Abstract
Background
Gene duplication has had a major impact on genome evolution. Localized (or tandem) duplication resulting from unequal crossing over and whole genome duplication are believed to be the two dominant mechanisms contributing to vertebrate genome evolution. While much scrutiny has been directed toward discerning patterns indicative of whole-genome duplication events in teleost species, less attention has been paid to the continuous nature of gene duplications and their impact on the size, gene content, functional diversity, and overall architecture of teleost genomes.
Results
Here, using a Markov clustering algorithm directed approach we catalogue and analyze patterns of gene duplication in the four model teleost species with chromosomal coordinates: zebrafish, medaka, stickleback, and Tetraodon. Our analyses based on set size, duplication type, synonymous substitution rate (Ks), and gene ontology emphasize shared and lineage-specific patterns of genome evolution via gene duplication. Most strikingly, our analyses highlight the extraordinary duplication and retention rate of recent duplicates in zebrafish and their likely role in the structural and functional expansion of the zebrafish genome. We find that the zebrafish genome is remarkable in its large number of duplicated genes, small duplicate set size, biased Ks distribution toward minimal mutational divergence, and proportion of tandem and intra-chromosomal duplicates when compared with the other teleost model genomes. The observed gene duplication patterns have played significant roles in shaping the architecture of teleost genomes and appear to have contributed to the recent functional diversification and divergence of important physiological processes in zebrafish.
Conclusions
We have analyzed gene duplication patterns and duplication types among the available teleost genomes and found that a large number of genes were tandemly and intrachromosomally duplicated, suggesting their origin of independent and continuous duplication. This is particularly true for the zebrafish genome. Further analysis of the duplicated gene sets indicated that a significant portion of duplicated genes in the zebrafish genome were of recent, lineage-specific duplication events. Most strikingly, a subset of duplicated genes is enriched among the recently duplicated genes involved in immune or sensory response pathways. Such findings demonstrated the significance of continuous gene duplication as well as that of whole genome duplication in the course of genome evolution.
Keywords: Gene duplication, Whole genome duplication, Teleost species, Tandem duplication
Background
Three main mechanisms are believed to generate gene duplications; unequal crossing over, retrotransposition, and chromosomal (genome) duplication [1,2]. Of these, localized (or tandem) duplication resulting from unequal crossing over and genome duplication are believed to be the two dominant mechanisms contributing to vertebrate genome evolution [3,4]. Much energy has been devoted to the examination and modeling of the whole genome duplication events believed to have shaped vertebrate genomes. Over four decades ago, Ohno (1970) suggested that two rounds of large-scale gene duplication had occurred early in vertebrate evolution. Sequencing analysis of Hox gene clusters from a spectrum of vertebrate species provided critical evidence in support of Ohno’s hypothesis [5-8] and indicated, in turn, an additional round of fish-specific genome duplication (FSGD) prior to the divergence of most teleost species [9-13]. Additional evidence supporting FSGD has been garnered from studies of pufferfish, Takifugu rubripes and Tetraodon nigroviridis. In these studies, hundreds of genes and gene clusters are present in duplicate in teleost fish but possessing only single copy in other vertebrates, illustrating fish-specific duplication of syntenic regions between humans and fish [14-16]. Ongoing examination of gene families across vertebrate evolution continues to provide general support for the three rounds of genome duplication (3R) hypothesis [17-22] in teleost fish.
By contrast, far less energy has been expended in understanding the larger and, arguably, more complicated landscape of gene duplication across model fish genomes and examining how genomes have been shaped and sized by gene duplication forces. Tandem duplication, in particular, is now recognized as a powerful, fast-acting evolutionary mechanism in the generation and expansion of gene families [4], accounting for greater than 10% of human genes [23]. Tandemly-arrayed genes (TAGs) are critical zones of adaptive plasticity, forming the building blocks for sensitive immune, reproductive, and sensory responses [24-26]. However, their extent and impact on teleost genome architecture has been routinely overlooked in the search for broader genome duplication patterns.
While many teleost fish species are in advanced stages of genome sequencing and assembly, only four species currently possess well-annotated genomes with chromosomal-anchored sequence information allowing extensive analysis of gene duplication—zebrafish, Danio rerio, medaka, Oryzias latipes, green spotted pufferfish, T. nigroviridis, and stickleback, Gasterosteus aculeatus. These fish, however, represent an interesting cross section of teleost diversity, with genomes differing in size from 342 Mb in pufferfish to 1.5 Gb in zebrafish, and with great variations in effective population sizes and generation intervals ranging from 7 weeks to 2 years. Differences in life history may reasonably be expected to impact patterns of gene duplication and retention. According to the neutral theory of molecular evolution [27] a new paralogous allele, if selectively neutral, has a probability of 1/2 N (where N is effective population size) of being fixed in a diploid population, with fixation occurring, on average, over 4 N generations. Differences in population size and generation interval among the teleost model species may also impact the extent and effectiveness of positive selection as seen previously in comparisons of duplicated genes between human and mouse [28].
Several recent studies have highlighted exceptional features of the zebrafish genome. These include reports of significantly higher rates of evolution in conserved noncoding elements [29], the largest numbers of tandemly-arrayed duplicates among all surveyed vertebrate species [4], and the highest average duplication rate of all lineages in the vertebrate tree (9.04 duplications/Ma [30]). Our own research has previously revealed a potentially related phenomenon of lower levels of alternative splicing when compared to other teleost species [31] and has explored the extensive nature of tandem duplications within some zebrafish gene families, e.g. cc chemokines [32]. Indeed, the particularities of the zebrafish genome have led many studies to use the more canonical pufferfish and medaka genomes in testing genome and gene duplication models and theories. The zebrafish genome may be perceived to represent some of the genome architecture of a large number of vertebrate species given its location on a portion of the tree of life within Cyprinidae with over 2,400 extant species. However, huge diversities exist in this group of freshwater fishes. For instance, the genome of common carp (Cyprinus carpio) is believed to have gone through additional round of whole genome duplication. Therefore, in terms of gene duplication, the common carp genome could be drastically different from the architecture of the zebrafish genome. Detailed examination and comparative analysis of the nature and impact of duplications in the zebrafish genome may only provide some reference for gene duplication analysis in related species.
To study the nature and extent of duplication among teleost species, here, we used a Markov clustering dynamic programming algorithm to arrange gene duplicates within the four model fish genomes into sets. Further analyses based on set size, duplication type, synonymous substitution rate (Ks), and gene ontology emphasize shared and lineage-specific patterns of genome evolution via duplication. Most strikingly, our analyses confirm the extraordinary duplication and retention rate of recent duplicates in zebrafish and their likely role in the expansion of the zebrafish genome.
Results
Duplicated gene sets among four model teleost species
Unigene sets gathered from the Ensembl databases of the four teleost fish were used for self-BLAST (all vs. all) followed by Markov clustering dynamic programming utilizing chromosomal coordinates as implemented in the program MCScan [33]. As shown in Table 1, a total of 3,991, 2,584, 2,669, and 2,020 duplicated gene sets were identified from zebrafish, medaka, stickleback, and green spotted pufferfish (Tetraodon), respectively. Based on chromosomal positions and relationships, the duplication sets were divided into three non-exclusive types: tandem duplication, inter-chromosomal duplication (non-tandem) and intra-chromosomal duplication (non-tandem). Definitions for the duplication types were as follows: 1) tandem duplication: duplicated gene copies located within 10 kb of one another (pairwise); 2) Intra-chromosomal duplication (Non-tandem): duplicated gene copies located on the same chromosome with a distance of greater than 10 kb between all members; and 3) Inter-chromosomal duplication (Non-tandem): duplicated gene copies located on different chromosomes. A portion of the duplicated sets combined several duplication types (e.g., duplicate set members present in both tandem and inter-chromosomal arrangements; Table 1). Inter-chromosomal duplications were the most prevalent among the three types across all four teleost species, accounting for around 80% of duplication sets and indicating the importance of genome-level duplication events in shaping teleost genome architecture. Intra-chromosomal and tandem duplication were the second and third most prevalent types, respectively. Zebrafish had the highest percentage of sets within these latter two categories, 47%, compared with 33.9%, 35.5%, and 38.6% in medaka, stickleback, and Tetraodon, respectively. In addition, zebrafish differed noticeably from medaka, stickleback, and Tetraodon in average duplication set size, with 4.3 genes per duplication set compared to 5.4 genes per set in the three other species.
Table 1.
Summary of gene duplications in four teleost model species
| Zebrafish | Medaka | Stickleback | Tetraodon | |
|---|---|---|---|---|
|
Genes |
26,842 |
18,027 |
19,178 |
14,038 |
|
Duplication sets |
3,991 |
2,584 |
2,669 |
2,020 |
|
Average duplication set size (gene number) |
4.3 |
5.4 |
5.4 |
5.4 |
|
Inter-chromosomal duplication sets |
3,109 (77.9%) |
2,249 (87.0%) |
2,262 (84.8%) |
1,645 (81.4%) |
|
Intra-chromosomal duplication sets |
1,264 (31.7%) |
614 (23.8%) |
573 (21.5%) |
477 (23.6%) |
|
Tandem duplication sets |
612 (15.3%) |
260 (10.1%) |
373 (14.0%) |
303 (15.0%) |
| Mixed duplication sets | 994 | 539 | 539 | 405 |
Duplication sets reflect groups of putatively paralogous genes clustered together by MCScan. Duplication type classifications are non-exclusive in some cases (Methods) due to multiple duplication types being found in some sets. The number of these “mixed” type sets is listed below for each species.
Duplication set size prevalence differs between zebrafish and other teleost species
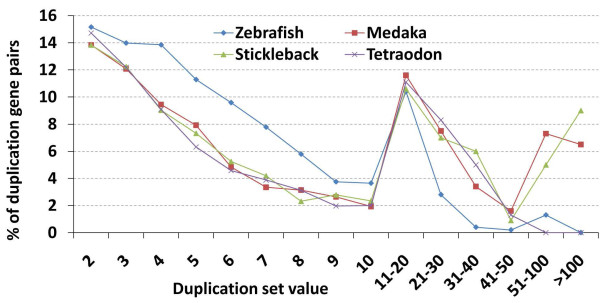
To better understand the distribution of duplicated genes within the four model teleost species, we examined the number of genes on a percentage basis found within duplication sets of varying size. While the relationship between duplication set size and percentage of duplicated genes was similar among the four species (Figure 1; Additional file 1: Table S1.), zebrafish again was the outlier, showing a pattern of more numerous small-scale duplications (set sizes 2–10). This pattern was consistent with our observation of smaller average set size in zebrafish, as was the larger number of duplications found in set sizes greater than 20 in medaka, stickleback, and Tetraodon.
Figure 1.
The distribution of duplicated genes from the four model teleost species across varying duplication set sizes
Lineage-specific patterns of duplication events among four teleost species
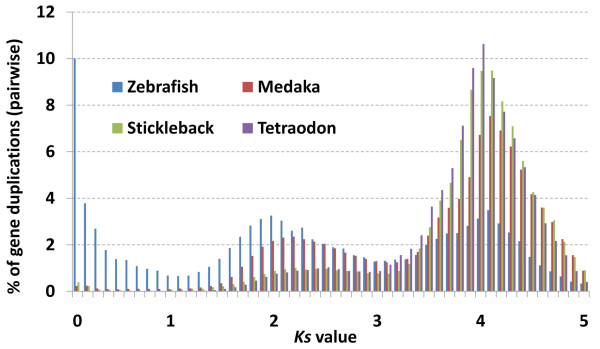
We next asked whether the observed prevalence of small duplication sets in zebrafish reflected a faster evolutionary rate in the species as manifested in its duplicated genes. To answer the question, we first examined the mutational distance between the duplicated genes (pairwise) of each species using Ks, a measure of the number of substitutions per synonymous site. We again noted a strikingly different Ks distribution in zebrafish when compared with the three other model species (Figure 2). Over 24.4% of duplicated genes in zebrafish had Ks values of ≤1.0 compared to 1.3%, 0.97%, and 0.05% of duplicated genes in medaka, stickleback and Tetraodon, respectively.
Figure 2.
The distribution of duplicated genes (pairwise comparisons) from the four model teleost species across varyingKsvalues
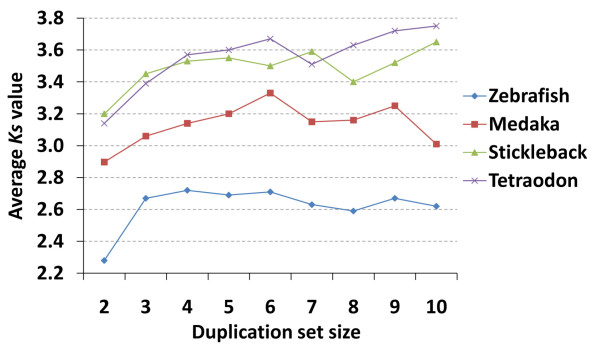
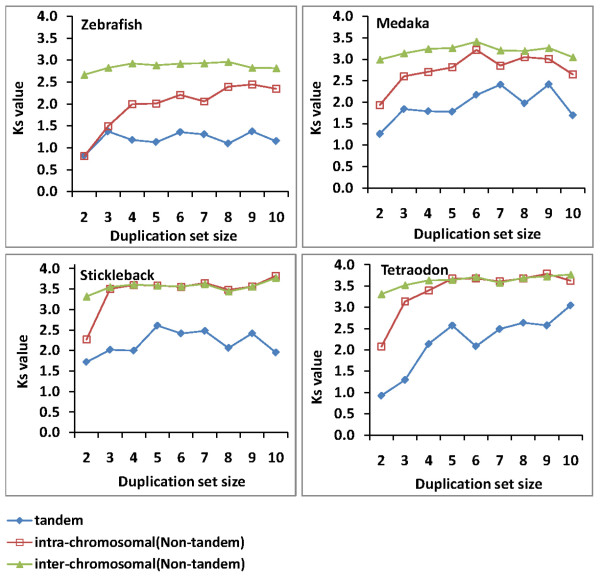
To determine whether the abundance of small duplicate sets in zebrafish may be explained by recent evolution (low Ks) of these genes, we calculated average Ks values for each duplicated set size in the size ranges where zebrafish has a greater percentage of duplicated genes (set size 1–10; Figure 3). Indeed, Ks values in these sets are markedly lower in zebrafish than in medaka, stickleback, and Tetraodon. Interestingly, while a clear positive correlation existed between duplication set size and Ks value in stickleback and Tetraodon, this pattern was obscured in medaka and not apparent in zebrafish.
Figure 3.
The relationship between duplication set size and averageKsvalue of duplicated genes from the four teleost species
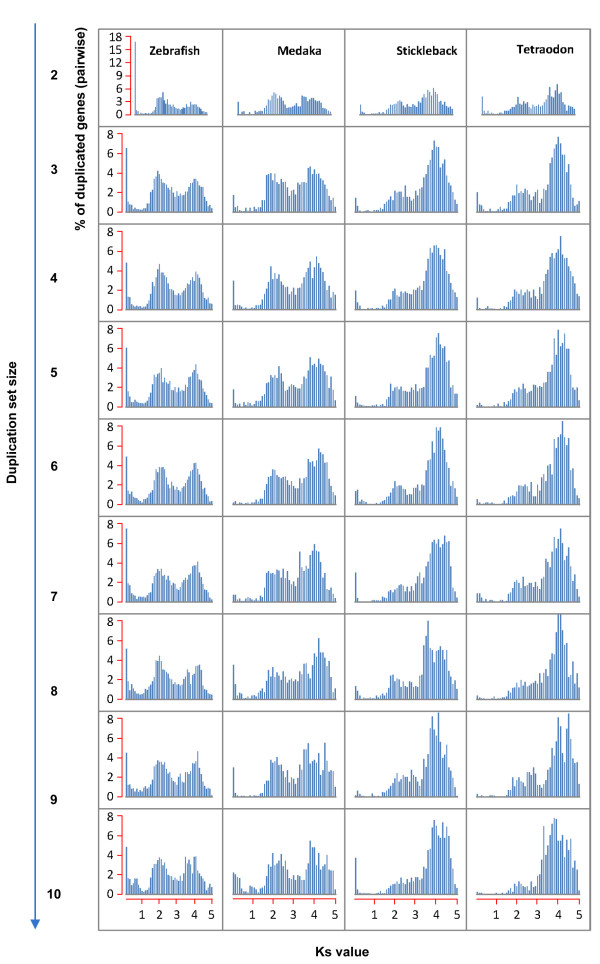
The relationship between Ks and set size was even more evident when the duplicated set sizes were analyzed separately and individual pairwise Ks values were plotted (Figure 4). As seen previously, zebrafish has an abundance of low Ks (Ks <1) duplicate pairs at all the studied set sizes when compared with the other three species. However, several other interesting patterns were evident in this analysis. Zebrafish and medaka maintain two roughly proportional peaks of Ks values (approximate mean values of Ks = 2 and Ks = 4), indicating two broad age (divergence level) categories of duplicated genes in these species, irrespective of duplicate set number. In contrast, a single major peak (mean Ks = 4) was observed in stickleback and Tetraodon, with a much smaller Ks peak (Ks = 2) appearing to generally diminish with increasing set size. The Ks distributions of stickleback and Tetraodon are particularly striking in their similarity to one another and suggest a dramatically diminished role for recent duplications in shaping these species’ genomes when compared with zebrafish and medaka.
Figure 4.
The distribution of duplicated genes (pairwise comparisons) across increasingKsvalues for each duplication set size (2 to 10)
Tandem duplications are predominant among small, recent gene duplications in zebrafish
We next asked whether the large numbers of small, recent duplications observed in zebrafish were evenly distributed across duplication types or whether they were biased toward a particular type. As seen in Figure 5, tandem gene duplicates had the lowest Ks values in each species irrespective of duplication set size. Tandem duplicates from zebrafish had the lowest Ks values observed in any species with little perceptible increase in mutational distance across the analyzed duplicated set sizes. Intra-chromosomal duplicates in zebrafish and medaka had intermediate Ks values between tandem and inter-chromosomal duplication with an upward trend correlated with increasing duplication set size. By contrast, Ks values for intra-chromosomal duplicates in stickleback and Tetraodon were virtually indistinguishable from those of inter-chromosomal duplicates in duplication sets of size ≥3. These patterns again point to the static nature of these genomes, with diminished retention and/or minimal levels of recent intra-chromosomal or tandem duplication activity to shape their genome architecture.
Figure 5.
AverageKsvalues for varying duplication set sizes and among the three different duplication types in the four model teleost species
Functional bias of recent (low Ks) duplicates in zebrafish
In order to determine whether the expansion of recent, retained duplicates in zebrafish has contributed to the diversification of genes mediating particular physiological functions in the species, we carried out gene ontology analysis on the duplicated gene sets with Ks values ≤1.0. This Ks range comprises the duplicated set with the most striking expansion when compared with the three other teleost models (Figures 3 and 5). Three GO terms were enriched among these duplicates when compared to the larger set of duplicated zebrafish genes (Additional file 2: Table S2.)—MHC protein complex, olfactory receptor activity, and antigen processing and presentation. Similar enrichment was not detected in the other three species, precluded in part by their small set sizes in this Ks range. The enriched categories, critical for immune and sensory capabilities, strongly suggest a functional bias in mechanisms of duplication and retention in zebrafish and further point to the importance of lineage-specific patterns of duplication in genome evolution and species diversification.
Discussion
Gene duplication has been described as an opportunity to explore forbidden evolutionary space [2], the idea that duplicated genes operating under temporary conditions of relaxed selection provide the raw material for evolution of new gene functions. While whole-genome duplication events are critical in shaping broader genome architecture, gene duplication, particularly tandem events, represent more recent, and potentially, adaptive signatures of evolution [34] which are expected to differ among vertebrate lineages [23,35]. Indeed [36], using zebrafish as their model, and others have shown evidence that evolutionary rates of duplicated genes in teleost fish far outstrip those of the mouse lineage. These differences, aside from adaptive consequences, can have profound effects on the degree of shared ancestry and synteny among vertebrate genomes. For example, only 50% of duplicated genes in zebrafish, and 70% in Tetraodon, have their origin in 1R/2R WGD events, compared to over 80% in mammalian, avian, and amphibian lineages. The remaining fraction comes from FSGD and species-specific events [30]. Clearly, patterns of teleost gene duplication deserve closer scrutiny to better understand how this process continues to shape genome evolution. Therefore, here we examined the nature and extent of gene duplication in four model teleosts, zebrafish, medaka, stickleback and Tetraodon.
Our approach divided duplicated genes into sets based on duplication type and captured larger gene families as well as smaller, recent duplications. From the onset of our analysis, zebrafish stood out from the other three model species by most measures, with a larger percentage of sets involved in tandem and intra-chromosomal arrangements and numerous small duplication sets (Table 1, Figure 1). Our analysis of the mutational distance between duplicate pairs (Ks) across the teleost species (Figure 2), however, produced the most striking illustration of different patterns of duplication and retention. Over 24% of duplicate pairs in zebrafish had Ks values of ≤1.0 compared to around 1% or less in the other three species. These results are supported by previous studies which noted high evolutionary rates and duplicate retention rates in zebrafish [29,30]. The abundance of low Ks duplicate pairs in zebrafish may stem from a greater number of birth events or fewer gene loss events among young duplicates. Although homogenization through gene conversion is a possibility [2,37,38], the low Ks values are mostly associated with tandem duplicates, suggesting recent gene duplications.
Our approach focused on surveying the broader architecture of duplication in the teleost genomes rather than relying on cross-species phylogenetic analysis for identification of orthologous relationships. Our analyses are limited, therefore, in distinguishing between rapid lineage specific gains in zebrafish and excessive gene loss in other teleosts for particular duplicate sets. The bias in the low Ks duplicate pairs in zebrafish toward tandem duplication (Figure 5) provides support for these being recent duplication events. Close to 65% of these zebrafish duplicate pairs with Ks ≤ 1.0 are found in tandem arrangements compared with ~15% of total duplicated sets (data not shown). In addition, gene ontology analysis revealed a bias in these duplicates toward physiological functions previously associated with rapid evolution and adaptation [28,39,40]. Indeed, the enriched categories (olfactory receptors, MHC) are well known for their rapid diversification through duplication, recombination, and gene conversion [39,41,42]. Taken together, our results suggest strikingly rapid evolution and high retention of recent duplicates in zebrafish in a manner likely to result in specialization of immune and sensory mechanisms.
The differences observed in Ks distributions among the four teleost species (Figures 3 and 5) raised several intriguing questions for further research: What is the effect of life history on the genome architecture of fish, and is there a link between genome size and duplication rate/retention rate in fish? Shiu et al. (2006) examined similar lineage-specific patterns when comparing human and mouse duplicates, suggesting that the larger population size and shorter generation interval in murine species could account for more effective natural selection and retention of duplicated genes. In the four investigated teleost genomes, zebrafish and medaka share similar life history patterns, generation intervals of 7–9 weeks and large effective population sizes, and similar Ks distributions (excluding Ks <1.0). In contrast, Tetraodon and stickleback, with generation intervals of 1–2 year and smaller effective population sizes, had a notable absence of young (low Ks) duplicates and shared remarkably similar Ks distributions (Figure 5) across their duplicated genes. These patterns of duplication rate and retention have been explored in the light of population size using genome sequence information in invertebrates [43] and previously, on a more theoretical basis [44,45]. Previous observations of correlations between spontaneous duplication/deletion rates and effective population size and increasing retention of linked (tandem) duplicates at intermediate population sizes appear to support such a connection between life history and duplication profiles as suggested by our data. Another pattern deserving further attention as additional teleost genomes become available is a potential association between duplication timing/retention rates and genome size. Based on the limited data available from the four model genomes here, patterns of duplication rate (especially as reflected by those pairs with Ks ≤ 1.0) reflect genome size with zebrafish with the largest genome at 1.5 Gb, followed by medaka (700 Mb), stickleback (446 Mb) and Tetraodon (342 Mb). The drastically differing patterns of duplicate formation and retention as detected here and by Blomme (2006) may be reflected in evolution of non-coding elements as well [29] and, together, could contribute to significantly higher genic content and associated genome size, as observed in zebrafish [46].
The observed differences in age of duplicated genes as reflected in Ks values could also result from errors in genome sequence assemblies of medaka, stickleback and Tetraodon. As these genomes were sequenced using the shotgun approaches, sequence assembly could have underestimated the segmental duplicated genes. In other words, the most similar paralogues could have been assembled as one gene while they are truly two or more genes in the genome. In this scenario, the missing segmental duplications do affect the assessment of the age of duplications [47]. However, this problem cannot be easily addressed. In order to determine if such a possibility could have caused the major differences in Ks values between zebrafish and the other three fish species, we conducted simulations using zebrafish chromosome 1. The whole genome sequence assembly of zebrafish chromosome 1 was “segmented” into 500 bp pieces and then de novo assembly was conducted using a 10X sequence coverage. In this assembly, a large number of contigs were obtained, 37,396 contigs. Apparently, the large numbers of contigs were resulted from interspersed repetitive segments, most notably the TC1-like transposons. We then mapped the assembled contigs in silico to the reference genome sequence of zebrafish chromosome 1. Over 99.7% of these assembled contigs were mapped to chromosome 1 sequences, suggesting that the “shotgun” approach did not affect the identification of paralogs. Therefore, we believe that the differences in Ks values were likely not caused by sequence assembly errors in medaka, stickleback and Tetraodon although all these genomes were sequenced using whole genome shotgun sequencing.
Previously, we highlighted the low levels of alternative splicing detected from zebrafish (17% of mapped genes) compared with the other model teleost species [31]. By contrast, the compact genome of Tetraodon showed alternative splicing in 43% of mapped genes. In that study, an inverse correlation between genome size and alternative splicing was observed. Researchers have previously suggested an inverse relationship between rates of gene duplication and alternative splicing in animals [48] and, more recently, in plants [49] based on single gene or gene family investigations. Our previous analysis of alternative splicing combined with our present examination of gene duplication in the same teleost species appears to support this connection on a genome scale. Further study is warranted to investigate whether the recent duplicates of zebrafish can provide the functional repertoire generated through alternative splicing in other, smaller teleost genomes.
Our findings indicate that varying rates of gene duplication and retention can have a dramatic impact on the ancestry and architecture of teleost genomes and contribute to functional diversification and divergence of important physiological processes. These patterns may be reflective of differences in life history across the teleost radiation and may ultimately influence genic content and genome size. Further analyses of the genomes of additional, key teleosts (i.e. catfish, carp) in the near future will allow us to test these theoretical relationships and analyze the particularities of the zebrafish genome in the context of more recently diverged species.
In Brown’s paper, the Copy number variation elements (CNVE) appeared to be consistent with extensive population substructuring (i.e., local adaption) among zebrafish population, with 4,199 (69%) of the identified CNVEs unique to one strain and only 457 (7.5%) CNVEs are common to all four groups [50]. Given this large amount of genome variation among zebrafish populations, analysis of genomes from additional zebrafish populations may reveal differences in gene copy numbers within a given duplication set. This would be of great interest in helping to establish the rate of gene birth in zebrafish. However, only the reference genome sequences were available for the present analysis. In addition, large differences of gene copy number variations have been mostly associated with anonymous genomic segments, not protein-encoding genes.
Conclusions
We have analyzed gene duplication patterns and duplication types among the available teleost genomes and found that a large number of genes were tandemly and intrachromosomally duplicated, suggesting their origin of independent and continuous duplication. This is particularly true for the zebrafish genome. Further analysis of the duplicated gene sets indicated that a significant portion of duplicated genes in the zebrafish genome were of recent, lineage-specific duplication events. Most strikingly, a subset of duplicated genes is enriched among the recently duplicated genes involved in immune or sensory response pathways. Such findings demonstrated the significance of continuous gene duplication as well as that of whole genome duplication in the course of genome evolution.
Methods
Gene set and duplicated gene search
The zebrafish, medaka, stickleback, and Tetraodon protein sequences used in this study were obtained from Ensembl (www.ensembl.org; Ensembl Gene 63; Zv9 for zebrafish, HdrR for medaka, BROAD S1 for stickleback, and TETRAODON 8.0 for Tetraodon) were used for the gene duplication analysis. Sequences annotated as unknown, random, and mitochondrial were removed, and only genes with known chromosome location were kept. For all genes with overlapping chromosomal locations, shorter genes were discarded and the longest coding form kept following similar methods used previously [23,34]. Following filtering, there were 26,842 genes in the zebrafish genome, 18,027 genes in the medaka genome, 19,178 genes in the stickleback genome, and 14,038 genes in the Tetraodon genome (Table 1). These genes then were used for all-against-all blastp searches [51] using the BLOSUM62 matrix and the SEG filter to mask regions of low compositional complexity [52]. Next, all the gene pairs were sorted by gene name and a filter script was used to remove all the redundant pairs, including self matches and multiple matches. These unique and sorted BLAST results were used as the input of MCscan [33]. MCscan is based on a Markov cluster algorithm which retrieves multiple chromosomal regions using dynamic programming based on the similarity matrix generated from previous BLAST results. The default parameter was used (‘mul (0.4343), ceil (200)’) to generate the prerequisite .mcl file for MCscan. For the generated duplication sets, we examined the chromosomal locations of the family members for the following duplication type categories.
Duplication categories
The copies of the duplicated gene sets may reside on the same chromosome (intra-chromosomal) or on different chromosomes (inter-chromosomal). Based on the locations and arrangements of the duplicated gene copies, we classified the duplicated genes into the following three categories: 1) Tandem duplication: duplicated gene copies are located next to each other on the same chromosome within a distance of less than 10 kb; 2) Intra-chromosomal duplication (Non-tandem): duplicated gene copies are located on the same chromosome with a distance of greater than 10 kb between all set members; and 3) Inter-chromosomal duplication (Non-tandem): duplicated gene copies are located on different chromosomes.
Synonymous substitution (Ks) mutation rates for gene pairs
For each pair of homologs, their protein sequences were aligned with CLUSTALW [53] and their protein alignment converted to DNA alignment with PAL2NAL [54]. The Ks values were calculated using the PAML software package [55]. The Nei-Gojobori algorithm [56] was implemented in the PAML package.
Gene ontology calculation for gene pairs
Gene ontology enrichment was calculated using goatools [33]. The resulting data structure is based on a directed acyclic graph (DAG) which can be easily traversed from leaf to root. The over-representation and under-representation of certain GO terms were analyzed based on Fisher’s exact test. Also several multiple corrections were implemented including Bonferroni, Sidak, and false discovery rate. The latest version (Jun. 6th, 2011) obo-formatted file was downloaded from Gene Ontology website (http://geneontology.org).
Sequence simulation
The zebrafish chromosome 1 (Zv9) was downloaded from Ensembl database and then it was segmented into 500 bp pieces using CLC bio assembly simulator [57]. De novo assembly was conducted with 10-fold chromosome coverage using CLC Genomics Workbench.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JL conceived the study, carried out the bioinformatics analysis, and was involved in drafting the manuscript. EP conceived the study and was involved in drafting the manuscript. HT conceived the study. JL carried out the bioinformatics analysis. ZL conceived the study, carried out the bioinformatics analysis, and was involved in drafting the manuscript carried out the bioinformatics analysis. All authors read and approved the final manuscript for publication.
Supplementary Material
Table S1. Duplication set size distribution in four teleost species. Non-bracketed number reflects the number of duplication sets of the listed set size, while the bracketed percentage reflects the percentage of duplicated genes found in the listed set size as represented in Figure 1.
Table S2. Gene ontology enrichment in zebrafish duplicate pairs with low Ks values (Ks ≤ 1.0).
Contributor Information
Jianguo Lu, Email: jzl0004@auburn.edu.
Eric Peatman, Email: peatmer@auburn.edu.
Haibao Tang, Email: tanghaibao@gmail.com.
Joshua Lewis, Email: jtl0003@auburn.edu.
Zhanjiang Liu, Email: zliu@acesag.auburn.edu.
Acknowledgements
Thanks to Qing Yang for providing data analysis programming, Huseyin Kucuktas for useful discussions. This project was supported by Agriculture and Food Research Initiative.
Competitive Grant no. 2009-35205-05101 and 2010-65205-20356 from the USDA National Institute of Food and Agriculture
References
- Zhang JZ. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- Hurles M. Gene duplication: the genomic trade in spare parts. PLoS Biol. 2004;2:900–904. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Pan D, Zhang LQ. Tandemly arrayed genes in vertebrate genomes. Comp Funct Genomics. 2008;2008:1–11. doi: 10.1155/2008/545269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994. pp. 125–133. [PubMed]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z. et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:1700–1708. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL. et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Meyer A. Revisiting recent challenges to the ancient fish-specific genome duplication hypothesis. Curr Biol. 2001;11:R1005–R1007. doi: 10.1016/s0960-9822(01)00610-8. [DOI] [PubMed] [Google Scholar]
- Vandepoele K, De Vos W, Taylor JS, Meyer A, Van de Peer Y. Major events in the genome evolution of vertebrates: Paranome age and size differ considerably between ray-finned fishes and land vertebrates. Proc Natl Acad Sci U S A. 2004;101:1638–1643. doi: 10.1073/pnas.0307968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A, Koh EGL, Chia JM, Brenner S, Aparicio S, Venkatesh B. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol. 2004;21:1146–1151. doi: 10.1093/molbev/msh114. [DOI] [PubMed] [Google Scholar]
- Crow KD, Stadler PF, Lynch VJ, Amemiya C, Wagner GP. The “fish-specific” Hox cluster duplication is coincident with the origin of teleosts. Mol Biol Evol. 2006;23:121–136. doi: 10.1093/molbev/msj020. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A. et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Ravi V, Venkatesh B. Rapidly evolving fish genomes and teleost diversity. Curr Opin Genet Dev. 2008;18:544–550. doi: 10.1016/j.gde.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Winkler C, Schafer M, Duschl J, Schartl M, Volff JN. Functional divergence of two zebrafish midkine growth factors following fish-specific gene duplication. Genome Res. 2003;13:1067–1081. doi: 10.1101/gr.1097503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Schartl M, Volff JN. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol Biol. 2007;7:74. doi: 10.1186/1471-2148-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li Q, Lu H, Chen H, Guo YQ, Cheng HH, Zhou RJ. Fish specific duplication of Dmrt2: characterization of zebrafish Dmrt2b. Biochimie. 2008;90:878–887. doi: 10.1016/j.biochi.2008.02.021. [DOI] [PubMed] [Google Scholar]
- Kassahn KS, Dang VT, Wilkins SJ, Perkins AC, Ragan MA. Evolution of gene function and regulatory control after whole-genome duplication: comparative analyses in vertebrates. Genome Res. 2009;19:1404–1418. doi: 10.1101/gr.086827.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188:799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam HK, Ferguson MM, Danzmann RG. Whole genome duplication: challenges and considerations associated with sequence orthology assignment in Salmoninae. J Fish Biol. 2011;79:561–574. doi: 10.1111/j.1095-8649.2011.03030.x. [DOI] [PubMed] [Google Scholar]
- Shoja V, Zhang LQ. A roadmap of tandemly arrayed genes in the genomes of human, mouse, and rat. Mol Biol Evol. 2006;23:2134–2141. doi: 10.1093/molbev/msl085. [DOI] [PubMed] [Google Scholar]
- Rizzon C, Ponger L, Gaut BS. Striking similarities in the genomic distribution of tandemly arrayed genes in Arabidopsis and rice. PLoS Comput Biol. 2006;2:989–1000. doi: 10.1371/journal.pcbi.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. A role for gene duplication and natural variation of gene expression in the evolution of metabolism. PLoS One. 2008;3(3):e1838. doi: 10.1371/journal.pone.0001838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Aa LM, Levraud JP, Yahmi M, Lauret E, Briolat V, Herbomel P, Benmansour A, Boudinot P. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge, London: Cambridge University Press; 1983. [Google Scholar]
- Shiu SH, Byrnes JK, Pan R, Zhang P, Li WH. Role of positive selection in the retention of duplicate genes in mammalian genomes. Proc Natl Acad Sci U S A. 2006;103:2232–2236. doi: 10.1073/pnas.0510388103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 2011;28:1205–1215. doi: 10.1093/molbev/msq304. [DOI] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7(5):R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JG, Peatman E, Wang WQ, Yang Q, Abernathy J, Wang SL, Kucuktas H, Liu ZJ. Alternative splicing in teleost fish genomes: same-species and cross-species analysis and comparisons. Mol Genet Genomics. 2010;283:531–539. doi: 10.1007/s00438-010-0538-3. [DOI] [PubMed] [Google Scholar]
- Peatman E, Liu ZJ. CC chemokines in zebrafish: evidence for extensive intrachromosomal gene duplications. Genomics. 2006;88:381–385. doi: 10.1016/j.ygeno.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Tang HB, Wang XY, Bowers JE, Ming R, Alam M, Paterson AH. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008;18:1944–1954. doi: 10.1101/gr.080978.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. The temporal distribution of gene duplication events in a set of highly conserved human gene families. Mol Biol Evol. 2003;20:154–161. doi: 10.1093/molbev/msg017. [DOI] [PubMed] [Google Scholar]
- Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res. 2004;14:1248–1257. doi: 10.1101/gr.2400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Laudet V. Evolutionary rates of duplicate genes in fish and mammals. Mol Biol Evol. 2001;18:681–683. doi: 10.1093/oxfordjournals.molbev.a003849. [DOI] [PubMed] [Google Scholar]
- Ohta T. Gene conversion and evolution of gene families: an overview. Genes. 2010;1:7. doi: 10.3390/genes1030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan JP, Grimwood J, Danke J, Schmutz J, Dickson M, Amemiya CT, Myers RM. Coelacanth genome sequence reveals the evolutionary history of vertebrate genes. Genome Res. 2004;14:2397–2405. doi: 10.1101/gr.2972804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon D, Glusman G, Pilpel Y, Khen M, Gruetzner F, Haaf T, Lancet D. Primate evolution of an olfactory receptor cluster: diversification by gene conversion and recent emergence of pseudogenes. Genomics. 1999;61:24–36. doi: 10.1006/geno.1999.5900. [DOI] [PubMed] [Google Scholar]
- Chen M, Peng Z, He S. Olfactory receptor gene family evolution in stickleback and medaka fishes. Sci China Life Sci. 2010;53:257–266. doi: 10.1007/s11427-010-0025-4. [DOI] [PubMed] [Google Scholar]
- Böhme J, Högstrand K. Timing and effects of template number for gene conversion of histocompatibility complex genes in the mouse. Hereditas. 1997;127:11–18. doi: 10.1111/j.1601-5223.1997.00011.x. [DOI] [PubMed] [Google Scholar]
- Amadou C, Younger RM, Sims S, Matthews LH, Rogers J, Kumanovics A, Ziegler A, Beck S, Lindahl KF. Co-duplication of olfactory receptor and MHC class I genes in the mouse major histocompatibility complex. Hum Mol Genet. 2003;12:3025–3040. doi: 10.1093/hmg/ddg317. [DOI] [PubMed] [Google Scholar]
- Lipinski KJ, Farslow JC, Fitzpatrick KA, Lynch M, Katju V, Bergthorsson U. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr Biol. 2011;21:306–310. doi: 10.1016/j.cub.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Hou Y, Lin S. Distinct gene number-genome size relationships for eukaryotes and non-eukaryotes: gene content estimation for dinoflagellate genomes. PLoS One. 2009;4:e6978. doi: 10.1371/journal.pone.0006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X, Jiang Z. et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C. Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat Rev Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Chung JD, Fu X, Johnson VE, Ranjan P, Booth SL, Harding SA, Tsai CJ. Alternative splicing and gene duplication differentially shaped the regulation of isochorismate synthase in Populus and Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:22020–22025. doi: 10.1073/pnas.0906869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KH, Dobrinski KP. et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci. 2012;109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC, Federhen S. Statistics of local complexity in amino-acid-sequences and sequence databases. Comput Chem. 1993;17:149–163. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. ClustalW - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Knudsen B, Forsberg R, Miyamoto M. A computer simulator for assessing the different challenges and strategies of de novo sequence assembly. Genes. 2010;1:263–282. doi: 10.3390/genes1020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Duplication set size distribution in four teleost species. Non-bracketed number reflects the number of duplication sets of the listed set size, while the bracketed percentage reflects the percentage of duplicated genes found in the listed set size as represented in Figure 1.
Table S2. Gene ontology enrichment in zebrafish duplicate pairs with low Ks values (Ks ≤ 1.0).