Abstract
Background
Bdellovibrio bacteriovorus HD100 must regulate genes in response to a variety of environmental conditions as it enters, preys upon and leaves other bacteria, or grows axenically without prey. In addition to “housekeeping” sigma factors, its genome encodes several alternate sigma factors, including 2 Group IV-RpoE-like proteins, which may be involved in the complex regulation of its predatory lifestyle.
Results
We find that one sigma factor gene, bd3314, cannot be deleted from Bdellovibrio in either predatory or prey-independent growth states, and is therefore possibly essential, likely being an alternate sigma 70. Deletion of one of two Group IV-like sigma factor genes, bd0881, affects flagellar gene regulation and results in less efficient predation, although not due to motility changes; deletion of the second, bd0743, showed that it normally represses chaperone gene expression and intriguingly we find an alternative groES gene is expressed at timepoints in the predatory cycle where intensive protein synthesis at Bdellovibrio septation, prior to prey lysis, will be occurring.
Conclusions
We have taken the first step in understanding how alternate sigma factors regulate different processes in the predatory lifecycle of Bdellovibrio and discovered that alternate chaperones regulated by one of them are expressed at different stages of the lifecycle.
Background
Bdellovibrio bacteriovorus HD100 must regulate genes in response to a variety of environmental conditions as it enters, digests, and leaves other Gram-negative bacteria, or when it grows axenically without prey [1-3]. Discrete waves of enzymes digesting different prey contents are required so that predatory enzymes do not act on each other, as the Bdellovibrio changes from a non-replicating “attack-phase” outside the prey, to a growing and replicating state inside prey. The B. bacteriovorus HD100 genome encodes several potential sigma factors for RNA polymerase which may contribute to such organised waves of gene regulation [4]. The Bdellovibrio bacteriovorus HD100 genome has several predicted “housekeeping” sigma factors: gene bd0242 encoding an RpoD sigma 70 sigma factor; gene bd3318, encoding a FliA-like sigma factor and gene bd0843 encoding an RpoN-like sigma factor. In addition, there are two homologues of genes predicted to encode Group IV-RpoE-like sigma factors, bd0881 (product predicted at 162 amino-acids) and bd0743 (product predicted at 206 amino-acids). Further, gene bd3314 is predicted to encode a larger sigma factor homologue (predicted at 373 amino-acids) with sigma 70 homology.
RpoE-like sigma factors in other bacteria mediate gene expression in response to changes in host/external environment and bacteria with mutations in rpoEs can be defective in virulence or other host interactions [5]. Bd0881 and Bd0743 predicted proteins show significant homology (28.6% and 31.8% identity respectively) to the rpoE gene product of E. coli which encodes a sigma factor of the ECF type that is responsive to extra-cytoplasmic, periplasmic events; RpoE in E. coli is sequestered at the inner membrane by an RseA RseB pair of proteins, until inducing-events, in the shape of abnormally folded proteins in the periplasm, cause it to be released and active [6]. The Bdellovibrio genome, like that of other delta-proteobacteria, does not contain rseAB genes, suggesting that the RpoE-like sigma factors encoded by bd0881 and bd0743 belong more generally to the Group IV-type sigma factors. Unlike some members of this group, the Bdellovibrio genes lack the typical downstream co-transcribed gene encoding a product with homology to an anti-sigma factor. Indeed the genes (bd0745 and bd0882) that are immediately downstream of bd0743 and bd0881 are unique to the Bdellovibrio genome, with no other significant homologues in other bacteria.
We hypothesised that the regulatory functions of alternate Group IV sigma factors might be diverse and important in the Bdellovibrio lifestyle, where prey-interaction versus prey-independent axenic growth brings with it many different challenges to the cell, including outer membrane insults, and a need for a great deal of de novo protein synthesis. Thus we used directed mutagenesis with kanamycin cartridge insertion, to test if inactivation of the three sigma factor genes bd3314, bd0881 and bd0743, affected viability and to determine what their regulatory roles in the Bdellovibrio axenic and predatory lifestyles may be. We find that one is likely essential, one is involved in regulating predatory processes and one is involved in repression of different components of the GroESEL chaperone complex, which themselves may have different roles in the predatory lifecycle.
Results
Transcriptional studies and bioinformatics show the operon structures for bd0743 and bd0881
RT-PCR on total RNA taken from predatory growth conditions demonstrated that adjacent genes bd0880bd0881 and bd0882 are co-transcribed in an operon, but that bd0743 was not co-transcribed with the adjacent gene bd0745. This differs to the situation for Group IV sigma factors in other bacteria where the downstream gene usually encodes an anti-sigma-factor [7]. Alignment of the RpoE protein from E. coli with the predicted gene products from bd0743 and bd0881 gave another indication that these Bdellovibrio proteins may have different roles from that of E. coli RpoE. Amino acids known to bind the −35 recognition site in E. coli differ in Bd0743 and Bd0881 as illustrated in Table 1 and Figure 1, suggesting that these sigma factors may recognise different sequences to those of E. coli and also to each other. Additionally bd0881 is conserved in the genome of Bacteriovorax marinus, a marine Bdellovibrio-like bacterium but bd0743 does not have a strong homologue in that genome. These data were provided by BLAST analysis hosted by the Wellcome Trust Sanger Institute and can be obtained from http://www.sanger.ac.uk/cgi-bin/blast/submitblast/b_marinus.
Table 1.
amino acid composition of −35 recognition sites of theBdellovibriosigma factor gene products compared toE. coliRpoE[8]
| -35 recognition site amino acids inE. coli RpoE | Corresponding amino acid in Bd0743 | Corresponding amino acid in Bd0881 |
|---|---|---|
|
R149 |
R |
F |
|
Y156 |
F* |
L |
|
E157 |
N |
K |
|
P166 |
P |
P |
|
G168 |
D |
G |
|
T169 |
T |
T |
|
R171 |
K* |
K* |
|
S172 |
A |
S |
|
R173 |
A |
R |
|
F175 |
M |
S |
|
R176 |
K* |
L |
| R178 | R | R |
Many of the residues comprising the −35 recognition site of E. coli RpoE (bold) are not conserved in B. bacteriovorus HD100 (shown as non-bold), suggesting that these RpoE-like proteins may recognise different DNA consensus sequences and correlating with the lack of classical E. coli RpoE consensus sequences in promoters in the B. bacteriovorus HD100 genome. (* = conservative substitution)
Figure 1.
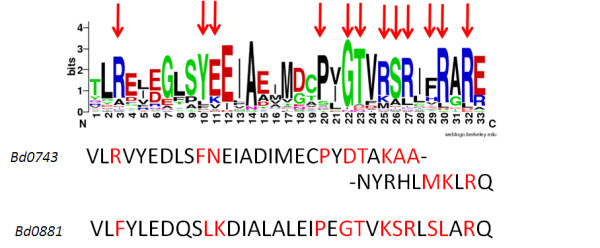
Sequence LOGO showing DNA binding region of RpoEs [8]. The first 35 sequences annotated as rpoE in the NCBI database were entered into the Weblogo program (http://weblogo.berkeley.edu/) using default parameters. The red arrows indicate the residues known to bind DNA in E. coli. The residues highlighted in red on the Bdellovibrio sequences show those that align to these using the ClustalW program and indicate that these are different from most RpoEs and each other, suggesting that they may well bind to different DNA motifs. There is also a 4 residue insertion in the Bd0743 sequence relative to the other sequences.
Inactivation of sigma factor genes suggests that bd3314 may be essential
Kanamycin resistant cassettes were inserted into the bd0743bd0881 and bd3314 genes to disrupt their coding sequences, and knockout mutants were screened for as described previously [9]. Viable knock-out mutants capable of predation were obtained for bd0743 and bd0881, but they could not be obtained for bd3314, despite extensive screening in axenic (prey-independent) and predatory conditions well beyond the bacterial numbers from which the other two mutants were isolated, suggesting that Bd3314 may be essential (a total of 287 isolates were screened from 4 separate conjugation experiments, yielding only bd3314 merodiploids, compared to 10 and 29 isolates yielding 6 and 1 knock-out mutants for bd0743 and bd0881 respectively). Bd3314 is larger than the other RpoE-like sigma factors (predicted 373 amino acids compared to 162 and 206) with homology to regions 1.2, 2, 3 and 4 of sigma 70 and so this may be acting as an alternative sigma 70 factor guiding the transcription of housekeeping genes which would explain why generating a knock-out mutant was not obtained. Top hits from a BLAST search for Bd3314 are sigma-70 genes from many delta-proteobacteria, (outwith the predatory Bdellovibrio) further supporting its possible role as an alternative sigma 70 protein. Some hits from BLAST were annotated as RpoH, but Bd3314 is unlikely to be RpoH as it lacks the “RpoH box” conserved in these proteins [10]. Further studies on the groups of genes it regulates is beyond the scope of this manuscript, but it is likely that as Bd3314 is conserved in other delta-proteobacteria, including many non-predatory bacteria, it may not have a specialised predatorily associated function.
Luminescent prey assay shows less efficient predation by a Bdellovibrio bd0881 knockout strain
Both the ΔBd0743 and ΔBd0881 knockout strains were able to grow predatorily but a predation efficiency assay [9] using luminescent prey cells showed that the ΔBd0881 mutant was less efficient at predation upon E. coli than the ΔBd0743 mutant and the wild-type control (Figure 2). For any given ratio of E. coli to Bdellovibrio, the ΔBd0881 strain took longer to reduce light emitted from the luminescent E. coli to half of its maximum, and hence took longer to kill the prey. An extra sum of squares F test carried out using the GraphPad Prism 5 software showed that this difference was significant (P < 0.0001). This suggests that Bd0881 controls, or optimises, the transcription of some genes involved in the predatory lifestyle while Bd0743 does not and thus Bd0881 is the first experimentally identified Bdellovibrio transcriptional regulator of predation genes. Axenic, prey-independent growth of both mutants was not significantly different from wild-type and heat shock (at 42°C for 10 min) did not reduce viability suggesting that they are not acting as typical alternate sigma32-like factors.
Figure 2.
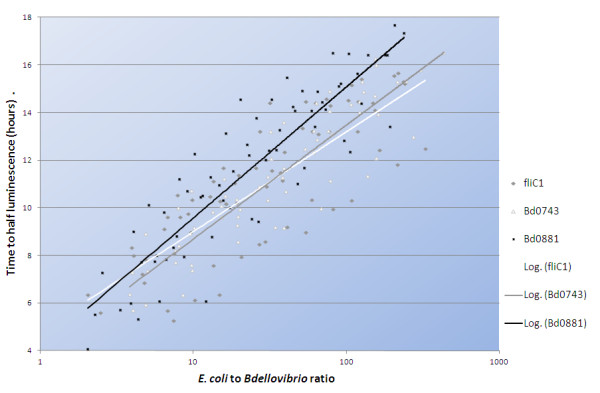
Predation efficiency assay using luminescent prey shows reduced efficiency for the ΔBd0881 mutant. Predatory efficiency plot showing log10 initial ratios of prey to predator against time to reach half of starting luminescence for the strains. Equivalent numbers of the ΔBd0881 mutant Bdellovibrio killed the prey cells more slowly than ΔBd0743 or kanamycin resistant “reconstituted wild-type”, fliC1 merodiploid strain.
Previously we have shown [11] that predatory efficiency in liquid media can be affected by the swimming speed of the Bdellovibrio which affects how efficiently they enter areas where they collide with prey. Interestingly, the transcription of the bd3052 fliC5 flagellin gene was found, by RT-PCR on attack phase Bdellovibrio RNA, (Figure 3) to be significantly down regulated in the ΔBd0881 mutant compared to the ΔBd0743 mutant and the wild type (WT) HD100 under heat shock conditions. This suggests that Bd0881 may have some role in regulating the expression of fliC5, altering protein composition and thus rigidity and/or the lengths of flagellar filaments in Bdellovibrio.
Figure 3.
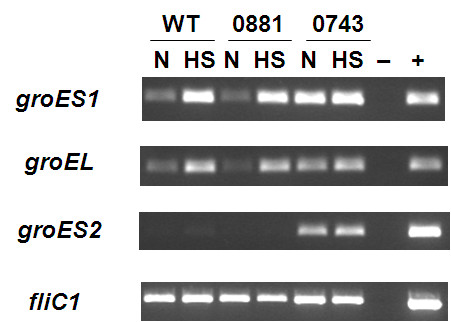
RT-PCR showing relative levels of transcription of chaperonin and flagellin genes in total RNA from attack phaseBdellovibrio, under normal and heat-shocked conditions. RT-PCR with transcript specific primers was carried out on matched concentrations of RNA (matched by Nanodrop spectrophotometer readings) from wild-type and mutant attack-phase Bdellovibrio including samples subjected to heat shock (42°C for 10 minutes). Total RNA samples from :-WT- wild-type HD100 attack phase, N- non-heat shocked 29°C, HS- heat shocked at 42°C for 10 minutes, 0881- ΔBd0881 attack phase, 0743- ΔBd0743 attack phase, Lane 7- no template negative control, Lane 8- HD100 genomic DNA positive control. “No reverse transcriptase” controls were performed for each template and were negative for DNA contamination (data not shown). The abundant transcript produced using primers designed to anneal to the fliC1 gene acts as a positive control by showing that there was ample total RNA in all samples.
A comparison of the flagellar lengths of the two strains versus WT, at the exact same growth conditions, revealed that the flagellar filaments of ΔBd0881 were slightly but significantly (P = 0.0026), shorter than those in wild type Bdellovibrio. In contrast, those in ΔBd0743 were longer (P = 0.0016) than the wild type (Figure 4A). We have previously shown [11] that fliC5 deletion shortens flagella and that ΔfliC5 flagellar mutants swim more slowly and prey less efficiently on E. coli in the luminescent prey assay. Interestingly, when we compared the swimming speeds of the two strains (Figure 4B) we found that the ΔBd0881 cells swam significantly (P = 0.044) but only slightly faster than the wild type, however, surprisingly both swam significantly (P < 10-5) faster than the ΔBd0743 strain despite it having longer flagellar filaments. Thus having a changed flagellin composition in the ΔBd0743 mutant strains produced a longer flagellum but either it had a “flaccid” wave form structure that produced less torque and thus swimming speed, or the ΔBd0743 mutation affected its complement of motor proteins so that the longer flagellum in this strain rotated slower than the wild type. We couldn’t test this by antibody-tethering cells by their flagella to glass slides because the flagella are sheathed with an outer membrane. The effect was a subtle one as the speeds were only slightly altered and the flagella waveforms did not appear to be grossly altered by electron microscopy.
Figure 4.
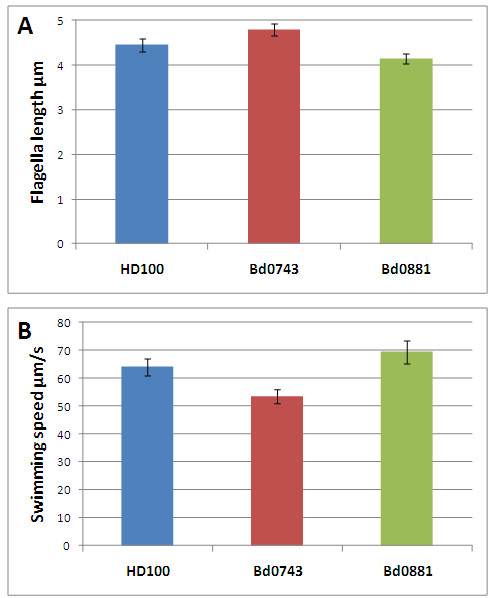
Lengths of flagella and swimming speeds of the mutants and wild-type. A- Flagellar length of wild type and sigma factor mutants measured from electron micrographs, error bars show 95% confidence intervals. B- Speeds of wild type and mutant predatory strains measured by the Hobson Bactracker, error bars show 95% confidence intervals.
To look for any evidence of association between RpoE-like sigma factor proteins and motility gene expression, we firstly measured the transcription of the 3 motA genes in ΔBd0881 and ΔBd0743, but found no difference compared to wild type (data not shown). This led us to conclude that Bd0881 does not act at motor regulation and does not produce faster rotating but shorter flagella.
We next tested whether there was an association between the transcriptional expression profiles of the rpoE-like genes and flagellar genes, measuring this by RT-PCR in total RNA from across the predatory cycle (Figure 5). We found that the expression patterns for bd0743 and bd3314 were constitutive but the expression pattern of bd0881 was similar to that seen for the key fliC3 gene of Bdellovibrio[11]; fliC3 is the only flagellin gene (from 6 fliCs) whose expression is crucial to flagellar synthesis, and its repression prevents motility of Bdellovibrio[6].
Figure 5.
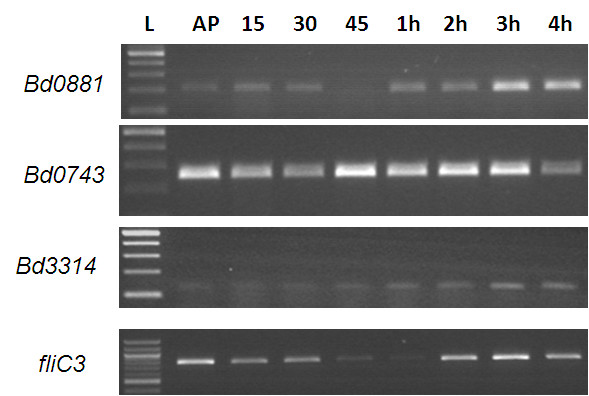
Expression patterns ofrpoE-like genes compared tofliC3in total RNA taken from across the predatory cycle studied by RT-PCR. RT-PCR with transcript-specific primers on total RNA prepared from identical numbers of B. bacteriovorus HD100 predator synchronously invading an E. coli S17-1 prey culture, with samples taken as the predatory infection, and Bdellovibrio development proceeds across a time course. L- NEB 100 bp ladder, AP- attack-phase 15–15 minutes predation, 30–30 minutes predation, 45–45 minutes predation 1-4 h: 1,2,3,4 hours predation respectively. Controls of no template, no reverse transcriptase, E. coli S17-1 only RNA as template and B. bacteriovorus HD100 genomic DNA were carried out. Primers designed to bd0743 give a product in every sample, thus act as a positive control for the RNA, validating the lack of expression in some of the samples. A similar expression pattern was seen for bd0881 and fliC3.
Our results showed that expression of bd0881 was all but abolished at 45 min to 1 hour after Bdellovibrio addition to prey, and resumed later in the predatory cycle, before prey lysis, as shown in Figure 4 alongside expression of the critical fliC3 gene. The expression of the fliC3 gene initially drops early in the predatory cycle, then resumes as the Bdellovibrio are nearing septation and flagella are synthesised prior to prey lysis and progeny escape from the prey cell debris into liquid cultures. Thus the similarity in expression patterns of fliC3 gene and bd0881 during predation may imply that Bd0881 protein is involved in regulatory events to do with the timepoints where flagella are being synthesised, i.e. around septation, but the fact that ΔBd0881 mutants are not immotile shows that Bd0881 is not required for the “all or nothing” induction of the fliC3 gene expression itself.
RT-PCR reveals regulation of chaperone genes by Bd0743
RT-PCR was used to study the expression of GroE chaperone protein genes in wild-type and sigma-factor knockout Bdellovibrio strains, as chaperone genes are typically RpoE-regulated in other bacteria, although no obvious E. coli RpoE- like consensus sequence was seen upstream of them in the B. bacteriovorus HD100 genome. Other bacteria induce expression of GroE protein chaperones upon heat shock (typically experimentally 42°C) in order to deal with misfolded proteins [12]. Furthermore, over-expression of chaperones can aid the expression of high levels of proteins in cells [13] including situations where addition of phage–encoded GroES proteins modify the size of protein that the bacterial chaperone can fold, to assemble large phage capsid proteins [14]. The Bdellovibrio genome has, in addition to the bd0097bd0099 groES groEL genes, a second homologue, bd3349, of groES (here designated groES2 versus groES1 for bd0097), so we investigated the expression of all these genes by RT-PCR using matched amounts of RNA from wild-type and sigma-factor mutant Bdellovibrio, treated in attack phase, at different temperatures (29°C and heat-shock 42°C for 10 mins; Figure 3) using methods previously described [15]. In wild-type Bdellovibrio, as is the case in many other bacteria, groES1EL expression was low at normal Bdellovibrio growth temperature (29°C) and expression was induced at a higher level under heat shock (42°C). This situation was the same for wild type and the ΔBd0881 mutant indicating that the Bd0881 sigma factor is not involved in this heat shock event. In the ΔBd0743 mutant, however, groES1EL expression was de-repressed, even in non-heat shock conditions suggesting that the Bd0743 sigma factor controls, directly or indirectly, the repression of groES1EL under normal temperature conditions. The viability of the ΔBd0743 cells was not affected under predatory growth conditions as determined by plaque assay indicating that this GroE deregulation does not severely affect the cells during laboratory culturing.
The second chaperone gene groES2 (bd3349) was expressed at a very low level, in attack phase cells of in the wild-type and ΔBd0881 mutant, under both normal and heat shock conditions,(Figure 3); suggesting that possibly it is not normally part of the heat shock response and may have a different role outside. In the ΔBd0743 mutant, however, groES2 expression was de-repressed in both normal and heat shock conditions, again implying that this sigma factor controls the expression of repressors of chaperone gene expression.
Recent work [16] has demonstrated different roles for multiple chaperone groEL gene products in the predatory Myxococcus including differential roles in predation and so it is possible that a similar situation exists with the duplicate groES gene products of Bdellovibrio. The groES2 gene was annotated in the B. bacteriovorus HD100 genome as encoding a 224 amino acid protein, but closer inspection reveals that a more likely start codon is at the methionine at base pair position 322 within this orf as the region before this, in the old annotation, includes lots of repetitive sequence. Using this start codon, the predicted protein of 117 amino acids has 34% identity and 62% similarity with the predicted (100 amino-acid) GroES protein of E. coli, and this 117aa region only of Bd3349 GroES2 is homologous to all predicted GroES sequences of delta-proteobacteria which give the highest BLAST homology scores for the Bd3349 protein. RT-PCR primers for groES2 were designed to anneal to RNA encoding this orf and transcription of both groES genes was monitored in RNA extracted over a wild type predatory time-course of B. bacteriovorus HD100 preying upon E. coli (Figure 6). This showed that groES1 was upregulated early at 15 minutes upon Bdellovibrio contact with prey cells and when the Bdellovibrio were growing within prey, remaining constitutively expressed throughout the predatory cycle. In contrast groES2 was not expressed early but was upregulated later, at 2–4 hours in the predation cycle when Bdellovibrio were beginning to septate and lyse prey. Although there are more Bdellovibrio present at this stage of the predatory cycle as a result of replication within the prey, the upregulation is unlikely to solely be a result of this as groES2 is not expressed at all in earlier stages of the cycle and so its induction here is significant. RT-PCR was also performed on matched amounts of RNA derived from 3 different host-independent strains derived from each sigma-factor mutant and a control wild-type (Figure 7) and revealed that groEL, groES1 and groES2 were all expressed at similar levels in each of the mutants in axenic, prey-independent (HI) growth. As (HI) host-independently growing Bdellovibrio populations include a mixture of attack phase and filamentous growth stage cells, it is not surprising that all of the chaperones are expressed in these cells.
Figure 6.
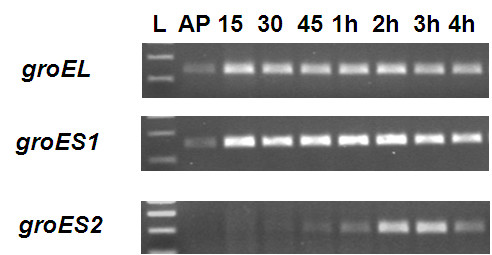
Transcriptional expression patterns of the threeBdellovibriochaperonin genes across the wild type predatory cycle. RT-PCR with transcript-specific primers was performed on total RNA prepared from identical volumes of B. bacteriovorus HD100 predator with E. coli S17-1 prey infection culture as the predatory infection proceeds across a time course. L- NEB 100 bp ladder, AP- attack-phase 15–15 minutes predation, 30–30 minutes predation, 45–45 minutes predation 1-4 h: 1,2,3,4 hours predation respectively. Controls of no template, no reverse transcriptase, E. coli S17-1 only RNA as template and bacteriovorus HD100 genomic DNA were carried out. Primers designed to bd0097 groES1 and bd0099 groEL give a product in every sample, thus act as a positive control for the RNA, validating the lack of expression of bd3349 groES2 in the earlier part of the infectious cycle.
Figure 7.
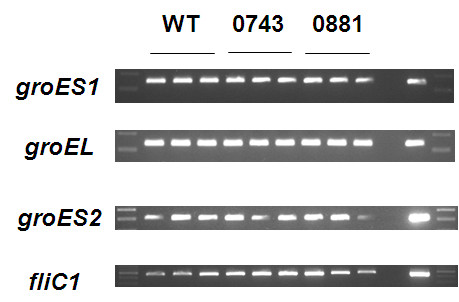
Transcriptional expression patterns of the threeBdellovibriochaperonin genes during axenic Host-Independent growth. RT-PCR with transcript specific primers was carried out on matched concentrations of RNA (matched by Nanodrop spectrophotometer readings) from axenically grown Host-Independent Bdellovibrio. Three independently isolated strains of each sigma factor mutant and each host-independent (HI) wild-type were used to account for HI strain-to strain variation. L- NEB 100 bp ladder –ve - no template negative control + ve- HD100 genomic DNA positive control.
Conclusions
We have shown that of three B. bacteriovorus HD100 sigma factor genes with at least partial rpoE homology, one- bd3314, is likely essential for Bdellovibrio cell life and cannot be deleted. bd0881 and bd0743 can be deleted with the Bdellovibrio retaining the ability to grow predatorily or prey-independently.
In the case of ΔBd0881 the predatory efficiency was reduced, despite the flagellar motility of the mutant being slightly increased, (despite a slight but statistically significant shortening of flagellar filament length) thus the change in predation efficiency may not be due to motility changes but regulation of other predatory genes. The bd0881 gene has an expression pattern across the predatory cycle that is similar to that of the flagellin genes whose expression is required for Bdellovibrio motility. That bd0881 expression is turned off and then resumes at a similar time to flagellin gene expression, during the predatory cycle, implies that Bd0881 may have a role associated with pre-septation developmental maturation of Bdellovibrio around the time that flagella are being built in newly dividing cells. However the Bd0881 sigma factor does not directly regulate the expression of fliC flagellin or mot flagellar motor genes themselves.
Surprisingly, predatory efficiency was not affected in our cultures by the slower swimming speed of the ΔBd0743 sigma factor mutant; this is probably indicative of sufficient mixing of predator and prey at close quarters in lab conditions. The slight increase in flagellar length in ΔBd0743 mutants is likely to have come with the incorporation of a higher percentage of a less rigid flagellin in the flagella causing a less efficient “bow wave” and this may account for the slower swimming. In both the ΔBd0743 and ΔBd0881 mutants, small but significant changes in swimming speed were paradoxically associated with changes apparently in the wrong direction in flagellar length. This shows that it is not simply flagellar length that governs the thrust produced by flagellar propellers. In previous studies on the six different flagellins that are incorporated into the flagellar propeller of Bdellovibrio[11,17], we found that different flagellin compositions of a single Bdellovibrio flagellum are possible, and that in the case of a fliC4 mutant, for example, an apparently wild type-length flagellum gave a lower swimming speed than wild type [11] suggesting an altered filament rigidity. As flagellar filament growth, in a bacterium with six flagellins, is a post-transcriptionally highly controlled process involving diverse chaperones and gate keepers at the base of the flagellum allowing different subunits to be added into the growing flagellum [18] we cannot expect to tell anything meaningful about these small changes of swimming speed from simple studies of flagellar filament gene expression, so we have decided to leave this aspect of the investigation at this point.
In looking at chaperonin expression regulation by B. bacteriovorus HD100 sigma factors, we found that, in contrast to bd0881, deletion of which had no effect, the product of gene bd0743 acts more like the heat shock sigma factor RpoE of other bacteria and represses (directly or indirectly) the level of expression of chaperonin genes groES1 groEL (bd0097 and bd0099) in non-heat shock conditions and the level of expression of the groES2 (bd3349) gene under both heat-shock and non-heat-shock conditions. These data and the finding that the groES2 gene is normally expressed in wild type Bdellovibrio only during the late stages of predation (2–4 hours) when the Bdellovibrio are septating and preparing to lyse the exhausted prey bdelloplast, may suggest that a modified chaperonin complex involving GroES2 is used in Bdellovibrio protein expression and folding that occurs at this point. Ascertaining why this is the case requires more chaperone-specific experimentation, beyond the scope of this study and mutagenesis of bd3349 is underway. That the majority of GroES residues shown to interact with GroEL in E. coli[19] are conserved or have conserved substitutions in both of the GroES1 and GroES2 homologues of B. bacteriovorus HD100 supports the idea that they form genuine alternative chaperonin complexes, making GroEL protein folding chambers with different GroES “lids”. It is a tantalising possibility that Bdellovibrio has a requirement for a modified chaperonin complex for the folding of unusual Bdellovibrio proteins required for late-stage prey lysis or Bdellovibrio attack phase cell maturation. The Bd0743-controlled, late-stage expression of groES2 is a possible mechanism for this. Although the (reannotated) Bdellovibrio groES2 gene product is larger at 117 amino-acids than the bd0097 groES1 gene product which is 100 amino-acids, there is no significant additional homology (above that for GroES1) between Bdellovibrio GroES2 and the bacteriophage T4 Gp31 GroES-like protein (data not shown). The bacteriophage T4 Gp31 GroES-like protein allows formation of a larger protein folding chamber for unusual phage capsid protein Gp23 to fold. Bdellovibrio, being a bacterium rather than a phage, does not have any homologues of this protein, so any analogous alternative role for GroES2 in Bdellovibrio protein folding awaits the outcomes of further mutagenesis studies.
Methods
Strains and growth conditions
A list of bacterial strains used in this study is presented in Table 2. E. coli was grown on YT media overnight (about 16 hours) with 50 μg ml-1 kanamycin sulphate as appropriate. Host dependent, predatory Bdellovibrio were grown in liquid prey lysate cultures in Ca/HEPES buffer or on YPSC double agar overlays as described elsewhere [20].
Table 2.
List of strains used in this study
| Strain | Description | Reference |
|---|---|---|
|
E. coli S17-1 |
thi,pro,hsdR-,hsdM+,recA; integrated plasmid RP4-Tc::Mu-Kn::Tn7 |
[21] |
|
E. coli DH5α |
F’ endA1 hsdR17 (rk-mk-) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ(lacIZYA-argF) U169 deoR (ϕ80dlacΔ(lacZ)M15) |
[22] |
|
E. coli S17-1: pZMR100 |
Plasmid vector used to confer Kmr on S17-1 & DFB225 that are being used as prey for KmrBdellovibrio strains |
[23] |
|
Bdellovibrio bacteriovorus HD100 |
Wild-type |
[4] |
|
Bdellovibrio bacteriovorus fliC1 merodiploid |
Kmr derivative of HD100 merodiploid for fliC1 |
[24] |
|
Bdellovibrio bacteriovorus bd0743 |
HD100 bd0743::aphII |
This study |
| Bdellovibrio bacteriovorus bd0881 | HD100 bd0881::aphII | This study |
RNA isolation and RT-PCR
Total RNA was isolated with modifications of the Promega SV total isolation kit described previously [11]. Heat shock was carried out by incubating 20 ml of prey-dependent Bdellovibrio in 50 ml centrifuge tubes at 29°C, then transferring to a 42°C water bath (with a control transferred to a 29°C water bath) for 10 minutes before adding 5 ml 5% phenol 95% ethanol (v/v) and proceeding with RNA extraction. Plaque enumeration confirmed that this heat treatment had no significant affect on cell viability. RT-PCR was carried out with the Qiagen one-step RT-PCR kit according to the manufacturer’s instructions as described elsewhere [25]. Primers used are shown in Table 3.
Table 3.
List of primers used in this study
| Primer | Sequence | Use |
|---|---|---|
| fliC3RTF |
ATGCTCAGAGAGTTCTCTGG |
fliC3 RT-PCR |
| fliC3RTR |
AATGACTTGTTCAAGAGTCC |
fliC3 RT-PCR |
| fliC5RTF |
GCTCAACGTAACTTGGTCGG |
fliC5 RT-PCR |
| fliC5RTR |
AGCCGATCAGCTTAAGAGCC |
fliC5 RT-PCR |
| bd0881RTF |
CGCAAGGAAGAAGTCAGTCC |
bd0881 RT-PCR |
| bd0881RTR |
CAGGCTTAAACGGGATTTCA |
bd0881 RT-PCR |
| bd0743RTF |
GCTCTTTTTCCGAACTCGTG |
bd0743 RT-PCR |
| bd0743RTR |
TACAGCCAATTGCACATCGT |
bd0743 RT-PCR |
| Bd3314RTF |
GGATTCGCGGCTATATTCAA |
bd3314 RT-PCR |
| Bd3314RTR |
TGGCATCCAGAGCTTCTTTT |
bd3314 RT-PCR |
| fliC1RTF |
GCATCTATCGCAGCACAACG |
fliC1 RT-PCR |
| fliC1RTR |
CCGTCGAGTCGGCATCAAAT |
fliC1 RT-PCR |
| Bd743-F |
GAAATTCTTGAAGCCATGACCAATGCG |
Cloning bd0743 |
| Bd743-R |
CGGGATCCGAGTGGCCTCTGGATTCG |
Cloning bd0743 |
| Bd881-F2 |
CGGAATTCTGGTCGCAAGAATATCTGCC |
Cloning bd0881 |
| Bd881-R2 |
GCTCTAGAATGACTCCAAGCTGGTTGGC |
Cloning bd0881 |
| Bd3314-F |
GCTCTAGACAGAAAGGAAACGACGCAC |
Cloning bd3314 |
| Bd3314-R | GCTCTAGAGCTTAGGGGTTCTGTATAA | Cloning bd3314 |
Gene knock-out and luminescent prey assay
Kanamycin resistance cassettes were inserted into the rpoE-like sigma factor genes of Bdellovibrio, as described elsewhere [9,11]. Primers used are listed in Table 3. Luminescent prey assays (with E. coli S17-1 containing the plasmid pCL100) were carried out as described elsewhere [9,11] except using a Fluostar Optima machine and the final enumeration data were expressed as Bdellovibrio per E. coli. An extra sum of squares F test carried out using the GraphPad Prism 5 software was carried out to show significance.
Electron microscopy and flagella filament length analysis
Bdellovibrio cells were incubated for 24 hours in a predatory culture before being placed on a carbon formvar grid (Agar Scientific), and stained with 0.5% uranyl acetate pH 4.0 as described previously [17]. Cells were imaged using a JEOL JEM1010 transmission electron microscope. Flagellar lengths were measured to the nearest 0.01 μm for an average of 50 cells per strain, error bars show the 95% CI around the mean for each sample as described previously [17]. Student’s t-test was carried out to determine significance of results.
Hobson BacTracker analysis of bdellovibrio swimming speeds
The swimming speed of each Bdellovibrio strain was analysed using Hobson BacTracker (Hobson Tracking Systems, Sheffield, United Kingdom) exactly as described in [24], including the use of the lower run speed limit of 15 μm/s to reduce the influence of Brownian motion, and accidental tethered-cell-body rotation, on the speed outputs. Cells were pre-grown for 24 hours in a typical 10 ml predatory culture with E. coli S17-1 as prey under the same conditions as for the electron microscopic analysis above. Student’s t-test was carried out to determine significance of results.
Abbreviations
Rpo, RNA polymerase; E. coli, Escherichia coli; ECF, Extracytoplasmic Function; RT-PCR, Reverse Transcriptase Polymerase Chain Reaction; BLAST, Basic Local Alignment Search Tool.
Competing interests
The authors declare that they have no competing interests.
Authors contributions
RES designed the experiments and co-authored the manuscript. CL performed the RT-PCR and luminescence assays and co-authored the manuscript, RT constructed the mutants and performed RT-PCR, LH performed the electron microscopy and speed measurements. All authors read and approved the final manuscript
Contributor Information
Carey Lambert, Email: carey.lambert@nottingham.ac.uk.
Rob Till, Email: rob.till@nottingham.ac.uk.
Laura Hobley, Email: l.hobley@dundee.ac.uk.
R Elizabeth Sockett, Email: liz.sockett@nottingham.ac.uk.
Acknowledgements
The authors thank Marilyn Whitworth for technical assistance and thank Dr Peter Lund at Birmingham University for helpful suggestions for future GroES2 work. This research was supported by Wellcome Trust grant AL077459 and by Human Frontier Science Programme Grant RGP52/2005.
References
- Varon M, Shilo M. Interaction of Bdellovibrio bacteriovorus and host bacteria. J Bacteriol. 1968;95(3):744–753. doi: 10.1128/jb.95.3.744-753.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. In: The Prokaryotes. 2. Schleifer KH, editor. Springer, New York; 1991. The genus Bdellovibrio. [Google Scholar]
- Shilo M, Bruff B. Lysis of Gram-negative bacteria by host-independent ectoparasitic Bdellovibrio bacteriovorus isolates. J Gen Microbiol. 1965;40:317–328. doi: 10.1099/00221287-40-3-317. [DOI] [PubMed] [Google Scholar]
- Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A. et al. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science. 2004;303(5658):689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- Heusipp G, Schmidt MA, Miller VL. Identification of rpoE and nadB as host responsive elements of Yersinia enterocolitica. FEMS Microbiol Lett. 2003;226(2):291–298. doi: 10.1016/S0378-1097(03)00613-X. [DOI] [PubMed] [Google Scholar]
- Ades SE. Regulation by destruction: design of the sigmaE envelope stress response. Curr Opin Microbiol. 2008;11(6):535–540. doi: 10.1016/j.mib.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Mishra MN, Kumar S, Gupta N, Kaur S, Gupta A, Tripathi AK. The extra-cytoplasmic function sigma factor (RpoE) cotranscribed with its cognate anti-sigma factor confers tolerance to NaCl, ethanol and methylene blue in Azospirillum brasilense Sp7. Microbiology. 2011;157(4):988–999. doi: 10.1099/mic.0.046672-0. [DOI] [PubMed] [Google Scholar]
- Lane WJ, Darst SA. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4(9):e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Smith MCM, Sockett RE. A Novel assay to monitor predator–prey interactions for Bdellovibrio bacteriovorus 109 J reveals a role for methyl-accepting chemotaxis proteins in predation. Environ Microbiol. 2003;5(2):127–132. doi: 10.1046/j.1462-2920.2003.00385.x. [DOI] [PubMed] [Google Scholar]
- Nakahigashi K, Yanagi H, Yura T. Isolation and sequence analysis of rpoH genes encoding sigma 32 homologs from Gram negative bacteria: conserved mRNA and protein segments for heat shock regulation. Nucleic Acids Res. 1995;23(21):4383–4390. [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Evans KJ, Till R, Hobley L, Capeness M, Rendulic S, Schuster SC, Aizawa S, Sockett RE. Characterizing the flagellar filament and the role of motility in bacterial prey-penetration by Bdellovibrio bacteriovorus. Mol Microbiol. 2006;60(2):274–286. doi: 10.1111/j.1365-2958.2006.05081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisbert E, Yura T, Rhodius VA, Gross CA. Convergence of molecular, modeling, and systems approaches for an understanding of the Escherichia coli heat shock response. Microbiol Mol Biol Rev. 2008;72(3):545–554. doi: 10.1128/MMBR.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Aggarwal N, Batra P, Mishra S, Chaudhuri TK. Co-expression of chaperonin GroEL/GroES enhances in vivo folding of yeast mitochondrial aconitase and alters the growth characteristics of Escherichia coli. Int J Biochem Cell Biol. 2006;38(11):1975–1985. doi: 10.1016/j.biocel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457(7225):107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert C, Chang CY, Capeness MJ, Sockett RE. The first bite–profiling the predatosome in the bacterial pathogen Bdellovibrio. PLoS One. 2010;5(1):e8599. doi: 10.1371/journal.pone.0008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Y, Zhang CY, Zhang WY, Jiang DM, Wu ZH, Liu H, Li YZ. Myxococcus xanthus viability depends on groEL supplied by either of two genes, but the paralogs have different functions during heat shock, predation, and development. J Bacteriol. 2010;192(7):1875–1881. doi: 10.1128/JB.01458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida Y, Hobley L, Lambert C, Fenton AK, Sockett RE, Aizawa S. Roles of multiple flagellins in flagellar formation and flagellar growth post bdelloplast lysis in Bdellovibrio bacteriovorus. J Mol Biol. 2009;394(5):1011–1021. doi: 10.1016/j.jmb.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulds-Pain A, Birchall C, Aldridge C, Smith WD, Grimaldi G, Nakamura S, Miyata T, Gray J, Li G, Tang J. et al. Flagellin redundancy inCaulobacter crescentusand its implications for flagellar filament assembly. J Bacteriol. 2011;193(11):2695–2707. doi: 10.1128/JB.01172-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass I, Horovitz A. Mapping pathways of allosteric communication in GroEL by analysis of correlated mutations. Proteins. 2002;48(4):611–617. doi: 10.1002/prot.10180. [DOI] [PubMed] [Google Scholar]
- Lambert C, Sockett RE. Laboratory maintenance of Bdellovibrio. Curr Protoc Microbiol. 2008. pp. 7B 2.1–7B 2.13. Chapter 7. [DOI] [PubMed]
- Simon R, Preifer U, Puhler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983;9:184–191. [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Rogers M, Ekaterinaki N, Nimmo E, Sherratt D. Analysis of Tn7 transposition. Mol Gen Genet. 1986;205(3):550–556. doi: 10.1007/BF00338097. [DOI] [PubMed] [Google Scholar]
- Morehouse KA, Hobley L, Capeness M, Sockett RE. Three motAB Stator Gene Products in Bdellovibrio bacteriovorus Contribute to Motility of a Single Flagellum during Predatory and Prey-Independent Growth. J Bacteriol. 2011;193(4):932–943. doi: 10.1128/JB.00941-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans KJ, Lambert C, Sockett RE. Predation by Bdellovibrio bacteriovorus HD100 requires type IV pili. J Bacteriol. 2007;189(13):4850–4859. doi: 10.1128/JB.01942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]


