Background: APOBEC3A is a myeloid-specific interferon-inducible DNA C to U deaminase implicated in innate immunity.
Results: APOBEC3A also elicits MeC to T editing activity in vitro with deoxy-oligonucleotides and in vivo with transfected plasmids.
Conclusion: APOBEC3A accommodates both normal and larger DNA cytosine substrates.
Significance: The developmental specialization and broader substrate range of APOBEC3A may be an evolutionary adaptation for physiological function in foreign DNA restriction.
Keywords: DNA Enzymes, DNA Methylation, Innate Immunity, Interferon, RNA Editing, DNA Deamination, DNA Demethylation, Foreign DNA Restriction
Abstract
Multiple studies have indicated that the TET oxidases and, more controversially, the activation-induced cytidine deaminase/APOBEC deaminases have the capacity to convert genomic DNA 5-methylcytosine (MeC) into altered nucleobases that provoke excision repair and culminate in the replacement of the original MeC with a normal cytosine (C). We show that human APOBEC3A (A3A) efficiently deaminates both MeC to thymine (T) and normal C to uracil (U) in single-stranded DNA substrates. In comparison, the related enzyme APOBEC3G (A3G) has undetectable MeC to T activity and 10-fold less C to U activity. Upon 100-fold induction of endogenous A3A by interferon, the MeC status of bulk chromosomal DNA is unaltered, whereas both MeC and C nucleobases in transfected plasmid DNA substrates are highly susceptible to editing. Knockdown experiments show that endogenous A3A is the source of both of these cellular DNA deaminase activities. This is the first evidence for nonchromosomal DNA MeC to T editing in human cells. These biochemical and cellular data combine to suggest a model in which the expanded substrate versatility of A3A may be an evolutionary adaptation that occurred to fortify its innate immune function in foreign DNA clearance by myeloid lineage cell types.
Introduction
Although the methyltransferase enzymes that add methyl groups to cytosines are well defined, comparatively little is known about the opposing mechanism of demethylation (1–3). Proposals include direct demethylation by as yet unidentified enzymes, dilution by DNA replication (without maintenance methylation), modification of the methyl group by TET (ten-eleven translocation) proteins followed by excision and repair, or enzymatic deamination followed by canonical base excision repair (2–11). In support of the latter mechanism, the DNA cytosine deaminase, AID,5 has been shown in vitro to catalyze the conversion of MeC to T (12–14) and has been implicated in demethylation in mouse germ and stem cells (15, 16). The critical nature of modified MeC nucleobases (products of oxidation and/or deamination) is supported by the embryonic lethality of thymine DNA glycosylase (TDG) null mice (5, 17).
AID, however, cannot be the sole factor contributing to genomic DNA demethylation because AID-deficient animals are viable (18). The stark phenotypic difference between TDG and AID null animals may be due to overlapping function with TET enzymes, as yet unidentified enzymes, and/or related polynucleotide cytosine deaminase family members. Mice have two related enzymes, APOBEC1 and APOBEC3 (A3), whereas humans have eight, APOBEC1 and an expanded A3 repertoire: A3A, A3B, A3C, A3D, A3F, A3G, and A3H (19, 20). All mammals also express APOBEC2 and APOBEC4, which are more distant relatives with functions in cardiac development and as yet unknown processes, respectively (21–23).
MeC deamination activity may be dispensable for the established physiological functions of most polynucleotide cytosine deaminase family members. AID deaminates antibody gene variable and switch region DNA cytosines to initiate the distinct processes of somatic hypermutation and class switch recombination, respectively (24, 25). Most of the reported AID hotspots occur outside of potentially methylatable cytosine-guanine (CpG) dinucleotide motifs in antibody gene DNA (26). APOBEC1 edits a specific cytosine in the APOB mRNA to generate a shorter form of the encoded protein (27). Several A3 enzymes edit retroviral cDNA cytosines within the confines of a cytoplasmic capsid-encased structure, which is presumably not accessible to nuclear methyltransferase enzymes (28, 29).
However, several A3s and most prominently A3A are also capable of editing transfected plasmid DNA substrates and triggering their clearance from human cells (30–32). A specialized role for A3A in foreign DNA restriction is supported by its phagocytic/myeloid lineage-restricted expression and its strong IFN inducibility (30, 33–35). Interestingly, although foreign DNA substrates are readily deaminated, the genomic DNA of A3A-induced phagocytes does not appear to be susceptible to the same mechanism (30). The differential susceptibility of foreign DNA and self-DNA suggests that some process, such as cytosine methylation, may be affording protection to nuclear DNA.
To shed additional light on these areas, we investigated the substrate specificity and kinetics of two human enzymes, A3A and A3G. A3A readily deaminates both C and MeC single-stranded DNA substrates, whereas A3G is less efficient and appears to have an exclusive preference for normal C. Our studies are consistent with a biological role for A3A in foreign DNA restriction, where a broader substrate range may be advantageous for triggering the clearance of a larger number of foreign DNA substrates.
EXPERIMENTAL PROCEDURES
Plasmids
pcDNA3.1-A3A-mycHis and A3G-mycHis vectors were reported (30, 36). Standard PCR cloning and site-directed mutagenesis techniques were used to construct derivatives.
Proteins
A3A-mycHis6, A3G-mycHis6, or their derivatives were expressed in HEK293T cells and purified using nickel affinity purification techniques (30, 36). Escherichia coli UDG, UDG inhibitor, and M.SssI were purchased from New England Biolabs, and Methanobacterium thermoautotrophicum TDG from Trevigen.
Deamination Assays
In vitro deamination assays with partially purified proteins were performed in 25 mm HEPES, pH 7.4, 100 mm NaCl, and 0.1% Triton X-100. UDG, UDG inhibitor, or TDG was added as indicated. Most reactions with UDG or UDG inhibitor were done concurrently with A3 treatment. In kinetics assays where reaction time was critical, the reactions were flash frozen in liquid nitrogen and stopped by rapid heating to 95 °C for 10 min and then followed by UDG treatment at 37 °C. The reaction rates were determined by quantifying substrate and product bands at each time point and performing Michaelis-Menten analysis. Substrate concentration at each time point was calculated by multiplying the initial concentration by the ratio of the density of the substrate band divided by the sum of the substrate and product bands (12). Reactions with TDG were performed by adding complement to 3-fold molar excess to fluorescently labeled substrate and TDG or TDG dilution buffer followed by 12 h at 42 °C. Following A3 and glycosylase treatment, NaOH was added to a final concentration of 100 mm, and the reactions were heated for 10 min at 95 °C. The reactions were then mixed with 2× denaturing PAGE loading dye and run on 15 or 20% TBE-urea polyacrylamide gels. The gels were imaged with a Fuji FLA-5000 scanner and quantified with Fuji ImageGauge version 4.1. The 43-mer substrate for A3A was 5′-ATTATTATTATT(C or MeC)TAATGGATTTATTTATTTA TTTATTTATTT-(6FAM). For A3G, the substrate was 5′-ATTATTATTATTC(C or MeC)AATGGATTTATTTATTTATTTATTTATTT-(6FAM). In vitro deamination assays with lysates (see below for cells) were done as described (30).
Cellular Experiments
THP1 cells were obtained from Dr. Andrea Cimarelli (Ecole Normale Supérieure de Lyon). Control and A3A knockdown pools were obtained by transduction with pLKO-based lentiviral constructs (Open Biosystems) followed by puromycin resistance selection. Specific A3A knockdown was verified by immunoblotting (below) and quantitative PCR (30, 37). Foreign DNA restriction assays were done by transfecting mock or M.SssI-treated pEGFP-N3 (Clontech) into mock or IFNα-treated (300 units/ml Universal Type I INFα; PBL Interferon Source) THP1 cells as described (30). Transfections were performed with Transit LT1 (Mirus Bio). After 48 h, total cellular DNA was prepared, and foreign DNA was recovered by PCR cloning into pJet1.2 (Fermentas) and analyzed by three-dimensional PCR sequencing. A fraction of the cells was used for anti-A3A immunoblotting with a rabbit polyclonal antibody generated by immunization and boosting with a C-terminal peptide CPFQPWDGLEEHSQALSGRLRAILQNQGN (Epitomics Custom Rabbit Polyclonal Production Service). Each rabbit was immunized with 0.5 mg of peptide conjugated to keyhole limpet hemocyanin, followed by boosts of 0.25 mg after 3 and 5 weeks. Two further boosts of ovalbumin-conjugated peptide were injected after 7 and 9 weeks. Bleeds were collected 12 days after the fourth and fifth injections (about 9 and 11 weeks). A3A-transfected HEK293T cell lysates were screened by immunoblotting to identify positive sera (detected using a goat anti-rabbit IgG-800CW secondary antibody; LiCor Odyssey II). Positive bleeds were used for immunofluorescent microscopy of A3A-eGFP transfected HeLa cells (detected using a goat anti-rabbit FITC secondary antibody; Jackson ImmunoResearch).
Immunofluorescent Microscopy Studies
Endogenous A3A was up-regulated by treating THP1 cells or primary human monocytes with IFNα (300 units/ml Universal Type I INFα; R & D Systems) or phorbol 12-myristate 13-acetate (20 ng/ml; Sigma). Primary CD14+ monocytes were purified by negative selection (Stem Cell Tech) from fresh peripheral blood mononuclear cells obtained from Memorial Blood Center (St. Paul, MN). Purity was confirmed by flow cytometry (>70% pure). The cells were fixed with 4% paraformaldehyde, stained with rabbit anti-human A3A (above) and goat anti-rabbit IgG-TRITC (Jackson ImmunoResearch), and imaged with a DeltaVision deconvolution microscope (Applied Precision) (30, 38).
RESULTS
A3A Is a Single-stranded DNA Cytosine Deaminase
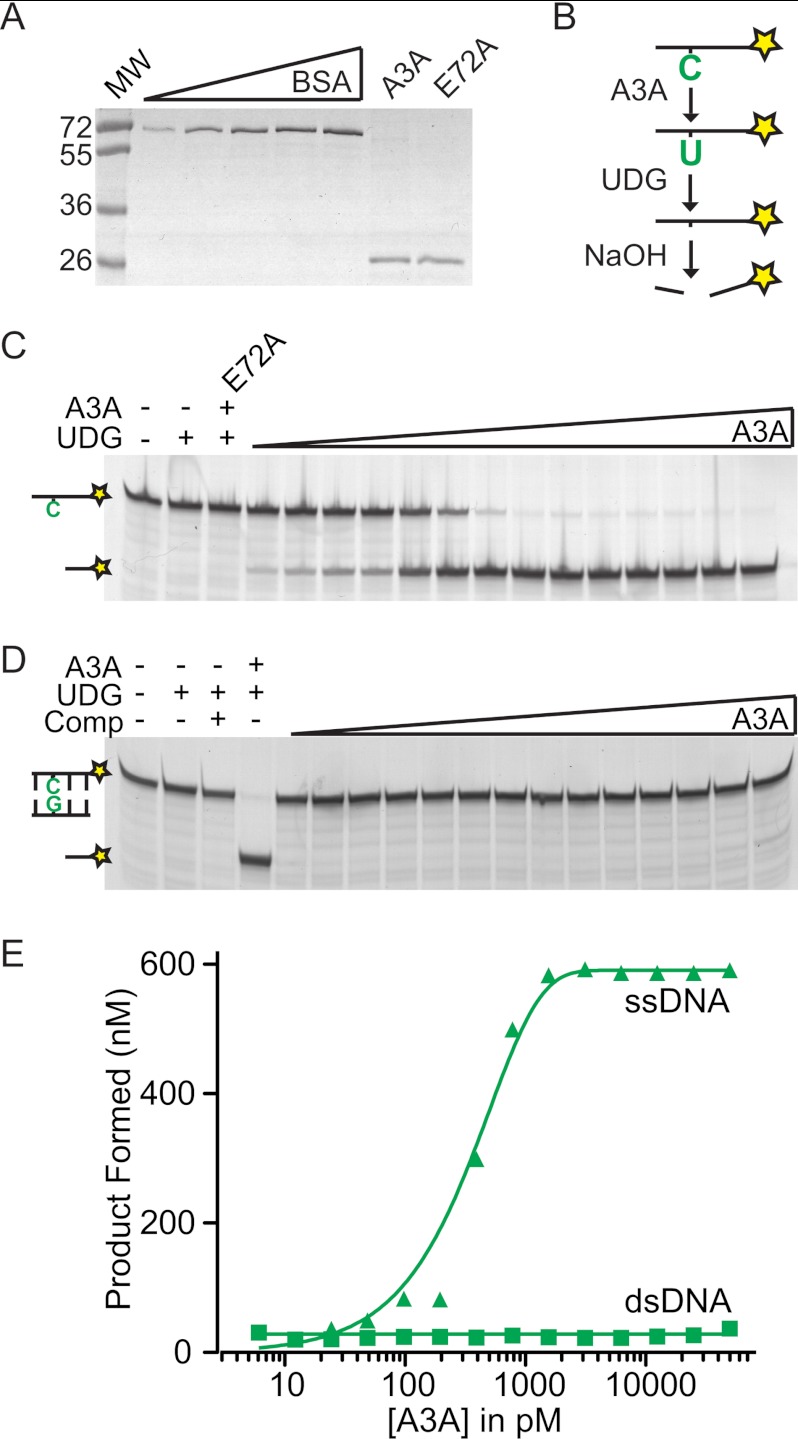
Prior studies with whole cell or HIV particle lysates containing A3A were used to infer that this enzyme prefers single-stranded ssDNA over dsDNA substrates (30, 33, 34). To revisit substrate specificity questions and perform kinetic studies, we partially purified human A3A from HEK293T cells. A catalytic mutant A3A-E72A was prepared in parallel to control for potentially undesirable co-purifying activities such as nucleases. A3A or its derivatives comprised >90% of each enzyme preparation as assessed by densitometry of Coomassie Blue-stained SDS-PAGE gels (Fig. 1A). A3A DNA deaminase activity was measured with a gel-based fluorescence assay, in which a cytosine-containing ssDNA is converted into a shorter product by deamination, uracil excision, and hydrolytic cleavage (36) (Fig. 1B). The deamination step is rate-limiting because the excision enzyme uracil-DNA glycosylase (UDG) is present in excess.
FIGURE 1.
A3A is a potent single strand DNA deaminase. A, Coomassie gel image of A3A-mycHis and its catalytic mutant in comparison with 0.2–1 μg of BSA. B, assay schematic in which a cytosine in a fluorescently labeled (asterisk) ssDNA substrate is deaminated to uracil and then converted to a shorter reaction product by UDG and NaOH. C, gel image showing the relationship between A3A (2-fold dilutions from 50 nm) and ssDNA deaminase activity with 200 nm substrate in 60 min. Parallel control reactions had no enzymes, UDG only, or 50 nm A3A-E72A. D, reaction series as in C, but dsDNA was used as substrate by preincubating with 1.5-fold excess complement. Parallel control reactions had no enzymes, UDG plus/minus complement, or 50 nm A3A with complement. E, quantification of the data in C and D, indicating the amount of product formed as a function of enzyme concentration. MW, molecular weight.
Human A3A is active with 3 nm able to fully deaminate 67-fold excess substrate ssDNA within 60 min (Fig. 1, C and E). Other products were not detected in the A3A reactions, and the substrate ssDNA remained unchanged in the A3A-E72A reactions, demonstrating that the enzyme preparations are free of detectable contaminating endo- or exonuclease activities (Fig. 1C and supplemental Fig. S1). Moreover, substrate ssDNA cytosines could be protected from A3A-dependent deamination by preincubating with 1.5-fold excess complement and converting potential substrate into dsDNA (Fig. 1, D and E). A small amount of deamination product was apparent at higher A3A concentrations, which may be due to incomplete annealing of the complementary strand, DNA breathing, small amounts of a co-purifying DNA unwinding activity such as a helicase and/or reduced levels of a potentially relevant dsDNA deaminase activity. Although more work will be needed to address every possibility, these studies extend prior work by demonstrating that A3A has a >1000-fold preference for a target cytosine in the context of ssDNA over dsDNA.
A3A Also Catalyzes Efficient ssDNA MeC to T Deamination
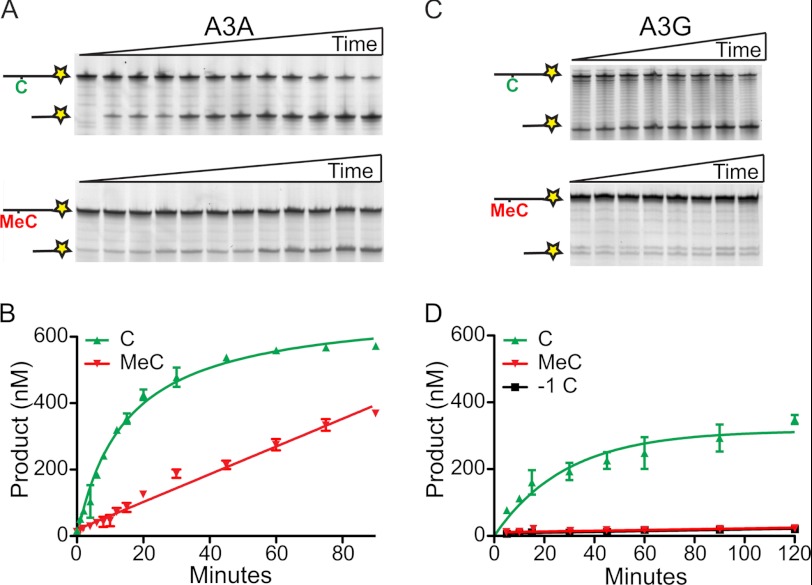
A3A is strongly induced in human CD14 positive phagocytic cells by IFNα and other innate immune agonists such as CpG DNA (30, 33–35, 39). Whether cytosine methylation, a common DNA modification in both prokaryotes and eukaryotes, has any influence on the deamination activity of A3A is of interest. A 90-min time course was done by incubating 3 nm A3A and a 200-fold excess of C or MeC containing 43-mer substrate. At each time point, the reactions were stopped by snap freezing in liquid N2, and at the end of the time course all of the reactions were mixed with 3-fold excess complementary DNA. UDG efficiently excises U from ssDNA and dsDNA substrates (40, 41), so the inclusion of a complementary strand does not affect this step of the assay. As above, A3A showed high activity on normal C ssDNA substrate, converting 93% to product in 45 min (green labels in Fig. 2, A and B).
FIGURE 2.
A3A is also a potent MeC to T deaminase. A, representative gels of an activity time course of 3 nm A3A on 600 nm oligonucleotide with a single cytosine (top gel) or MeC (bottom gel). B, quantification by densitometry of data in A and two independent experiments (means ± S.D. with error smaller than the symbol in most instances). C, representative gels of an activity time course of 100 nm A3G against 600 nm substrate ssDNA with either CC (top gel) or CMeC (bottom gel). D, quantification by densitometry of data in C and two independent experiments (means ± S.D. with error smaller than the symbol in most instances). Quantification of the −1 C in the sequence is also indicated, demonstrating that this C is not deaminated by A3G (black symbols mostly hidden).
The resulting deamination products were quantified by densitometry and used to determine kinetic parameters (12). This analysis yielded a reaction velocity (Vmax) of 45 ± 2.5 nm/min, a catalytic efficiency (Kcat) of 15 min−1, and a Km of 230 ± 30 nm. Similar kinetic values were obtained using Michaelis-Menten methodology (supplemental Fig. S2).
Under the same reaction conditions, apart from using TDG as the glycosylase, 3 nm A3A also elicited strong MeC to T deaminase activity (red labels in Fig. 2, A and B). Parallel control reactions demonstrated that the catalytic glutamate E72 is essential and that TDG addition (but not UDG) is required to convert T/G mispairs into abasic sites (supplemental Fig. S1). Compared with the matched normal C containing ssDNA substrate, A3A is ∼10-fold less efficient against MeC with a Vmax of 4.2 nm/min and a Kcat of 1.4 min−1. The linear relationship between substrate concentration and reaction time indicated that the MeC to T reaction is occurring at maximal velocity. These data combine to demonstrate that human A3A possesses both C to U and MeC to T ssDNA deamination activities, with the former being more efficient. The various enzymes, preparation methods, and substrates used in previous studies complicate quantitative comparisons with our work here on A3A, but see below for direct comparisons with A3G and supplemental Table S1 for a comprehensive list of prior biochemical work.
The Related Enzyme A3G Prefers Normal Single-stranded DNA Cytosines
A3A is 73% similar to the catalytic C-terminal domain of A3G. To test whether MeC to T deamination is a general activity of other A3 subfamily members, we performed a series of biochemical experiments with purified A3G on a substrate modified to contain an additional C adjacent to the target C to accommodate the strong preference of A3G for deaminating the second C in 5′-CC dinucleotide motifs (42–45). Although this modification created two potential target sites, it was essential because A3G elicits little activity on ssDNA substrates with a single target cytosine flanked on the 5′ side by A, G, or T (data not shown).
Using two-stage reactions, as described above, 100 nm A3G is required to achieve 67% deamination of the 600 nm substrate oligonucleotide in 120 min (i.e., 33-fold more enzyme and 2-fold more time are required to achieve lower product yields; Fig. 2, C and D). As expected, A3G preferentially deaminates the second cytosine in the 5′-CC motif. Quantifying early reaction products yielded a reaction velocity (Vmax) of 8.3 nm/min and a Kcat of 0.08 min−1. The appearance of a faint, slower migrating product indicated comparatively little alternative 5′-CC to UC product. Parallel reactions with A3A and the same 5′-CC ssDNA substrate showed that A3A was ∼10-fold more active than A3G (assuming the fraction of active enzyme in each preparation is similar; data not shown).
In contrast, full-length A3G or the C-terminal half of A3G (analogous in size to the mono-domain A3A protein) is unable to deaminate MeC to T under the same reaction conditions (Fig. 2, C and D, and supplemental Fig. S3). Low levels of putative C and MeC deamination products are evident in every lane, but these levels are most likely due to substrate impurities (U at the −1 position and/or T at the optimal A3G deamination target position). In contrast, A3A readily converts both the normal C and the adjacent MeC base in the same ssDNA substrate into U and T products, respectively (supplemental Fig. S3). Thus, despite strong amino acid identity with A3A, human A3G fails to deaminate MeC to T in ssDNA. It is premature to conclude that A3G cannot ever promote this reaction, especially in vivo where other proteins may be influential, but our head to head biochemical comparisons do not lend support to such a possibility.
Obvious Structural Differences between A3A and A3G Fail to Explain MeC to T Activity
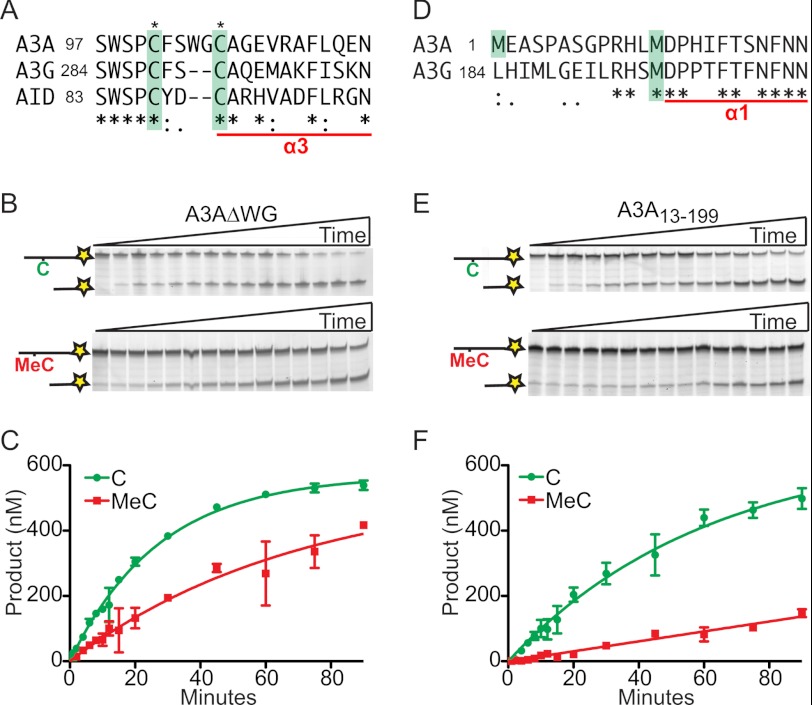
To try to explain the contrasting MeC deamination activity of A3A and A3G, the amino acids within or near the catalytic center of the enzyme were compared. One major difference is the number of amino acids that separate the zinc-coordinating cysteines (Fig. 3A). These cysteines are separated by four amino acids in A3A and A3B, but by only two residues in A3G and other family members (19, 20). To determine whether the larger intercysteine distance is responsible for the difference in activity, we deleted the WG residues in A3A, purified the resulting enzyme from HEK293T cells, and performed a series of deamination experiments (gel image of partially purified proteins in supplemental Fig. S4). This A3A deletion mutant was almost as active as the wild type enzyme using both normal C and MeC ssDNA substrates (Fig. 3, B and C). Therefore, the intercysteine distance is not responsible for the ability of A3A to accommodate MeC substrates.
FIGURE 3.
A3A mutants retain C to U and MeC to T activity. A, alignment of the zinc-binding motif from A3A, A3G, and AID. Asterisks and green boxes indicate zinc-binding cysteines. A3G α3 is underlined. B, representative gels showing a time course of 3 nm A3AΔWG on 600 nm substrate ssDNA with either a single C (top) or MeC (bottom). C, quantification of three independent assays with A3AΔWG against C and MeC (mean ± S.D.). D, alignment A3A N-terminal residues and the corresponding A3G region. The methionines discussed in the text are highlighted. A3G α1 is underlined. E, representative gels indicating the activity of 3 nm A3A13–199 on 600 nm C oligonucleotide (top) or MeC oligonucleotide (bottom). F, quantification of three independent assays with A3A13–199 against C and MeC ssDNA substrates (mean ± S.D.).
Another major difference between A3A and A3G is an N-terminal segment that occurs prior to each minimal catalytic domain of the enzyme (30, 34). A3A has 12 residues prior to Met-13 (homologous to A3G M197), whereas A3G has an additional entire (but inactive) 196-residue deaminase domain prior to residue Met-197 (Fig. 3D). A3G Met-197 and other residues within this region make chemical contacts with active site residues and promote C to U deaminase activity (46). Interestingly, two isoforms of A3A are clearly expressed in primary IFNα-stimulated monocytes, one starting at M1 and the other at M13 (Refs. 30 and 34 and below). To test the possibility that the unique N-terminal extension of A3A governs MeC to T deamination activity, we purified the shorter A3A13–199 isoform from HEK293T cells and assayed its activity (gel image of partially purified proteins in supplemental Fig. S4). In multiple independent experiments A3A13–199 is about 5-fold less active than the full-length enzyme, with maximum velocities of 9.4 nm/min for C and 1.6 nm/min for MeC ssDNA substrates (Fig. 3, E and F). However, because both normal C and MeC rates decreased, the N-terminal extension seems to be a determinant of enzyme efficiency rather than substrate specificity.
Endogenous A3A Deaminates Both Normal C and MeC in Transfected Foreign DNA Substrates
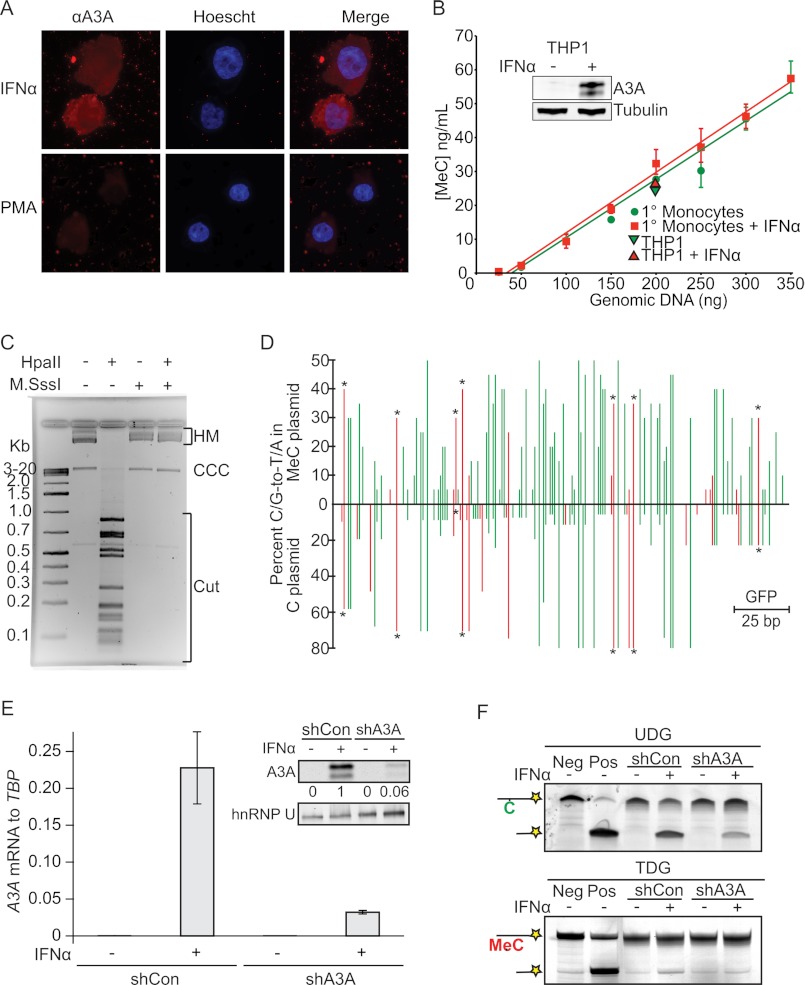
Epitope-tagged A3A has shown a cell-wide localization consistent with many possible biological functions (30, 33, 47) (supplemental Fig. S5). Using a new polyclonal antiserum, we found that endogenous A3A can be detected after IFNα treatment of the monocytic cell line THP1 or primary human monocytes (Fig. 4A and supplemental Fig. S5). To ask whether A3A alters the methylation state of nuclear DNA, ELISAs were used to quantify MeC levels of genomic DNA prepared from THP1 cells and primary monocytes treated with IFNα. Although IFNα caused a >100-fold increase in both A3A mRNA and protein levels, no detectable differences in overall MeC levels were found (Fig. 4B). These results indicate that genomic MeC residues are not generally accessible to A3A-mediated deamination.
FIGURE 4.
Endogenous A3A deaminates both normal C and MeC foreign DNA substrates. A, representative image of endogenous A3A localization in IFNα-treated THP1 cells. B, genomic MeC levels in IFNα-treated THP1 cells and primary monocytes. The inset shows a representative immunoblot of A3A with tubulin as a loading control. C, agarose gel image of MeC and normal C plasmid DNA substrates treated as shown. HM, CCC, and Cut indicate higher mass, covalently closed circular, and cleaved plasmid species, respectively. D, hypermutation profiles of MeC and normal C plasmid substrates. Each analysis consisted of >18 independent sequences (GFP nucleotides 516–720) and >380 C/G to T/A mutations. Red and green bars indicate mutations within CpG and other cytosine dinucleotide motifs, respectively. Asterisks denote highly mutated 5′-TCG sites. E, quantification of A3A mRNA and protein levels in mock or IFNα-treated THP1 cells stably expressing control (shCon) or A3A knockdown (shA3A) short hairpin RNAs. F, ssDNA C (top panel) and MeC (bottom panel) deaminase activity of extracts from cells in E. Parallel negative (Neg) and positive (Pos) control reactions used enzyme buffer minus and plus partially purified A3A, respectively. Oligonucleotide impurity is responsible for the low level of background product. PMA, phorbol 12-myristate 13-acetate.
We previously proposed that A3A is a critical effector protein of an IFN-inducible foreign DNA clearance mechanism (30). Foreign DNA can be microbial, but it can also be human DNA located outside of its normal cellular context. Therefore, as part of this innate defense mechanism, we reasoned that it may be advantageous for A3A to accommodate both normal C and MeC residues. To test this possibility in living cells, we generated normal plasmid DNA (unmodified at CpG), treated it with the CpG methyltransferase M.SssI, and transfected either normal C or MeC containing plasmid into IFNα-treated THP1 cells. This procedure resulted in plasmid DNA that was fully methylated at CpG dinucleotides as demonstrated by resistance to cleavage by the methyl-sensitive restriction enzyme HpaII (Fig. 4C). 48 h post-transfection, PCR was used to recover segments of the remaining plasmid DNA for sequence analysis. High frequencies of C/G to T/A mutations were evident in both the nonmethylated and the fully methylated substrates, and nearly half of these mutations occurred at A3A-preferred 5′-TC dinucleotides (30, 34) (Fig. 4D). Importantly, knockdown experiments demonstrated that both the normal C and MeC ssDNA deaminase activity in IFNα-stimulated THP1 cell extracts is dependent upon endogenous A3A (Fig. 4, E and F). It is unlikely that another enzyme in cells or cell extracts removes or modifies the methyl group prior to A3A-dependent DNA deamination because this hypothetical enzyme would also have to be IFN-inducible or activatable because MeC to T editing is only observed in extracts from IFNα-treated cells. Such an activity would also have to prefer 5′-TC dinucleotides, and it would have to be non-rate-limiting so that A3A would have sufficient time to access and deaminate any demethylated C residues. Taken together, our data show that foreign MeC (and normal C) DNA is highly susceptible to deamination by A3A in living cells, whereas genomic DNA does not appear to be affected.
DISCUSSION
DNA cytosine methylation and demethylation are important for numerous biological processes. Several models for genomic DNA demethylation have been proposed (see the Introduction). Recent reports have demonstrated roles for mammalian TET proteins in converting MeC residues into bulkier substrates for TDG-mediated base excision repair, thereby replacing MeC with normal C using the nonmodified DNA strand as a base excision repair template (6–9). A more controversial mechanism involves AID, APOBEC1, and/or APOBEC3-catalyzed deamination of MeC to T, which in the simplest scenario results in a T/G mispair that is recognized by TDG and repaired to normal C by downstream base excision repair pathway enzymes (4, 12–16).
Here, we provide evidence that human A3A is both a potent C to U and MeC to T ssDNA deaminase. A similar conclusion was reached using an E. coli-based mutation assay during revision of this manuscript (Ref. 48 and supplemental Table S1). Because A3A expression is interferon-inducible and developmentally confined to myeloid/phagocytic lineage cell types, we propose a model in which the expanded substrate capacity of A3A may be advantageous for physiological function in foreign DNA restriction. An expanded substrate range may benefit the host because it would facilitate the recognition and clearance of a broader number of foreign DNA substrates, including microbial as well as out of context self-DNA, for instance, from apoptotic or necrotic cells.
Although the catalytic domain of human A3G is 73% similar (65% identical) to A3A, we found that this related enzyme strongly prefers normal cytosines in ssDNA (also supported by Ref. 48; supplemental Table S1). This specificity is consistent with its role in retrovirus and retrotransposon restriction, where the retroelement ssDNA is located within the confines of the capsid. A3G specifically enters this structure during retroelement assembly and acts on newly sythesized ssDNA, which has not yet been integrated into the genome and subjected to methyltransferases. Therefore, the lack of activity on MeC substrates may not pose a challenge for the biological function of A3G in restriction of retroelements and viruses.
The A3G catalytic domain crystal structure (Protein Data Bank 3IR2; Ref. 49) and an A3A homology model offer little insight into the expanded substrate capability of A3A, and virtual docking even suggests that A3G may be able to accommodate MeC within its active site (supplemental Fig. S6). Thus, high resolution structures of A3A in the absence and presence of substrate C- and MeC-containing ssDNA may be necessary to fully explain the observed activity and substrate differences.
Another unresolved problem is fully reconciling the in vitro and the in vivo editing data. A3A has a strong preference for cytosines and methyl-cytosines in ssDNA molecules, yet transfected double-stranded plasmids are readily edited in living cells. It is conceivable, although unlikely given the strong biochemical preference shown in Fig. 1, that A3A could have an intrinsic capacity to deaminate dsDNA cytosines. However, more likely, one or more of many possible nucleic acid enzymes in cells may serve to open the DNA for A3A (e.g., a helicase or a polymerase).
This is the first study to provide evidence that DNA demethylation may have a physiological function beyond normal chromosomal processes in human cells. Given the present knowledge that A3A is conserved in primates, it is possible that the specialized innate immune function described here in foreign DNA restriction may be similarly conserved. This function may be sufficient to justify the maintenance of a promiscuous and highly active deaminase in primates despite the potential threat of creating chromosomal DNA lesions. Our studies here demonstrate that MeC alone is insufficient to protect DNA from deamination by A3A. Indeed, A3A overexpression in heterologous cell types, such as HEK293, is highly toxic (data not shown and Ref. 32). This potential hazard may be mitigated in primates by confining A3A expression to terminally differentiated cells in the myeloid lineage and also by as yet undefined processes that serve to protect chromosomal DNA and/or promote the targeting of the foreign DNA restriction mechanism.
Acknowledgment
We thank Andrea Cimarelli for THP1.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI064046 and P01 GM091743. This work was also supported by Seed Grant 11.54 from the University of Minnesota Academic Health Center.

This article contains supplemental Table S1 and Figs. S1–S6.
- AID
- activation-induced cytidine deaminase
- MeC
- methylcytosine
- C
- cytosine
- TDG
- thymine DNA glycosylase
- A3A
- APOBEC3A
- CpG
- cytosine-guanine.
REFERENCES
- 1. Feng S., Jacobsen S. E., Reik W. (2010) Epigenetic reprogramming in plant and animal development. Science 330, 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bhutani N., Burns D. M., Blau H. M. (2011) DNA demethylation dynamics. Cell 146, 866–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nabel C. S., Manning S. A., Kohli R. M. (2012) The curious chemical biology of cytosine. Deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem. Biol. 7, 20–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gehring M., Reik W., Henikoff S. (2009) DNA demethylation by DNA repair. Trends Genet. 25, 82–90 [DOI] [PubMed] [Google Scholar]
- 5. Cortázar D., Kunz C., Selfridge J., Lettieri T., Saito Y., MacDougall E., Wirz A., Schuermann D., Jacobs A. L., Siegrist F., Steinacher R., Jiricny J., Bird A., Schär P. (2011) Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423 [DOI] [PubMed] [Google Scholar]
- 6. He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., Xu G. L. (2011) Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333, 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., Zhang Y. (2011) Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wossidlo M., Nakamura T., Lepikhov K., Marques C. J., Zakhartchenko V., Boiani M., Arand J., Nakano T., Reik W., Walter J. (2011) 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat. Commun. 2, 241. [DOI] [PubMed] [Google Scholar]
- 10. Hajkova P., Jeffries S. J., Lee C., Miller N., Jackson S. P., Surani M. A. (2010) Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science 329, 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fritz E. L., Papavasiliou F. N. (2010) Cytidine deaminases. AIDing DNA demethylation? Genes Dev. 24, 2107–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morgan H. D., Dean W., Coker H. A., Reik W., Petersen-Mahrt S. K. (2004) Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues. Implications for epigenetic reprogramming. J. Biol. Chem. 279, 52353–52360 [DOI] [PubMed] [Google Scholar]
- 13. Bransteitter R., Pham P., Scharff M. D., Goodman M. F. (2003) Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc. Natl. Acad. Sci. U.S.A. 100, 4102–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larijani M., Frieder D., Sonbuchner T. M., Bransteitter R., Goodman M. F., Bouhassira E. E., Scharff M. D., Martin A. (2005) Methylation protects cytidines from AID-mediated deamination. Mol. Immunol. 42, 599–604 [DOI] [PubMed] [Google Scholar]
- 15. Popp C., Dean W., Feng S., Cokus S. J., Andrews S., Pellegrini M., Jacobsen S. E., Reik W. (2010) Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463, 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhutani N., Brady J. J., Damian M., Sacco A., Corbel S. Y., Blau H. M. (2010) Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortellino S., Xu J., Sannai M., Moore R., Caretti E., Cigliano A., Le Coz M., Devarajan K., Wessels A., Soprano D., Abramowitz L. K., Bartolomei M. S., Rambow F., Bassi M. R., Bruno T., Fanciulli M., Renner C., Klein-Szanto A. J., Matsumoto Y., Kobi D., Davidson I., Alberti C., Larue L., Bellacosa A. (2011) Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 19. LaRue R. S., Andrésdóttir V., Blanchard Y., Conticello S. G., Derse D., Emerman M., Greene W. C., Jónsson S. R., Landau N. R., Löchelt M., Malik H. S., Malim M. H., Münk C., O'Brien S. J., Pathak V. K., Strebel K., Wain-Hobson S., Yu X. F., Yuhki N., Harris R. S. (2009) Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 83, 494–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Conticello S. G. (2008) The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sato Y., Probst H. C., Tatsumi R., Ikeuchi Y., Neuberger M. S., Rada C. (2010) Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J. Biol. Chem. 285, 7111–7118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vonica A., Rosa A., Arduini B. L., Brivanlou A. H. (2011) APOBEC2, a selective inhibitor of TGFβ signaling, regulates left-right axis specification during early embryogenesis. Dev. Biol. 350, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rogozin I. B., Basu M. K., Jordan I. K., Pavlov Y. I., Koonin E. V. (2005) APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle 4, 1281–1285 [DOI] [PubMed] [Google Scholar]
- 24. Longerich S., Basu U., Alt F., Storb U. (2006) AID in somatic hypermutation and class switch recombination. Curr. Opin Immunol. 18, 164–174 [DOI] [PubMed] [Google Scholar]
- 25. Di Noia J. M., Neuberger M. S. (2007) Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 [DOI] [PubMed] [Google Scholar]
- 26. Rada C., Di Noia J. M., Neuberger M. S. (2004) Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol Cell 16, 163–171 [DOI] [PubMed] [Google Scholar]
- 27. Blanc V., Davidson N. O. (2003) C-to-U RNA editing. Mechanisms leading to genetic diversity. J. Biol. Chem. 278, 1395–1398 [DOI] [PubMed] [Google Scholar]
- 28. Malim M. H. (2009) APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos Trans. R. Soc. Lond. B Biol. Sci. 364, 675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Albin J. S., Harris R. S. (2010) Interactions of host APOBEC3 restriction factors with HIV-1 in vivo. Implications for therapeutics. Expert Rev. Mol. Med. 12, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stenglein M. D., Burns M. B., Li M., Lengyel J., Harris R. S. (2010) APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bulliard Y., Narvaiza I., Bertero A., Peddi S., Röhrig U. F., Ortiz M., Zoete V., Castro-Díaz N., Turelli P., Telenti A., Michielin O., Weitzman M. D., Trono D. (2011) Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J. Virol. 85, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Landry S., Narvaiza I., Linfesty D. C., Weitzman M. D. (2011) APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12, 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H., Lilley C. E., Yu Q., Lee D. V., Chou J., Narvaiza I., Landau N. R., Weitzman M. D. (2006) APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16, 480–485 [DOI] [PubMed] [Google Scholar]
- 34. Thielen B. K., McNevin J. P., McElrath M. J., Hunt B. V., Klein K. C., Lingappa J. R. (2010) Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J. Biol. Chem. 285, 27753–27766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koning F. A., Newman E. N., Kim E. Y., Kunstman K. J., Wolinsky S. M., Malim M. H. (2009) Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83, 9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li M., Shandilya S. M., Carpenter M. A., Rathore A., Brown W. L., Perkins A. L., Harki D. A., Solberg J., Hook D. J., Pandey K. K., Parniak M. A., Johnson J. R., Krogan N. J., Somasundaran M., Ali A., Schiffer C. A., Harris R. S. (2012) First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem. Biol. 7, 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Refsland E. W., Stenglein M. D., Shindo K., Albin J. S., Brown W. L., Harris R. S. (2010) Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues. Implications for HIV-1 restriction. Nucleic Acids Res. 38, 4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lackey L., Demorest Z. L., Land A. M., Hultquist J. F., Brown W. L., Harris R. S. (2012) APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 419, 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peng G., Lei K. J., Jin W., Greenwell-Wild T., Wahl S. M. (2006) Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203, 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B. (1977) DNA N-glycosidases. Properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 252, 3286–3294 [PubMed] [Google Scholar]
- 41. Stivers J. T., Pankiewicz K. W., Watanabe K. A. (1999) Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry 38, 952–963 [DOI] [PubMed] [Google Scholar]
- 42. Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 43. Nowarski R., Britan-Rosich E., Shiloach T., Kotler M. (2008) Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 15, 1059–1066 [DOI] [PubMed] [Google Scholar]
- 44. Chelico L., Pham P., Calabrese P., Goodman M. F. (2006) APOBEC3G DNA deaminase acts processively 3′ → 5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 13, 392–399 [DOI] [PubMed] [Google Scholar]
- 45. Iwatani Y., Chan D. S., Wang F., Maynard K. S., Sugiura W., Gronenborn A. M., Rouzina I., Williams M. C., Musier-Forsyth K., Levin J. G. (2007) Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35, 7096–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harjes E., Gross P. J., Chen K. M., Lu Y., Shindo K., Nowarski R., Gross J. D., Kotler M., Harris R. S., Matsuo H. (2009) An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., Cullen B. R. (2006) Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wijesinghe P., Bhagwat A. S. (2012) Efficient deamination of 5-methylcytosines in DNA by human APOBEC3A, but not by AID or APOBEC3G. Nucleic Acids Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shandilya S. M., Nalam M. N., Nalivaika E. A., Gross P. J., Valesano J. C., Shindo K., Li M., Munson M., Royer W. E., Harjes E., Kono T., Matsuo H., Harris R. S., Somasundaran M., Schiffer C. A. (2010) Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]