Background: The C-type lectin RegIIIβ has bactericidal activity against certain Gram-negative bacteria. The mechanism had remained unclear.
Results: RegIIIβ binds to lipid A through a loop motif, which is also required for the bactericidal activity.
Conclusion: Bacterial binding of RegIIIβ to lipid A is critical for its bactericidal activity against Gram-negative bacteria.
Significance: This study provides novel insights into the bactericidal mechanism and recognition specificity of RegIIIβ.
Keywords: Antimicrobial Peptides, Bacteria, Carbohydrate Binding Protein, Lectin, Lipopolysaccharide (LPS), Bactericidal Activity, Carbohydrate Recognition Specificity
Abstract
RegIIIβ is a member of the C-type lectin family called RegIII. It is known to bind peptidoglycan, and its bactericidal activity shapes the interactions with commensal and pathogenic gut bacteria. However, little is known about its carbohydrate recognition specificity and the bactericidal mechanism, particularly against Gram-negative bacteria. Here, we show that RegIIIβ can bind directly to LPS by recognizing the carbohydrate moiety of lipid A via a novel motif that is indispensable for its bactericidal activity. This bactericidal activity of RegIIIβ could be inhibited by preincubation with LPS, lipid A, or gentiobiose. The latter is a disaccharide composed of two units of β-(1→6)-linked d-glucose and resembles the carbohydrate moiety of lipid A. Therefore, this structural element may form a key target site recognized by RegIIIβ. Using point-mutated RegIIIβ proteins, we found that amino acid residues in two structural motifs termed “loop 1” and “loop 2,” are important for peptidoglycan and lipid A binding (Arg-135, Asp-142) and for the bactericidal activity (Glu-134, Asn-136, Asp-142). Thus, the ERN motif and residue Asp-142 in the loop 2 are of critical importance for RegIIIβ function. This provides novel insights into the carbohydrate recognition specificity of RegIIIβ and explains its bactericidal activity against Gram-negative bacteria.
Introduction
The gastrointestinal tracts of mammals represent highly complex ecosystems colonized with very dense microbial communities called “microbiota.” These communities are composed of several hundred different bacterial species (1, 2). However, the mechanisms shaping the community structure of the microbiota are still not well understood. Antimicrobial proteins produced by intestinal epithelial cells such as α-defensins can affect the composition of gut microbiota and limit opportunistic invasion by symbiotic bacteria (3, 4). In addition, antimicrobial proteins also play a pivotal role in protecting from enteropathogenic infections (4, 5).
The bactericidal RegIII (regenerating gene family protein III) lectin family plays a crucial role in maintenance of microbiota homeostasis with commensal bacteria and protection of the host from pathogens (6–11). The matured RegIII lectins have a molecular mass of ∼16 kDa and are secreted from Paneth cells and epithelial cells of the intestine into gut lumen (6, 12). The murine intestine expresses two RegIII family members, RegIIIβ and RegIIIγ (13). Production of RegIIIβ and RegIIIγ is dramatically increased in response to bacterial colonization and pathogenic infection (6, 8, 12). Similarly, human hepatocarcinoma intestine pancreas/pancreatitis-associated protein (HIP/PAP),3 a human homolog for RegIIIγ, is expressed in the small intestine, and strong HIP/PAP expression has been shown in patients with inflammatory bowel disease (14, 15). Hence, RegIII proteins are thought to contribute substantially to the bacteria-host interaction in the gut.
The different RegIII lectins have distinct spectra of antimicrobial activity. RegIIIγ and HIP/PAP recognize peptidoglycan and display bactericidal activity against Gram-positive bacteria, whereas Gram-negative bacteria are not affected. Presumably, this is attributable to the topology of the Gram-negative cell envelope where the LPS-loaded outer membrane shields the access of RegIIIγ and HIP/PAP to the peptidoglycan (6). In contrast, despite sharing a high degree of sequence similarity to HIP/PAP and RegIIIγ, RegIIIβ can kill not only Gram-positive bacteria, but also some certain Gram-negative bacterial species. For example, RegIIIβ was found to kill Clostridium butyricum, Lactobacillus reuteri, and different Escherichia coli strains but not Enterococcus faecalis, Lactobacillus murinus, and Salmonella typhimurium (8). The mechanism explaining the activity against some Gram-negative species (but not others) had remained unknown.
As a structural feature, RegIII lectins have two distinct loop regions called loop 1 and loop 2 (16, 17). In HIP/PAP, and possibly also in RegIIIγ, a canonical EPN tripeptide motif in loop 1 was shown to be essential for carbohydrate recognition and bactericidal activity (18). Although the canonical motif is also found in loop 1 of RegIIIβ, roles of the motif in RegIIIβ-mediated carbohydrate recognition and bactericidal activity have remained unknown. Furthermore, little is known about the carbohydrate recognition specificity of RegIIIβ and how this protein kills bacteria.
Here, we show that the bactericidal effect of RegIIIβ against Salmonella typhimurium, an enteropathogenic Gram-negative bacterium that causes self-limiting diarrhea in humans and diverse animal models, including mice (19), is dependent on the bacterial growth phase. RegIIIβ kills S. typhimurium in logarithmic but not in stationary growth phase. The bactericidal activity correlates with the ability to bind to the bacterial surface. We finally show that RegIIIβ binds to LPS by recognizing the carbohydrate moiety of lipid A via a novel motif in the loop 2 region. Moreover, we found that Asp-142 is also involved in lipid A binding and the bactericidal activity of RegIIIβ. Our results provide new insights into the carbohydrate recognition specificity of the RegIII lectin family and explain why RegIIIβ is bactericidal for Gram-negative bacteria.
EXPERIMENTAL PROCEDURES
Bacterial Strains
S. typhimurium SL1344 was used in this study (20). Bacteria were cultured in LB broth under mild aeration. Specifically, the bacteria were grown in 1-cm diameter glass test tubes in 3 ml of LB broth on a rotating wheel (160 rpm) at 37 °C overnight, diluted 1:200 into a 100-ml Erlenmeyer flask (20 ml of LB broth), incubated at 37 °C in an orbital shaker (160 rpm) and grown to the indicated OD.
Reagents
Insoluble peptidoglycan from Bacillus subtilis, mannan from S. cerevisiae, dextran from Leuconostoc spp., lipopolysaccharides (LPS) from S. typhimurium, LPS Ra from S. typhimurium, LPS Re from S. minnesota, 3-deoxy-d-manno-oct-2-ulosonic acid, (Kdo; ammonium salt), lipid A from S. minnesota, chitobiose, chitotriose, and gentiobiose were purchased from Sigma-Aldrich. Soluble peptidoglycan from E. coli was purchased from InvivoGen (San Diego, CA).
Purification of Recombinant RegIIIβ Proteins
Recombinant untagged RegIIIβ and its point-mutated variants were prepared as described previously (8). Briefly, E. coli expressing RegIIIβ were lysed by sonication, and then the resulting inclusion bodies including RegIIIβ were purified. The purified RegIIIβ inclusion bodies were solubilized in denaturing buffer containing guanidine-HCl and then subjected to refolding buffer containing arginine-HCl to refold the RegIIIβ protein. Finally, the refolded RegIIIβ was purified by dialysis in binding buffer (25 mm MES, pH 6.0, 25 mm NaCl). Point mutations were introduced into RegIIIβ using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies, Inc., Santa Clara, CA) with specific primers harboring the desired mutations. The primer sequences were: for E114Q, 5′-GACCCTGGGTGCGCAGCCGAATGGCGGCG-3′ (primer 1) and 5′-CGCCGCCATTCGGCTGCGCACCCAGGGTC-3′ (primer 2); for D108N, 5′-CTGGATCGGTCTGCATAATCCGACCCTG-3′ (primer 1) and 5′-CCAGGGTCGGATTATGCAGACCGATCCAG-3′ (primer 2); for E134Q, 5′-ATGAACTATTTCAACTGGCAGCGTAATCCGAGCACCGCG-3′ (primer 1) and 5′-CGCGGTGCTCGGATTACGCTGCCAGTTGAAATAGTTCAT-3′ (primer 2); for R135T, 5′-GAACTATTTCAACTGGGAAACTAATCCGAGCACCGCGCTG-3′ (primer 1) and 5′-CAGCGCGGTGCTCGGATTAGTTTCCCAGTTGAAATAGTTC-3′ (primer 2); for N136A, 5′-GTTATGAACTATTTCAACTGGGAACGTGCGCCGAGCACCGCGCT-3′ (primer 1) and 5′-AGCGCGGTGCTCGGCGCACGTTCCCAGTTGAAATAGTTCATAAC-3′ (primer 2); and for D142A, 5′-GAGCACCGCGCTGGCGCGTGCGTTTTGCGG-3′ (primer 1) and 5′-CCGCAAAACGCACGCGCCAGCGCGGTGCTC-3′ (primer 2). The mutations were confirmed by DNA sequencing.
In Vitro Killing Assay
The in vitro killing assay was performed as described previously (8). In brief, bacteria grown at various growth phases were washed and resuspended in binding buffer (25 mm MES, pH 6.0, 25 mm NaCl) at a density of 1–3 × 106 cfu/ml. The diluted bacterial suspension was exposed to 10 μm RegIIIβ or 0.7 μm polymyxin B at 37 °C for 30 min. Bacteria were then plated on selective LB media. The recovered cfu were normalized for the original cfu of the inoculum, thus yielding the bacterial “survival” (in %). For the competitive in vitro killing assay, we used bacteria from the mid-logarithmic growth phase. Preincubation steps were performed for 10 min at 37 °C, as indicated.
Bacterial Binding Assay
Fifty microliter aliquots (1.0 × 107 cfu) of S. typhimurium grown up to the logarithmic (or the stationary) growth phase were pelleted by centrifugation, washed with binding buffer (25 mm MES, pH 6.0, 25 mm NaCl), suspended in binding buffer, and incubated with 10 μg of RegIIIβ (in 50 μl of total volume) for 15 min at 37 °C. The reaction mixtures were centrifuged at 6000 × g for 5 min, and the resulting supernatant was isolated, mixed with SDS-PAGE sample buffer, and boiled for 5 min. The pellet was washed once with binding buffer and boiled for 5 min in SDS-PAGE sample buffer. The samples were subjected to SDS-PAGE and analyzed by Coomassie Brilliant Blue (CBB) staining.
Gel Filtration Chromatography
Gel filtration chromatography was carried out using a Superdex 75 (10/300) column, which was equilibrated in running buffer (25 mm MES, pH 6.0, 125 mm NaCl). One hundred and fifty micrograms of RegIIIβ and/or 3 mg of LPS were mixed, incubated for 30 min at 37 °C in binding buffer, and applied to the column at flow rate of 0.5 ml/min. The elution profile was analyzed by monitoring the absorbance at 280 nm and by SDS-PAGE and Western blot analyses of the fractions using an affinity-purified anti-RegIIIβ (8) and an anti-LPS (Salmonella O antiserum Factor 5, Group B; Difco, Detroit, MI) antibodies.
Lipid A Binding Assay Using Polymyxin B-agarose
To generate “lipid A-coated” beads, 100 μg of lipid A (Sigma) were preincubated with polymyxin B-agarose (Sigma). Twenty-five μg of RegIIIβ was added to the resulting resin in a total volume of 0.5 ml of binding buffer. After rotation for 2 h at 4 °C, the resin was washed five times with 0.5 ml of wash buffer (25 mm MES, pH 6.0, 50 mm NaCl). Bound protein was eluted by boiling the resin in SDS-PAGE sample buffer (50 mm Tris-HCl, pH 6.8, 2% (w/v) SDS, 0.1% (w/v) bromphenol blue, 10% (v/v) glycerol, 100 mm 2-mercaptoethanol) and then resolved by SDS-PAGE through 15% acrylamide gels and analyzed by CBB.
Peptidoglycan Binding Assay
Ten μg of RegIIIβ or its point-mutated variants were added to 50 μg of insoluble peptidoglycan from B. subtilis (Sigma) in a total volume of 200 μl of binding buffer (25 mm MES, pH 6.0, 25 mm NaCl) and incubated for 2 h at 4 °C on a rotational mixer (12 rpm). After the incubation, centrifugation at 6000 × g for 5 min was carried out to sediment insoluble peptidoglycan. The supernatant was concentrated via filter centrifugation to reduce the volume. The pellet (insoluble peptidoglycan plus bound material) was washed twice in binding buffer, and the bound protein was eluted by boiling the peptidoglycan pellet in 2× SDS-PAGE sample buffer (100 mm Tris-HCl, pH 6.8, 4% (w/v) SDS, 0.2% (w/v) bromphenol blue, 20% (v/v) glycerol, 200 mm 2-mercaptoethanol). The samples were subjected to SDS-PAGE through 15% SDS-acrylamide gels and analyzed by CBB staining. For inhibition experiments, the RegIIIβ (10 μg) was preincubated for 30 min at 37 °C with 100 μg of soluble peptidoglycan (InvivoGen, San Diego, CA). After the preincubation, 50 μg of insoluble peptidoglycan was added, and binding was analyzed as described above.
Statistical Analysis
Statistical analysis was performed using the Mann Whitney U test or unpaired Student's t test. p < 0.05 was considered to be statistically significant.
RESULTS
Growth Phase-dependent Bactericidal Effect of RegIIIβ
We have shown that the C-type lectin RegIIIβ has bactericidal effects against certain Gram-positive and Gram-negative bacteria in vitro (8). Moreover, our findings have suggested that RegIIIβ can promote S. typhimurium gut infection by eliminating the competing gut microbiota.
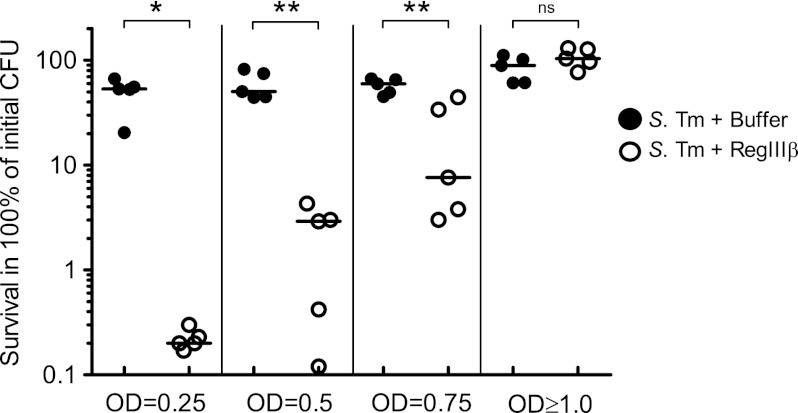
To analyze the bactericidal mechanism of RegIIIβ in more detail, we have analyzed killing of S. typhimurium cells taken from distinct bacterial growth phases in vitro. Similar to our previous data (8), wild-type S. typhimurium (SL1344) grown up to the stationary phase (A600 ≥ 1.0) was resistant to RegIIIβ (Fig. 1). In contrast, S. typhimurium cells from the early- (A600 = 0.25), the mid- (A600 = 0.5), or the late-logarithmic growth phase (A600 = 0.75) were susceptible to RegIIIβ (Fig. 1). These results establish that the RegIIIβ-mediated bactericidal effect against S. typhimurium is dependent on the bacterial growth phase. Thus, S. typhimurium from the early- or the mid-logarithmic growth phase can be employed to study how RegIIIβ acts on Gram-negative bacteria.
FIGURE 1.
RegIIIβ-mediated bactericidal effect is dependent on the bacterial growth phase. The S. typhimurium (S. Tm) wild-type strain (SL1344) was grown to early logarithmic growth phase (A600 = 0.25), middle logarithmic growth phase (A600 = 0.5), late logarithmic growth phase (A600 = 0.75), or stationary growth phase (A600 ≥ 1.0) and incubated with purified recombinant RegIIIβ (10 μm) for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. Assays were done at least in five independent experiments. Bar shows the median. *, p < 0.05; **, p < 0.01; ns, not significant (Mann-Whitney U test).
The Ability of RegIIIβ to Bind Gram-negative Bacteria Correlates with Its Bactericidal Activity
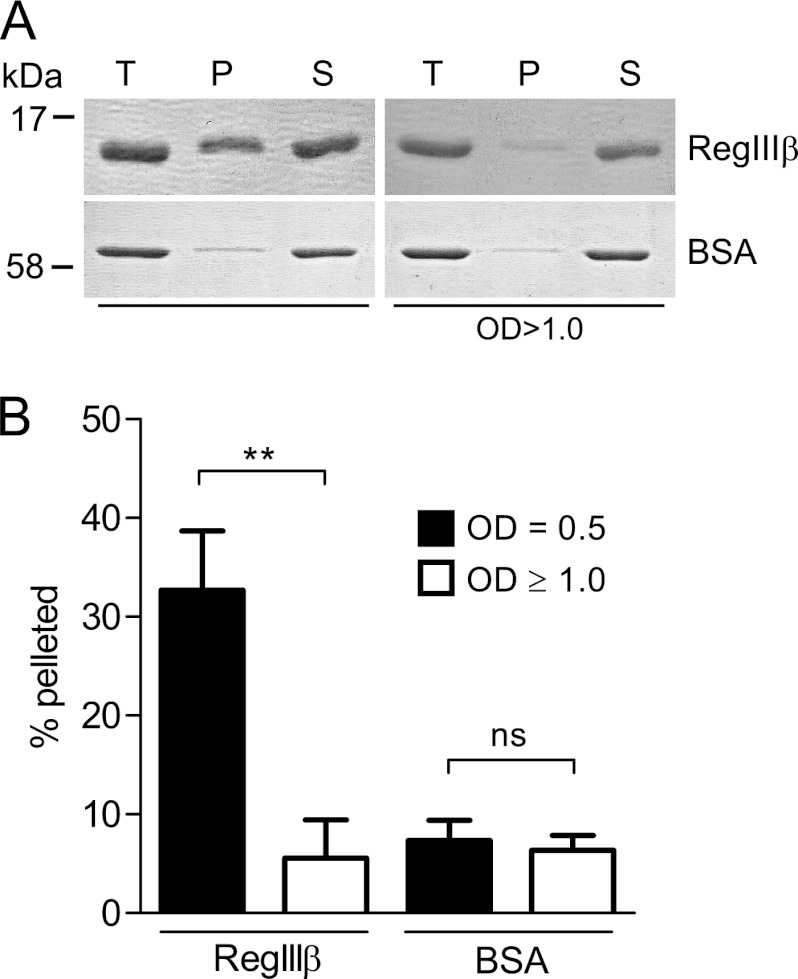
Binding to bacterial surface structures (i.e. peptidoglycan) is essential for the antibacterial activity of RegIII lectins such as HIP/PAP (18). This correlation has not yet been established for RegIIIβ. Therefore, we compared the binding ability of RegIIIβ to S. typhimurium grown to mid-logarithmic or to stationary growth phase. RegIIIβ was bound more efficiently to S. typhimurium from the mid-logarithmic growth phase than to bacteria from the stationary phase (Fig. 2, A and B). In contrast, BSA did not bind to S. typhimurium from either growth phase. The results indicate that the binding capacity of RegIIIβ to S. typhimurium correlates with its bactericidal activity.
FIGURE 2.
Binding of RegIIIβ to S. typhimurium from the middle logarithmic or from the stationary growth phase. A, bacteria were grown up to the middle logarithmic growth phase or the stationary growth phase (1.0 × 107 cfu) and incubated with RegIIIβ (10 μg) at 37 °C for 15 min (T, total fraction). The RegIIIβ bound or unbound to bacteria was separated by centrifugation (P, pellet; S, supernatant) and then analyzed by SDS-PAGE and CBB staining. B, quantified protein levels (%) of the pellet fractions were determined by defining the relative intensity of the total fraction as 100%. The error bars represent the S.D. of the mean from three independent experiments. **, p < 0.01; ns, not significant (unpaired Student's t test).
Insoluble Peptidoglycan and Mannan Can Block the Bactericidal Effect of RegIIIβ
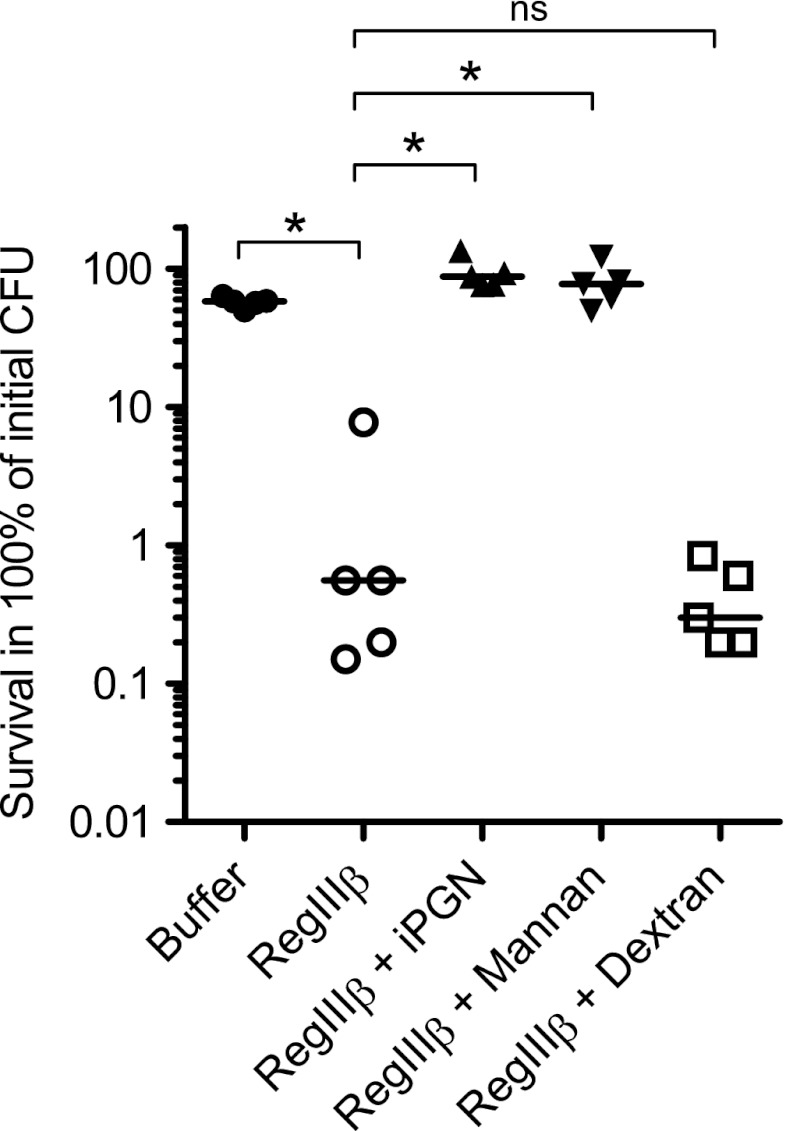
Earlier work has demonstrated that RegIIIβ can bind to peptidoglycan (18). This result also may imply that preincubation with peptidoglycan should inhibit the bactericidal effect of RegIIIβ. To test this hypothesis, RegIIIβ was preincubated with insoluble peptidoglycan (10 min, 37 °C; “Experimental Procedures”) before adding it to mid-logarithmic phase S. typhimurium. Preincubation with insoluble peptidoglycan from B. subtilis (supplemental Fig. 1A) completely abolished the RegIIIβ-mediated bactericidal effect (Fig. 3). A similar inhibitory effect was observed upon preincubation with mannan (supplemental Fig. 1B), another known substrate for RegIIIβ binding, whereas dextran (no binding to RegIIIβ) (supplemental Fig. 1C) had no the inhibitory effect (Fig. 3). Thus, preincubation with known substrates for RegIIIβ can inhibit its bactericidal activity against Gram-negative Salmonella spp., presumably by inhibiting RegIIIβ-binding to the bacterial surface.
FIGURE 3.
Preincubation with carbohydrate substrates inhibits the RegIIIβ-mediated bactericidal effect. Before mixing with bacteria, insoluble peptidoglycan (iPGN) from B. subtilis (160 μg/ml), mannan (1.6 mg/ml), or dextran (1.6 mg/ml) were incubated with purified RegIIIβ (10 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) grown up to the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. At least five independent experiments were performed. The bar shows the median. *, p < 0.05; ns, not significant (Mann-Whitney U test).
The Effect of LPS on RegIIIβ-mediated Bactericidal Activity
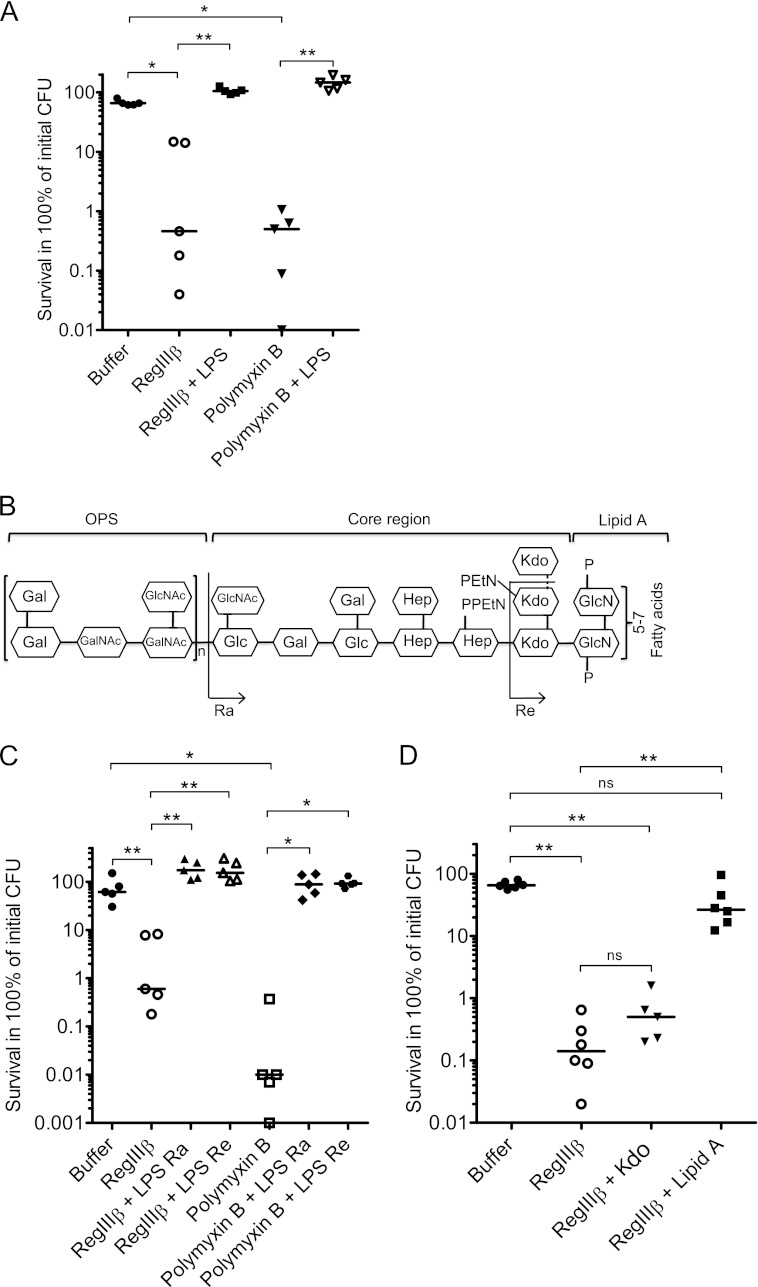
In Gram-negative bacteria such as S. typhimurium, the peptidoglycan is normally not accessible to proteins of >600 Da, as it is shielded by the outer membrane. The outer leaflet of the outer membrane is composed mostly of LPS, a large, complex glycolipid. We thus hypothesized that RegIIIβ may initially interact with LPS and that this may represent a critical step in its bactericidal effect on Gram-negative bacteria. To test this hypothesis, we investigated whether LPS preincubation may inhibit RegIIIβ-mediated killing of S. typhimurium. Indeed, preincubation of RegIIIβ with LPS (10 min, 37 °C; “Experimental Procedures”) completely abolished the bactericidal activity of RegIIIβ (Fig. 4A).
FIGURE 4.
Preincubation with LPS or lipid A inhibits the RegIIIβ-mediated bactericidal effect. A, the effect of LPS on RegIIIβ-mediated killing. Before mixing with bacteria, LPS from S. typhimurium (1.6 μm) was incubated with purified RegIIIβ (10 μm) or polymyxin B (0.7 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. B, chemical structure of LPS from S. typhimurium. LPS rough Ra and Re forms are indicated. Gal, galactose; GalNAc, N-acetyl-galactosamine; Glc, glucose; GlcNAc, N-acetylglucosamine; Hep, heptose; EtN, ethanolamine; GlcN, glucosamine; P, phosphate; n, the number of repeating units may vary from 0 to ∼50. C, the effect of rough LPS mutants Ra and Re on RegIIIβ-mediated killing. LPS Ra from S. typhimurium (38.4 μm) or LPS Re from S. minnesota (60.8 μm) was incubated with purified RegIIIβ (10 μm) or polymyxin B (0.7 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C, and bacterial survival was quantified by dilution-plating. D, the effect of Kdo and lipid A on RegIIIβ-mediated killing. Kdo (62.4 mm) or lipid A from S. minnesota (40 μm) was incubated with purified RegIIIβ (10 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C and quantified by dilution plating. Assays were repeated at least five times. Bars show the median. *, p < 0.05; **, p < 0.01; ns, not significant (Mann-Whitney U test).
Formally, the total RegIIIβ concentration was ∼3 times lower than that of the LPS in this assay. This might suggest that one LPS molecule might bind more than one RegIIIβ molecule. However, an alternative explanation is equally plausible. The bacterial binding assay (Fig. 2) indicated that, at least in some preparations, only one-third of the RegIIIβ protein does bind to the LPS (and/or other bacterial surface structures). The “inactivity” of the remaining two-thirds of the protein might be explained by incomplete refolding and/or by inhibitory bacterial molecules (LPS, murein, other inhibitors) which were carried over during the purification procedure, thus reducing the active RegIIIβ concentration. Thus, the LPS and the active fraction of the RegIIIβ might be present at approximately equimolar ratios in the experiment shown in Fig. 4A. Therefore, we cannot draw any firm conclusions on the stoichiometry of the RegIIIβ-LPS complex from these data.
Polymyxin B was used as a control. This cationic antimicrobial antibiotic is well known to bind LPS (21, 22), to disrupt the bacterial membrane, and thus to kill Gram-negative bacteria. Similar to RegIIIβ, the bactericidal effect of polymyxin B was inhibited by LPS preincubation (Fig. 4A). Furthermore, we wanted to exclude that micelle formation affected LPS-mediated inhibition. Triton X-100 did not alter the effect of RegIIIβ on Salmonella viability and did not affect the inhibition of RegIIIβ by LPS (supplemental Fig. 2). These results suggest that RegIIIβ may indeed bind to LPS and that this interaction may be critical for the bactericidal effect of RegIIIβ.
Lipid A Is Responsible for Counteracting the RegIIIβ-mediated Bactericidal Effect
LPS consists of three structural domains, the O-saccharide antigen, the core oligosaccharide, and the lipid A (Fig. 4B). To identify the structural domain required for inhibiting RegIIIβ-mediated bacterial killing, we performed a series of inhibition experiments using truncated versions of LPS. Rough LPS Ra lacks the O-specific polysaccharide (OPS, O-antigen), the Re core lacks the OPS and most residues of the core oligosaccharide, and lipid A lacks the OPS and the entire core oligosaccharide (Fig. 4B). Preincubation with LPS Ra and Re inhibited RegIIIβ-mediated killing (Fig. 4C). Control experiments were performed with polymyxin B. Similar to RegIIIβ, polymyxin B-mediated killing was inhibited by preincubation with LPS Ra or LPS Re (Fig. 4C). These results suggest that neither the OPS nor an intact core oligosaccharide are necessary for inhibiting the bactericidal activity of RegIIIβ.
To this point, it had remained unclear whether the two Kdo residues remaining on the LPS Re or the lipid A moiety itself might be involved. Therefore, we performed additional inhibition experiments. Preincubation with lipid A, but not with Kdo, inhibited the RegIIIβ-mediated bactericidal activity (Fig. 4D). We also tested the effect of dimethyl sulfoxide (DMSO) as DMSO served as the solvent for lipid A. However, DMSO had no effect on the bactericidal activity (data not shown). These results indicate that lipid A is the LPS moiety inhibiting the RegIIIβ-mediated bactericidal activity and imply that RegIIIβ may bind directly to lipid A.
Direct Interaction of RegIIIβ with LPS
To further substantiate the direct interaction of RegIIIβ with LPS, we employed a gel filtration chromatography assay. In this assay, LPS and/or RegIIIβ were preincubated for 30 min in binding buffer and run on a Superdex 75 (10/300) gel filtration column, and the elution profile was analyzed by photometry (A280) and Western blot of the eluted fractions using an anti-RegIIIβ antibody. In the absence of LPS, RegIIIβ eluted at fraction numbers 18–24, suggesting a molecular mass of ∼24 kDa (Fig. 5, A and B). This “apparent” molecular mass was slightly higher than the molecular mass of RegIIIβ (16 kDa). Similar observations have been reported for HIP/PAP (23). Overall, these observations indicated that RegIIIβ mainly exists in a monomeric state.
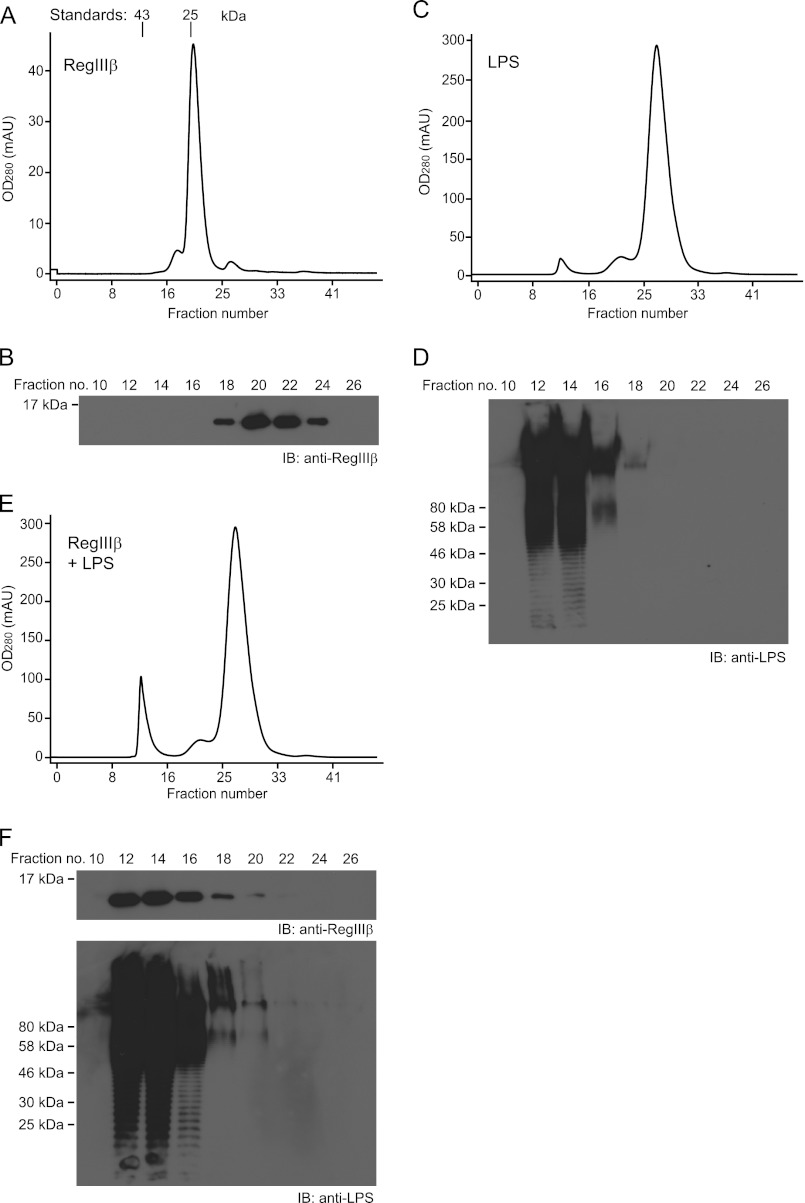
FIGURE 5.
Direct interaction of RegIIIβ with LPS. A, C, and E, gel filtration chromatography of RegIIIβ (A), LPS (C), and mixture of RegIIIβ and LPS (E). B, D, and F, detection of RegIIIβ and LPS in the eluate fractions of A and C and E by Western blot (IB) analysis using anti-RegIIIβ and anti-LPS antibodies. Standard proteins are indicated in A: ovalbumin (43 kDa) and chymotrypsinogen (25 kDa). mAU, milliabsorbance units.
In the absence of RegIIIβ, LPS yielded three peaks as observed by photometry. However, only the first peak harbored LPS as indicated by Western blotting (fractions 12–16; Fig. 5, C and D). It remains unclear whether the peaks observed around fractions number 21 and 27 might be attributable to a protein contamination in the LPS purchased from Sigma, a crude LPS extracted via the chloroform method (see also below). Nevertheless, the co-incubation of RegIIIβ with LPS resulted in a shift of the RegIIIβ peak (from fractions 18–24) into fractions 12–16, as indicated by photometry and by Western blotting (Fig. 5, E and F). These results strongly suggest that RegIIIβ binds directly to LPS.
To further substantiate this and to exclude effects from unknown contaminants, we repeated this assay using pure LPS prepared as described in Ref. 24. As expected, preincubation with the pure LPS completely inhibited the bactericidal activity of RegIIIβ (data not shown). In the gel filtration chromatography assay, LPS from this preparation yielded virtually no peaks around fractions 21 or 27 (data not shown). However, the pure LPS retained the capacity to shift RegIIIβ into fractions 12–16. This lends further support to the notion that RegIIIβ may bind directly to LPS.
Preincubation with Gentiobiose, a Disaccharide Composed of Two β-(1→6)-Linked Units of nd-Glucose, Inhibits the RegIIIβ-mediated Bactericidal Effect
Published data and our new findings have established that peptidoglycan and lipid A may represent target structures for RegIIIβ. Both target molecules include a disaccharide composed of modified glucosamines connected by β-(1→4)- or β-(1→6)-linkages, respectively (supplemental Fig. 3, A and B). Therefore, we hypothesized that RegIIIβ might recognize these disaccharide moieties. If so, preincubation with disaccharides resembling those found in peptidoglycan or lipid A should inhibit RegIIIβ-mediated killing.
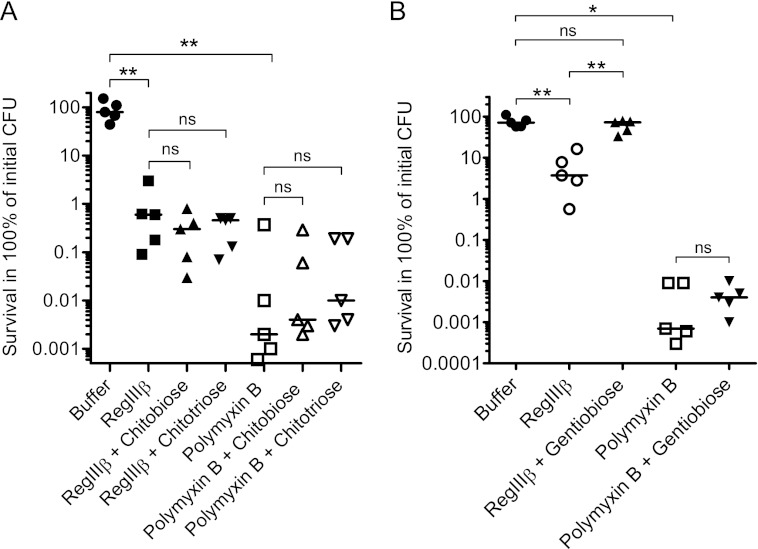
Chitobiose is a dimer composed of two units of N-acetylglucosamine and resembles the carbohydrate structure of peptidoglycan (supplemental Fig. 3A). Preincubation with chitobiose had no effect on the RegIIIβ-mediated bactericidal effect (Fig. 6A). The same held true for chitotriose, a trisaccharide composed of β-(1→4)-linked N-acetylglucosamine (supplemental Fig. 3A). Similarly, neither chitobiose nor chitotriose inhibited the polymyxin B-mediated bacterial killing activity (Fig. 6A). Thus, the carbohydrate moiety of peptidoglycan alone seems insufficient for binding by RegIIIβ.
FIGURE 6.
Preincubation with gentiobiose inhibits the bactericidal effect of RegIIIβ. A, the effect of chitobiose and chitotriose on RegIIIβ-mediated killing. chitobiose (7.7 mm) or chitotriose (5.1 mm) was incubated with purified RegIIIβ (10 μm) or polymyxin B (0.7 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. B, the effect of gentiobiose on RegIIIβ-mediated killing. Gentiobiose (9.3 mm) was incubated with purified RegIIIβ (10 μm) or polymyxin B (0.7 μm) for 10 min at 37 °C. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase was incubated with the preincubated mixture for 30 min at 37 °C, and bacterial survival was quantified by dilution-plating. Assays were done in at least five independent experiments. The bar shows the median. **, p < 0.01; ns, not significant (Mann-Whitney U test).
Gentiobiose is a disaccharide composed of β-(1→6)-linked d-glucose residues and resembles the carbohydrate structure of lipid A (supplemental Fig. 3B). Preincubation with gentiobiose blocked the RegIIIβ-mediated bactericidal activity (Fig. 6B). In contrast, gentiobiose had no effect on the polymyxin B-mediated killing, indicating that the inhibitory effect is specific for RegIIIβ. Collectively, these results suggest that the carbohydrate moiety of lipid A represents a key determinant sufficient for targeting by RegIIIβ and suggested that this interaction is critical for the bactericidal effect of this antimicrobial protein.
The EPN and DPT Motifs of Loop 1 Are Not Required for Binding to Peptidoglycan, Lipid A, or for the Bactericidal Activity of RegIIIβ
In the human protein HIP/PAP, the canonical EPN motif (in particular amino acid Glu-114) was shown to be of critical importance for binding to peptidoglycan and for the bactericidal activity against Gram-positive bacteria (18). This EPN motif is also conserved in RegIIIβ and RegIIIγ (Fig. 7A and supplemental Fig. 4). However, it has remained unclear whether EPN contributes to the biological activity of RegIIIβ.
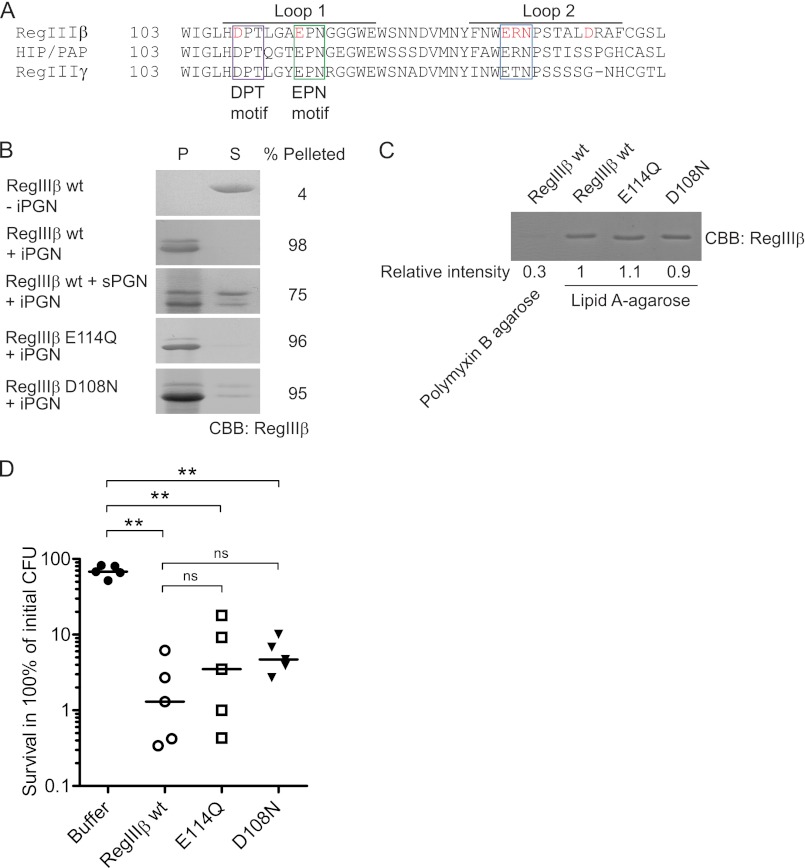
FIGURE 7.
Loop 1 of RegIIIβ is dispensable for binding to both peptidoglycan and lipid A and the bactericidal activity. A, alignments of HIP/PAP from human and RegIIIβ and RegIIIγ from mouse. Loop 1 and 2 of the long loop region were indicated. Loop 1 of all displayed homologs has an EPN and a DPT motif, indicated by magenta or green boxes, respectively. Loop 2 of RegIIIβ and HIP/PAP has a putative ERN motif, whereas RegIIIγ has a putative motif ETN, indicated by a blue box. Amino acid residues examined by targeted mutagenesis in this study are indicated in red. B, peptidoglycan binding assays of the loop 1 mutants of RegIIIβ. Ten μg of wild-type RegIIIβ (WT) or point mutated RegIIIβ E114Q or D108N proteins were added to 50 μg of insoluble peptidoglycan (iPGN) from B. subtilis. After incubation, the mixture was pelleted by centrifugation. The resulting pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE and CBB staining. For binding competition, 100 μg of soluble peptidoglycan (sPGN) from E. coli was preincubated with RegIIIβ. Percent of band intensity of the pellet fraction was calculated using NIH ImageJ software. C, lipid A binding assays of the loop 1 mutants of RegIIIβ. Purified wild-type RegIIIβ (wt), or point mutated RegIIIβ E114Q or D108N proteins were incubated with lipid A-immobilized polymyxin B-agarose. After washing the agarose, the bound RegIIIβ proteins were eluted by boiling in SDS-PAGE sample buffer and then subjected to SDS-PAGE and analyzed by CBB staining. The band intensity was calculated using ImageJ software by defining the relative intensity of wild-type RegIIIβ bound to the lipid A-immobilized agarose as “1”. D, in vitro killing assay of the loop 1 mutants of RegIIIβ. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth phase (A600 = 0.5) was incubated with purified recombinant RegIIIβ WT or E114Q or D108N (10 μm) for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. Assays were done at least in five independent experiments. The bar shows the median. **, p < 0.01; ns, not significant (Mann-Whitney U test).
To address this issue, we generated an E114Q point-mutated RegIIIβ protein and analyzed possible functional defects. First, we performed the binding assay to insoluble peptidoglycan (see “Experimental Procedures”). (Note that some peptidoglycan-associated proteolytic activity facilitates N-terminal cleavage of RegIIIβ (6, 25); this results in a slightly decreased molecular weight but does not affect target binding.) As shown in Fig. 7B, WT RegIIIβ bound efficiently to the insoluble peptidoglycan as it partitioned (almost exclusively) into the pellet fraction. If pre-incubated with soluble peptidoglycan, a significant fraction of RegIIIβ was retained in the supernatant fraction (Fig. 7B). This validated the assay and confirmed the binding specificity of RegIIIβ to peptidoglycan. Surprisingly, the E114Q variant of RegIIIβ bound with equivalent efficiency as the WT protein (Fig. 7B).
In a second assay, we analyzed the binding of the E114Q variant to lipid A. For this purpose, we coated polymyxin B-agarose beads with lipid A and performed a pulldown assay (see “Experimental Procedures”). Also in this assay, the E114Q variant of RegIIIβ bound with equivalent efficiency as the WT protein (Fig. 7C).
In a third approach, we tested the bactericidal effect of RegIIIβ E114Q. In contrast to the mutant phenotype reported for HIP/PAP (HIP/PAP-E114Q), RegIIIβ E114Q retained the bactericidal activity of the WT protein (Fig. 7D). These results indicate that the EPN motif is not required for either peptidoglycan or lipid A recognition and for the killing of Gram-negative bacteria by RegIIIβ.
Recently, a second motif (DPQ) was discovered in the loop 1 region of RegIV, another human Reg family lectin (26). RegIIIβ displays a similar sequence (DPT) in loop 1 (Fig. 7A and supplemental Fig. 4), suggesting that this motif might play a role in the interaction with bacterial target structures or the antimicrobial activity. To analyze the role of the DPT sequence, we generated RegIIIβ D108N. However, RegIIIβ D108N did show wild type activity in all three functional assays (Fig. 7, B–D). These results indicate that neither the EPN nor the DPT motif were required for substrate recognition or the bactericidal activity of RegIIIβ.
Residue Arg-135 of Loop 2 Is Required for Peptidoglycan and Lipid A Binding
Loop 2 of RegIIIβ features an ERN tripeptide (Fig. 7A and supplemental Fig. 4). The similarity to the EPN motif (i.e. loop 1 of HIP/PAP and RegIIIβ) prompted us to explore whether the ERN might contribute to substrate recognition or the bactericidal activity of RegIIIβ. For this purpose, we generated three variants of RegIIIβ, i.e. E134Q, R135T, and N136A. In binding assays to peptidoglycan or lipid A, RegIIIβ R135T was attenuated, whereas RegIIIβ E134Q and RegIIIβ N136A displayed wild-type activity (Fig. 8, A and B). These results indicate that Arg-135 contributes significantly to target binding by RegIIIβ.
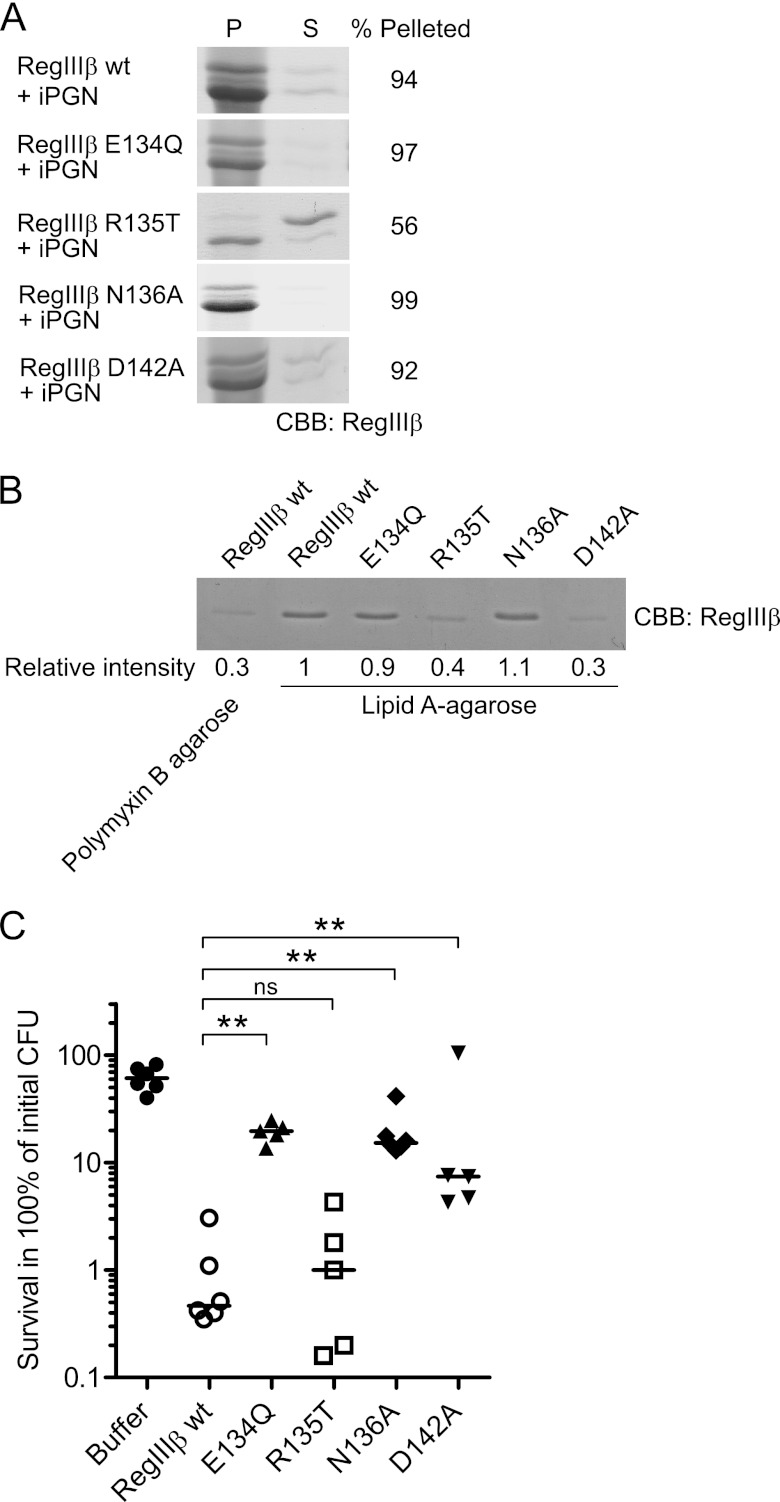
FIGURE 8.
Loop 2 is essential for substrate recognition of peptidoglycan and lipid A and for the bactericidal activity of RegIIIβ. A, peptidoglycan binding assays of the loop 2 mutants of RegIIIβ. 10 μg of RegIIIβ proteins, wild-type RegIIIβ (WT), E134Q, R135T, N136A, and D142A point mutants in loop 2 were added to 50 μg of insoluble peptidoglycan (iPGN) from B. subtilis. After incubation, the mixture was pelleted by centrifugation. The resulting pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE and CBB staining. Percent of band intensity of the pellet fraction was calculated using ImageJ software. B, lipid A binding assays of the loop 2 mutants of RegIIIβ. Purified wild-type RegIIIβ or point mutated RegIIIβ E134Q, R135T, N136A, or D142A were incubated with lipid A-immobilized polymyxin B-agarose. After washing the agarose, the bound RegIIIβ proteins were eluted by boiling in SDS-PAGE sample buffer and then subjected to SDS-PAGE and analyzed by CBB staining. The band intensity was calculated using ImageJ software. The relative intensity of wild-type RegIIIβ bound to the lipid A-immobilized agarose was defined as “1”. C, in vitro killing assay of the Loop 2 mutants of RegIIIβ. S. typhimurium wild-type strain (SL1344) from the middle logarithmic growth (A600 = 0.5) was incubated with purified recombinant RegIIIβ wild-type E134Q, R135T, N136A, or D142A (10 μm) for 30 min at 37 °C, and bacterial survival was quantified by dilution plating. Assays were performed in at least five independent experiments. The bar shows the median. **, p < 0.01; ns, not significant (Mann-Whitney U test).
RegIIIβ Mutants E134Q and N136A Have a Reduced Bactericidal Activity
To clarify the role of the ERN motif in the bactericidal activity of RegIIIβ, we performed bacterial killing assays with RegIIIβ E134Q, R135T, or N136A. The R135T mutant had bactericidal activity similar to wild-type RegIIIβ protein (Fig. 8C). In contrast, the bactericidal activity of RegIIIβ E134Q and N136A was significantly impaired (Fig. 8C). Thus, Glu-134 and Asn-136 seem crucial for the bactericidal effect of RegIIIβ. Together with the data presented above, these results indicate that ERN represents a novel motif and implicates loop 2 in the bactericidal activity of RegIIIβ.
Asp-142 of Loop 2 Is Required for Lipid A Binding and Bactericidal Activity
In several C-type lectins, substrate binding is facilitated via asparagine or glutamine residues and/or the COOH group of aspartic or glutamic acid (16, 27, 28). Loop 2 of RegIIIβ also features such a residue (Fig. 7A and supplemental Fig. 4). To address involvement of this Asp-142 amino acid residue, we generated RegIIIβ D142A. In binding assays, RegIIIβ D142A was impaired in binding to lipid A, but not to peptidoglycan (Fig. 8, A and B). Furthermore, RegIIIβ D142A displayed a reduced bactericidal activity (Fig. 8C). These results indicate that the residue Asp-142 of loop 2 contributes to substrate recognition and bactericidal activity of RegIIIβ.
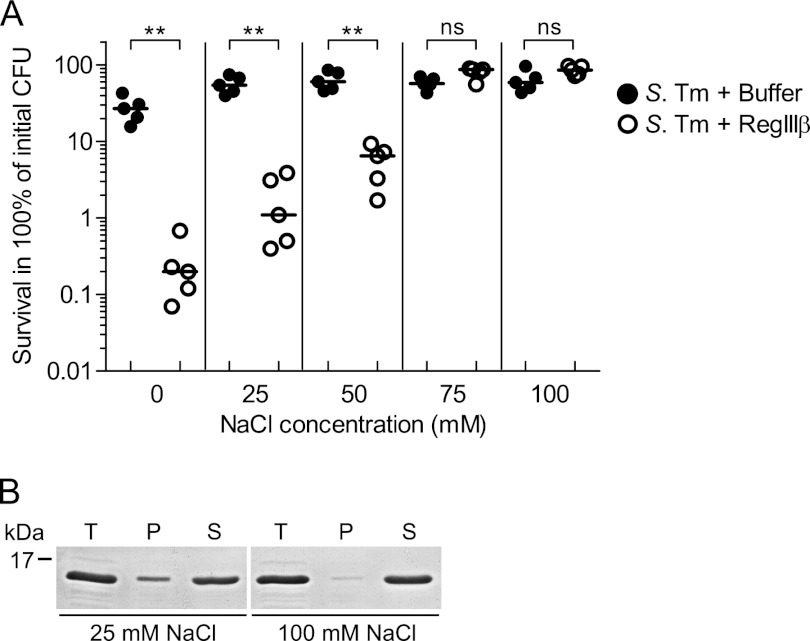
Salt Concentrations of ≥75 mm NaCl Alleviate Binding and Antimicrobial Activity of RegIIIβ
Our results from the binding assays suggest that polar, hydrogen, or ionic interactions might contribute to the binding of RegIIIβ to peptidoglycan and to LPS. These bonds should be sensitive to the salt concentration of the buffer. To test this hypothesis, we performed the in vitro killing assay using the binding buffers with increasing NaCl concentrations. In the presence of low NaCl concentrations (0, 25, 50 mm), RegIIIβ had bactericidal activity (Fig. 9A). In contrast, no bactericidal activity was detected at 75 and 100 mm NaCl.
FIGURE 9.
Salt concentration dependence of RegIIIβ-mediated killing. A, in vitro killing assay in the presence of various concentration of NaCl. S. typhimurium (S. Tm) wild-type strain (SL1344) from the middle logarithmic growth phase (A600 = 0.5) was incubated with purified recombinant RegIIIβ (10 μm) for 30 min at 37 °C in binding buffer containing the indicated concentration of NaCl (0, 25, 50, 75, and 100 mm), and bacterial survival was quantified by dilution plating. Assays were done in at least five independent experiments. The bar shows the median. **, p < 0.01; ns, not significant (Mann-Whitney U test). B, the effect of NaCl on binding of RegIIIβ to bacteria. Bacteria from the middle logarithmic growth phase (1.0 × 107 cfu) were incubated in binding buffer containing 25 mm NaCl or 100 mm NaCl and RegIIIβ (10 μg) at 37 °C for 15 min (T, total fraction). The fraction of RegIIIβ bound to bacteria was recovered by centrifugation (P, pellet; S, supernatant), and binding was analyzed by SDS-PAGE and CBB staining.
In a second approach, we compared the binding capacity of RegIIIβ to mid-log phase S. typhimurium in the presence of low (25 mm) or high (100 mm) concentrations of NaCl. In the presence of 100 mm NaCl, RegIIIβ bound with reduced efficiency (Fig. 9B; compare proteins in the pellet fractions “P”). Together, these results indicate that the ionic strength of the medium has a significant impact on the biological activity of RegIIIβ.
DISCUSSION
We have previously shown that RegIIIβ can contribute to Salmonella-induced colitis by eliminating competing gut microbiota (8). This was attributed to the susceptibility of many commensals to RegIIIβ-mediated killing, whereas S. typhimurium was found to be resistant in vitro (8). However, our results shown here reveal that S. typhimurium is not resistant per se. Instead, resistance depends on the bacterial growth phase: S. typhimurium taken from early- to mid-logarithmic growth phase are killed by RegIIIβ (this work), whereas bacteria from the stationary growth phase are resistant (as shown previously in Ref. 8). The susceptibility of S. typhimurium from the logarithmic growth phase correlated with enhanced binding to RegIIIβ, suggesting that killing might be attributable to the mode and/or the efficiency of RegIIIβ binding. Here, we have found that LPS is (besides peptidoglycan) a key target of RegIIIβ in the Gram-negative bacterial cell envelope, and we determined the molecular domains of LPS and of RegIIIβ, which are required for binding and for the antimicrobial activity against mid-log phase S. typhimurium.
Historically, RegIIIβ was found to bind peptidoglycan (18). However, in the context of the Gram-negative cell envelope, peptidoglycan should be shielded from antimicrobial peptides present in the surrounding environment. Therefore, we have hypothesized that RegIIIβ may also bind to LPS, as this complex lipid features a large number of carbohydrate residues and makes up the bulk of the outer leaflet of the outer membrane. Indeed, we found that RegIIIβ directly binds to the lipid A moiety of LPS and that this is an important step in the bactericidal activity against S. typhimurium. Most likely, the carbohydrate moiety of lipid A represents the main structural feature recognized by RegIIIβ, as gentiobiose (but not chitobiose or chitotriose), which mimics the carbohydrate backbone of lipid A, can protect S. typhimurium from RegIIIβ-mediated killing.
Is there a functional link between the binding of RegIIIβ to LPS and to peptidoglycan? Despite all progress, we still cannot answer this important question. One may consider at least three alternative scenarios: (i) “LPS-mediated access to peptidoglycan”. RegIIIβ binding to LPS may compromise the outer membrane integrity. This facilitates access of additional RegIIIβ molecules to the peptidoglycan in a second step. (ii) “Parallel access”. RegIIIβ might (at least initially) access the periplasm through pre-formed (growth-related) gaps in the outer membrane. Thus, RegIIIβ may target LPS and peptidoglycan at the same time. (iii) “Two-step-mechanism”. Alternatively, the very same RegIIIβ molecules, which initially bind to the LPS, might go on to penetrate the outer membrane and access the periplasm where they bind to peptidoglycan. Deciphering these steps will be an important topic for future research.
Similarly, the mechanism allowing RegIIIβ access to lipid A is not entirely clear. According to our findings of the growth phase-dependent killing effect of RegIIIβ, it would be conceivable if lipid A (and also maybe peptidoglycan) was accessible to RegIIIβ at the septum or the growth zone, i.e. features which may expose lipid A to access by RegIIIβ and which exist only in growing bacteria, but not in stationary phase cultures. Furthermore, the OPS production by S. typhimurium has been shown to depend on the bacterial growth phase, i.e. S. typhimurium grown up to the stationary phase has higher expression of OPS than bacteria from the logarithmic growth phase (29). Our previous data also have shown that the OPS-deficient S. typhimurium is more susceptible to RegIIIβ than the wild-type strain (8). It is conceivable that the rigid LPS layer limits the access of RegIIIβ to lipid A, especially in S. typhimurium grown up to the stationary phase. Revealing the mechanism underlying the growth phase-dependent bactericidal activity of RegIIIβ will be an important topic for future work.
RegIIIβ seems to employ different moieties for target binding than other RegIII family proteins. Lectins of the RegIII family (except for RegIIIα) display an EPN motif in loop 1. In the case of HIP/PAP, this motif was shown to be required for carbohydrate/peptidoglycan recognition (18). In contrast, the EPN motif in loop 1 of RegIIIβ is dispensable for peptidoglycan recognition and for its bactericidal activity. Instead, our results indicate that the carbohydrate recognition and the bactericidal activity (as well as lipid A binding) of RegIIIβ rely on a novel motif in loop 2, the ERN tripeptide, and residue Asp-142. This may resemble to some extent the carbohydrate recognition by mannose-binding lectin and asialoglycoprotein receptor. In these C-type lectins, EPN or QPD motifs of the long loop 2 mediate the recognition specificity to GlcNAc and mannose or Gal, respectively (27). The EPN and the QPD motif act as hydrogen bond donors (asparagine and glutamine) or acceptors (glutamic acid and aspartic acid) during carbohydrate recognition (16). In conclusion, the mode of carbohydrate recognition and bactericidal activity of RegIIIβ differs from that of HIP/PAP and RegIIIγ. It is tempting to speculate that these differences may explain the distinct antimicrobial spectra of the different RegIII family members.
Which target structures are recognized by the RegIII lectins? Peptidoglycan-binding was observed for HIP/PAP, RegIIIγ, and RegIIIβ, but not for RegIIIα (6, 18 and in this study). This was a first indication that substrate specificities do differ between the different RegIII family members. If binding is observed, the carbohydrate backbone of the peptidoglycan seems to represent the most important target structure. This is indicated by affinity assays and the inhibition of bacterial killing using chitin, a β-(1→4)-linked N-acetyl-glucosamine polymer (6, 18). However, the individual RegIII proteins seem to differ in their requirement for target oligomerization. The bactericidal activity of HIP/PAP is inhibited by chitobiose (a β-(1→4)-linked N-acetyl-glucosamine dimer) (6), whereas RegIIIγ-mediated killing effect is blocked by chitotetraose, but not by chitobiose (6). Similarly, we found that the bactericidal activity of RegIIIβ is not inhibited by chitobiose or chitotriose.
Besides the carbohydrate backbone, at least some of the RegIII family proteins also seem to recognize the linker peptides. Weak binding affinities of l-Ala-d-Glu-mDAP and l-Ala-d-Glu-l-Lys to HIP/PAP have been demonstrated by NMR (18). This binding does not seem to involve loop 1 (i.e. Glu-114, Glu-118), suggesting that different moieties of HIP/PAP are responsible for binding to the carbohydrate and the peptide moieties of peptidoglycan (18). Currently, we do not know whether RegIIIβ binding may also involve an additional epitope such as the linker peptides of the peptidoglycan. Such a requirement is known for peptidoglycan recognition proteins, which recognize a muramyl-tripeptide (30–32). Overall, the current data support the notion that different antimicrobial lectins of the RegIII family do differ in their target preference and in their affinities toward monomeric or oligomeric peptidoglycan units.
In addition to peptidoglycans, RegIIIβ can also bind to LPS. Again, the carbohydrate moiety seems to be the key binding determinant of this target molecule as demonstrated via gentiobiose inhibition assays. In this case, a carbohydrate dimer was sufficient for detectable binding. Future work will have to determine whether/how the carbohydrate binding site of RegIIIβ may allow discriminating between different carbohydrate moieties. The second residue (Arg-135) of the ERN motif affected both, binding to lipid A and to peptidoglycan and thus might constitute part of a “common” recognition mechanism.
The bactericidal activity of RegIIIβ is quite sensitive to NaCl concentrations of >50 mm. So far, we do not know whether this effect is attributable to the osmolarity or to the shielding of ionic/polar interactions. In the gut lumen, the total osmolarity is thought to range around the equivalent of 300 mm NaCl (33). However, in this environment, NaCl is only one of many components contributing to the total osmolarity. Based on these observations, it is tempting to speculate that salt concentration or osmolarity might represent a mechanism for controlling RegIIIβ activity in the gut. This will be an important topic for future work.
In summary, we propose a novel bacterial target for RegIIIβ, identify a novel functional motif involved in target recognition, and propose a mode of action that differs in important aspects from that of the other well characterized members of the RegIII family. Specifically, RegIIIβ was found to bind LPS by recognizing the carbohydrate moiety of lipid A via ERN and Asp-142 of loop 2. Our results provide novel insights into the carbohydrate recognition specificity and bactericidal mechanism of the RegIII lectin family. This will enable detailed structural studies aiming to clarify the bonding patterns explaining carbohydrate recognition by RegIIIβ. Such information may empower systematic approaches for developing new antimicrobial agents.
Acknowledgments
We are grateful to Markus Aebi, Markus Künzler, and Mikael Sellin for discussions and critical comments on this manuscript. We also thank Toshihiko Kitajima for assistance with gel filtration chromatography and helpful discussion.
This work was supported by Grant ETH-08-08-3 from the ETH Zurich Research Foundation (to W.-D. H.).

This article contains supplemental Figs. 1–4.
- HIP/PAP
- human hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- CBB
- Coomassie Brilliant Blue
- OPS
- O-specific polysaccharide.
REFERENCES
- 1. Dethlefsen L., McFall-Ngai M., Relman D. A. (2007) An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449, 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ley R. E., Lozupone C. A., Hamady M., Knight R., Gordon J. I. (2008) Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Salzman N. H., Underwood M. A., Bevins C. L. (2007) Paneth cells, defensins, and the commensal microbiota: A hypothesis on intimate interplay at the intestinal mucosa. Semin. Immunol. 19, 70–83 [DOI] [PubMed] [Google Scholar]
- 4. Ganz T. (2003) Defensins: Antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 5. Wilson C. L., Ouellette A. J., Satchell D. P., Ayabe T., López-Boado Y. S., Stratman J. L., Hultgren S. J., Matrisian L. M., Parks W. C. (1999) Regulation of intestinal α-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117 [DOI] [PubMed] [Google Scholar]
- 6. Cash H. L., Whitham C. V., Behrendt C. L., Hooper L. V. (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brandl K., Plitas G., Schnabl B., DeMatteo R. P., Pamer E. G. (2007) MyD88-mediated signals induce the bactericidal lectin RegIII γ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med. 204, 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stelter C., Käppeli R., König C., Krah A., Hardt W. D., Stecher B., Bumann D. (2011) Salmonella-induced mucosal lectin RegIIIβ kills competing gut microbiota. PLoS One 6, e20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vaishnava S., Yamamoto M., Severson K. M., Ruhn K. A., Yu X., Koren O., Ley R., Wakeland E. K., Hooper L. V. (2011) The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dessein R., Gironella M., Vignal C., Peyrin-Biroulet L., Sokol H., Secher T., Lacas-Gervais S., Gratadoux J. J., Lafont F., Dagorn J. C., Ryffel B., Akira S., Langella P., Nùñez G., Sirard J. C., Iovanna J., Simonet M., Chamaillard M. (2009) Toll-like receptor 2 is critical for induction of Reg3β expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 58, 771–776 [DOI] [PubMed] [Google Scholar]
- 11. Keilbaugh S. A., Shin M. E., Banchereau R. F., McVay L. D., Boyko N., Artis D., Cebra J. J., Wu G. D. (2005) Activation of RegIIIβ/γ and interferon γ expression in the intestinal tract of SCID mice: An innate response to bacterial colonization of the gut. Gut 54, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaishnava S., Behrendt C. L., Ismail A. S., Eckmann L., Hooper L. V. (2008) Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. U.S.A. 105, 20858–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Narushima Y., Unno M., Nakagawara K., Mori M., Miyashita H., Suzuki Y., Noguchi N., Takasawa S., Kumagai T., Yonekura H., Okamoto H. (1997) Structure, chromosomal localization and expression of mouse genes encoding type III Reg, RegIII α, RegIII β, RegIII γ. Gene 185, 159–168 [DOI] [PubMed] [Google Scholar]
- 14. Christa L., Carnot F., Simon M. T., Levavasseur F., Stinnakre M. G., Lasserre C., Thepot D., Clement B., Devinoy E., Brechot C. (1996) HIP/PAP is an adhesive protein expressed in hepatocarcinoma, normal Paneth, and pancreatic cells. Am. J. Physiol. 271, G993–1002 [DOI] [PubMed] [Google Scholar]
- 15. Ogawa H., Fukushima K., Naito H., Funayama Y., Unno M., Takahashi K., Kitayama T., Matsuno S., Ohtani H., Takasawa S., Okamoto H., Sasaki I. (2003) Increased expression of HIP/PAP and regenerating gene III in human inflammatory bowel disease and a murine bacterial reconstitution model. Inflamm. Bowel Dis. 9, 162–170 [DOI] [PubMed] [Google Scholar]
- 16. Zelensky A. N., Gready J. E. (2005) The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217 [DOI] [PubMed] [Google Scholar]
- 17. Ho M. R., Lou Y. C., Lin W. C., Lyu P. C., Huang W. N., Chen C. (2006) Human pancreatitis-associated protein forms fibrillar aggregates with a native-like conformation. J. Biol. Chem. 281, 33566–33576 [DOI] [PubMed] [Google Scholar]
- 18. Lehotzky R. E., Partch C. L., Mukherjee S., Cash H. L., Goldman W. E., Gardner K. H., Hooper L. V. (2010) Molecular basis for peptidoglycan recognition by a bactericidal lectin. Proc. Natl. Acad. Sci. U.S.A. 107, 7722–7727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kaiser P., Diard M., Stecher B., Hardt W. D. (2012) The streptomycin mouse model for Salmonella diarrhea: Functional analysis of the microbiota, the pathogen's virulence factors, and the host's mucosal immune response. Immunol. Rev. 245, 56–83 [DOI] [PubMed] [Google Scholar]
- 20. Hoiseth S. K., Stocker B. A. (1981) Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 [DOI] [PubMed] [Google Scholar]
- 21. Morrison D. C., Jacobs D. M. (1976) Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry 13, 813–818 [DOI] [PubMed] [Google Scholar]
- 22. Moore R. A., Bates N. C., Hancock R. E. (1986) Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob. Agents Chemother. 29, 496–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cash H. L., Whitham C. V., Hooper L. V. (2006) Refolding, purification, and characterization of human and murine RegIII proteins expressed in Escherichia coli. Protein Expr. Purif. 48, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Castro C., Parrilli M., Holst O., Molinaro A. (2010) Microbe-associated molecular patterns in innate immunity: Extraction and chemical analysis of Gram-negative bacterial lipopolysaccharides. Methods Enzymol. 480, 89–115 [DOI] [PubMed] [Google Scholar]
- 25. Mukherjee S., Partch C. L., Lehotzky R. E., Whitham C. V., Chu H., Bevins C. L., Gardner K. H., Hooper L. V. (2009) Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J. Biol. Chem. 284, 4881–4888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ho M. R., Lou Y. C., Wei S. Y., Luo S. C., Lin W. C., Lyu P. C., Chen C. (2010) Human RegIV protein adopts a typical C-type lectin fold but binds mannan with two calcium-independent sites. J. Mol. Biol. 402, 682–695 [DOI] [PubMed] [Google Scholar]
- 27. Drickamer K. (1992) Engineering galactose-binding activity into a C-type mannose-binding protein. Nature 360, 183–186 [DOI] [PubMed] [Google Scholar]
- 28. Drickamer K. (1999) C-type lectin-like domains. Curr. Opin. Struct. Biol. 9, 585–590 [DOI] [PubMed] [Google Scholar]
- 29. Bravo D., Silva C., Carter J. A., Hoare A., Alvarez S. A., Blondel C. J., Zaldívar M., Valvano M. A., Contreras I. (2008) Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J. Med. Microbiol. 57, 938–946 [DOI] [PubMed] [Google Scholar]
- 30. Kim M. S., Byun M., Oh B. H. (2003) Crystal structure of peptidoglycan recognition protein LB from Drosophila melanogaster. Nat. Immunol. 4, 787–793 [DOI] [PubMed] [Google Scholar]
- 31. Guan R., Roychowdhury A., Ember B., Kumar S., Boons G. J., Mariuzza R. A. (2004) Structural basis for peptidoglycan binding by peptidoglycan recognition proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 17168–17173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chang C. I., Chelliah Y., Borek D., Mengin-Lecreulx D., Deisenhofer J. (2006) Structure of tracheal cytotoxin in complex with a heterodimeric pattern-recognition receptor. Science 311, 1761–1764 [DOI] [PubMed] [Google Scholar]
- 33. Gupta S., Chowdhury R. (1997) Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65, 1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]