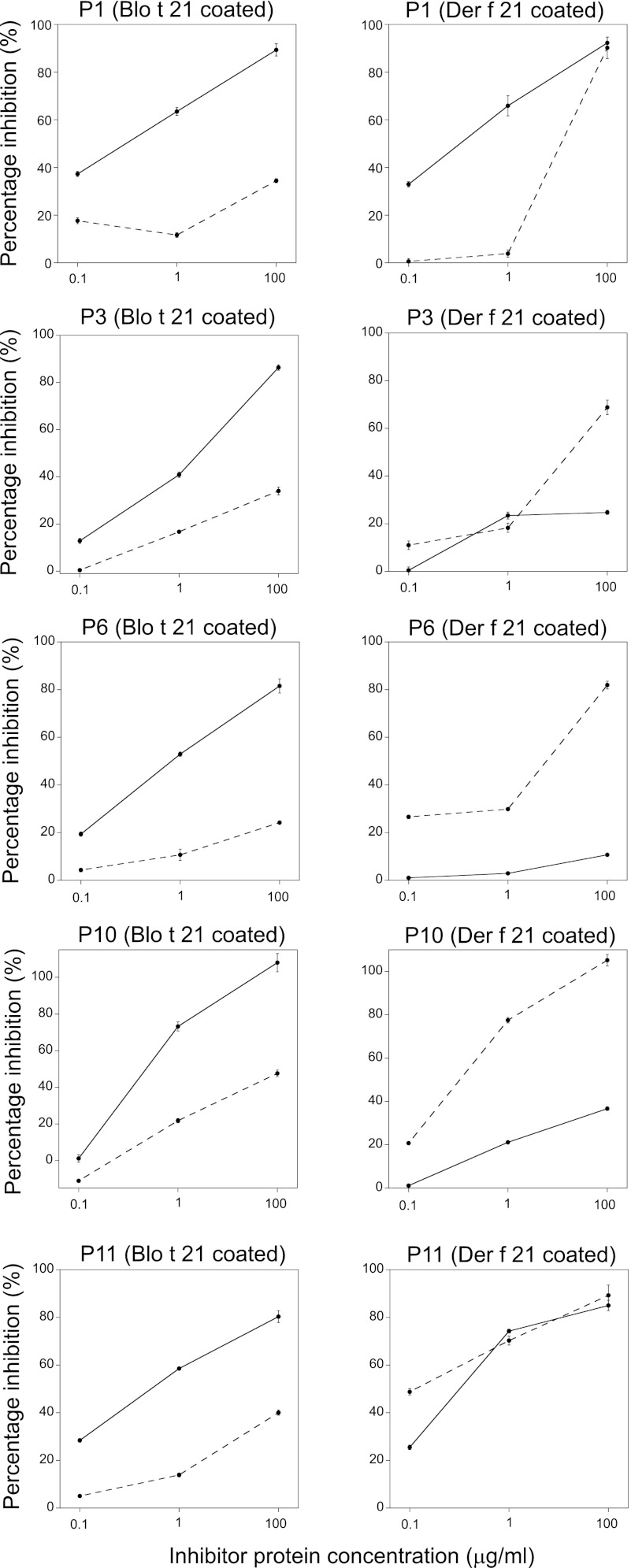
Sera from five allergic patients were used for a cross-inhibition IgE binding assay between Blo t 21 and Der f 21. Based on the data from an inhibitor concentration of 100 μg/ml, Blo t 21 could inhibit an average of 89.1% (range, 80.3–108%) of IgE binding to itself, but it could only inhibit an average of 49.9% (range, 10.7–92.3% for five patients) of IgE binding to Der f 21 (Fig. 6). On the other hand, Der f 21 could inhibit, on average, 87.1% (range, 68.8–105.2% for five patients) of IgE binding to itself, whereas it only inhibited 36.0% (range, 24.2–47.6%) of IgE binding to Blo t 21 (Fig. 6). In certain cases, e.g., in patients P1 and P11, Blo t 21 could inhibit IgE binding to Der f 21 to a similar degree as Der f 21 inhibiting IgE binding to itself. These results suggest that, although both Blo t 21 and Der f 21 belong to the group 21 allergen, they have only a low to moderate degree of cross-reactivity because they are from different species of dust mites.
FIGURE 6.
Cross-inhibition of IgE binding between Blo t 21 and Der f 21. Cross-inhibition experiments of IgE binding were conducted using sera from five patients preadsorbed with 0.1, 1, or 100 μg/ml of Blo t 21 (solid line) or Der f 21 (dotted line). The percentage of inhibition of IgE binding were determined with plates coated with either Blo t 21 or Der f 21 and using patient sera preadsorbed with different concentrations of either Blo t 21 or Der f 21. The average from duplicates of a single experiment was plotted with their standard deviations.
DISCUSSION
Blo t 5 is a well characterized major allergen of B. tropicalis (4), but because of gene duplication, a paralogue, Blo t 21, exists (5). The occurrence of this paralogue in a group of allergens is not common, and Blo t 21 is the first case to be reported. Although both Blo t 5 and Blo t 21 are among the most allergenic groups of allergens in B. tropicalis (7), as yet, the exact physiological function of these allergens is unknown. The multimeric state of Der p 5 in crystal contains a hydrophobic pocket that may be involved in binding of ligand molecules (10); whether binding of the ligand will affect allergenicity remains to be determined. More cases are needed to assess whether there is a relationship between the allergenicity of a protein and the chance occurrence of a paralogue.
Here, we report the first structure of a group 21 allergen from dust mite. The three-dimensional NMR structure of Blo t 21 shows that it consists of three anti-parallel α-helices assembled in a helical bundle resembling that of Blo t 5 (8, 30) and Der p 5 (10). Like many other allergens, both group 5 and 21 allergens have a high percentage of charged residues. The sequence identities of Blo t 21 to group 5 allergens, such as Blo t 5 (40.7%) and Der p 5 (44.4%), are low, suggesting that it may represent a novel group of allergens with disparate IgE epitope(s) compared with group 5 allergens. It was previously reported that Blo t 5 and Blo t 21 have only a low to moderate degree of cross-reactivity (5). The sequence identity of Blo t 21 to other identified group 21 allergens, such as Der p 21 (38.1%) and Der f 21 (38.9%) from Dermatophagoides mite, is also low. However, the sequence identity between Der p 21 and Der f 21 is very high (70.6%), suggesting that B. tropicalis and Dermatophagoides mites represent different major sources of dust mite allergens (4).
Despite the similar overall folding of Blo t 21 and Blo t 5, a closer examination of their structures shows dissimilar local arrangements of charged residues at the L2 loop region and on helix α3. The four identified, putative IgE epitope residues of Blo t 5, i.e., Glu-76, Asp-81, Glu-86, and Glu-91, are also conserved in Blo t 21, corresponding to residues Glu-74, Asp-79, Glu-84, and Glu-89. These four residues are arranged in a continuous stretch in Blo t 5, forming a linear IgE-binding epitope at this loop region. However, in Blo t 21, residues Glu-74, Asp-79, and Glu-84 form a cluster on one side of the protein, whereas residue Glu-89 is separated from these residues and forms a new epitope with Asp-96 on the opposite side of the protein. This arrangement may allow binding of two IgE antibodies for cross-linking of the IgE receptors, FcϵRI. In Blo t 5, residue Asp-96 of Blo t 21 is replaced with a residue of the opposite charge, Lys-98, although in Der p 5, residue Glu-74 of Blo t 21 is replaced with a hydrophobic residue, Leu-73. Blo t 21 and Blo t 5 seem to have conserved as well as unique IgE epitope residues located at the L2 loop region and helix α3. These subtle differences in composition and the distribution of charges in this discrete region may determine the specificity of IgE binding and may account for the difference in the IgE binding specificities of Blo t 21, Blo t 5, and Der p 5. In Der f 21, all five of the IgE epitope residues of Blo t 21 are conserved, with the exception of residue Asp-96 of Blo t 21, which is replaced with residue Glu-99 in Der f 21. We speculate that the discrepancy in the IgE binding specificities of Blo t 21 and Der f 21 may be due to differences in other residues adjacent to the charged residues in this region. We also cannot rule out the possibility of other potential IgE binding sites at other regions of the protein. A detailed structure and IgE epitope mapping of Der f 21 will be required to explain the variances in IgE binding between Blo t 21 and Der f 21.
Mutation of residue Lys-107 of Blo t 21 causes a significant drop in IgE binding in more than 50% of patients. Residue Lys-107 is well isolated from the charged residues at the L2 loop region but lies very close to the region previously described as the IgE epitope of Blo t 5 that was based on inhibition of IgE binding using a monoclonal antibody targeting the L1 loop region (30). IgE epitopes that are further apart may also allow easier cross-linking of FcϵRI. Therefore, it would be reasonable to suggest that Lys-107 and the residues around Lys-107 may represent a common IgE-binding epitope between Blo t 21 and Blo t 5. However, because Lys-107 is replaced with residue Gln-109 in Blo t 5, this may in fact represent a unique IgE epitope in Blo t 21. Cross-inhibition experiments between Blo t 5 and the K107A mutant of Blo t 21 will be needed to answer this. The combination of K107A mutation with the 4A mutant of Blo t 21 may further reduce IgE binding and constitute a better hypoallergen for immunotherapy, provided that the mutant is stable for expression and purification.
In this study, we showed that Blo t 21 has only a low to moderate cross-reactivity with Blo t 5 and could therefore represent a novel group of major allergens in B. tropicalis. Another study on the characterization of Der p 21 also showed that it lacks IgE cross-reactivity with Der p 5 and represents a new allergen in D. pteronyssinus (6). Both studies suggest that group 21 is a novel group of allergens with only moderate sequence identity to group 5 allergens, even though the overall structures are highly similar. Other than the low to moderate cross-reactivity between group 5 and group 21 allergens, the cross-reactivity of the same group 21 allergens from B. tropicalis and Dermatophagoides mites that have moderate sequence identities is also low to moderate, e.g., sequence identities between Blo t 21 and Der f 21. This low to moderate cross-reactivity among the same group of allergens with moderate sequence identities from B. tropicalis and Dermatophagoides mites is also observed between Blo t 5 and Der p 5 (12), between Blo t 1 and Der p 1 (14), and between Blo t 10 and Der p 10 (11). The results presented in this study are in agreement with the notion that B. tropicalis is a major source of dust mite allergens in the tropics and subtropics that is distinct from Dermatophagoides mites (4).
This work was supported by Biomedical Research Council Grant 04/1/21/19/315, National Medical Research Council of Singapore Grant NMRC/1150/2008, and Ministry of Education Tier 2 Grant MOE 2011-T2-2-002.

This article contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (code 2LM9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
F. T. Chew and Y. F. Gao, unpublished results.
- TOCSY
- total correlation spectroscopy
- PBS-T
- 1 × phosphate buffered saline with 0.05% Tween-20.
REFERENCES
- 1. Chew F. T., Lim S. H., Goh D. Y., Lee B. W. (1999) Sensitization to local dust-mite fauna in Singapore. Allergy 54, 1150–1159 [DOI] [PubMed] [Google Scholar]
- 2. Chua K. Y., Cheong N., Kuo I. C., Lee B. W., Yi F. C., Huang C. H., Liew L. N. (2007) The Blomia tropicalis allergens. Protein Pept. Lett. 14, 325–333 [DOI] [PubMed] [Google Scholar]
- 3. Tsai W. J., Liu C. H., Chen S. T., Yang C. Y. (2003) Identification of the antigenic determinants of the American cockroach allergen Per a 1 by error-prone PCR. J. Immunol. Methods 276, 163–174 [DOI] [PubMed] [Google Scholar]
- 4. Yi F. C., Shek L. P., Cheong N., Chua K. Y., Lee B. W. (2006) Molecular cloning of Blomia tropicalis allergens. A major source of dust mite allergens in the tropics and subtropics. Inflamm. Allergy Drug Targets 5, 261–266 [DOI] [PubMed] [Google Scholar]
- 5. Gao Y. F., Wang D. Y., Ong T. C., Tay S. L., Yap K. H., Chew F. T. (2007) Identification and characterization of a novel allergen from Blomia tropicalis. Blo t 21. J. Allergy Clin. Immunol. 120, 105–112 [DOI] [PubMed] [Google Scholar]
- 6. Weghofer M., Dall'Antonia Y., Grote M., Stöcklinger A., Kneidinger M., Balic N., Krauth M. T., Fernández-Caldas E., Thomas W. R., van Hage M., Vieths S., Spitzauer S., Horak F., Svergun D. I., Konarev P. V., Valent P., Thalhamer J., Keller W., Valenta R., Vrtala S. (2008) Characterization of Der p 21, a new important allergen derived from the gut of house dust mites. Allergy 63, 758–767 [DOI] [PubMed] [Google Scholar]
- 7. Kidon M. I., Chiang W. C., Liew W. K., Ong T. C., Tiong Y. S., Wong K. N., Angus A. C., Ong S. T., Gao Y. F., Reginald K., Bi X. Z., Shang H. S., Chew F. T. (2011) Mite component-specific IgE repertoire and phenotypes of allergic disease in childhood. The tropical perspective. Pediatr. Allergy Immunol. 22, 202–210 [DOI] [PubMed] [Google Scholar]
- 8. Chan S. L., Ong T. C., Gao Y. F., Tiong Y. S., Wang D. Y., Chew F. T., Mok Y. K. (2008) Nuclear magnetic resonance structure and IgE epitopes of Blo t 5, a major dust mite allergen. J. Immunol. 181, 2586–2596 [DOI] [PubMed] [Google Scholar]
- 9. Naik M. T., Chang C. F., Kuo I. C., Yu T., Fang P. J., Chua K. Y., Huang T. H. (2007) Complete 1H, 13C and 15N resonance assignments of Blo t 5, a major mite allergen from Blomia tropicalis. J. Biomol. NMR 38, 189. [DOI] [PubMed] [Google Scholar]
- 10. Mueller G. A., Gosavi R. A., Krahn J. M., Edwards L. L., Cuneo M. J., Glesner J., Pomés A., Chapman M. D., London R. E., Pedersen L. C. (2010) Der p 5 crystal structure provides insight into the group 5 dust mite allergens. J. Biol. Chem. 285, 25394–25401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. García Robaina J. C., Sánchez Machín I., Fernández-Caldas E., Iraola Calvo V., Vázquez Moncholi C., Bonnet Moreno C., de la Torre Morín F. (2003) Skin tests and conjunctival and bronchial challenges with extracts of Blomia tropicalis and Dermatophagoides pteronyssinus in patients with allergic asthma and/or rhinoconjunctivitis. Int. Arch. Allergy Immunol. 131, 182–188 [DOI] [PubMed] [Google Scholar]
- 12. Chew F. T., Yi F. C., Chua K. Y., Fernandez-Caldas E., Arruda L. K., Chapman M. D., Lee B. W. (1999) Allergenic differences between the domestic mites Blomia tropicalis and Dermatophagoides pteronyssinus. Clin. Exp. Allergy 29, 982–988 [DOI] [PubMed] [Google Scholar]
- 13. Kuo I. C., Cheong N., Trakultivakorn M., Lee B. W., Chua K. Y. (2003) An extensive study of human IgE cross-reactivity of Blo t 5 and Der p 5. J. Allergy Clin. Immunol. 111, 603–609 [DOI] [PubMed] [Google Scholar]
- 14. Cheong N., Soon S. C., Ramos J. D., Kuo I. C., Kolatkar P. R., Lee B. W., Chua K. Y. (2003) Lack of human IgE cross-reactivity between mite allergens Blo t 1 and Der p 1. Allergy 58, 912–920 [DOI] [PubMed] [Google Scholar]
- 15. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 16. Goddard T. D., Kneller D. G. (2008) SPARKY 3, University of California, San Francisco, CA [Google Scholar]
- 17. Wittekind M., Mueller L. (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. B101, 201–205 [Google Scholar]
- 18. Grzesiek S., Bax A. (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 19. Grzesiek S., Anglister J., Bax A. (1992) Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N enriched proteins by isotopic mixing of 13C magnetization. J. Magn. Reson. B101, 114–119 [Google Scholar]
- 20. Clore G. M., Bax A., Driscoll P. C., Wingfield P. T., Gronenborn A. M. (1990) Assignment of the side-chain 1H and 13C resonances of interleukin-1 beta using double- and triple-resonance heteronuclear three-dimensional NMR spectroscopy. Biochemistry 29, 8172–8184 [DOI] [PubMed] [Google Scholar]
- 21. Wishart D. S., Sykes B. D. (1994) The 13C chemical-shift index. A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 22. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 23. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 24. Nederveen A. J., Doreleijers J. F., Vranken W., Miller Z., Spronk C. A., Nabuurs S. B., Güntert P., Livny M., Markley J. L., Nilges M., Ulrich E. L., Kaptein R., Bonvin A. M. (2005) RECOORD. A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59, 662–672 [DOI] [PubMed] [Google Scholar]
- 25. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 27. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272. [DOI] [PubMed] [Google Scholar]
- 28. Sippl M. J., Wiederstein M. (2008) A note on difficult structure alignment problems. Bioinformatics 24, 426–427 [DOI] [PubMed] [Google Scholar]
- 29. Sippl M. J. (2008) On distance and similarity in fold space. Bioinformatics 24, 872–873 [DOI] [PubMed] [Google Scholar]
- 30. Naik M. T., Chang C. F., Kuo I. C., Kung C. C., Yi F. C., Chua K. Y., Huang T. H. (2008) Roles of structure and structural dynamics in the antibody recognition of the allergen proteins. An NMR study on Blomia tropicalis major allergen. Structure 16, 125–136 [DOI] [PubMed] [Google Scholar]
- 31. Caraballo L., Mercado D., Jiménez S., Moreno L., Puerta L., Chua K. Y. (1998) Analysis of the cross-reactivity between BtM and Der p 5, two group 5 recombinant allergens from Blomia tropicalis and Dermatophagoides pteronyssinus. Int. Arch. Allergy Immunol. 117, 38–45 [DOI] [PubMed] [Google Scholar]
- 32. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL. A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 33. DeLano W. L. (2002) PyMOL, DeLano Scientific, San Carlos, CA [Google Scholar]
- 34. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]